Abstract
Objective
Plasma kynurenine/tryptophan (KT) ratio, a biomarker of indoleamine 2,3-dioxygenase-1 (IDO) activity, is a strong independent predictor of mortality in HIV-infected Ugandans initiating antiretroviral therapy (ART) and may play a key role in HIV pathogenesis. We performed a genome-wide study to identify potential host genetic determinants of KT ratio in HIV-infected ART-suppressed Ugandans.
Design/Methods
We performed genome-wide and exome array genotyping and measured plasma KT ratio during the initial 6-12 months of suppressive ART in Ugandans. We evaluated >16 million SNPs in association with log10KT ratio using linear mixed models adjusted for cohort, gender, pregnancy, and ancestry.
Results
Among 597 Ugandans, 62% were female, median age was 35, median baseline CD4+ count was 135 cells/mm3, and median baseline HIV-1 RNA was 5.1 log10copies/mL. Several polymorphisms in candidate genes TNF, IFNGR1, and TLR4 were associated with log10KT ratio (P<5.0×10−5). An intergenic polymorphism between CSPG5 and ELP6 was genome-wide significant, while several others exhibited suggestive associations (P<5.0×10−7), including genes encoding protein tyrosine phosphatases (PTPRM, PTPRN2) and the vitamin D metabolism gene, CYP24A1. Several of these SNPs were associated with markers of inflammation, coagulation, and monocyte activation, but did not replicate in a small U.S. cohort (N=262; 33% African-American).
Conclusions
Our findings highlight a potentially important role of IFN-γ, TNF−α, and toll-like receptor signaling in determining IDO activity and subsequent mortality risk in HIV-infected ART-suppressed Ugandans. These results also identify potential novel pathways involved in IDO immunoregulation. Further studies are needed to confirm these findings in treated HIV-infected populations.
Keywords: HIV, kynurenine, tryptophan, antiretroviral therapy, genome-wide association study
INTRODUCTION
Despite effective antiretroviral therapy (ART), HIV-infected individuals experience higher rates of mortality and aging-associated diseases than HIV-uninfected controls [1, 2]. Markers of immune activation predict clinical progression of disease, but even with ART suppression, immune activation levels fail to normalize [3, 4]. To date, the specific biologic pathways driving persistent immune activation and its relationship to mortality remain unclear.
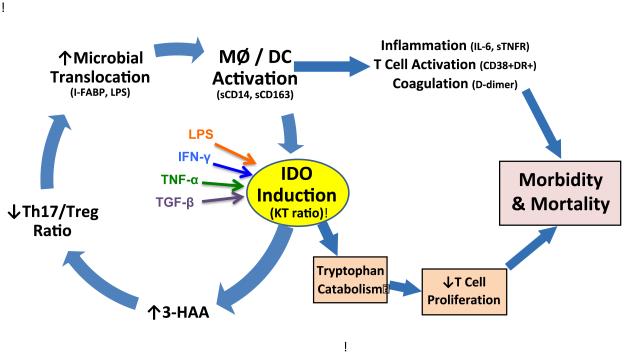
One proposed mechanism is the indoleamine 2,3-dioxygenase (IDO)-induced tryptophan catabolism pathway, which can be quantified as the ratio of kynurenine to tryptophan (KT) in plasma. IDO plays a critical role in autoimmune disorders [5], cancer [5], and fetal tolerance during pregnancy [5]. IDO is a key immunoregulatory enzyme that converts tryptophan to kynurenine [6]. Bacterial molecules such as lipopolysaccharide (LPS) and pro-inflammatory cytokines such as IFN-γ, TNF-α, and TGF-β promote increased IDO activity (Figure 1) [7, 8]. Several IDO-induced catabolites have been shown to decrease T cell proliferation and function [9], and may compromise mucosal integrity, thereby increasing bacterial translocation and systemic immune activation [10]. These downstream effects may contribute to increased morbidity and mortality even during ART suppression [11].
Figure 1.
Schematic illustrating the potential contribution of IDO to ongoing immune dysfunction and increased risk of morbidity and mortality during treated HIV Infection. Abbreviations: IDO, indoleamine 2;3-dioxygenase; KT, kynurenine/tryptophan; 3-HAA, 3-hydroxyanthranilic acid; MΦ, macrophage; DC, dendritic cell; i-FABP, intestinal fatty acid binding protein; LPS, lipopolysaccharide; IFN-γ, interferon gamma; TNF-α , tumor necrosis factor alpha; TGF-β, transforming growth factor beta; sCD14, soluble CD14; sCD163, soluble CD163; IL-6, interleukin-6; sTNFR, soluble tumor necrosis factor receptor.
Asymptomatic untreated HIV-infected participants have higher KT ratios than HIV-uninfected controls, while HIV-infected participants with progression to AIDS have the highest KT ratios [12]. Though IDO activity declines with ART, IDO has also been associated with a significant increase in mortality risk in Ugandans, both during untreated and treated HIV disease [11]. In a subsequent analysis of multiple immune markers in this same Ugandan cohort, KT ratio was the strongest independent risk factor for mortality, even after adjusting for other markers of inflammation (IL-6), monocyte activation (sCD14), and CD4+ or CD8+ T cell activation (%CD38+HLA-DR+) [13]. KT ratio has also been shown to predict mortality in ART-suppressed North Americans [3, 4].
We performed a genome-wide association study (GWAS) of KT ratio in HIV-infected Ugandans initiating ART. The goal of the study was to identify potential targets for intervention – perhaps in genes other than IDO itself - that may naturally (and presumably safely) influence IDO immunoregulation. We performed both a candidate gene (based on known factors that influence the IDO pathway) and genome-wide analysis. We found that single nucleotide polymorphisms (SNPs) in TNF, IFNGR1, and TLR4 and SNPs in two additional pathways – one involving protein tyrosine phosphatase signaling and another related to vitamin D metabolism – are strongly associated with IDO activity. Several of the SNPs identified from these analyses were also associated with markers of monocyte activation (sCD14, sCD163) and inflammation (IL-6, D-dimer), additional biomarkers previously associated with increased risk of mortality during treated HIV disease. These findings provide further support that variation at these loci may play an important role in influencing the IDO pathway, as well as other interrelated immunologic pathways.
