Abstract
The unique mental abilities of humans are rooted in the immensely expanded and folded neocortex, which reflects the expansion of neural progenitors, especially basal progenitors including basal radial glia (bRGs, also called outer RGs) and intermediate progenitor cells (IPCs). Here, we show that constitutively active Shh signaling expanded bRGs and IPCs and induced folding in the otherwise smooth mouse neocortex, whereas the loss of Shh signaling decreased the number of bRGs and IPCs and the size of the neocortex. SHH signaling was strongly active in the human fetal neocortex but not in the mouse embryonic neocortex, and blocking SHH signaling in human cerebral organoids decreased the number of bRGs. Mechanistically, Shh signaling increased the initial generation and self-renewal of bRGs as well as increasing IPC proliferation. Thus, robust SHH signaling in the human fetal neocortex may contribute to bRG and IPC expansion and neocortical growth and folding.
The neocortex, a six-layered structure covering mammalian brains, computes high-order sensory, motor, and cognitive processes. During evolution, the neocortex has expanded dramatically and folded in certain species. The immense expansion of the neocortex in humans has made possible the complex behavior, cognition, and intellect that are quintessential to humans. Neocortical expansion and folding reflect an increase in the number of neurons, especially upper-layer neurons, and are thus dependent on the number and proliferative capacity of neural progenitors1–5. The primary neural progenitors are radial glia (RGs), whose cell bodies reside in the ventricular zone (VZ) at the apical side of the developing brain and are hence called ventricular RGs (vRGs) or apical RGs (aRGs). The aRGs generate neurons directly or via intermediate progenitor cells (IPCs) that occupy the subventricular zone (SVZ). Newborn neurons migrate along radial processes of RGs through the intermediate zone (IZ) to the cortical plate (CP) to form neuronal layers.
A neurogenic area basal to the archetypal SVZ was recently identified in the brains of monkeys and humans6,7. This area is called the outer SVZ (oSVZ) and is thought to be responsible for the expansion of the neocortex, especially the upper-layer neurons, in primates6,8. The primary neural progenitors in the oSVZ are outer RGs (oRGs, also called basal RGs [bRGs]) that are detached from the ventricle but maintain some aRG features, including radial processes9,10. bRGs are also present in other species including the mouse10–15; however, bRGs are greatly expanded in species with large and folded brains, especially humans, and their expansion is thought to underlie the complexity of the human brain1–5,11. Yet, little is known about the molecular pathways that lead to the expansion of bRGs and to neocortical growth and folding.
Sonic Hedgehog (Shh) signaling regulates brain development from early patterning to adult neurogenesis. Defective SHH signaling causes holoprosencephaly, a defect in the separation of the brain hemispheres that mostly results from abnormal patterning. Notably, patients with mild holoprosencephaly often have smaller-than-normal brains at birth (microcephaly)16, suggesting that mechanisms affecting early patterning also affect neurogenesis and brain size. Indeed, mutations in SHH and PATCHED1, an SHH receptor gene, were found in microcephaly patients who were not holoprosencephalic17,18. In mice, defective Shh signaling causes defective proliferation of IPCs and microcephaly19; however, the role of SHH signaling in human neocortical development beyond patterning is unknown.
Results
Elevated Shh signaling induces cortical growth and folding
To study the role of Shh signaling in neocortical development without affecting early patterning, we expressed a constitutively active Smoothened (SmoM2), an activator of Shh signaling, in aRGs and their progenies from embryonic day 13.5 (E13.5) by using a GFAP promoter–driven Cre. GFAP::Cre; SmoM2fl/+ mice (SmoM2 mutants) had cortices larger than those of controls (Fig. 1). Remarkably, SmoM2 mutants consistently developed folding in the cingulate cortex and showed increased cell density in the upper layers of the cingulate and medial cortices (Fig. 1b). SmoM2 mutants normally expressed layer-specific markers (Fig. 1c). The density of deep-layer neurons expressing Tbr1 (Tbr1+) was slightly decreased in SmoM2 mutants, but Satb2+ upper-layer (layer II and III) neurons in the cingulate and medial cortices were denser in SmoM2 mutants than in controls (Fig. 1c,e). Notably, white matter extended into the induced gyri (Fig. 1b), suggesting that the upper-layer neurons in the folded area had axonal projections. The Satb2+ neuronal density was unchanged in the laterally located sensory cortex (data not shown), which showed no folding. These results suggest that increased Satb2+ upper-layer neurons induce folding in the cingulate cortex.
Figure 1. SmoM2 induces neocortical expansion and folding.
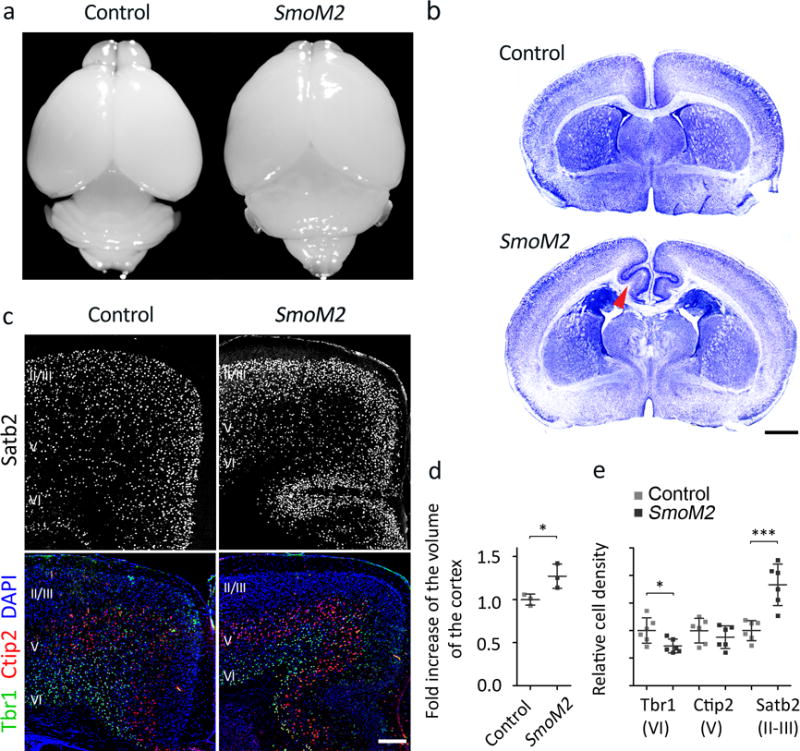
a. Control and GFAP::Cre; SmoM2fl/+ (SmoM2 mutant) brains at P10. b. Nissl staining of control and SmoM2 brain sections at P7. The arrowhead indicates white matter extended into the induced gyrus. Scale bar = 1 mm. Pictures represent at least 5 repeats. c. Expression of layer-specific markers: Satb2 (layers II–IV), Ctip2 (official name Bcl11b, red, layer V), Tbr1 (green, layer VI), and DAPI (blue). Scale bar = 0.2 mm. Pictures represent at least 3 repeats. d. Cortical volume calculated from serial MRI scans of P7 control and SmoM2 mutants. Results were normalized to fold change compared to controls. Two tailed unpaired t-test, P = 0.0380, n = 3 mice per group. We assumed normal distribution although the number of samples was too small for normality test. F test for variance, P = 0.3510, F(2, 2) = 4.697. e. Quantification of cell density in the cingulate and medial cortex. Two tailed unpaired t-test, P = 0.0006 (Satb2), P = 0.3634 (Ctip2), P = 0.0250 (Tbr1); t(10) = 4.877 (Satb2), t(10) = 0.9522 (Ctip2), t(10) = 2.635 (Tbr1); n = 9 brain slices from 3 mice per group. All data have passed Kolmogorov–Smirnov (KS) test for normality (P > 0.1) and F test for variance P = 4.255 (Satb2), P = 0.8609 (Ctip2), P = 0.2070 (Tbr1); F(5, 5) = 0.1380 (Satb2), F(5, 5) = 1.179 (Ctip2), F(5,5) = 3.385 (Tbr1). *P < 0.05; ***P < 0.001. Error bars represent standard deviation.
Elevated Shh signaling expands bRGs and IPCs
To understand upper-layer neuronal expansion, we examined neural progenitors at E16.5, when mostly upper-layer neurons are generated. Consistent with the mitogenic effect of Shh on IPCs19, IPCs expressing Tbr2 (Eomes) significantly increased in the SmoM2 mutants, especially in the SVZ, where most IPCs proliferate (Fig. 2a,c and Supplementary Fig. 1). In controls, Tbr2+ cells were spread to the IZ, where newborn neurons maintain Tbr220. Although the selective expansion of IPCs alone can cause the cortex to grow even larger than in SmoM2 mutants, such selective expansion does not induce denser upper layers or folding in mice21, suggesting that additional mechanisms are involved in folding.
Figure 2. SmoM2 expands IPCs and bRGs (oRGs).
