Summary
When sulfur compounds are scarce or difficult to process, E. coli adapts by inducing the high-level expression of sulfur-compound importers. If cystine then becomes available, the cystine is rapidly overimported and reduced, leading to a burgeoning pool of intracellular cysteine. Most of the excess cysteine is exported, but some is adventitiously degraded, with the consequent release of sulfide. Sulfide is a potent ligand of copper and heme moieties, raising the prospect that it interferes with enzymes. We observed that when cystine was provided and sulfide levels rose, E. coli became strictly dependent upon cytochrome bd oxidase for continued respiration. Inspection revealed that low-micromolar levels of sulfide inhibited the proton-pumping cytochrome bo oxidase that is regarded as the primary respiratory oxidase. In the absence of the back-up cytochrome bd oxidase, growth failed. Exogenous sulfide elicited the same effect. The potency of sulfide was enhanced when oxygen concentrations were low. Natural oxic-anoxic interfaces are often sulfidic, including the intestinal environment where E. coli dwells. We propose that the sulfide resistance of the cytochrome bd oxidase is a key trait that permits respiration in such habitats.
Introduction
Contemporary organisms have inherited from the primordial world a metabolism that depends in many ways upon sulfur chemistry. The chemical properties of sulfur are far more varied than those of most biological elements. Sulfur is redox active at physiological potentials, it is a potent nucleophile even when exposed to bulk solution, it binds transition metals tightly, and it exhibits flexible coordination geometry. Universal aspects of metabolism exploit these qualities. For example, because organic sulfur can be oxidized from its sulfhydryl form to its sulfenic and disulfide forms, cysteine residues can sense and scavenge reactive oxygen species (Zheng et al., 1998; Poole, 2007) and provide stabilizing crosslinks to secreted proteins. The reversible formation of thioesters is central to energy conservation during glycolysis and pyruvate dissimilation. Sulfur ligands coordinate iron-sulfur clusters, adhere molybdenum to pterins, and organize zinc fingers. Particularities of sulfur bond angles tune the reactivities of thiamine and biotin and enable the group-transfer activity of S-adenosylmethionine.
Due to this reliance upon sulfur chemistry, all organisms must acquire substantial sulfur in order to grow: the total sulfur-atom concentration inside E. coli is ~0.13 M (calculated from Neidhardt and Umbarger, 1996). To meet this demand, microorganisms express a suite of transporters and pathways that allow them to convert a wide variety of inorganic and organic sulfur molecules to cysteine. The intracellular pool of cysteine then serves as the central reservoir of sulfur atoms: in addition to its direct incorporation into polypeptides, cysteine is a precursor of methionine and the direct donor of sulfur atoms for the synthesis of multiple cofactors. When environmental sources of sulfur dwindle, bacteria respond by strongly inducing the synthesis of each of its sulfur-compound importers, effectively turning the bacterium into a sulfur sponge (Kredich, 1996). In the model bacterium Escherichia coli, sulfur deficiency is sensed by the transcription factor CysB (Kredich, 1992).
E. coli dwells in aerobic and microaerobic habitats in which extracellular cysteine is chemically oxidized by oxygen. Thus cystine—the disulfide form of cysteine—is likely to be the most abundant form of organic sulfur. Indeed, the bacterium expresses two forms of cystine importer: TcyP, a high-flux importer driven by membrane potential, and TcyJLN, a lower-flux but higher-affinity system driven by ATP hydrolysis (Imlay et al., 2015). Both systems are strongly induced by CysB when sulfur levels decline. Together they enable E. coli to grow at full speed even when environmental cystine concentrations fall as low as five nanomolar.
However, the use of cystine as a sulfur source raises an unusual problem in transport control. Because disulfide bonds can be quickly transferred to protein thiols, cystine must be reduced as quickly as it enters the cell. Glutaredoxin systems do so very efficiently, and even during rapid cystine import only cysteine is detected inside the cell (Imlay et al., 2015). A consequence, however, is that cystine importers cannot be controlled by product inhibition. In contrast, most other transporters slow down when the accumulated product occupies their active sites. Indeed, when sulfur-poor E. coli cultures encounter even 5 micromolar cystine, the unfettered action of its induced TcyP causes the rate of cystine import to exceed the sulfur demand of the cell by as much as 50-fold (Imlay et al., 2015). Cysteine pools swell more than 10-fold. Excess cysteine is problematic, as it inhibits a key enzyme in isoleucine synthesis (Harris, 1981) and promotes Fenton chemistry (Park & Imlay, 2003). E. coli responds to this dilemma by exporting the excess cysteine (Imlay et al., 2015). The continuous cycle of cystine import, reduction, and export is energetically expensive, increasing the demand for ATP and NADPH by an estimated 10% and 100%, respectively. This futile cycle continues for an hour or longer, until the membrane content of TcyP declines through cell division. Salmonella and Lactobacilli also excrete excess cysteine in this situation, suggesting that this dilemma is widespread (Turner et al., 1999, Imlay et al., 2015). It seems odd that cystine importers did not evolve to be allosterically inhibited by the growing cysteine pool, but perhaps this option is precluded by the structural similarities between cysteine and serine.