MATERIALS AND METHODS
Study participants
Participants were sampled from the UARTO and ARKS cohorts in Uganda. UARTO includes HIV-infected individuals initiating first ART at a rural clinic in Mbarara. ARKS includes HIV-infected participants with mild-to-moderate cutaneous Kaposi’s sarcoma (KS) at a specialty clinic in Kampala. The primary ART regimen included two nucleoside reverse transcriptases (e.g., zidovudine/lamivudine, stavudine/lamivudine, or tenofovir/emtricitabine) plus a non-nucleoside reverse transcriptase (e.g., nevirapine or efavirenz). Participants were included if they had sustained plasma HIV RNA <400 copies/mL (the lower limit of detection of an earlier Amplicor assay [Roche]; the same cut-off was applied to a more recent Cobas Taqman assay [Roche] to maintain consistency), KT ratio at month 6 and/or 12 of ART, and genotype data. ARKS participants were excluded for active untreated opportunistic infections or mucocutaneous KS requiring chemotherapy. All participants provided written informed consent. This research was approved by the institutional review boards at each site and the University of California, San Francisco.
Phenotype and covariate data
Demographic and clinical data were collected at screening and follow-up visits approximately every three months. Data included self-reported pregnancy status and symptom reporting, as well as fasting morning laboratory measures (e.g., plasma HIV-1 RNA, CD4+ T cell counts, and archival of peripheral blood mononuclear cells) as described previously [11]. Plasma KT ratio was measured in cryopreserved plasma using LC-MS/MS [14] at pre-ART and approximately 6 and 12 months of ART. Mortality and follow-up were tracked via a limited vital status ascertainment program as described previously [15]. Cause of death was classified only in broad categories (e.g., illness, trauma, suicide, or related to childbirth). For this study, deaths due to trauma, injury, or suicide were excluded. Self-reported diagnoses of tuberculosis (TB) and/or TB medication history were adjudicated by reviewing clinical records and laboratory smear/culture results [16]. These data were only available for UARTO since TB diagnosis was an exclusion criterion for the ARKS study [16].
Genotyping and quality control
Genome-wide genotyping was performed at the RIKEN Center for Genomic Medicine using the Human Omni Express Bead Chip (Illumina, San Diego, California), including over 700,000 SNPs. Additional whole exome genotyping of over 200,000 coding putative functional SNPs was performed at the University of California, San Francisco using the Axiom Whole Exome Array Chip (Affymetrix, Santa Clara, California). A total of 928 participant samples were genotyped with the exome array and of these, 642 were also genotyped using a genome-wide array (Supplementary Figure 1). After excluding samples due to ineligibility (lack of viral suppression), inadequate genotyping quality (<95% call rate), genetic relatedness (IBD>0.125), and heterozygosity (±2 SD), 788 samples passed quality control for exome and/or genome-wide array data. Among these 788 samples with quality-controlled genetic data, 597 also had existing KT ratio data (at month 6 and/or 12 of ART). We generated principal components using Eigensoft to include as covariates for ancestry in linear mixed effects models (http://genetics.med.harvard.edu/reich/Reich_Lab/Software.html) [17].
Genome-wide SNP imputation
Quality-controlled genotyping data were used to impute additional genotypes using IMPUTE2 (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html). We performed pre-phasing to estimate haplotypes prior to imputation, using SHAPEIT (https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html). Only imputed SNPs with imputation quality score ≥0.80 were included in the final dataset.
Association Analyses
We fit linear mixed effects regression models to test for associations between genotypes (coded as allele dosages) and log10KT ratios during month 6 and 12 of ART suppression. We restricted all analyses to ART-suppressed timepoints since SNPs associated with plasma KT ratio prior to sustained ART suppression would mostly reflect genetic predictors of HIV viral load setpoint or CD4+ T cell count in these chronically HIV-infected participants initiating therapy. We also did not include HIV RNA levels or CD4+ T cell counts as covariates in the models as these factors could potentially be intermediates or colliders in a causal pathway between host genetics and plasma KT ratio. Only SNPs with minor allele frequencies (MAF)>1% were included in the analyses. The final model included covariates for gender, pregnancy, and cohort (to account for KS diagnosis[16] and other unmeasured cohort-specific differences), as well as the first ten principal components to control for population stratification.
We performed candidate gene analyses on 17,307 SNPs in the following genes (including 100 kilobases flanking regions): IDO1, IDO2, IFNG, IFNGR1, IFNGR2, TNF, TNFRSF1A, TGFB1, TGFBR1, TGFBR2, TGFBR3, IFNA1, IFNAR1, IFNB1, TLR4, TLR7, TLR8, and TLR9. For the candidate gene analyses, we used a Bonferroni-adjusted significance threshold of P<5×10−5, based on the number of SNPs tested. For the genome-wide analysis, we considered suggestive (P<5.0×10−7) as well as genome-wide significant (P<5.0×10−8) SNPs [18]. This is especially important in light of our limited sample size, and allows for further investigation of potentially biologically relevant SNPs. SNPs exhibiting suggestive or significant associations with KT ratios were then tested for association with additional biomarkers (e.g., IL-6, D-dimer, etc.) using linear mixed regression.
Replication Analysis
We performed validation analyses among participants in the AIDS Clinical Trials Group (ACTG) studies NWCS 279 and NWCS 329 who had both GWAS [19] and KT ratio data at one year of ART suppression [4]. Linear regressions were adjusted for ancestry, batch (for KT ratio measurements), case-control status (occurrence of non-AIDS defining events) [4], and gender.
RESULTS
Among 597 participants, 43% in ARKS and 70% in UARTO were female, the median age was 35 years, median baseline CD4+ T cell count was 135 cells/mm3, and median HIV RNA was 5.1 log10copies/mL. At month 12 of ART, the median CD4+ T cell count was 266 cells/mm3, and all participants had HIV RNA ≤2.6 log10copies/mL (Table 1). Median KT ratios in UARTO and ARKS were similar (63 versus 66 nM/μM, P=0.93). Variability (R2) in KT ratio at month 6 was 3.7% and at month 12 was 3.0%, and there was a statistically significant decline in plasma KT ratio from month 6 to 12 of ART (69 to 60 nM/μM, P<0.0001) overall, as well as within individuals (P<0.0001), consistent with previously published data [11].