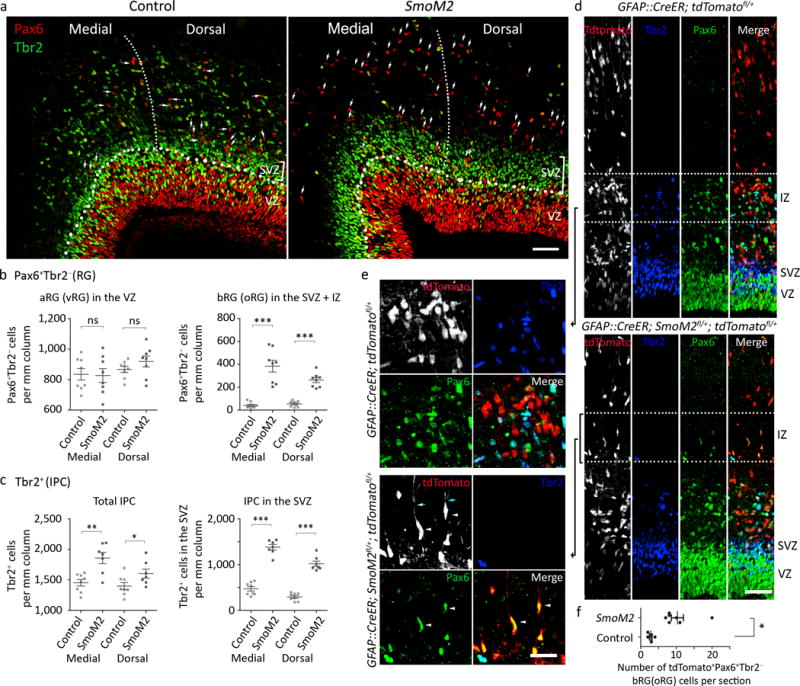
a. E16.5 cortices labeled for an RG marker, Pax6 (red), and an IPC marker, Tbr2 (green). The thin dotted line demarcates the medial (M) and dorsal (D) cortices. The thick dotted line marks the boundary between the VZ and SVZ. The Pax6+ Tbr2− cells separated from the VZ by bands of Tbr2+ IPC cells were counted as bRGs (oRGs; white arrows indicate examples). Pictures represent at least 6 repeats. b, c. Quantification of Pax6+ Tbr2− RGs and Tbr2+ IPCs at E16.5 Two tailed unpaired t-test (aRG, IPC, SVZ IPC, equal variance) or two tailed unpaired t-test with Welch’s correction (bRG, unequal variance), P = 0.7888 (aRG_M), P = 0.2648 (aRG_D), P = 0.0003 (bRG_M), P = 0.0001 (bRG_D), P = 0.0017 (IPC_M), P = 0.0458 (IPC_D), P = 0.0000 (SVZ IPC_M), P = 0.0000 (SVZ IPC_D); t(14) = 0.2730 (aRG_M), t(14) = 1.162 (aRG_D), t(7)=6.503 (bRG_M), t(9)=8.780 (bRG_D), t(13)=3.926 (IPC_M), t(13)=2.208 (IPC_D), t(13)=11.92 (SVZ IPC_M), t(13)=11.52 (SVZ IPC_D). All data have passed Kolmogorov–Smirnov (KS) test for normality P > 0.1. bRG did not pass F test for equal variance, P = 0.6628 (aRG_M), P = 0.1556 (aRG_D), P = 0.0003 (bRG_M), P = 0.0369 (bRG_D), P = 0.2378 (IPC_M), P = 0.6396 (IPC_D), P = 0.7069 (SVZ IPC_M), P = 0.1388 (SVZ IPC_D); F(7, 7) = 1.408 (aRG_M), F(7, 7) = 3.127 (aRG_D), F(7, 7) = 28.04 (bRG_M), F(7, 7) = 5.603 (bRG_D), F (6, 7) = 2.596 (IPC_M), F (6, 7) = 1.441 (IPC_D), F (6, 7) = 1.336 (SVZ IPC_M), F (6, 7) = 3.350 (SVZ IPC_D). N = 8 brain slices from 3 mice per group d, e. E16.5 cortices labeled with Pax6 (green) and Tbr2 (blue) after tamoxifen injection at E13.5. The dotted lines in (d) indicate areas enlarged in (e). f. Quantification of bRGs (tdTomato+ Pax6+ Tbr2−). Two tailed unpaired t-test with Welch’s correction, P = 0.0051, t(6) = 4.300. All data have passed Kolmogorov–Smirnov (KS) test for normality, P > 0.1. F test for equal variance, P = 0.0059, F (6, 4) = 3.350. N = 5 (control) and 7 (SmoM2) brain slices from 3 mice per group. ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001. IZ, intermediate zone; SVZ, subventricular zone; VZ, ventricular zone. Scale bars = 50 μm in (a, d), 25 μm in (e). Error bars represent standard error of the mean.
The density of Pax6+ Tbr2− aRGs was unchanged in SmoM2 mutants; however, Pax6+ Tbr2− cells were significantly increased outside the VZ in the SmoM2 cortex (Fig. 2a,b). Notably, Pax6+ Tbr2− cells increased significantly more in the medial than in the dorsal part of the neocortex (1.6 fold, Mann Whitney test, P = 0.0261, Sum of rank = 89.50, 46.50, Mann-Whitney U = 10.50, standard error = 0.216, 0.096, n = 8) reflecting SmoM2 expression in a high-medial to low-lateral gradient in SmoM2 mutants (Supplementary Fig. 1c, top). The Pax6+ Tbr2− cells outside the VZ also expressed Sox2 (Supplementary Fig. 2a), a transcription factor expressed by RGs. The expression pattern (Pax6+ Sox2+ Tbr2−) and location of these cells were reminiscent of those of bRGs. Indeed, most Sox2+ cells outside the VZ had processes that were positive for the RG markers RC2 and Glast, confirming their bRG identity (Supplementary Fig. 2a). In SmoM2 mutants, bRGs showed diverse morphology, with basal, apical, or bipolar processes being oriented radially or tangentially, similar to bRGs in monkeys and ferrets22,23, and some had a growth cone–like structure, as in human and ferret bRGs23 (Supplementary Fig. 2a).
To test whether SmoM2 increased bRGs cell-autonomously, we induced sparse recombination and labeled the recombined cells with tdTomato in GFAP::CreER; SmoM2fl/+; tdTomatofl/+ mice by injecting tamoxifen at E13.5 (Fig. 2d,e). At E16.5, tdTomato+ bRGs were significantly increased in GFAP::CreER; SmoM2fl/+; tdTomatofl/+ mice compared to controls, indicating cell-autonomous functioning of SmoM2 (Fig. 2d–f). As in monkeys22, bRGs showed diverse morphology, with bipolar bRGs being the most abundant (Supplementary Fig. 2b). Thus, SmoM2 cell-autonomously increased bRGs that resembled those in gyrencephalic species.
Elevated Shh signaling keeps bRGs and IPCs in the cell cycle
Manipulating Trnp1, PDGFD signaling, or ARHGAP11B increases bRGs in mice via delamination of aRGs24–26. However, SmoM2 increased the number of bRGs without decreasing that of aRGs (Fig. 2b), similar to the initial bRG expansion in humans1. Thus, SmoM2 may have expanded bRGs by increasing their self-renewal, production, or both. To test if SmoM2 increased self-renewal, we investigated the fraction of self-renewed bRGs remaining in the cell cycle 24 h after the previous S phase by injecting BrdU at E15.5 and EdU at E16.5 1.5 h before collecting embryos (Fig. 3a). At E16.5, the proliferation of the IPCs (EdU+) but not that of the aRGs and bRGs, was increased in SmoM2 mutants; however, the proportions of bRGs and IPCs remaining in the cell cycle 24 h after the previous S phase (EdU+ BrdU+ cells) were significantly higher in SmoM2 mutants than in controls (Fig. 3a,b). bRGs migrate basally before dividing; thus the more they divide and self-renew, the further they move from the ventricular surface9,12. Consistent with the increase in BrdU+ EdU+ bRGs, bRGs distributed more basally in SmoM2 mutants than in controls (Fig. 3c), indicating increased self-renewal of bRGs in SmoM2 mutants.
Figure. 3. SmoM2 expands bRGs (oRGs) by increasing their self-renewal and changing the aRG (vRG) division angle toward bRG production.
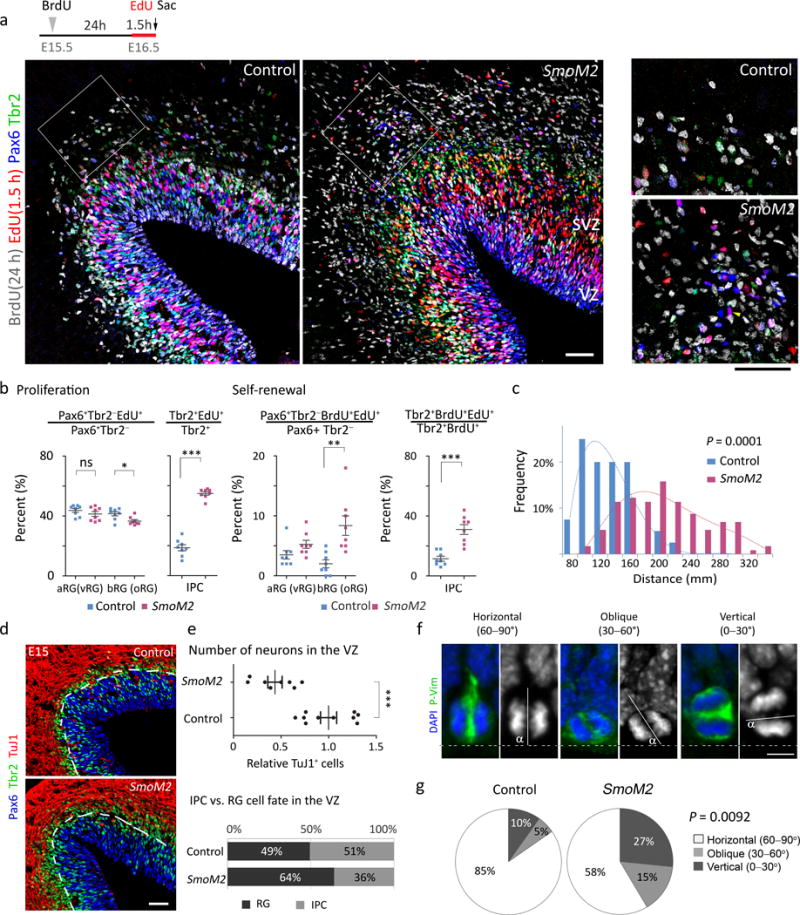
a. The experimental scheme and E16.5 cortices labeled for Pax6 (blue), Tbr2 (green), BrdU (grey), and EdU (red). The boxed areas are enlarged in the right. Pictures represent at least 3 repeats. b. Quantification of proliferation (EdU+) and self-renewal (EdU+ BrdU+) of RGs and IPCs. All the data collected from 8 brain slices from 3 mice per group passed Kolmogorov–Smirnov (KS) test for normality, P > 0.1. For proliferation studies, two tailed unpaired t-test was used, for aRG P = 0.2680, t(14)=1.154; for bRG, P = 0.0103, t(14)=2.960; for IPC, P = 0.0001, t(14)=15.75. F test for equal variance, for aRG, P = 0.5533, F(7, 7) = 1.594; for bRG, P = 0.3659, F(7, 7) = 2.045; for IPC, P = 0.1751, F(7, 7) = 2.963. For self-renewal of bRG, two tailed unpaired t-test with Welch’s correction was used, P = 0.0056, t(9) = 3.618; F test for variance P = 0.0274, F(7, 7) = 6.245. For self-renewal of IPC, two tailed unpaired t-test was used, P = 0.0001, t(14) = 5.321; F test for variance P = 0.0869, F(7, 7) = 4.013. c. Distribution of the distances of the EdU+ bRGs from the ventricular surface. Mann Whitney test, P = 0.0001, Sum of ranks = 1392, 10540, Mann-Whitney U = 571.5, n = 40 for control and n = 114 for SmoM2. N = 8 brain slices from 3 mice per group. d, e. E15.5 cortices labeled for TuJ1 (blue), Pax6 (green), and Tbr2 (red) (d) and quantifications (e). Pictures represent at least 3 repeats. For relative density of TuJ1 in the VZ, Mann Whitney test, P = 0.0006, Sum of ranks = 134, 37, Mann-Whitney U = 1.000, n = 10 and 8 for control and SmoM2 brain slices from 3 pairs of mice. For ratio of RG and IPC in the VZ, Mann Whitney test, P = 0.0021, Sum of ranks = 28, 77, Mann-Whitney U = 0.000, n = 7 brain slices from 3 mice per group. f, g. Mitotic aRGs at E14.5 labeled for phospho-vimentin (P-Vim, green) and DAPI (blue) (f) and quantification of the division angle (g). We blindly analyzed 101 cells (SmoM2 mutants) and 71 cells (controls) from 3 pairs of mice. In f, α indicates the angle between the apical surface (dotted line) and the plane of division (solid line). The divisions are termed ‘horizontal’ if 60° < α ≤ 90°, ‘oblique’ if 30° < α ≤ 60°, and ‘vertical’ if 0° ≤ α ≤ 30°. Mann Whitney test, P = 0.0092, Sum of ranks = 6937, 7769, Mann-Whitney U = 2719. ns, P > 0.05; *P < 0.05; **P < 0.005; ***P < 0.001. Error bars represent standard error of the mean. The scale bars represent 50 μm (a, d) and 5 μm (f).