In studying this process, we noticed that during the period of excessive cystine import, E. coli cultures emanate the odor of hydrogen sulfide. (The term sulfide is used here to denote both H2S and HS−, as the pKa of H2S is 6.9.) Like cysteine, sulfide is chemically reactive and potentially problematic. It exhibits a substantial affinity for heme and for soft metals, including those in proteins (Wu et al., 2015, Vitvitsky et al., 2015, Bolic et al., 2015, Nagy, 2015). Due to its tiny size, sulfide cannot easily be physically excluded from their active sites. Therefore, we anticipated that during periods of cystine uptake, endogenous sulfide might threaten the activities of heme or copper enzymes. How does E. coli cope with what must be a common event?
In this study we examined the impact of cystine influx, and of the consequent evolution of sulfide, upon cytochrome oxidases, the most critical heme and copper enzymes in this cell. We learned that when cysteine pools swell, some of the cysteine is adventitiously degraded to sulfide, and this sulfide is abundant enough to potently inhibit the energy-conserving cytochrome bo oxidase. This action does not block growth, however, because the respiratory flux shifts to the cytochrome bd oxidase, a copper-free enzyme. Although the latter enzyme does not pump protons, its action enables aerobic metabolism and growth to continue. A similar series of events is likely to occur when E. coli encounters exogenous sulfide, a common solute in oxic/anoxic interfaces. A key role of cytochrome bd oxidase may be to enable respiration in that situation.
Results
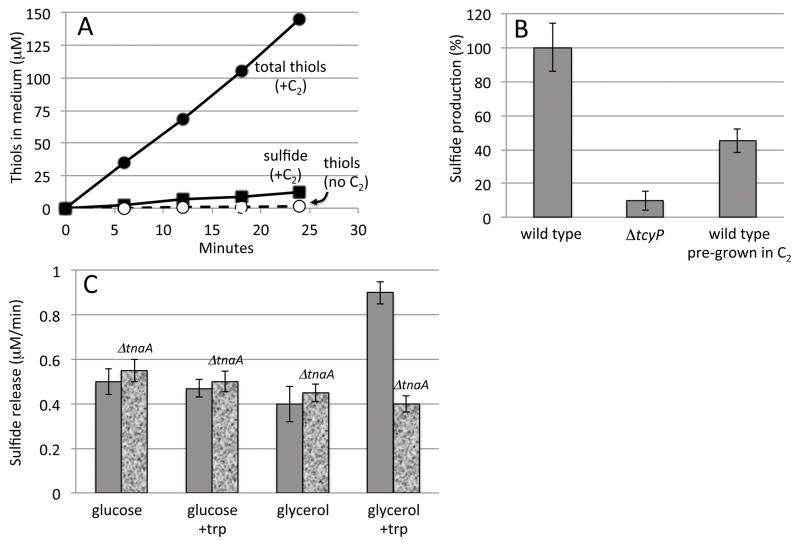
Sulfate is traditionally employed as the sulfur source in defined lab growth medium. It is not the sulfur source that E. coli prefers, however, as the CysB transcription factor is only fully deactivated when cystine is provided (Imlay et al., 2015). When cystine was added to wild-type cells growing in sulfate medium, thiol compounds were immediately and continuously excreted (Fig. 1A). Calculations (Experimental Procedures) demonstrate that the rate of thiol excretion was approximately 50 times the total sulfur demand of the cell.
Figure 1. Cystine stimulates rapid cysteine and sulfide excretion.
A. Total thiol excretion (circles) and hydrogen sulfide release (squares) from wild-type (MG1655) cells. Cells were cultured in glycerol medium with sulfate as the sole sulfur source. Solid symbols: 0.1 mM cystine (C2) was added at time zero. Open symbols: no addition. B. Rate of hydrogen sulfide release after cystine addition. Bar 3 represents sulfide release by cells that had been pre-cultured in medium containing 0.5 mM cystine. Time courses are representative of at least three independent experiments; error bars in all figures represent the standard deviation from three experiments. C. Wild-type and tryptophanase mutant (ΔtnaA) cells were cultured in glucose or glycerol medium, with 5 mM tryptophan supplements where indicated. After washing, sulfide production was measured immediately upon the addition of 0.5 mM cystine.