Table 1.
Characteristics of HIV-infected individuals initiating first ART in the ARKS and
| Characteristics | ARKSa
(N =177) |
UARTO (N = 420) |
|---|---|---|
| Age | 39 (29-40) | 35 (29-40) |
| Female | 76 (43%) | 292 (70%) |
| Baseline CD4+ count (cells/mm3) | 116 (25-250) | 137 (80-206) |
| Proximal CD4+ count (cells/mm3)b | 273 (171-465) | 257 (175-257) |
| Baseline HIV RNA (log10copies/mL) | 5.2 (4.9 – 5.5) | 5.1 (4.1 – 5.5) |
| Proximal HIV RNA (log10copies/mL) | ≤ 2.6 | ≤ 2.6 |
| Plasma KT ratio (nM/μM)c | 63 (50 – 93) | 66 (50 – 86) |
Abbreviations: ART, antiretroviral therapy; ARKS, Anti-Retrovirals for Kaposi’s Sarcoma; UARTO, Uganda AIDS Rural Treatment Outcomes cohort; KT, kynurenine/tryptophan ratio.
Median and interquartile range shown except for gender (number and percent).
“Proximal” refers to the most recent CD4+ T cell count or HIV RNA relative to the most recent KT ratio measurement.
Plasma KT ratio during month 6-12 of ART.
By principal components analyses using 1000 Genomes reference data, our Ugandan population was most genetically related to Luhyans in Kenya (Supplementary Figure 2). Q-Q plots of the discovery genome-wide association results gave an inflation factor of λ=1.04, indicating that the results are not confounded by genetic ancestry (Supplementary Figure 3).
Candidate gene SNPs and plasma KT ratio
Several SNPs in the candidate genes TNF, IFNGR1, and TLR4 were associated with KT ratio at the Bonferroni-adjusted significance level of P<5.0×10−5, conferring between a 1.11 to 1.71-fold increase in KT ratio per increase in minor allele copy (Table 2). These included SNPs in the 5’ and 3’ regions of TNF, a SNP in IFNGR1, and a SNP in TLR4. SNPs strongly associated with KT ratio included rs17200810 (3’ of TNF), which localizes to a region predicted to bind to several transcription factors, and rs276565 (5’ of IFNGR1) and rs270148 (intron 2 of TLR4), which are in active H3K27Ac histone mark regulatory regions (Supplementary Figure 4). The SNP in the IFNGR1 region also lies 100 kb upstream of IL20RA and 43 kb downstream of IL22RA2.
Table 2.
Association of candidate gene polymorphisms with KT ratioa.
| SNPb | Exp(Beta)c | 95% CI | P | Location | MAF | Gene |
|---|---|---|---|---|---|---|
| rs17200810d | 1.65 | (1.35, 2.02) | 1.41×10−6 | 3’ | 0.02 | TNF |
| rs34451538 | 1.50 | (1.26, 1.79) | 3.90×10−6 | 5’ | 0.03 | TNF |
| rs276565e | 1.11 | (1.06, 1.16) | 5.30×10−6 | 5’ | 0.28 | IFNGR1f |
| rs114064880 | 1.71 | (1.36, 2.16) | 5.33×10−6 | 5’ | 0.01 | TNF |
| rs11575838 | 1.68 | (1.34, 2.11) | 7.13×10−6 | 3’ | 0.01 | TNF |
| rs2770148g | 1.15 | (1.08, 1.23) | 2.94×10−5 | Intron 2 | 0.12 | TLR4 |
Abbreviations: KT, kynurenine/tryptophan ratio; SNP, single nucleotide polymorphism; CI, confidence interval; MAF, minor allele frequency.
Linear mixed effects regression models of log10plasma KT ratio were adjusted for gender, pregnancy, cohort (ARKS vs. UARTO), and the first ten principal components.
Genotyped or imputed single nucleotide polymorphism.
Fold change in KT ratio (nM/μM) per copy of minor allele.
Genotyped SNP, multiple transcription factors predicted to bind to this loci (e.g. IRF4, JUND; RUNX).
Genotyped SNP, in active H3K27Ac mark regulatory region.
This SNP also lies 100 kb upstream of IL20RA and 43 kb downstream of IL22RA2.
In linkage disequilibrium (LD) with genotyped SNP, rs1413088.
Genome-wide SNPs and plasma KT ratio
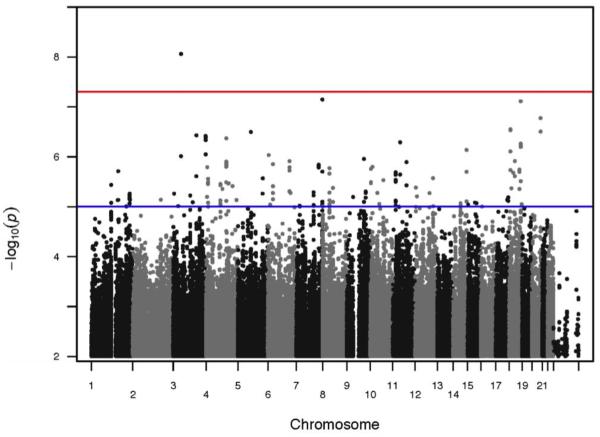
Suggestive and significant SNPs from the genome-wide analysis were estimated to increase KT ratio by 1.20 to 1.61-fold per increase in minor allele copy (Table 3). An intergenic SNP (rs56185965, 5’ of CSPG5 and 3’ of ELP6) was significantly associated with KT ratio at the genome-wide level (P<5.0×10−8), while several SNPs exhibited suggestive associations (Figure 2). Among the latter, two SNPs (rs6950107 and rs75257475) were in genes encoding two distinct protein tyrosine phosphatases; these SNPs localize to predicted transcription factor-binding sites for proteins involved in activator protein 1 (AP-1) signaling. A third SNP (rs13041834) was 67 kb upstream of CYP24A1, a gene encoding a key enzyme involved in vitamin D metabolism, and lies in an H3K27Ac histone mark-enriched region (Table 3, Supplementary Figure 5). This SNP also lies 16 kb downstream of BCAS1 (Breast Carcinoma Amplified Sequence 1), a gene that is overexpressed in breast cancer cell lines [20], and 18 kb downstream of MIR4756, a small non-coding RNA gene, or microRNA, of unknown clinical significance [21].