Elevated Shh signaling shifts aRG division to produce bRGs
Multiple types of bRGs and IPCs, including apical IPCs, are all descendants of aRGs12,13,22,27,28. Therefore, we investigated whether aRGs produced more bRGs in SmoM2 mutants. To compare the direct progenies of aRGs and exclude IPCs and neurons generated from IPCs, we compared cells in the VZ but excluded the SVZ, where IPCs were actively proliferating (Fig. 3b and Supplementary Fig. 3d). At E15.5, RGs constituted 49% of the cells in the control VZs and 64% of the cells in the SmoM2 VZs (Fig. 3d,e and Supplementary Fig. 3a). The percentage of IPCs (Tbr2+ Pax6+ or −) and the number of neurons (TuJ1+) were significantly reduced in SmoM2 VZs. Thus, SmoM2-expressing aRGs produced more RGs at the expense of IPCs and neurons, which may explain the decrease in Tbr1+ neurons (Fig. 1e). Despite this, aRGs were not increased at E16.5 in SmoM2 mutants, whereas bRGs were markedly increased (Fig. 2b), suggesting that the additional RGs in the mutant VZ at E15.5 became bRGs.
To investigate the mechanism by which the cell composition changed in the SmoM2 VZ, we investigated the division angles of aRGs. aRGs dividing on an axis horizontal to the ventricular surface (horizontal division) mostly produce neurons or IPCs, whereas those dividing vertically or obliquely produce bRGs13,27. Vertical and oblique divisions were remarkably increased in SmoM2 mutants, as compared to controls (42% vs. 15%) (Fig. 3f,g). Furthermore, RGs dividing in the VZ but away from the ventricular surface were significantly increased in SmoM2 mutants (Supplementary Fig. 3b–d). RGs dividing non-apically in the VZ produce bRGs23,29 and may also represent bRGs in transit to the SVZ after generation at the ventricular surface; bRGs are also found in the inner SVZ and VZ11,14,23. Thus, SmoM2 increased the number of bRGs by shifting aRG division toward bRG production. SmoM2 also greatly increased the number of proliferating IPCs (Fig. 3b and Supplementary Fig. 3b,d), resulting in their expansion in the SVZ despite their decreased production from aRGs (Fig. 3e). These results suggest that the expansion of bRGs through increased production and self-renewal, along with an increased number of IPCs, led to neocortical expansion and folding in SmoM2 mutants.
Fate analysis of progenies from aRGs expressing SMOM2
To confirm that SmoM2 shifted aRG division toward bRG production at the expense of IPCs and neurons, we transduced aRGs sparsely with retroviruses expressing GFP either alone or with SMOM2 (a constitutively active human SMO) at E13.5 and examined the fates of GFP+ cells in transduced clones that had at least two cells. At E15, 46% of clones expressing GFP alone contained only IPCs and/or neurons without RGs (IPC/N clones), a further 46% contained one aRG with IPC/N (aRG + IPC/N), and 8% contained two aRGs with or without IPC/N (2aRGs + IPC/N) (Fig. 4). Similar to what is observed in SmoM2 mutants (Fig. 3e), SMOM2 transduction increased clones containing aRGs to 67% (56% aRG+IPC/N and 11% 2aRG+IPC/N) at the expense of IPC/N clones (33%). Remarkably, by E16, the proportion of aRG-containing clones decreased to the control level (47%), concomitant with a marked increase in the proportion of bRG-containing clones to 23%, which was much higher than that observed in control clones (5%). At E16, among clones containing IPCs, the number of IPCs per clone was higher in SMOM2-transduced clones than in control clones (1.18 vs. 1.36, P = 0.0158, by Mann Whitney test, Sum of ranks = 4390, 1938, Mann-Whitney U = 1118). Together with the results of SmoM2 mutant analyses, these results indicate that elevated Shh signaling shifts aRG division toward bRG production at the expense of IPCs and neurons and increases the proliferative divisions of IPCs.
Figure. 4. Retrovirus expressing SMOM2 promotes bRG production at the clonal level.
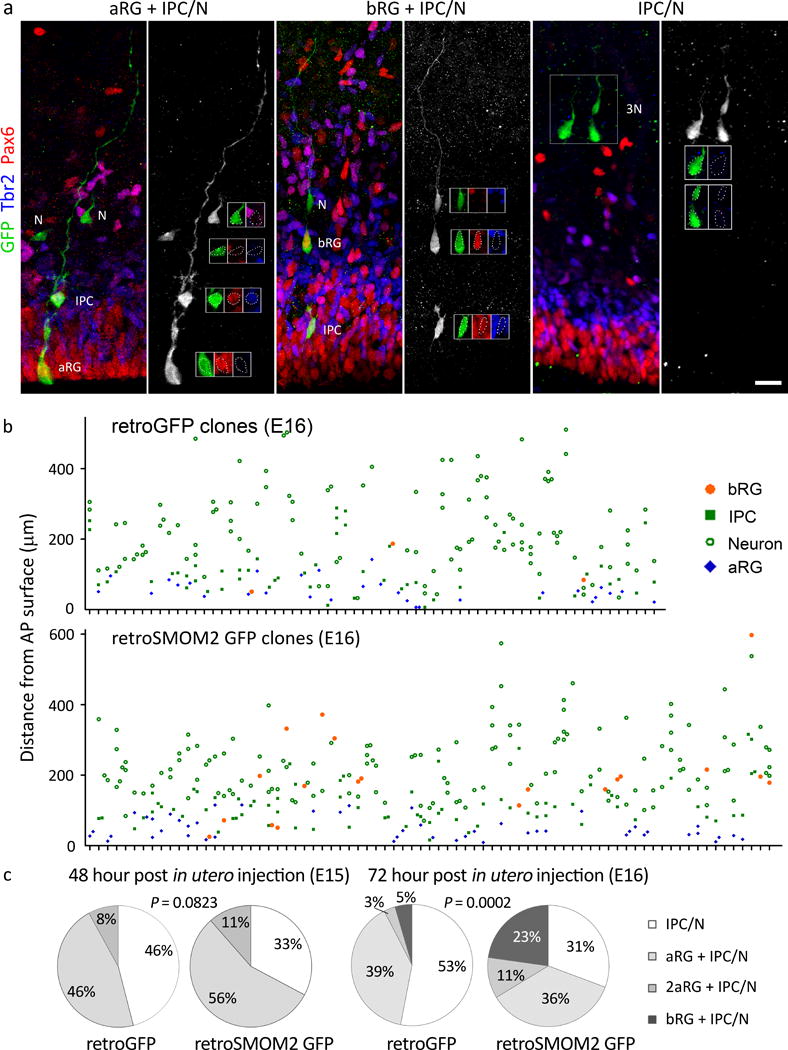
a. Micrographs showing examples of different types of clones. Cortices were labeled with GFP (green), Pax6 (red), and Tbr2 (blue) to determine the cell fate 48 h (E15) or 72 h (E16) after the in utero intraventricular injection of GFP or SMOM2 GFP retroviruses at E13. Clones were defined as follows: IPC/N clones contained IPCs and/or neurons but no RGs; aRG+IPC/N clones had one aRG and IPC/N; 2aRG+IPC/N clones had two aRGs with or without IPC/N; and bRG clones had at least one bRG and other cell types. Scale bar = 20 μm. b. Composition of individual clones at E16. Each vertical column of cells above a mark on the X-axis represents a single clone. c. Distribution of clone types. At E15, we found 23 IPC/N, 23 aRG+IPC/N, and 4 2aRG+IPC/N clones in control, and 20 IPC/N, 34 aRG+IPC/N, and 7 2aRG+IPC/N clones in SMOM2. At E16, we found 35 IPC/N, 26 aRG+IPC/N, 2 2aRG+IPC/N, and 3 bRG clones in control, and 31 IPC/N, 36 aRG+IPC/N, 11 2aRG+IPC/N, and 23 bRG clones in SMOM2. Two sided Fisher’s exact test, P = 0.0823 at E15 (50 GFP clones, 61 SMOM2 GFP clones), or Chi-square test, P = 0.1659, Chi-square (2) = 3.593 at E15; Chi-square test, P = 0.0002, Chi-square (2) = 17.61 at E16 (66 GFP clones, 101 SMOM2 GFP clones).