Previous mass spectroscopic analysis determined that the majority of excreted thiol compounds were cysteine (Imlay et al., 2015). However, direct assay for sulfide—which would have been lost from mass spectrometry samples—demonstrated that sulfide is also generated (Fig. 1). Sulfide was released from cells only after cystine was provided, and its release depended upon the presence of TcyP (Fig. 1B). TcyP is the predominant cystine importer; it exhibits an apparent KM for cystine of 2 μM (Imlay et al., 2015), so that low micromolar cystine is sufficient to cause the full rate of thiol excretion that was observed. In the experiments reported here, higher cystine concentrations were used only so that the external cystine was not depleted during the period of the experiments. Notably, cells that were grown continuously in cystine released substantially less cysteine and sulfide than did those to which cystine was abruptly added (Fig. 1B). Prior work showed that when cells grow in cystine-containing medium, CysB is deactivated, tcyP transcription falls to basal levels, and the rate of cystine import declines (Imlay et al., 2015). Therefore, cysteine and sulfide excretion is maximal in the generation or two that immediately follow an increase in cystine availability.
The release of sulfide from cysteine can be accidental
We hoped to identify the endogenous source of sulfide during cystine import. A substantial number of E. coli enzymes can exhibit cysteine desulfidase activity in vitro: 3-mercaptopyruvate:cyanide sulfurtransferase (SseA), O-acetylserine sulfhydrylases A and B (CysK and CysM), cystathionine β-lyases A and B (MetC and MalY), and IscS (Awano et al., 2005, Smith et al., 2001). Importantly, most of these enzymes have other recognized functions in vivo, indicating that their action upon cysteine is adventitious. Mutants that lacked these various enzymes were tested, but none of them were substantially diminished in the rate at which they generated sulfide during cystine import (data not shown). Either the sulfide production is due to the collective action of all these enzymes, or else another enzyme with especially high activity has escaped notice.
The literature suggests that when cysteine is incubated with extracts from cells grown on LB medium, the primary sulfide source is tryptophanase (Awano et al., 2005). This enzyme exhibits parallel activities upon both substrates:
| (1) |
| (2) |
The transcription of tnaA, encoding tryptophanase, is positively regulated by Crp bound to cyclic AMP, and it requires the unusual binding of free tryptophan to a ribosome/leader-peptide complex (Cruz-Vera & Yanofsky, 2008). The implication is that the enzyme is synthesized only if abundant tryptophan is available to be used as a carbon source. LB matches that condition, but our Trp-free glycerol medium did not. Indeed, we found that the addition of cystine to cells grown on glycerol triggered tryptophanase-driven sulfide release only if tryptophan had been provided in the pre-culture (Fig. 1C). If glucose (a PTS substrate) was present or if tryptophan was absent, tryptophanase did not contribute to sulfide release. The tnaA regulatory scheme indicates that the physiological role of tryptophanase is indeed tryptophan degradation and that its activity upon cysteine is accidental.
The kinetic properties of the enzyme fit this view. The chemical mechanism of the pyridoxal phosphate-dependent elimination reaction fails to discriminate against cysteine, as Vmax values differ only two-fold (Newton et al., 1965). The adventitious reaction is assisted because sulfide is an especially excellent leaving group. The small size of cysteine enables it to occupy the tryptophan binding site, but it does so loosely, so that the KM value for cysteine (11 mM) is much higher than that of tryptophan (0.33 mM). Under our standard growth conditions the intracellular cysteine levels are ~ 0.2 mM (Park & Imlay, 2003), and so degradation by tryptophanase will be minimal. Only when cystine is over-imported into the cell does the cysteine concentration rise to millimolar levels (Park & Imlay, 2003), approaching the KM of the enzyme and triggering degradation at a rate that creates a sulfide hazard.
The endogenous sulfide impairs cytochrome bo oxidase
Thiol compounds bind with substantial avidity to soft metals and hemes and therefore are potentially inhibitory to enzymes that use these cofactors (Nies, 2003, Wu et al., 2015, Vitvitsky et al., 2015, Bolic et al., 2015, Nagy, 2015). Zinc has a moderate affinity for thiols, and zinc enzymes can be inhibited when moderate concentrations of cysteine or sulfide coordinate and perhaps extract the metal (Martin & Maricle, 2015, Gu & Imlay, 2013). However, the softest biological metal is copper. E. coli synthesizes only a handful of copper enzymes: the respiratory cytochrome bo oxidase, copper-zinc superoxide dismutase, monoamine oxidase, and multicopper oxidase. E. coli also has relatively few heme enzymes (Table S3 in (Mancini & Imlay, 2015)), of which the cytochrome bo oxidase and a partner cytochrome bd oxidase are the only ones that provide an essential activity under standard growth conditions. Cytochrome bo oxidase is a four-subunit transmembrane complex that catalyzes the transfer of four electrons from two molecules of reduced ubiquinone to molecular oxygen in conjunction with the pumping of four protons into the periplasm (Abramson et al., 2000). This proton translocation contributes to the energy yield of respiration. During the reaction the bo oxidase binds molecular oxygen between its CuB copper atom and heme o3 cofactor. Cytochrome bo oxidase is similar in active-site structure to the mitochrondrial cytochrome c oxidase. The latter enzyme differs most substantially in that it receives electrons from cytochrome c rather than ubiquinone and in that it uses an a-type heme in its binuclear cluster.