Table 3.
Association of polymorphisms with plasma KT ratio from genome-wide analysisa.
| SNPb | Exp(Beta)c | 95% CI | P | Location | MAF | Nearest Gene(s) |
|---|---|---|---|---|---|---|
| rs56185965 | 1.55 | (1.33, 1.79) | 8.73×10−9 | intergenic | 0.02 | CSPG5; ELP6 |
| rs6950107d | 1.44 | (1.26, 1.64) | 7.14×10−8 | intron 6 | 0.03 | PTPRN2 |
| rs17085469 | 1.20 | (1.13, 1.29) | 7.75×10−8 | 5’ | 0.10 | RP11-736G13.1 |
| rs13041834e | 1.17 | (1.10, 1.25) | 3.13×10−7 | intergenic | 0.12 | CYP24A1; BCAS1; MIR4756 |
| rs75257475f | 1.23 | (1.13, 1.32) | 2.95×10−7 | intron 1 | 0.07 | PTPRM |
| rs200564710 | 1.30 | (1.18, 1.44) | 3.20×10−7 | intergenic | 0.05 | HOMER1; PAPD4 |
| rs114598920 | 1.37 | (1.21, 1.54) | 3.73×10−7 | intron 1 | 0.03 | CLSTN2 |
| rs115718192 | 1.61 | (1.34, 1.93) | 4.13×10−7 | intergenic | 0.02 | TFRC; SDHAP1 |
| rs76101033 | 1.41 | (1.23, 1.61) | 4.29×10−7 | Intron 2 | 0.02 | RP11-6757F12.1 |
Abbreviations: KT, kynurenine/tryptophan ratio; SNP, single nucleotide polymorphism; CI, confidence interval; MAF, minor allele frequency.
Linear mixed effects regression models of log10plasma KT ratio were adjusted for gender, pregnancy, cohort (ARKS vs. UARTO), and the first ten principal components.
Genotyped or imputed single nucleotide polymorphism.
Fold change in KT ratio (nM/μM) per copy of minor allele.
SNP predicted to decrease binding to SMAD3.
SNP in active H3K27Ac mark region, in linkage disequilbrium (LD) with genotyped SNP, rs6064045.
SNP predicted to alter binding to FOSL2, in LD with 11 other SNPs with P<1×10−7, including genotyped SNP rs1695225.
Figure 2.
Manhattan plot for association analyses. Genome-wide association test results are shown as −log10transformed p-values for the linear mixed effects regression analyses of plasma log10KT ratio. Chromosomal location of SNPs is indicated on the x-axis. SNPs significantly (P<5.0×10−8) or suggestively (P<5.0×10−5) associated with plasma log10KT ratio are noted above the red or blue line, respectively.
Associations between selected SNPs and markers of inflammation and immune activation
To evaluate whether SNPs associated with KT ratio might be related to additional immunologic pathways that have been associated with increased morbidity/mortality during treated HIV disease [4, 11], we evaluated SNPs in relation to D-Dimer, IL-6, sCD163, sCD14, and CD4+ and CD8+ T cell activation levels during ART suppression (Supplementary Table 1). These biomarkers were only modestly correlated with each other (R=0.09 to 0.45), with the exception of CD4+ and CD8+ T cell activation (R=0.67) (Supplementary Figure 6). Several of the SNPs identified in the candidate gene analysis were associated with increased monocyte activation (sCD163 and sCD14) and increased IL-6 (Supplementary Table 1). SNPs identified in the genome-wide analysis were associated with sCD163, sCD14, D-Dimer, and IL-6 (Supplementary Table 2).
Associations between selected SNPs and mortality
There were too few deaths in the cohort (35 deaths over a median of 7 years of follow-up) to directly perform a genome-wide analysis of predictors of mortality [11]. Nonetheless, we were able to assess whether SNPs associated with KT ratio were also associated with mortality. We observed statistically significant associations with mortality and rs13041834 (adjusted HR=1.82, 95% CI=1.06, 3.13) and rs75257475 (adjusted HR=2.05, 95%CI=1.12, 3.76) (Supplemental Table 3, Supplementary Figure 7).
Sensitivity Analyses to Evaluate Potential Effect of Non-Genetic Factors During Early ART
We had limited data on non-genetic factors, such as co-infections. However, we were able to perform post-hoc analyses excluding 39 individuals with adjudicated TB diagnoses and found no significant change in the effect estimates - though exclusion of these participants reduced the sample size and significance of some associations (Supplementary Table 4). We also compared the associations between SNPs and KT ratios at month 6 and 12 of ART separately to evaluate potential non-genetic influences (i.e., inflammatory conditions) that might increase KT ratio during early, but not later, ART timepoints. Of the 597 participants, 586 had KT ratio measurements at month 6, and 538 had KT ratio measurements at month 12 of ART. The effect estimates were largely unchanged at month 12 compared to month 6 of ART (Supplementary Table 5).
Attempt to Replicate Findings in a Non-African Sample of Convenience
We lacked resources to be able to measure KT ratio in an additional African cohort. However, we had access to existing data from ACTG studies that included participants with genome-wide array [19] and KT ratio data (at one year of ART suppression) [4] (Supplemental Table 6). While this ACTG cohort was smaller than our Ugandan sample (262 versus 597 participants), only assessed KT ratio at a single timepoint (as opposed to two timepoints in our Ugandan study), and was comprised of 33% participants with African ancestry (Supplementary Table 6), we evaluated whether the SNPs associated with KT ratio in our Ugandan cohort might be replicated - but were unable to confirm these findings (Supplementary Tables 7 and 8).