SmoM2 induces folding outside the cingulate cortex
In SmoM2 mutants, folding was present in only the cingulate cortex. As GFAP::Cre induces recombination at E13.5 and SmoM2 was expressed in a high-medial to low-lateral gradient in SmoM2 mutants, we used Nestin::Cre and Nestin::CreER, which can induce recombination earlier than E13.5 and did not show such a gradient (Supplementary Fig. 1c), to test whether SmoM2 could induce folding outside the cingulate cortex. Nestin::Cre induces recombination at E10.5. Only a few Nestin::Cre; SmoM2fl/+ mutants survived to birth, and these survivors had folding in both the cingulate cortex and other cortical areas (Supplementary Fig. 4a). The cortical layering was not disrupted in the folded area (Supplementary Fig. 4b). Inducing recombination moderately at E12.5 in Nestin::CreER; SmoM2fl/+ mutants avoided early lethality and induced folding outside the cingulate cortex (Supplementary Fig. 4c). Thus, SmoM2 could induce folding outside the cingulate cortex.
Cortical progenitor specification in SmoM2 mutants
In contrast to our results, previous studies showed that elevating Shh signaling earlier in Nestin::Cre; Gli3Xt/fl, Nestin::Cre; Ptch1fl/fl, or Emx1::Cre; Sufufl/fl mice disrupted the cortical progenitor specification and decreased the number of IPCs and upper-layer neurons30–32. The difference in the results is probably due to the different timing and efficiency of Shh signaling activation, because 1) Yabut et al. found no specification defects in GFAP::Cre; Sufufl/fl or GFAP::Cre; Smofl/+ mutants32, 2) overexpression of Shh in the neocortex at E13.5 increased IPCs33, and 3) we obtained far fewer than expected number of Nestin::Cre; SmoM2fl/+ mutants, suggesting that the few survivors are escapees that may have undergone inefficient recombination. In Emx1::Cre; Sufufl/fl mice, cortical progenitors ectopically expressed Dlx2 and Ascl1 (Mash1), which are ventral forebrain progenitor markers32; however, their expression was unchanged in our SmoM2 mutants at E14.5, by which time the aRG division mode had already shifted to produce bRGs (Supplementary Fig. 5a). At E16.5, cells expressing Ascl1 and Dlx2 appeared in SmoM2 mutant cortices (Supplementary Fig. 5b); however, the majority of neural progenitors were negative for Ascl1 and Dlx2, unlike those in Emx1::Cre; Sufufl/fl mice32. Notably, a subpopulation of human cortical progenitors express ASCL1, and human bRGs directly produce ASCL1+ cortical progenitors9. Thus, the specification of cortical progenitors in our SmoM2 mutants was largely unaffected and was somewhat similar to that of human cortical progenitors.
SmoM2 requires cilia and Gli2 to induce neocortical folding
To test whether cilia-dependent canonical Shh signaling was required for folding, we ablated the primary cilia from SmoM2-expressing cells by removing Kif3a, an essential ciliogenic gene. The loss of cilia in GFAP::Cre; SmoM2fl/+; Kif3afl/fl mice blocked neocortical expansion and folding (Supplementary Fig. 6a), as did removing Gli2, a transcription factor essential to the Shh-induced transcriptional program, in GFAP::Cre; SmoM2fl/+; Gli2fl/fl mice (data not shown). Therefore, transcriptional regulation through primary cilia and Gli2 was required for SmoM2-driven neocortical expansion and folding.
Loss of Smo decreases IPCs, bRGs, and the neocortex
Because elevated Shh signaling expands bRGs and the neocortex, we investigated whether endogenous Shh signaling affected the bRG population by deleting Smo in GFAP::Cre; Smofl/fl mice. Smo mutants had abnormally small brains (Supplementary Fig. 6b) and significantly fewer IPCs and bRGs but not aRGs (Fig. 5a–c). Remarkably, the loss of Smo decreased the proportion of aRGs dividing vertically or obliquely (Fig. 5d). Consequently, Smo mutants had more Tbr1+ deep-layer neurons and fewer Satb2+ upper-layer neurons than did control mice—the exact opposite of what is found in SmoM2 mutants (Fig. 5e vs. Fig. 1e). Thus, endogenous Shh signaling was required to expand IPCs, bRGs, upper-layer neurons, and the neocortex.
Figure. 5. Smo is required to expand IPCs, bRGs, and upper-layer neurons.
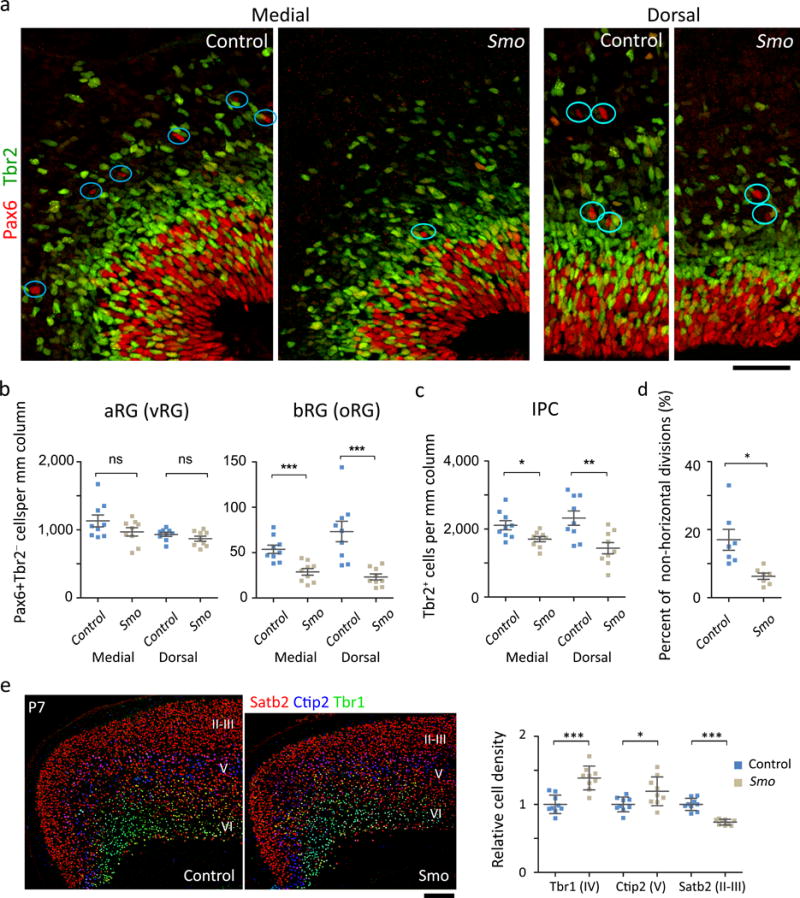
a. E16.5 cortices labeled for Pax6 (red) and Tbr2 (green). The circles indicate examples of Pax6+ Tbr2− bRGs. Scale bar = 50 μm. We analyzed 9 sections from 3 mice per group. b, Quantification of RGs. Mann Whitney test, for medial aRG, P = 0.5076, Sum of ranks = 93.50, 77.50, Mann-Whitney U = 32.50; for dorsal aRG, P = 0.3401, Sum of ranks = 97, 74, Mann-Whitney U = 29.00; for medial bRG, P = 0.0005, Sum of ranks = 122, 49, Mann-Whitney U = 4.000; for dorsal bRG P = 0.0002, Sum of ranks =124, 47, Mann-Whitney U = 2.000. c. Quantification of IPCs. Mann Whitney test, for medial IPC, P = 0.0188, Sum of ranks = 112, 59, Mann-Whitney U = 14.00; for dorsal IPC, P = 0.0078, Sum of ranks = 115, 56, Mann-Whitney U = 11.00. d. Quantification of aRGs dividing non-horizontally (0° ≤ α ≤ 60°). We blindly analyzed 144 cells (Smo mutants) and 190 cells (controls) in 7 sections from 3 mice per group. Two tailed unpaired t-test with Welch’s correction, P = 0.0124, t(7) = 3.343; F test for variance, P = 0.0098, F(6, 6) = 11.18. e. Expression and quantification of layer-specific markers: Satb2 (red), Ctip2 (blue), and Tbr1 (green). Scale bar = 0.2 mm. Two tailed unpaired t-test: for Tbr1, P = 0.0000, t(16)=5.244; for Ctip2, P = 0.0271, t(16)=2.432; for Satb2, P = 0.0000, t(11)=7.947. N = 9 sections from 3 mice per group. All data have passed Kolmogorov–Smirnov (KS) test for normality, P > 0.1 and tested for equal variance by F test: P = 0.4990, F (8,8) = 1.642 (Tbr1), P = 0.0701, F (8,8) = 3.928 (Ctip2), P = 0.0418, F (8,8) = 4.719 (Satb2). ns, P > 0.05; *P < 0.05; **P < 0.005; ***P < 0.001. Error bars represent standard error of the mean (b-d) and standard deviation (e).
Active SHH signaling in the human fetal neocortex
Despite its critical roles in expanding basal progenitors and the neocortex, Shh signaling is minimally active in the embryonic mouse neocortex. Gli1, a faithful marker of strong Shh signaling, is highly expressed in the ganglionic eminence but is undetectable in the embryonic neocortex (Supplementary Fig. 7a). Consistently, LacZ or CreER knocked into the Gli1 locus label cells in the ganglionic eminence but not in the neocortex34. In contrast, GLI1 is highly enriched in human aRGs35. In the ferret cortex, Gli1 expression is significantly higher in the region of the VZ that gives rise to the thick oSVZ containing many bRGs than in the VZ region that gives rise to the thin oSVZ containing fewer bRGs36. Therefore, strong SHH signaling may contribute to bRG and neocortical expansion in humans. To compare SHH signaling activity in the developing neocortex of humans and mice, we performed RNAseq with RNAs from the medial or lateral neocortices of E14 mouse embryos and compared Gli1 levels in our data with GLI1 levels in human RNAseq data obtained from BrainSpan (brainspan.org), which also used RNA from macrodissected neocortical tissues. To compare Gli1 and GLI1 levels in the two species, we normalized their levels to the expression levels of RG markers, SOX2, NESTIN, and PAX6, which show similar expression patterns in human and mouse RGs35. Notably, GLI1 and Gli1 levels correlated positively with bRG numbers in each species. In the human fetal neocortex, GLI1 levels greatly increased between 9 and 12 postconception weeks (pcw), coincident with bRG expansion, and remained high through mid-gestation, when the oSVZ is the main neurogenic area9 (Fig. 6a and Supplementary Fig. 8a). In mice, the Gli1 level was higher in the lateral cortex than in the medial cortex, consistent with there being more bRGs in the lateral cortex than in the medial cortex12 (top two graphs in the right panel of Fig. 6a). Notably, the relative GLI1 levels in human fetuses were higher than were the relative Gli1 levels in mouse embryos (Fig. 6a and Supplementary Fig. 8b). Another RNAseq dataset generated from sorted RGs also showed higher GLI1 levels in human aRGs26 (Supplementary Fig. 8c). Consistent with the RNAseq data and in contrast to the mouse embryonic neocortex, in situ hybridization detected GLI1 expression in the human neocortical VZ (Fig. 6b) as well as in the ganglionic eminence (Supplementary Fig. 7b). The transcriptome and in situ data suggest that there is potent SHH signaling in the human fetal neocortex.