Sulfide can potently inhibit mitochondrial cytochrome c oxidase. The mechanism is complex and depends upon details of the active-site structure (see Discussion), and relatively resistant forms of the enzyme are found in a few higher organisms that routinely experience sulfide stress (Martin & Maricle, 2015, Pfenninger et al., 2013, Tobler et al., 2014). The impact of sulfide upon bacterial cytochrome bo oxidase has not been reported.
The simpler two-subunit cytochrome bd oxidase is an alternative ubiquinol oxidase that binds oxygen between its heme d and heme b595 moieties (Junemann, 1997). It does not pump protons, although by enabling electron flow it sustains proton translocation by upstream complexes of the respiratory chain. It has been proposed that the physiological role of cytochrome bd oxidase is to maximize respiration when oxygen levels are very low, since its affinity for oxygen is ~0.3 μM, whereas that of cytochrome bo oxidase is ~6 μM (Mason et al., 2009, D’mello et al., 1996). (For comparison: Air-equilibrated 37° C water contains 210 μM oxygen.) Indeed, synthesis of cytochrome bd oxidase is induced when oxygen levels fall (Tseng et al., 1996).
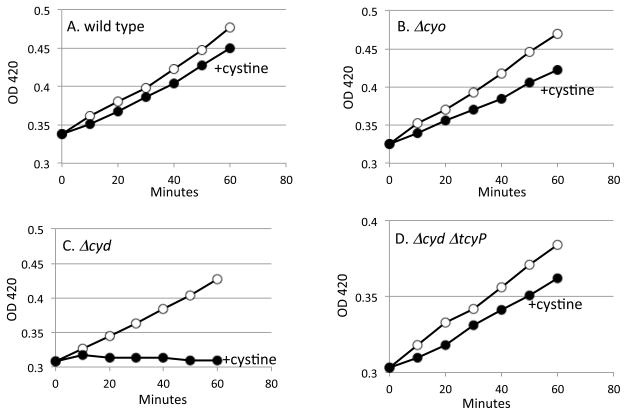
Because these ubiquinol oxidases rely upon solvent-exposed copper and heme moieties, we tested whether they continue to function during periods of cystine intoxication. In glycerol medium respiration is essential for growth. Both ΔcyoABCDE mutants (lacking cytochrome bo oxidase) and ΔcydAB mutants (lacking cytochrome bd oxidase) grew equally well in the base medium, as either of the two respiratory oxidases is sufficient. The addition of cystine only modestly slowed the growth of wild-type cells and of mutants lacking the cytochrome bo oxidase (Fig. 2A, B). However, cystine addition virtually stopped the growth of mutants that lacked cytochrome bd oxidase (Fig. 2C). The data suggest that upon cystine addition, cytochrome bo oxidase lost activity, but cytochrome bd oxidase did not. No other essential enzymes failed.
Figure 2. Continued growth after cystine addition requires cytochrome bd oxidase activity.
Open circles: no addition. Solid circles: cystine (0.5 mM) was added at time zero. Cells were cultured in glycerol medium supplemented with Ile. (A) Wild-type cells. (B) ΔcyoABCDE mutant. (C) ΔcydAB mutant. (d) ΔcydAB ΔtcyP mutant.
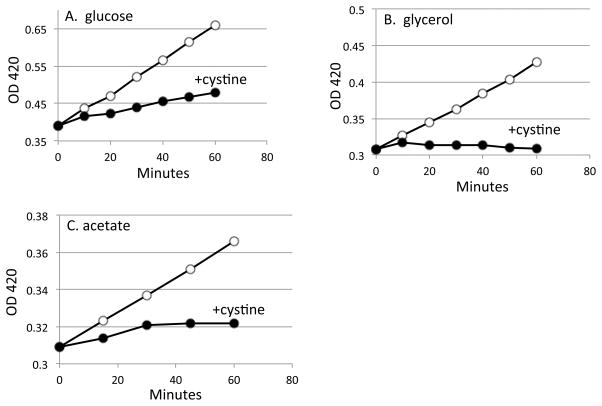
Respiration is required for energy production when glycerol and acetate are carbon sources, but glucose fermentation can allow slow growth to continue when respiration is impeded. Accordingly, the growth problem was reproduced in acetate medium but was slightly lessened in glucose medium (Fig. 3).
Figure 3. Cystine inhibits growth most potently in medium lacking a fermentable carbon source.