DISCUSSION
We performed a genome-wide study to identify potential host genetic determinants of KT ratio in HIV-infected ART-suppressed Ugandans. A previous study in SIV-infected rhesus macaques of direct IDO and CTLA-4 inhibition led to severe autoimmune necrotizing pancreatitis in animals [22]. However, several phase I and II cancer treatment trials of direct IDO inhibition are currently underway [23]. The goal of the present study was to identify pathways that may modify IDO activity during treated HIV disease, potentially identifying targets for interventions other than IDO itself.
The specific candidate genes assessed in the study were selected by a priori knowledge of factors that modulate IDO in published in vitro or animal model studies. IDO is upregulated in dendritic cells and phagocytes by pro-inflammatory cytokines including IFN-α, IFN-γ, TNF-α, and TGF-β [8, 24]. Translocation of microbial products, including LPS, may stimulate toll-like receptors, which then induce IDO activity [25]. The relative importance of these regulators in vivo during treated HIV infection, however, remains unclear.
We found that SNPs in TNF, IFNGR1, and TLR4 were strongly associated with KT ratio. Several of these SNPs localize to potential functionally relevant regions – e.g., rs17200810 (TNF) lies in a region predicted to bind to several transcription factors involved in host immunoregulation, including the response to HIV (e.g., RELA [26], IRF4 [27], JUND [28]). Both rs276565 (IFNGR1) and rs270148 (TLR4) lie in active H3K27Ac histone mark regulatory regions, loci that exhibit increased enhancer activity [29]. Interestingly, rs276565 (IFNGR1) also lies near IL20RA, which encodes for the IL−20 receptor (IL−20RA) and IL22RA2, which encodes for the soluble form of the IL-22 receptor (IL-22BP) (Supplementary Figure 4B). Both IL-20 and IL-22 are members of the IL-20 subfamily of cytokines, which plays a critical role in enhancing the host innate immune response such as during viral infection or with intestinal epithelial homeostasis [30].
In the genome-wide analysis, we identified additional pathways that might plausibly be involved in IDO immunoregulation. PTPRN2 and PTPRM are both protein tyrosine phosphatases (PTPs), which, with protein tyrosine kinases, rapidly modulate signaling processes to maintain cell proliferation, differentiation, and gene transcription [31, 32]. Based on ENCODE data, rs6950107 (PTPRN2) is predicted to decrease binding to SMAD3 [33], and rs75257475 (PTPRM) lies in a region that binds FOLS2 [34], a transcription factor that interacts with SMAD3 [35]. SMAD3 is a key intracellular effector of TGF-β signaling [36], and via formation of the AP-1 transcription factor complex [37] and binding to Foxp3 [38], plays a critical role in the development, function, and survival of regulatory T cells.
A second potential pathway identified from the genome-wide analysis is the vitamin D pathway. CYP24A1 (rs13041834) encodes the key enzyme catalyzing the metabolism of active vitamin D. In mouse models, vitamin D inhibits T cell proliferation [39], suppresses Th17 production [40], and induces regulatory T cell expansion [41, 42] - immunologic effects strikingly similar to those of IDO. Mice administered vitamin D also generate tolerogenic mature dendritic cells with enhanced TGFB and IDO mRNA expression, and increased IL-10 production with regulatory T cell expansion [43]. In HIV-infected ART-suppressed patients, vitamin D supplementation is associated with decreased T cell activation [44]. Of note, this SNP also lies downstream of BCAS1 and MIR4756. However, the clinical significance of these genes and their association with IDO signaling is unknown.
We identified one genome-wide significant SNP, but to our knowledge, neither chondroitin sulfate proteoglycan a5 (CSPG5) nor elongator acetyltransferase complex subunit (ELP6) are biologically relevant molecules in IDO signaling. The SNP does not lie in a putative regulatory site, polymorphisms closer to these two genes were not associated with KT ratio, and this SNP was not associated with other markers of immune dysfunction.
Our data are in contrast to a recent GWAS, which did not find any polymorphisms significantly associated with markers of immune activation and microbial translocation (sCD14 and i-FABP levels) [45]. However, only untreated HIV-infected individuals were included in the study, potentially making it more difficult to overcome additional variation due to (1) the extent of disease progression and (2) genetic and non-genetic contributors to viral load setpoint (factors that might confound the relationship between host genetics and sCD14). Participants were also restricted to those with higher CD4+ T cell counts, possibly excluding those with the most microbial translocation. Our study minimizes potential confounding by plasma HIV RNA levels by only evaluating ART-suppressed individuals. By sampling two different timepoints per participant, we also minimize within-subject variability and increase our power to detect an association with host genetic factors.
Our study has several limitations that deserve mention. First, we had limited data on non-genetic factors, such as co-infections. We were able to adjudicate potential TB diagnoses [16] and found little change in effect estimates after excluding these individuals. We also performed post-hoc analyses of SNPs in relation to month 6 and 12 KT ratios separately. These analyses did not demonstrate any bias towards a stronger relationship at month 6, which might suggest potential confounding by co-infection or other inflammatory conditions. Second, we had too few deaths in this cohort to directly perform a genome-wide analysis of mortality. Due to the lack of a dedicated mortality tracking system in Uganda, we also did not have information on the exact cause of death, e.g., AIDS-related death. However, we did identify two SNPs that were significantly associated with mortality, suggesting that the genetic determinants of this pathway may be clinically meaningful. Third, though we included nearly 600 participants, based on a median of two observations per participant, we only had 80% power to detect SNPs that explain approximately 5% of the variance in plasma log10KT ratio (assuming a log-additive model, two-tailed Type I error rate of 5.0×10−8 and MAF ≥1%). This difference would be equivalent to a 2.0-fold difference in KT ratio, corresponding to an estimated 3.7-fold increase in mortality risk [11]. Thus, it is likely that we lacked sufficient power, particularly for less common SNPs, to detect effect sizes that might be most clinically relevant. Finally, we did not have the resources to be able to measure KT ratio in an African replication cohort. We instead analyzed existing ACTG data but were unable to confirm the associations with KT ratio observed in our Ugandan study. Based on the above estimates of power and the small number of participants with African ancestry in the replication cohort, however, we cannot definitively exclude the associations observed in our Ugandan study at this time.