Figure. 6. Strong SHH signaling in human aRGs.
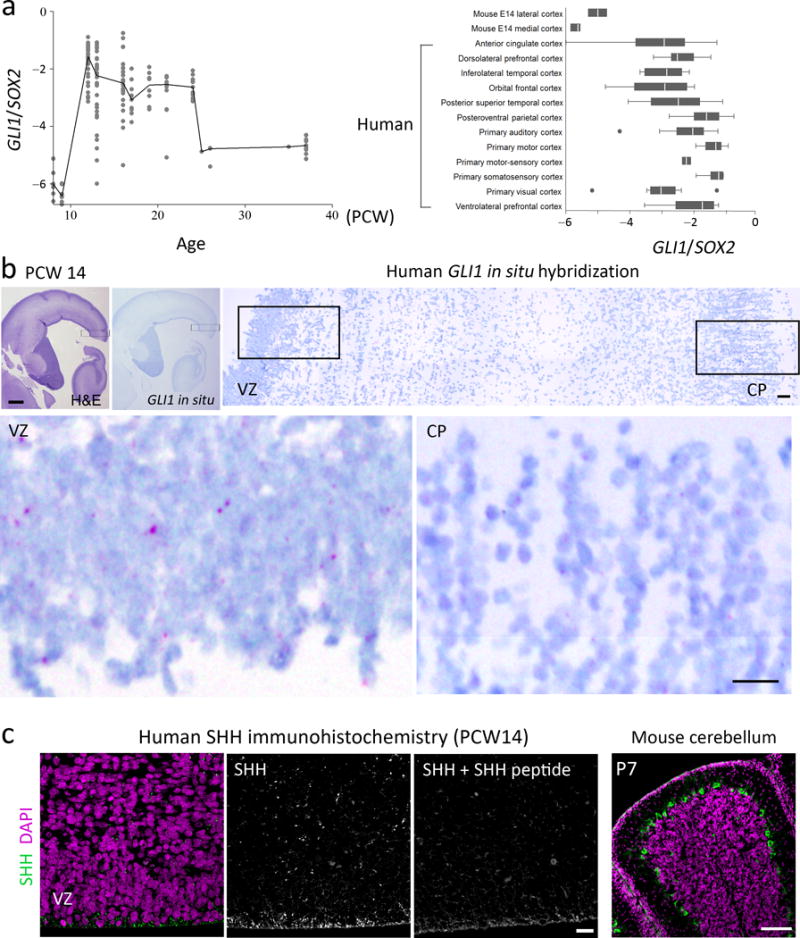
a. GLI1 expression during human fetal development (left) and a comparison of GLI1 and Gli1 expression levels in developing human and mouse cortices (right). b. H&E staining and in situ hybridization for GLI1 (purple dots) in the human fetal brain at 14 pcw. The boxed areas are enlarged below. Scale bars = 500 μm and 50 μm (enlarged images). Images represent results from 3 independent tissue samples. c. Human fetal brain at 14 pcw stained with anti-SHH antibody (green) and DAPI (purple). Co-incubation with SHH peptide blocks the strong SHH staining at the ventricular surface. This anti-SHH antibody specifically stained Purkinje cells in the mouse cerebellum, which express Shh. Scale bars = 200 μm for the rightmost figure in panel (c) and 20 μm for the others. Each figure represents at least 3 repeats on two different tissue samples.
Next, we investigated the source of SHH in human fetal brain. We detected SHH mRNA and SHH protein in the hypothalamic VZ (Supplementary Fig. 9a), which could secrete SHH into the ventricle. Consistently, SHH proteins were highly enriched at the ventricular surface of the neocortex (Fig. 6c). In mice, the loss of Smo but not of Shh in the cortex decreases cortical progenitor proliferation at E15.519, suggesting that Shh secreted from outside the cortex regulates cortical progenitor proliferation. Consistently, Shh is present in the embryonic cerebrospinal fluid and transventricularly delivered Shh regulates neural progenitor proliferation in the cerebellar VZ37.
SMO inhibitor decreases bRGs in human cerebral organoids
To investigate whether SHH signaling regulates the human bRG population, we employed a cerebral organoid model that recapitulates features of the developing human brain38. As reported, this model develops well-defined VZ- and SVZ-like structures and bRG-like cells (Supplementary Fig. 10a). To investigate whether SHH signaling affects the human aRG division angles, we treated organoids with either SANT1 (a SMO antagonist) or SAG (a SMO agonist). In contrast to mouse aRGs (Fig. 3g) but similar to human aRGs in slice culture27, more than half of the aRGs divided obliquely or vertically in the controls (Fig. 7a). Remarkably, SANT1 decreased the incidence of oblique and vertical division to 26%, similar to that in mouse aRGs, and subsequently decreased the number of PAX6+ TBR2− bRG-like cells outside the VZ, whereas neither effect was seen with SAG (Fig. 7a,b). This differential efficacy of SANT1 and SAG may reflect high levels of endogenous SHH signaling in the organoids. To test this, we examined the localization of SMO, which concentrates inside primary cilia upon activation39. Consistent with our hypothesis, SMO localized to 51% of the primary cilia in control aRGs, and SAG did not increase ciliary SMO localization, whereas SANT1 decreased it to 19% (Fig. 7c). Thus, human cerebral organoids exhibited intrinsic SHH signaling, and blocking this decreased oblique and vertical aRG divisions and the number of bRG-like cells. To test if blocking SHH signaling would affect the production of upper-layer neurons in organoids, we labeled a cohort of cells produced during SANT1 or DMSO (control) treatment with CldU. The proportion of CldU+ SATB2+ neurons in SATB2+ neurons was significantly decreased by SANT1 treatment (Supplementary Fig. 10b). Thus, blocking SHH signaling decreased the number of bRG and the production of SATB2+ upper-layer neurons in organoids.
Figure. 7. SHH signaling promotes bRG production in human cerebral organoids.
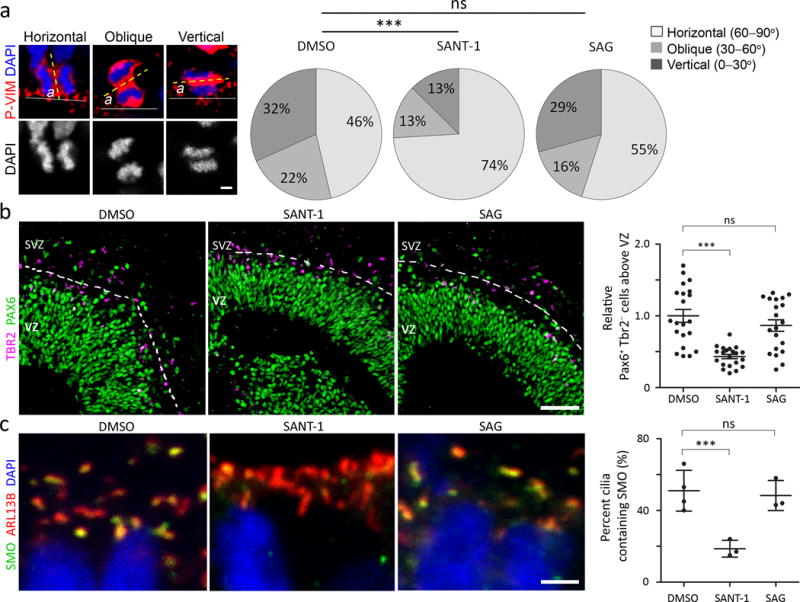
a. Examples of ‘horizontal’ (60° < α ≤ 90°), ‘oblique’ (30° < α ≤ 60°), and ‘vertical’ (0° ≤ α ≤ 30°) divisions of aRGs in cerebral organoids stained with phospho-vimentin (P-VIM, red) and DAPI (blue) and quantification of the division angles. We blindly analyzed 123 (DMSO), 133 (SAG), and 135 (SANT-1) cells from 15 organoids per group from 3 independent experiments. Mann Whitney test, DMSO vs. SANT-1, P = 0.0007, Sum of ranks = 14080, 19590, Mann-Whitney U = 6330; DMSO vs. SAG, P = 0.2681, Sum of ranks = 15336, 17817, Mann-Whitney U = 7586. Scale bar = 5 μm b. Human cerebral organoids labeled for PAX6 (green) and TBR2 (purple) and quantification of bRGs (PAX6+ TBR2− cells separated from dense PAX6+ cells by stretches of TBR2+ cells that are indicated by dotted lines). We analyzed 21 (DMSO), 20 (SAG), and 21 (SANT-1) VZ/SVZ structures from 11 (DMSO), 13 (SAG), and 15 (SANT-1) organoids from 3 independent experiments. Mann Whitney test, DMSO vs. SANT-1, P = 0.0000, Sum of ranks = 627.5, 275.5, Mann-Whitney U = 44.50; DMSO vs. SAG, P = 0.2565, Sum of ranks = 485, 376, Mann-Whitney U = 166.0. Scale bar = 50 μm. c. Apical surfaces of aRGs in organoids labeled for a cilia marker (ARL13B, red), SMO (green), and DAPI (blue) and quantification of cilia containing SMO. Scale bar = 2 μm. We examined 294 (DMSO), 292 (SANT-1), and 400 (SAG) cilia from 4 (DMSO), 3 (SANT-1), 3 (SAG) organoids from two independent experiments, of which 145 (DMSO), 53 (SANT-1), and 190 (SAG) cilia contained SMO. Two sided Fisher’s exact test, P = 0.0001 (DMSO vs. SANT-1), P = 0.6454 (DMSO vs. SAG). ns, P > 0.05; *P < 0.01; ***P < 0.001. Error bars represent standard deviation.
A recent study identified genes enriched in human bRGs40. Of those, we tested whether antibodies against PTPRZ1, TNC, ITGB5, or phospho-STAT3 can mark bRGs in human cerebral organoids and SmoM2 mutants and found that those did not preferentially mark bRGs in human cerebral organoids and SmoM2 mutants (data not shown).