Cystine (0.5 mM) was added to the ΔcydAB mutant at time zero. (A) Glucose medium. (B) Glycerol medium. (C) Acetate medium.
We considered the possibility that cystine might interfere with cytochrome bo oxidase function by transferring disulfide bonds to domains that are periplasmically exposed. However, the ΔcydAB mutant defect was alleviated in a tcyP mutant (Fig. 2D), indicating that toxicity required cystine influx into the cytoplasm.
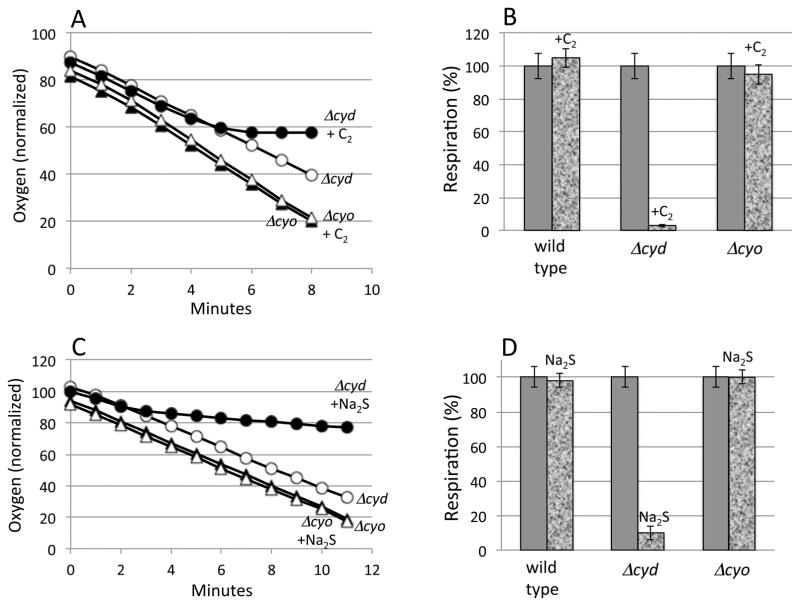
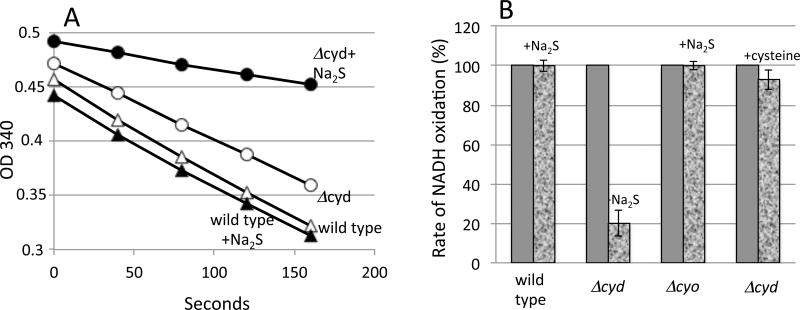
Oxygen consumption was monitored with a Clark-type electrode (Fig. 4A, B). As anticipated, cystine addition eliminated respiration in a ΔcydAB mutant that depended upon cytochrome bo oxidase for oxidase activity. It did not block the respiration of either the wild-type strain or the ΔcyoABCDE mutant, both of which retain cytochrome bd oxidase. Notably, although thiol excretion commenced immediately upon the addition of cystine (Fig. 1), there was a lag before respiration slowed (Fig. 4). This observation suggests that an inhibitor takes time to accumulate. Because the immediacy of cysteine efflux indicates that cysteine pools rise very rapidly, we inferred that sulfide rather than cysteine is the probable inhibitor. Indeed, the addition to the medium of 4 μM sulfide alone was sufficient to block respiration (Fig. 4C, D). Sulfide also generated the growth defect (Fig. S1).
Figure 4. Cystine and sulfide block cytochrome bo oxidase-dependent respiration, but cytochrome bd oxidase is unhindered.
(A, B) Cystine (0.5 mM, denoted C2) or (C, D) sodium sulfide (4 μM) was added to cultures in glycerol medium at time zero. Open symbols: no addition. Filled circles: cystine/sulfide added. Circles: ΔcydAB mutant. Squares: ΔcyoABCDE mutant. (B, D) Mean values +/− standard deviation of three replicate experiments. Respiration rates were measured 6–8 min after cystine addition.
Sulfide directly inhibits the cytochrome bo oxidase
Inverted membrane vesicles were prepared that contained intact respiratory chains. When NADH is added to these membranes, electrons flow through NADH dehydrogenases, ubiquinone, and cytochrome oxidases to molecular oxygen. The addition of cysteine did not diminish respiration by membranes that contained only cytochrome bo oxidase, but sulfide did inhibit (Fig. 5).
Figure 5. Sulfide directly inhibits cytochrome bo oxidase in isolated vesicles.