In conclusion, we identified several polymorphisms associated with KT ratio during treated HIV disease. Several of these SNPs lie in biologically plausible regions that could modulate the IDO pathway; these include polymorphisms in genes related to pro-inflammatory cytokines such as IFN-γ, TNF-α, and TGF-β, as well as genes involved in toll-like receptor signaling and vitamin D metabolism. While the SNP associations identified herein are plausible, in a small, primarily Caucasian cohort from a resource-rich region, we were unable to confirm these findings. The immunologic pathways predicting mortality among HIV-infected Ugandans may be different than those described in resource-rich settings; immune recovery (i.e., CD4+ T cell count) after ART initiation is significantly slower in resource-limited compared to resource-rich regions [15]. There may be non-genetic differences between regions that could plausibly affect KT ratio such as viral clade (e.g., A and D in Uganda versus B in the U.S.) and prevalent co-infections. Further studies are needed to confirm the role of these SNPs in IDO-mediated gut epithelial immunity, especially among African populations.
Supplementary Material
Supplementary Figure 1. Sample and SNP quality control selection steps to generate the final genotype and phenotype dataset.
Supplementary Figure 2. Principal components analysis for the genotyped and 1000 Genomes reference samples. The first three principal components are plotted. Each circle represents a sample from the genotyped cohorts (UARTO or ARKS) or from the 1000 Genomes reference samples. Abbreviations: LWK, Luhyans from Kenya; MKK, Masai from Kenya; YRI, Yorubans from Nigeria.
Supplementary Figure 3. Q-Q plots from genome-wide association analyses. Observed and expected −log10 p-value relationships are plotted. The red line shows the expected distribution assuming no inflation of statistics. The gray shaded region represents the 95% confidence interval. The genomic inflation factor (λ) is indicated.
Supplementary Figure 4. Regional visualization of significant and suggestive SNPs identified in the candidate gene analysis. The genomic region surrounding each SNP is annotated on the x-axis, and the −log10 p-values from the association analysis are shown on the y-axis. Circles represent genotyped SNPs while squares represent imputed SNPs. The purple shape at the center of the figure is the highlighted SNP of interest; surrounding circles/squares represent nearby SNPs. The colors of the neighboring SNPs indicate the degree of linkage disequilibrium (LD) with the highlighted SNP (red is strongest and dark blue is weakest degree of LD with the highlighted SNP). The recombination rate across the region is also illustrated. Figures were produced using LocusZoom (http://locuszoom.sph.umich.edu/locuszoom) with the 1000 Genomes African population (November 2014).
Supplementary Figure 5. Regional visualization of significant and suggestive SNPs identified in the genome-wide analysis. The genomic region surrounding each SNP is annotated on the x-axis, and the −log10 p-values from the association analysis are shown on the y-axis. Circles represent genotyped SNPs while squares represent imputed SNPs. The purple shape at the center of the figure is the highlighted SNP of interest; surrounding circles/squares represent nearby SNPs. The colors of the neighboring SNPs indicate the degree of linkage disequilibrium (LD) with the highlighted SNP (red is strongest and dark blue is weakest degree of LD with the highlighted SNP). The recombination rate across the region is also illustrated. Figures were produced using LocusZoom (http://locuszoom.sph.umich.edu/locuszoom) with the 1000 Genomes African population (November 2014).
Supplementary Figure 6. Correlations between log10KT ratio and log-transformed biomarkers of coagulation, inflammation, monocyte activation, and T cell activation during ART suppression.
Supplementary Figure 7. Kaplan-Meier survival curves of suggestive SNPs identified from the genome-wide analysis of KT ratio. Strata by minor allele copy number and the sample size contributing to each stratum are shown.
ACKNOWLEDGEMENTS
The authors wish to acknowledge the participation of all the study participants who contributed to this work as well as the clinical research staff of the UARTO and ARKS cohorts who made this research possible. This work was supported in part by the National Institutes of Health (R56AI100765, R21AI078774, K24MH087227, T32AA007240, R01MH054907, P30AI27763, P01AI076174, and U01GM061390), the Doris Duke Charitable Foundation (PWH, Clinical Scientist Development Award 2008047), the Sullivan Family Foundation, HIV Translational Research Training Grant (T32AI060530) (SAL), a Center for AIDS Research HIV Mentored Scientist Award (SAL), Bristol-Myers Squibb Virology Fellowship Award (SAL), and the NIH Pharmacogenomics Research Network-RIKEN Center for Genomic Medicine Global Alliance. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. SAL, PWH, and DLK conceived and designed the study. JNM, PWH, and HB coordinated collection, management, and quality control processes for the Ugandan cohort clinical data and specimens. DWH, PJM, and PWH coordinated collection of the ACTG replication cohort data. SAL, DLK, TM, and MK, coordinated generation of the genotyping data. YH conducted the laboratory KT ratio measurements. PWH coordinated the acquisition of additional biomarker measurements. SAL analyzed the data. JAM and JSW provided key data management and analysis support. SAL wrote the report. All authors provided critical feedback in finalizing the report. The authors would also like to acknowledge significant contributions by David R. Bangsberg, Yusuke Nakamura, Tricia H. Burdo, Kenneth C. Williams, Russell P. Tracy, Yap Boum, Miriam Laker-Oketta, Leslie W. Chinn, and Janine E. Micheli. LWC and JEM coordinated DNA sample collection and extraction for genome-wide genotyping and assisted with initial genetic data collection. DRB secured funding for and coordinated the design and operations of the UARTO cohort and YB oversaw its operations locally. MLO oversaw the local operations of the ARKS sutdy. YN coordinated the collection of the genotyping data at the RIKEN center. THB, KCW, and RPT generated the additional biomarker data.
Footnotes
The authors do not have a commercial or other association that might pose a conflict of interest.