Discussion
The prevalence of neocortical folding in many mammalian lineages, the presence of bRGs in all the mammalian species examined, and the selective expansion of bRGs in species with large and folded brains suggest that the mechanisms to induce bRG expansion and neocortical folding were present in a common ancestor of mammals and were subsequently selectively fortified or inactivated, giving rise to folded or smooth brains11. Our data suggest that Shh signaling has been central to the mechanisms underlying the evolutionary expansion of bRGs and neocortical growth and folding. Shh signaling promoted the expansion of mouse and human bRGs. Its activity was strong in human fetal cortex but not in mouse embryonic cortex, and elevating Shh signaling was sufficient to expand bRGs and induce neocortical folding in mice. Importantly, elevating Shh signaling expanded the IPCs as well as the bRGs. The expansion of both cell types probably contributes to the neocortical folding in SmoM2 mutants. The expansion of either progenitor in isolation is insufficient to induce folding21,41, whereas knockdown of Trnp1 or overexpression of ARHGAP11B induce folding after expanding both IPCs and bRGs24,26.
Shh signaling expanded bRGs and IPCs through distinct mechanisms. Shh signaling promoted the initial generation and self-renewal of bRGs but did not increase their proliferation rate. In contrast, Shh signaling decreased the generation of IPCs from aRGs but increased their proliferation and the proportion remaining in the cell cycle. Thus, Shh signaling offset the decreased generation of IPCs by promoting their proliferative divisions, leading to their great expansion in the SVZ. Similar to bRG expansion, the proliferative IPC division is characteristic of primates9,22; in rodents, IPCs mainly divide to form two neurons42–44. Thus, elevated Shh signaling elicited two developmental characteristics of large and folded brains, bRG expansion and proliferative IPC division, which are proposed to be necessary and sufficient for the evolution of an expanded and folded neocortex45.
The loss of Shh signaling in Smo mutants decreased the numbers of bRGs and IPCs. Previously, we showed that the loss of primary cilia resulted in no apparent defects in cortical development46. This seeming discrepancy is probably due to the fact that the loss of cilia results in the loss of both Gli activators and repressors, whereas the loss of Smo causes the loss of Gli activators and an increase in Gli repressors, leading to severer phenotypes than those resulting from the loss of cilia47–49. In the developing mouse neocortex, where Shh signaling activity is relatively weak, the loss of Gli activators in cilia mutants may be compensated for by the concomitant loss of Gli repressors. In contrast, the loss of Smo will result in the loss of activators and an increase in repressors, resulting in a pronounced decrease in signaling activity and in defective corticogenesis. We found strong SHH signaling activity in the human fetal cortex; thus, the loss of cilia and the resulting loss of Gli activators may strongly affect human corticogenesis. Notably, many of the genes mutated in congenital microcephaly are essential for the formation and function of centrioles and their associated structures including cilia50. Our findings suggest that defective SHH signaling contributes to congenital microcephaly.
In SmoM2 mutants, the folding occurred in the cingulate cortex medial to the area that showed the greatest increase in bRGs (the dorsomedial corner of the cortex). Similarly, Trnp1 knockdown or ARHGAP11B overexpression accomplished by in utero electroporation induced folding mostly in the cortex medial to the electroporated area that showed an increased number of bRGs24,26. It is unclear why the folding occurred preferentially in parts medial to the area with an increased number of bRGs. Neurogenesis proceeds from the lateral to medial cortex in mice. This lateral-to-medial gradient of formation and differentiation of neuronal layers may make those parts medial to the area that is interrupted by the increase in bRGs and neurons more amenable than lateral parts to structural changes. The physical constraint by the corpus callosum to limit lateral expansion of increased neurons may also have contributed to folding in the cingulate cortex in SmoM2 mutants.
The conserved genes including Ccnd1, Cdk4, Trnp1, PDGFD, and Pax6, and the human-specific genes, including ARHGAP11B, may have contributed to the evolutionary expansion and folding of the mammalian neocortex21,24–26,41. Shh signaling probably interacts with these and other unidentified genes to generate the complex and large mammalian neocortex. The SmoM2 mutants, being the first transgenic mouse model that expands both bRGs and IPCs and consistently develops folding in a defined cortical area, will be important in deciphering these interactions and the mechanisms underlying neocortical development and evolution.
METHODS
Mice
We used the following mouse strains: SmoM2flox (The Jackson Laboratory, JAX stock # 005130), Smoflox (JAX stock # 004526), tdTomatoflox (JAX stock # 007909), GFAP::Cre (JAX stock # 004600), Nestin::Cre (JAX stock # 003771), Nestin::CreERT2 (ref. 51), GFAP::CreER (ref. 52), and Kif3aflox (ref. 53). All mice were maintained in a mixed genetic background. To induce Cre-mediated recombination, we used females (2–6 month) carrying the flox alleles and males (2–9 month) carrying the Cre alleles for breeding. Timed pregnant CD-1 females were purchased from the Charles River Laboratories. All the mice were maintained at a 12/12 dark/light cycle and housed with maximum 5 mice of the same sex per cage. We used both sexes for experiments. All animal procedures were approved by the Institutional Animal Care and Use Committee of St Jude Children’s Research Hospital.
Human fetal brain samples
Formalin-fixed human fetal brain samples were obtained from the National Institutes of Health NeuroBioBank. The specimens were embedded in paraffin and sectioned coronally at a thickness of 5 μm. The St. Jude Children’s Research Hospital Institutional Review Board approved the use of fixed human fetal brain samples.
Thymidine analogues and tamoxifen injection
Solutions of 10 mg/mL bromodeoxyuridine (BrdU) (Sigma B-5002) and 2.5 mg/mL 5-ethynyl-2′-deoxyuridine (EdU) (Invitrogen A10044) were prepared in sterile 0.9% NaCl solution. BrdU (50 μg/g) and EdU (10 μg/g) were injected intraperitoneally into the dam mice at 24 h and 1.5 h, respectively, prior to euthanasia. Tamoxifen (Sigma T-5648) was dissolved in corn oil (Sigma C-8267) to a concentration of 20 mg/mL by rotation at 65°C for 1 h. Timed pregnant mice were injected intraperitoneally with tamoxifen at 1.5 mg/40 g at E12.5. For sparse labeling in the tdTomato reporter line, a dose of 0.5 mg/40 g was used.
Histologic processes
Embryonic brains were dissected out, fixed overnight in 4% paraformaldehyde (PFA) in PBS, cryoprotected for 24 h in 30% sucrose in PBS at 4°C, and embedded in OCT (Sakura Finetek). Tissue blocks were cryosectioned at a thickness of 12 μm and the sections were transferred to glass slides. Postnatal pups were perfused with 4% PFA. Their brains were dissected out, fixed overnight in 4% PFA at 4°C, embedded in paraffin, and sectioned at a thickness of 5 to 7 μm.
Immunohistochemical staining was performed as described previously48. EdU staining was performed using the Click-iT EdU Alexa Fluor® 488 kit in accordance with the manufacturer’s instructions. We used the following primary antibodies: anti-Arl13b (Proteintech 17711-1-AP, diluted 1:100), anti-Ascl1 (BD Pharmingen, 1:250; antigen retrieval in 10 uM Citric Acid Buffer ph 6.0 with steam for 10 minutes), anti-BrdU (Abcam ab1893, 1:500), anti-Ctip2 (Abcam ab18465, 1:500), anti-Dlx2 (ref. 54, a gift from Dr. J. L. Rubenstein, UCSF, 1:1000), anti-Itgb5 (Abcam ab15459, 1:50), anti-GLAST/EEA1 (Abcam ab416, 1:400), anti-Pax6 (Covance PRB-278P, 1:500; Development Studies Hybridoma Bank AB_528427, 1:200), anti-phospho-histone H3 Ser10 (Abcam 14955, 1:1,000), anti-phospho-Stat3 (Cell signaling #9145, 1:100), anti-phospho-Vimentin Ser55 (MBL International D076-3, 1:2,000), anti-PTPRZ1 (Sigma HPA015103, 1:250), anti-RC2 (Millipore MAB5740, 1:500), anti-Satb2 (Millipore ABE600, 1:2,000), anti-Shh (Abcam ab73958, 1:100), anti-Smo (Santa Cruz Biotechnology sc166685, 1:50), anti-Sox2 (Y-17) (Santa Cruz Biotechnology sc17320, 1:300), anti-Tbr1 (Millipore AB2261, 1:500), anti-Tbr2 (eBioscience 14-4875-82, 1:250; Abcam ab23345 1:250), anti-Tnc (Abcam ab108930, 1:500), and anti-TuJ1 (Covance MMS435P, 1:1,000). To block anti-SHH staining, we used recombinant human SHH N-terminus (R&D 1845-SH-025) at 20-fold molar excess of anti-SHH antibody. The following secondary antibodies were used at 1:400 dilutions and incubated for 2 h at room temperature or overnight at 4°C: Alexa Fluor® 488-conjugated donkey anti-rat IgG (Invitrogen, A-21208), goat anti-chicken IgY(Invitrogen, A-11039), goat anti-mouse IgG/IgM (Invitrogen, A-10684), donkey anti-mouse IgG (Invitrogen, A-21202); Alexa Fluor® 568-conjugated donkey anti-rabbit IgG (Invitrogen, A-10042), donkey anti-mouse IgG (Invitrogen, A-10037) and 647-conjugated donkey anti-goat IgG (Invitrogen, A-21447), donkey anti-rat IgG (Abcam, ab150155), Alexa Fluor® 405-conjugated donkey anti-sheep antibody (Abcam, ab175676), and DyLight™ 405-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch, 715-475-150). Coverslips were mounted on slides with Aqua-Poly/Mount (Polysciences).
Nissl staining was performed with cresyl violet (FD Neurotechnologies) according to the manufacturer’s instructions. Before Nissl staining, cryosections were dried overnight at room temperature then defatted in 50% chloroform in ethanol for 24 h.
In situ hybridization (ISH) was performed using an RNAscope® VS Assay (Advanced Cell Diagnostics) with a GLI1 probe (RNAscope VS Probe – Hs-GLI1; Advanced Cell Diagnostics, catalog no. 310996) and an SHH probe (RNAscope VS Probe – Hs-SHH; Advanced Cell Diagnostics, catalog no. 600956) in accordance with the manufacturer’s instructions.