Vesicles were prepared from wild-type, ΔcydAB, and ΔcyoABCDE strains. A. NADH oxidation was measured in vitro in the absence or presence of 8 μM sulfide. B. Normalized NADH oxidation rates. Where indicated, 8 μM sulfide or 2 mM cysteine were added.
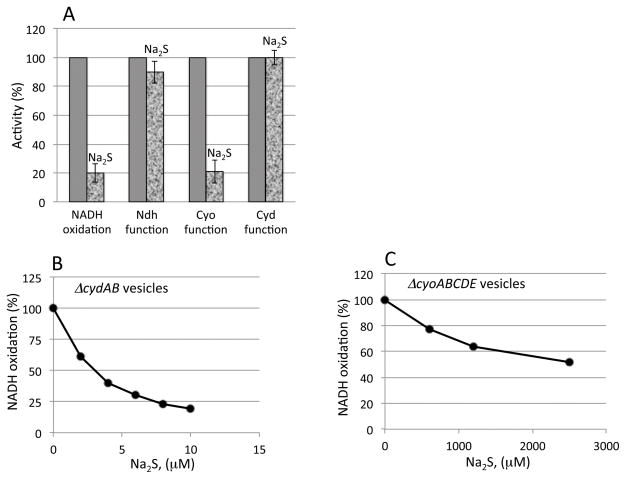
To confirm the site of inhibition, the NADH dehydrogenase and cytochrome oxidase activities were specifically assayed by measurement of NADH:plumbagin and dihydroubiquinone-1:oxygen oxidoreductase activities, respectively (Fig. 6A). Only the latter activity of ΔcydAB mutant membranes was blocked.
Figure 6. Low micromolar sulfide inhibits vesicular respiration only at the cytochrome bo oxidase site.
A) The NADH oxidase, NADH dehydrogenase, and quinone oxidase activities of membrane vesicles were each assayed in the absence or presence of 8μM sulfide. NADH oxidation requires function of both the dehydrogenase and oxidase complexes, whereas the NADH dehydrogenase and quinone oxidase activities are specific for those steps of the respiratory chain. (B and C) The sensitivities to sulfide of cytochrome bo oxidase (B) and cytochrome bd oxidase (C) were determined by assay of NADH oxidase activity in membrane vesicles expressing only one or the other oxidase.
We anticipated that the inhibition would occur through reversible binding of sulfide to the cytochrome bo oxidase active site rather than irreversible damage to the protein or metal cofactors. The notion fits the fact that doses of sulfide that partially inhibited the enzyme did so immediately but did not progressively diminish the activity any further (Fig. 5A). To test the reversibility of inhibition, sufficient sulfide was added to membranes to inhibit respiration to 15% of the control rate. The suspension was then bubbled for 1 min with nitrogen to remove sulfide, and then air was added. In this short time, respiration rebounded to 75% of the control rate, confirming that inhibition was quickly reversible. This result was replicated with intact cells: the removal of sulfide promptly restored full respiration (Fig. S2).
The sensitivities of the two cytochrome oxidases to sulfide were evaluated in air-saturated buffer. The cytochrome bo oxidase activity was progressively blocked by increasing levels of sulfide with half-inhibition occurring at ~ 5 micromolar sulfide (Fig. 6B, C). In contrast, a thousand-fold higher concentration was needed to inhibit the cytochrome bd oxidase.
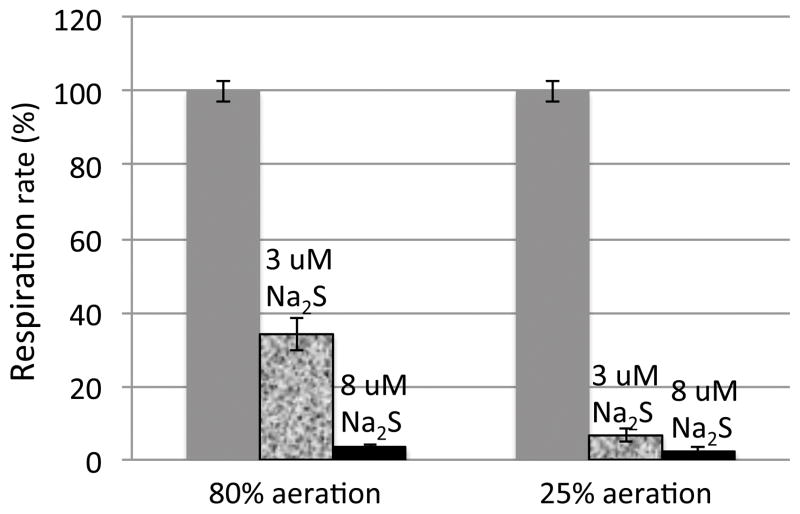
Because E. coli spends much of its time in hypoxic intestinal environments, it was pertinent to test whether sulfide inhibition is influenced by the oxygen concentration. Using the oxygen electrode to quantify both oxygen and respiration rate, we observed that a low concentration of sulfide was much more potent when the oxygen concentration was reduced (Fig. 7). Interestingly, inhibition occurred more readily at lower pH (Fig. S3), suggesting that H2S rather than HS− was the initial ligand. In fact, hemoglobin initially binds hydrogen sulfide in its H2S form (Vitvitsky et al., 2015), with deprotonation occurring subsequent to binding. It has also been proposed that cytochrome c oxidase may initially bind protonated hydrogen sulfide (Pietri et al., 2009). An alternative interpretation of our result is that low pH favors the dissociation from copper of the presumptive hydroxide anion that sulfide must displace.