REFERENCES
- 1.Lohse N, Hansen AB, Pedersen G, Kronborg G, Gerstoft J, Sorensen HT, et al. Survival of persons with and without HIV infection in Denmark, 1995-2005. Annals of internal medicine. 2007;146:87–95. doi: 10.7326/0003-4819-146-2-200701160-00003. [DOI] [PubMed] [Google Scholar]
- 2.Hunt PW. HIV and aging: emerging research issues. Current opinion in HIV and AIDS. 2014;9:302–308. doi: 10.1097/COH.0000000000000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunt PW, Sinclair E, Rodriguez B, Shive C, Clagett B, Funderburg N, et al. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. The Journal of Infectious Diseases. 2014;210:1228–1238. doi: 10.1093/infdis/jiu238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenorio AR, Zheng Y, Bosch RJ, Krishnan S, Rodriguez B, Hunt PW, et al. Soluble markers of inflammation and coagulation but not T-cell activation predict non-AIDS-defining morbid events during suppressive antiretroviral treatment. The Journal of Infectious Diseases. 2014;210:1248–1259. doi: 10.1093/infdis/jiu254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 6.Werner ER, Fuchs D, Hausen A, Jaeger H, Reibnegger G, Werner-Felmayer G, et al. Tryptophan degradation in patients infected by human immunodeficiency virus. Biol Chem Hoppe Seyler. 1988;369:337–340. doi: 10.1515/bchm3.1988.369.1.337. [DOI] [PubMed] [Google Scholar]
- 7.Stone TW, Darlington LG. Endogenous kynurenines as targets for drug discovery and development. Nature reviews. Drug discovery. 2002;1:609–620. doi: 10.1038/nrd870. [DOI] [PubMed] [Google Scholar]
- 8.Pallotta MT, Orabona C, Volpi C, Vacca C, Belladonna ML, Bianchi R, et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat Immunol. 12:870–878. doi: 10.1038/ni.2077. [DOI] [PubMed] [Google Scholar]
- 9.Baban B, Chandler PR, Sharma MD, Pihkala J, Koni PA, Munn DH, et al. IDO activates regulatory T cells and blocks their conversion into Th17-like T cells. Journal of immunology. 2009;183:2475–2483. doi: 10.4049/jimmunol.0900986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Favre D, Mold J, Hunt PW, Kanwar B, Loke P, Seu L, et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med. 2010;2:32ra36. doi: 10.1126/scitranslmed.3000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byakwaga H, Boum Y, 2nd, Huang Y, Muzoora C, Kembabazi A, Weiser SD, et al. The Kynurenine Pathway of Tryptophan Catabolism, CD4+ T-Cell Recovery, and Mortality Among HIV-Infected Ugandans Initiating Antiretroviral Therapy. The Journal of Infectious Diseases. 2014 doi: 10.1093/infdis/jiu115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huengsberg M, Winer JB, Gompels M, Round R, Ross J, Shahmanesh M. Serum kynurenine-to-tryptophan ratio increases with progressive disease in HIV-infected patients. Clin Chem. 1998;44:858–862. [PubMed] [Google Scholar]
- 13.Lee SA, Byakwaga H, Boum Y, Burdo TH, Haberer JE, Tracy RP, et al. Immunologic Pathways That Predict Mortality in HIV+ Ugandans Initiating ART; In the Program and Abstracts from the 22nd Conference on Retroviruses and Opportunistic Infections; Seattle, WA. 2015. Abstract # 317; 2015. [Google Scholar]
- 14.Huang Y, Louie A, Yang Q, Massenkoff N, Xu C, Hunt PW, et al. A simple LC-MS/MS method for determination of kynurenine and tryptophan concentrations in human plasma from HIV-infected patients. Bioanalysis. 2013;5:1397–1407. doi: 10.4155/bio.13.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng EH, Odeny TA, Lyamuya RE, Nakiwogga-Muwanga A, Diero L, Bwana M, et al. Estimation of Mortality among HIV-infected people on antiretroviral therapy treatment in east Africa: a sampling based approach in an observational, multisite, cohort study. The lancet. HIV. 2015;2:e107–e116. doi: 10.1016/S2352-3018(15)00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byakwaga H, Hunt PW, Laker-Oketta M, Glidden DV, Huang Y, Bwana BM, et al. The Kynurenine Pathway of Tryptophan Catabolism and AIDS-Associated Kaposi Sarcoma in Africa. Journal of acquired immune deficiency syndromes. 2015;70:296–303. doi: 10.1097/QAI.0000000000000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 18.Moskvina V, Schmidt KM. On multiple-testing correction in genome-wide association studies. Genetic epidemiology. 2008;32:567–573. doi: 10.1002/gepi.20331. [DOI] [PubMed] [Google Scholar]
- 19.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beardsley DI, Kowbel D, Lataxes TA, Mannino JM, Xin H, Kim WJ, et al. Characterization of the novel amplified in breast cancer-1 (NABC1) gene product. Experimental cell research. 2003;290:402–413. doi: 10.1016/s0014-4827(03)00353-7. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic acids research. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boasso A, Vaccari M, Fuchs D, Hardy AW, Tsai WP, Tryniszewska E, et al. Combined effect of antiretroviral therapy and blockade of IDO in SIV-infected rhesus macaques. Journal of immunology. 2009;182:4313–4320. doi: 10.4049/jimmunol.0803314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vacchelli E, Aranda F, Eggermont A, Sautes-Fridman C, Tartour E, Kennedy EP, et al. Trial watch: IDO inhibitors in cancer therapy. Oncoimmunology. 2014;3:e957994. doi: 10.4161/21624011.2014.957994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 25.O'Connor JC, Lawson MA, Andre C, Moreau M, Lestage J, Castanon N, et al. Lipopolysaccharide-induced depressive-like behavior is mediated by indoleamine 2,3-dioxygenase activation in mice. Mol Psychiatry. 2009;14:511–522. doi: 10.1038/sj.mp.4002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coiras M, Lopez-Huertas MR, Rullas J, Mittelbrunn M, Alcami J. Basal shuttle of NF-kappaB/I kappaB alpha in resting T lymphocytes regulates HIV-1 LTR dependent expression. Retrovirology. 2007;4:56. doi: 10.1186/1742-4690-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carbone A, Gloghini A, Larocca LM, Capello D, Pierconti F, Canzonieri V, et al. Expression profile of MUM1/IRF4, BCL-6, and CD138/syndecan-1 defines novel histogenetic subsets of human immunodeficiency virus-related lymphomas. Blood. 2001;97:744–751. doi: 10.1182/blood.v97.3.744. [DOI] [PubMed] [Google Scholar]