Human cerebral organoids
Cerebral organoids were developed using the protocol of Lancaster et al.38 with minor modifications. Briefly, H9 human embryonic stem cells (hESCs) were obtained from WiCell Research Institute and maintained in mTeSR™1 medium (Stemcell Technologies) according to the supplier’s instructions. To generate embryoid bodies (EBs), hESCs were suspended in low–basic fibroblast growth factor (bFGF) hESC medium consisting of DMEM/F12 (Life Technologies, 11330-032) supplemented with 10% knockout serum replacement (KOSR) (Life Technologies, 10828-028), 3% ES-quality FBS (Life Technologies, 10439-016), 1% GlutaMAX (Life Technologies, 35050-061), 1% MEM-NEAA (Life Technologies, 11140-050), 7 ppm (v/v) β-mercaptoethanol (Life Technologies, 21985-023), 4 ng/mL bFGF (Peprotech, 100-18B), and 50 μM Rho-associated kinase inhibitor (ATCC, ACS-3030) and seeded at 9,000 cells/150 μL in each well of a 96-well Lipidure®-Coat Plate (Gel Company, LCV96). The medium was changed every other day for 6 to 7 days, omitting the bFGF and ROCK inhibitor after day 4. When the EBs attained a diameter of approximately 500 μm, they were transferred to wells of a Costar® 24-well plate (1 EB/well) (Corning 3473). The EBs were fed every other day with neural induction medium consisting of DMEM/F12 supplemented with 1% N2 supplement (Life Technologies 17502-048), 1% GlutaMAX, 1% MEM-NEAA, and 1 μg/mL heparin for 4 to 5 days until neuroepithelial morphology became evident. The neuroepithelial aggregates were then embedded in a drop of Matrigel (Corning 356234). The embedded aggregates (n = 16) were grown in 6-mm dishes containing 5 mL of differentiation medium (50% DMEM/F12, 50% Neurobasal Medium, 0.5% N2 supplement, 1% B27 without vitamin A [Life Technologies, 12587-010], 0.025% [v/v] human insulin [Sigma, I9278], 3.5 ppm [v/v] β-mercaptoethanol, 1% GlutaMAX, 0.5% MEM-NEAA, and 1% penicillin-streptomycin) with constant shaking at 75 rpm for 4 days, with the medium being changed on the second day. Four days after differentiation, the tissue droplets were fed with differentiation medium containing B27 Supplement with vitamin A (Life Technologies 17504-044) and incubated for 4.5 weeks at 37°C in 5% CO2 with constant rotation at 75 rpm, with the medium being replenished every 3 days.
To investigate the effects of SHH signaling on bRG generation, cerebral organoids that had been differentiated for 5 weeks (6–10 organoids/well) were treated with 400 nM SAG, 400 nM SANT-1, or DMSO in differentiation medium with vitamin A for 3 days, with the medium being replenished once 24 h after the initial treatment.
To investigate the effect of SANT1 on upper-layer neuron production, organoids were treated with SANT1 (400 nM) or DMSO for 10 days from 29 days to 39 days after differentiation. To label a cohort of neurons produced during treatment, we treated organoids with CldU (3 μg/mL) for 48 h from 35 days to 37 days after differentiation. The organoids were fixed at 64 days after differentiation. We changed the medium every other day; thus, the organoids were treated with fresh SANT1 and DMSO every other day. The organoids were fixed in 4% formaldehyde for 15 min at 4°C, embedded in 7.5 % gelatin, and cryosectioned at a thickness of 20 μm.
Microscopy and data analysis
Images were acquired on a Zeiss780 microscope equipped with a Zen module. To quantify the cell number or density, Z-stacks of 4 to 5 optical sections at a step-size of 1 μm and a tiling of 4 to 6 images acquired using 20× objectives were combined for analysis. We used at least 3 mutant and 3 control brains for each group. Quantification and analysis were carried out using ImageJ, the image-processing module of Zen (Zeiss), and Photoshop (Adobe).
Determination of VZ, SVZ, aRGs, IPCs, bRGs: For all VZ/SVZ analyses, we stained sections for Pax6 (or Sox2) and Tbr2 to demarcate the VZ and the SVZ. We defined the VZ as the area lining the ventricle and containing dense Pax6+ (or Sox2+) Tbr2− nuclei up to an area where cells uniformly express Tbr2. We defined the SVZ as the second cell-dense area containing uniformly Tbr2+ cells above the VZ and below a cell-sparse area. We defined the IZ as the cell-sparse area between the SVZ and the CP, the cell-dense area lying beneath the pial surface. We defined aRGs as Pax6+ (or Sox2+) Tbr2− cells in the VZ and IPCs as cells expressing Tbr2 irrespective of Pax6 or Sox2 expression. We defined bRGs as Pax6+ (or Sox2+) Tbr2− cells present above the VZ. Only cells that were clearly separated from the zone of Pax6+ cells by dense Tbr2+ cells were considered to be bRGs.
Human organoid analyses: In organoids, we defined the VZ as the area lining the lumen and containing dense Pax6+ Tbr2− cells up to an area where the cells were sparser than in the VZ and formed stretches of Tbr2+ cells. In organoids, there were far fewer Tbr2+ cells than in mouse brain, but they formed thin but distinct bands above the VZ to form the SVZ. To quantify the bRG cells, we adopted similar criteria to those used for the mouse brain, i.e., Pax6+ Tbr2− cells above the VZ. A thin but continuous line of Tbr2+ cells formed the boundary between the VZ and SVZ. We observed little variation in the thickness of the VZ and SVZ between organoids in the same batch of organoids, though the sizes of the lumen varied substantially. For VZ/SVZ analyses, we only imaged the VZ/SVZs that were (1) not obliquely cut, (2) close to the edges of the organoids to ensure adequate drug access, (3) well separated from the neighboring VZ/SVZ, and (4) medial parts of the VZ/SVZ structures in serial sections.
Quantification of spindle orientations: To examine the division angles of vRGs, the cells were stained for P-Vim/PH3/DAPI and examined under 40× or 63× oil objectives. Only cells displaying clear mitotic figures and clear separation of the chromosomes by cytoplasm as revealed by P-Vim staining were included in the analysis. The angles of division were measured using ImageJ.
In utero intraventricular injection and clonal analysis
The plasmid vectors pBABE-GFP (originally a gift from William Hahn [Addgene plasmid # 10668]) and pBABE-SMOM2 were gifts from M.E. Hatley at St. Jude Children’s Research hospital. We inserted SMOM2 into EcoR1 and Sal1 sites of the pBABE GFP vector to make a pBABE SMOM2 GFP vector, where LTR drives SMOM2 expression and SV40 promoter/enhancer drives GFP expression. Replication-incompetent retroviruses were produced from 293T packaging cell lines. For in utero intraventricular injection, timed pregnant CD1 females (Charles River Laboratories) of gestation stage E13 were used. By using a Nanoject II Auto-Nanoliter injector (Drummond Scientific), approximately 0.5–0.7 μL of retroviral solution premixed with 1% Fast Green dye was delivered into the lateral ventricles of the embryonic brains via a glass pipette that was pre-beveled on a microgrinder (EG 401, NARISHIGE Groups) and had an opening of 2 to 3 μm. Glass pipette needles used for microinjection were pulled from glass capillaries (3.5″ Drummond 3-000-203-G/X, Drummond Scientific) by a P30 micropipette puller (Sutter instrument company). Dams were harvested 48 h (E15) or 72 h (E16) post injection. Coronal sections of 50 μm were collected and stained for GFP, Pax6, and Tbr2. Clones that were clearly separated from each other and were entirely included in the 50-μm thickness were imaged on the confocal microscope using Z-stacks at step-sizes of 1 μm. The GFP+ Pax6+ Tbr2−, GFP+ Tbr2+ (Pax6+ or Pax6−), and GFP+ Pax6− Tbr2− cells were defined as RGs, IPCs, and neurons, respectively. bRGs were determined as GFP+ Pax6+ Tbr2− cells located above the VZ, or those cells whose processes clearly had no AP surface attachment. Clones with at least 2 cells were used for analyses. The clones were classified as follows: IPC/N clones that contained IPCs and/or neurons but not RGs; aRG+IPC/N clones with one aRG and IPC/N; 2aRG+IPC/N clones with two aRGs with or without IPC/N; and bRG clones with at least one bRG and other cell types.
Magnetic resonance imaging (MRI)
To measure the volume of the neocortex, MRI was performed using a 7-T ClinScan animal MRI scanner (Bruker BioSpin MRI GmbH) equipped with a BGA-12S gradient insert and a 2-channel phased-array surface coil. Animals were anesthetized with isoflurane (1–2 % in O2) for the duration of the data acquisition. Turbo spin-echo protocols (TR/TE = 1900–2500/39–42 ms) were used to acquire T2-weighted images (sagittal, transverse, and coronal) with a matrix of 320 × 320, a field of view of 25 × 25 mm, and a slice thickness of 0.5 mm.
RNA isolation and sequencing
Total RNA was extracted from dissected medial or lateral cortices of E14.5 embryonic brains by using the RNeasy Micro Kit (Qiagen). A sequencing library was prepared using the TruSeq Stranded Total RNA Kit (Illumina) and sequenced with an Illumina HiSeq system. FASTQ sequences were mapped to the mm9 genome and counted with HTSEQ55 and the gene-level fragments per kilobase of transcript per million (FPKM) values were then computed. Exon junction data were extracted through the in-house RNApeg pipeline (M. Edmonson et al., unpublished data). Statistical analyses and visualizations were performed using Partek Genomics Suite 6.6 and Stata MP/11.2 software.
Cross-species RNAseq comparisons
We used RNAseq data from human fetal neocortices (8–37 pcw), obtained from BrainSpan (http://brainspan.org), to plot GLI1 expression levels over developmental periods. We used RNAseq data from early mid-trimester human fetuses at 12–19 pcw to compare the expression levels of GLI1 in humans and Gli1 in E14 mouse embryonic cortices. As an independent validation of the relative expression levels of GLI1 and Gli1, we also compared them by using RNAseq data from sorted RGs from the neocortices of human fetuses and mouse embryos (GSE65000) (ref. 26). Given the diverse sources, species, and methods of measurement, only the internally controlled relative expression could reasonably be compared. The FPKM data were log-start transformed [log2 FPKM = log(FPKM +2)/log(2)]. This variance-stabilizing transformation minimizes the effect of extremely low numbers in ratios arising from low signal strength or low read count. The resulting log ratios of GLI1 to the established RG marker genes PAX6, NES, and SOX2 were plotted by category with Stata MP\11.2.
qRT-PCR
Total RNA extracted from dissected medial cortices of E14.5 embryonic brains was reverse transcribed by Superscript III reverse transcriptase (Invitrogen). Quantitative PCR was performed on an Applied Biosystems 7900 quantitative PCR instrument using SYBR green. Transcript levels were normalized to the expression levels of Gapdh. Primer sequences are as follows:
Ascl1 F: CATCTCCCCCAACTACTCCA; Ascl1 R: CCAGCAGCTCTTGTTCCTCT;
Dlx2 F: AGCTACGACCTGGGCTACAC; Dlx2 R: TGGCTTCCCGTTCACTATTC;
Gapdh F: CGTCCCGTAGACAAAATGGT; Gapdh R: GAATTTGCCGTGAGTGGAGT.