Figure 7. Sulfide is a more potent inhibitor of cytochrome bo oxidase at low oxygen concentrations.
NADH oxidation rates were measured in ΔcydAB vesicles at 80% air saturation (170 μM O2) and 25% air saturation (50 μM O2).
The environmental sulfide concentration determines sulfide levels in the cytoplasm
The rate of diffusion across membranes must be considered in order to evaluate whether intracellular sulfide production immediately poisons the producing cell, or whether rapid egress will keep the cytoplasmic sulfide concentration low. As a charged species, the sulfide anion (HS−) cannot penetrate membranes at a significant rate. However, hydrogen sulfide (H2S) is substantially more hydrophobic than water and exhibits a twofold partition coefficient into hydrophobic phases relative to aqueous ones (Cuevasanta et al., 2012). Using these values, Cuevasanta et al. estimated the membrane permeability coefficient P of H2S to be 3 cm s−1, which is 1000-fold higher than that of water (P = 2–4 × 10− 3 cm s−1, (Fettiplace, 1978, Xiang & Anderson, 1994)). This value was consistent with experimental measurements of H2S diffusion across bilayer lipid membranes (P > 0.5 +/− 0.4 cm s−1 (Mathai et al., 2009).
The relationship between intracellular sulfide production and intracellular sulfide concentration can then be calculated. The rate of sulfide formation detected in Fig. 1 is an underestimate, because some H2S escaped from the cultures in these shaken flasks. Using cells transferred to a closed test tube (Procedures), we directly measured the rate of sulfide production inside cystine-treated E. coli to be approximately 10 mM min−1. Cell volume in minimal medium is 3.2 × 10−15 L cell−1 (Seaver & Imlay, 2001), so this rate of sulfide production = 6 × 10−19 mole sec−1 cell−1. The rate of H2S efflux can be calculated:
| (3) |
where A is the area of the cytoplasmic membrane (1.4 × 10−7 cm2 cell−1 (Seaver & Imlay, 2001)). For a cell in an infinitely dilute environment, where the external sulfide concentration is regarded as zero, the steady-state cytoplasmic H2S concentration can then be calculated by equating the rate of sulfide production with the rate of sulfide efflux. Despite the rapid rate of sulfide production, that calculation indicates an intracellular concentration of only 1.4 nM H2S. Since the pKa of sulfide = 6.89 (Mathai et al., 2009), the total sulfide concentration (H2S + HS−) would be 5 nM in a pH 7.4 cytoplasm. This concentration is far below the low-micromolar concentrations that poison cytochrome bo oxidase. For biological purposes sulfide can be regarded as equilibrating instantaneously across cell membranes, and toxic doses can be achieved only if the collective sulfide production by the microbial community drives the extracellular sulfide concentration to micromolar levels.
This analysis was tested by examination of the kinetics of respiratory inhibition in cystine-treated cells. Oxygen consumption ceased most quickly when cells were at higher densities during treatment (Fig. 8), in accordance with the faster accumulation of sulfide in the medium. Thus in nature very dilute sulfide-producing bacteria will be spared its toxicity. Denser bacteria in biofilms or microcolonies must contend with the sulfide that accumulates locally.
Figure 8. Cystine inhibits respiration more quickly if cell density is high.
Respiration rates were determined with an oxygen electrode using a single culture of cells diluted to 0.2 (left set of bars) or 0.06 OD600 (right set). Cystine (0.5 mM) was added in both cases at time zero.
Similarly, bacteria that enter a sulfide-rich environment will experience intracellular sulfide concentrations equivalent to that outside the cell. That expectation was borne out by the inhibition of respiration when sulfide was added to culture medium in Fig. 4C, D.