- 28.Kagnoff MF, Roebuck KA. Human immunodeficiency virus type 1 (HIV-1) infection and expression in intestinal epithelial cells: role of protein kinase A and C pathways in HIV-1 transcription. The Journal of Infectious Diseases. 1999;179(Suppl 3):S444–447. doi: 10.1086/314801. [DOI] [PubMed] [Google Scholar]
- 29.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutz S, Wang X, Ouyang W. The IL-20 subfamily of cytokines--from host defence to tissue homeostasis. Nature reviews. Immunology. 2014;14:783–795. doi: 10.1038/nri3766. [DOI] [PubMed] [Google Scholar]
- 31.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nature reviews. Molecular cell biology. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 32.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 33.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic acids research. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis C, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Sun D, Wang Y, Ren F, Pang S, Wang D, et al. FOSL2 positively regulates TGF-beta1 signalling in non-small cell lung cancer. PLoS One. 2014;9:e112150. doi: 10.1371/journal.pone.0112150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Feng XH, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 38.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nature immunology. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 39.Rigby WF, Noelle RJ, Krause K, Fanger MW. The effects of 1,25-dihydroxyvitamin D3 on human T lymphocyte activation and proliferation: a cell cycle analysis. Journal of immunology. 1985;135:2279–2286. [PubMed] [Google Scholar]
- 40.Cantorna MT, Yu S, Bruce D. The paradoxical effects of vitamin D on type 1 mediated immunity. Molecular aspects of medicine. 2008;29:369–375. doi: 10.1016/j.mam.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniel C, Sartory NA, Zahn N, Radeke HH, Stein JM. Immune modulatory treatment of trinitrobenzene sulfonic acid colitis with calcitriol is associated with a change of a T helper (Th) 1/Th17 to a Th2 and regulatory T cell profile. The Journal of pharmacology and experimental therapeutics. 2008;324:23–33. doi: 10.1124/jpet.107.127209. [DOI] [PubMed] [Google Scholar]
- 42.Cantorna MT, McDaniel K, Bora S, Chen J, James J. Vitamin D, immune regulation, the microbiota, and inflammatory bowel disease. Experimental biology and medicine. 2014;239:1524–1530. doi: 10.1177/1535370214523890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira GB, Gysemans CA, Demengeot J, da Cunha JP, Vanherwegen AS, Overbergh L, et al. 1,25-Dihydroxyvitamin D3 promotes tolerogenic dendritic cells with functional migratory properties in NOD mice. Journal of immunology. 2014;192:4210–4220. doi: 10.4049/jimmunol.1302350. [DOI] [PubMed] [Google Scholar]
- 44.Fabre-Mersseman V, Tubiana R, Papagno L, Bayard C, Briceno O, Fastenackels S, et al. Vitamin D supplementation is associated with reduced immune activation levels in HIV-1-infected patients on suppressive antiretroviral therapy. AIDS. 2014;28:2677–2682. doi: 10.1097/QAD.0000000000000472. [DOI] [PubMed] [Google Scholar]
- 45.Perkins MR, Bartha I, Timmer JK, Liebner JC, Wollinsky D, Gunthard HF, et al. The Interplay Between Host Genetic Variation, Viral Replication, and Microbial Translocation in Untreated HIV-Infected Individuals. The Journal of Infectious Diseases. 2015;212:578–584. doi: 10.1093/infdis/jiv089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Sample and SNP quality control selection steps to generate the final genotype and phenotype dataset.
Supplementary Figure 2. Principal components analysis for the genotyped and 1000 Genomes reference samples. The first three principal components are plotted. Each circle represents a sample from the genotyped cohorts (UARTO or ARKS) or from the 1000 Genomes reference samples. Abbreviations: LWK, Luhyans from Kenya; MKK, Masai from Kenya; YRI, Yorubans from Nigeria.
Supplementary Figure 3. Q-Q plots from genome-wide association analyses. Observed and expected −log10 p-value relationships are plotted. The red line shows the expected distribution assuming no inflation of statistics. The gray shaded region represents the 95% confidence interval. The genomic inflation factor (λ) is indicated.
Supplementary Figure 4. Regional visualization of significant and suggestive SNPs identified in the candidate gene analysis. The genomic region surrounding each SNP is annotated on the x-axis, and the −log10 p-values from the association analysis are shown on the y-axis. Circles represent genotyped SNPs while squares represent imputed SNPs. The purple shape at the center of the figure is the highlighted SNP of interest; surrounding circles/squares represent nearby SNPs. The colors of the neighboring SNPs indicate the degree of linkage disequilibrium (LD) with the highlighted SNP (red is strongest and dark blue is weakest degree of LD with the highlighted SNP). The recombination rate across the region is also illustrated. Figures were produced using LocusZoom (http://locuszoom.sph.umich.edu/locuszoom) with the 1000 Genomes African population (November 2014).
Supplementary Figure 5. Regional visualization of significant and suggestive SNPs identified in the genome-wide analysis. The genomic region surrounding each SNP is annotated on the x-axis, and the −log10 p-values from the association analysis are shown on the y-axis. Circles represent genotyped SNPs while squares represent imputed SNPs. The purple shape at the center of the figure is the highlighted SNP of interest; surrounding circles/squares represent nearby SNPs. The colors of the neighboring SNPs indicate the degree of linkage disequilibrium (LD) with the highlighted SNP (red is strongest and dark blue is weakest degree of LD with the highlighted SNP). The recombination rate across the region is also illustrated. Figures were produced using LocusZoom (http://locuszoom.sph.umich.edu/locuszoom) with the 1000 Genomes African population (November 2014).
Supplementary Figure 6. Correlations between log10KT ratio and log-transformed biomarkers of coagulation, inflammation, monocyte activation, and T cell activation during ART suppression.
Supplementary Figure 7. Kaplan-Meier survival curves of suggestive SNPs identified from the genome-wide analysis of KT ratio. Strata by minor allele copy number and the sample size contributing to each stratum are shown.