Statistics
Statistical analysis was performed using the GraphPad Prism software. All the statistical analyses were two tailed. Data analyzed by parametric statistical methods (unpaired t-test) were pre-tested for normality by Kolmogorov–Smirnov (K-S) test and for equal variance by F-test. For normal distribution data with unequal variance, an unpaired t-test with Welch’s correction was used. Non-parametric analysis including Mann-Whitney test, Chi-square test, and Fisher’s exact test were also used. P-values of less than 0.05 were considered significant. No statistical methods were used to predetermine the sample size, but our sample sizes are similar to those generally employed in the field. The mice were not randomized. The investigators were not blinded, except in the analyses of the division angles of mouse and human vRGs. No animal or data points were excluded from analysis.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank S. Baker at St. Jude Children’s Research Hospital for the GFAP::CreER and Nestin::CreER mice; L. S. Goldstein at the University of California San Diego for the Kif3afl/fl mice; M.E. Hatley at St. Jude Children’s Research hospital for the pBABE-GFP (originally a gift from William Hahn [Addgene plasmid # 10668]) and pBABE-SmoM2 vectors; J. L. Rubenstein and S. Pleasure at the University of California San Francisco for the Dlx2 antibody and the protocol for Ascl1 immunostaining, respectively; D. Finkelstein and J. Peng at St. Jude Children’s Research Hospital for help with the RNAseq analyses and hES cell culture, respectively. Human tissue was obtained from the NIH NeuroBioBank Brain at the University of Maryland, Baltimore, MD. We thank the staff of the Cell and Tissue Imaging Center, the Small Animal Imaging Center, the Hartwell Center for Bioinformatics and Biotechnology, and the Veterinary Pathology Core at St Jude Children’s Research Hospital for technical assistance. We thank S. Baker, X. Cao, M. Dyer, and K.A. Laycock for comments on the manuscript. Y.-G.H. is supported by NIH/NCI Cancer Center Core Support grant CA021765 (SJCRH), the Sontag Foundation Distinguished Scientist Award, Whitehall Foundation Research Grant, and ALSAC.
Footnotes
AUTHOR CONTRIBUTIONS
L.W. and Y.-G.H. designed and performed the experiments and wrote the manuscript. S.H. performed the in utero retroviral injections. Y.-G.H. conceived and supervised the study.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Lui JH, Hansen DV, Kriegstein AR. Development and Evolution of the Human Neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 3.Borrell V, Götz M. Role of radial glial cells in cerebral cortex folding. Current Opinion in Neurobiology. 2014;27:39–46. doi: 10.1016/j.conb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Sun T, Hevner RF. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci. 2014;15:217–232. doi: 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dehay C, Kennedy H, Kosik KS. The Outer Subventricular Zone and Primate-Specific Cortical Complexification. Neuron. 2015;85:683–694. doi: 10.1016/j.neuron.2014.12.060. [DOI] [PubMed] [Google Scholar]
- 6.Smart IHM, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cerebral cortex (New York, NY: 1991) 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zecevic N, Chen Y, Filipovic R. Contributions of cortical subventricular zone to the development of the human cerebral cortex. J Comp Neurol. 2005;491:109–122. doi: 10.1002/cne.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukaszewicz A, et al. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron. 2005;47:353–364. doi: 10.1016/j.neuron.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen DV, Lui JH, Parker PRL, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 10.Fietz SA, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nature Neuroscience. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 11.Reillo I, De Juan Romero C, García-Cabezas MÁ, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cerebral Cortex. 2011;21:1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Tsai J-W, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nature Neuroscience. 2011;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. Journal of Neuroscience. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García-Moreno F, Vasistha NA, Trevia N, Bourne JA, Molnár Z. Compartmentalization of cerebral cortical germinal zones in a lissencephalic primate and gyrencephalic rodent. Cerebral Cortex. 2012;22:482–492. doi: 10.1093/cercor/bhr312. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-Cerdeño V, et al. Comparative Analysis of the Subventricular Zone in Rat, Ferret and Macaque: Evidence for an Outer Subventricular Zone in Rodents. PLoS ONE. 2012;7:e30178. doi: 10.1371/journal.pone.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stashinko EE, et al. A retrospective survey of perinatal risk factors of 104 living children with holoprosencephaly. Am J Med Genet. 2004;128A:114–119. doi: 10.1002/ajmg.a.30070. [DOI] [PubMed] [Google Scholar]
- 17.Heussler HS, Suri M, Young ID, Muenke M. Extreme variability of expression of a Sonic Hedgehog mutation: attention difficulties and holoprosencephaly. Arch Dis Child. 2002;86:293–296. doi: 10.1136/adc.86.4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derwińska K, et al. PTCH1 duplication in a family with microcephaly and mild developmental delay. Eur J Hum Genet. 2009;17:267–271. doi: 10.1038/ejhg.2008.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komada M, et al. Hedgehog signaling is involved in development of the neocortex. Development. 2008;135:2717–2727. doi: 10.1242/dev.015891. [DOI] [PubMed] [Google Scholar]
- 20.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. Journal of Neuroscience. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nonaka-Kinoshita M, et al. Regulation of cerebral cortex size and folding by expansion of basal progenitors. The EMBO Journal. 2013;32:1817–1828. doi: 10.1038/emboj.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betizeau M, et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 23.Gertz CC, Lui JH, LaMonica BE, Wang X, Kriegstein AR. Diverse Behaviors of Outer Radial Glia in Developing Ferret and Human Cortex. Journal of Neuroscience. 2014;34:2559–2570. doi: 10.1523/JNEUROSCI.2645-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stahl R, et al. Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell. 2013;153:535–549. doi: 10.1016/j.cell.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 25.Lui JH, et al. Radial glia require PDGFD-PDGFRβ signalling in human but not mouse neocortex. Nature. 2014;515:264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Florio M, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 27.LaMonica BE, Lui JH, Hansen DV, Kriegstein AR. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nature Communications. 2013;4:1665. doi: 10.1038/ncomms2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gal JS, et al. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. Journal of Neuroscience. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilz GA, et al. Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nature Communications. 2013;4:2125. doi: 10.1038/ncomms3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Ge G, Uchida Y, Luu B, Ahn S. Gli3 Is Required for Maintenance and Fate Specification of Cortical Progenitors. Journal of Neuroscience. 2011;31:6440–6448. doi: 10.1523/JNEUROSCI.4892-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dave RK, et al. Sonic Hedgehog and Notch Signaling Can Cooperate to Regulate Neurogenic Divisions of Neocortical Progenitors. PLoS ONE. 2011;6:e14680. doi: 10.1371/journal.pone.0014680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yabut OR, Fernández G, Huynh T, Yoon K, Pleasure SJ. Suppressor of Fused Is Critical for Maintenance of Neuronal Progenitor Identity during Corticogenesis. CellReports. 2015;12:2021–2034. doi: 10.1016/j.celrep.2015.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shikata Y, et al. Ptch1-mediated dosage-dependent action of Shh signaling regulates neural progenitor development at late gestational stages. Developmental Biology. 2011;349:147–159. doi: 10.1016/j.ydbio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Yu W, Wang Y, McDonnell K, Stephen D, Bai CB. Patterning of ventral telencephalon requires positive function of Gli transcription factors. Developmental Biology. 2009;334:264–275. doi: 10.1016/j.ydbio.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 35.Johnson MB, et al. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nature Neuroscience. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Juan Romero C, Bruder C, Tomasello U, Sanz-Anquela JM, Borrell V. Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly. The EMBO Journal. 2015;34:1859–1874. doi: 10.15252/embj.201591176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang X, et al. Transventricular delivery of Sonic hedgehog is essential to cerebellar ventricular zone development. Proceedings of the National Academy of Sciences. 2010;107:8422–8427. doi: 10.1073/pnas.0911838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature Cell Biology. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 40.Pollen AA, et al. Molecular Identity of Human Outer Radial Glia during Cortical Development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong FK, et al. Sustained Pax6 Expression Generates Primate-like Basal Radial Glia in Developing Mouse Neocortex. Plos Biol. 2015;13:e1002217. doi: 10.1371/journal.pbio.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature Neuroscience. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 43.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyata T, et al. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 45.Lewitus E, Kelava I, Kalinka AT, Tomancak P, Huttner WB. An adaptive threshold in mammalian neocortical evolution. Plos Biol. 2014;12:e1002000. doi: 10.1371/journal.pbio.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong CK, et al. Primary cilia are required in a unique subpopulation of neural progenitors. Proceedings of the National Academy of Sciences. 2014;111:12438–12443. doi: 10.1073/pnas.1321425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han Y-G, Alvarez-Buylla A. Role of primary cilia in brain development and cancer. Current Opinion in Neurobiology. 2010;20:58–67. doi: 10.1016/j.conb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han Y-G, et al. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nature Neuroscience. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 49.Spassky N, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Developmental Biology. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu WF, Chahrour MH, Walsh CA. The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genom Human Genet. 2014;15:195–213. doi: 10.1146/annurev-genom-090413-025600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu G, et al. Pten deletion causes mTorc1-dependent ectopic neuroblast differentiation without causing uniform migration defects. Development. 2012;139:3422–3431. doi: 10.1242/dev.083154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chow LML, Zhang J, Baker SJ. Inducible Cre recombinase activity in mouse mature astrocytes and adult neural precursor cells. Transgenic Res. 2008;17:919–928. doi: 10.1007/s11248-008-9185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marszalek JR, et al. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 54.McKinsey GL, et al. Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron. 2013;77:83–98. doi: 10.1016/j.neuron.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.