Discussion
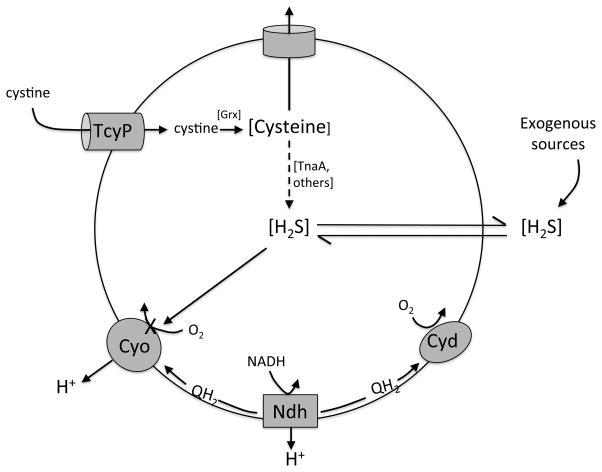
Respiration is essential for the growth of aerobic E. coli, but we have found that the primary cytochrome bo oxidase can be completely blocked by low-micromolar concentrations of hydrogen sulfide (Fig. 9). Such doses can accumulate through the inadvertent degradation of cysteine during periods of cystine import; they are also likely to be encountered in many natural habitats. In such circumstances respiration is sustained by diversion of electrons to the cytochrome bd oxidase. The sulfide resistance of the latter enzyme may be central to the role it plays in bacteria.
Figure 9. Sulfide physiology in E. coli.
Excessive import of cystine by TcyP enlarges cysteine pools. Most cysteine is excreted, but some is adventitiously degraded by tryptophanase and other enzymes. The sulfide is membrane permeant and accumulates in the local environment. Environmental sulfide also arises from sulfate-reducing bacteria in nearby anoxic zones. Intracellular sulfide inhibits the proton-pumping cytochrome bo oxidase, diverting respiratory flux through the non-pumping cytochrome bd oxidase.
Interaction of sulfide with cytochrome oxidases
The toxic effects of sulfide upon mammals has long been recognized (Reiffenstein et al., 1992), and at its core lies the inhibition of cytochrome c oxidase. The mechanism is more complex than for cyanide, nitric oxide, and carbon monoxide, which form reversible complexes with the active-site heme a3 (Pietri et al., 2011, Nicholls et al., 2013, Cooper & Brown, 2008). Sulfide can bind both the heme and copper moieties, it can do so in protonated or anionic form, and it can transfer electrons to them in acting as a substrate rather than inhibitor. Spectroscopic analysis of inhibited enzymes reveals that sulfide ultimately binds to both the iron and copper atoms (Hill et al., 1984), but it may be copper-mediated transfer of sulfide to the heme moiety that produces the reversibly inhibited structure at physiological doses of sulfide (Nicholls et al., 2013, Pietri et al., 2011). The local polypeptide has a hand in the avidity and fate of the bound complex, as residue-catalyzed deprotonation of the sulfide may enable a cycle of oxidase activity rather than inhibition (Pietri et al., 2011). Further, polypeptide changes that constrict the D pathway, a water-filled channel that delivers protons to the binuclear active site, appear to hinder sulfide entry and confer relative resistance (Pfenninger et al., 2013). Such changes were observed among fish that live in sulfidic waters and plants that must tolerate sulfide exudations from sediments during flooding. Intriguingly, a cytochrome c oxidase in a sulfate-reducing bacterium appeared to be tolerant of 0.25 mM sulfide, but the structural basis was not explored (Wu et al., 2015).
Evidently E. coli copes with sulfide not by evolving a resistant heme o3-CuB site but by providing the alternative cytochrome bd oxidase. It has been noted that this oxidase is modestly resistant to nitric oxide and cyanide, which are competitive inhibitors of oxygen binding (Mason et al., 2009). Its NO resistance may help respiration to persist when bacteria are confronted by NO that is released by phagocytes or that leaks from nitrate-reducing bacteria (Fu et al., 2013). The NO resistance does not arise from poor NO binding—the bd oxidase actually binds NO slightly (8-fold) more tightly than does the bo oxidase (Mason et al., 2009). Yet the bd oxidase binds oxygen with 20-fold higher affinity, so the net effect is that the IC50 of NO for the bd oxidase is three-fold higher. Similarly, the IC50 of cyanide for the bd oxidase about 10-fold higher than its IC50 for the bo oxidase Figure S4, which again suggests that the key difference stems from the difference in the affinities of the two enzymes for oxygen. These effects, however, are very small compared to the 1000-fold difference in sulfide IC50 values of the two enzymes (Fig. 6). We infer that sulfide binds much more poorly to the bd oxidase than to the bo oxidase, either because binding is weak or because it triggers sulfide oxidation rather than stable complex formation.
Interestingly, E. coli expresses a second bd-type oxidase (cytochrome bd-II oxidase, encoded by appBC). It is very poorly expressed under the aerobic, exponential-growth conditions employed in the experiments here, and its role in E. coli physiology is obscure. However, it can be induced in anoxic, stationary-phase cells (Atlung and Brondsted, 1994). We found that the cytochrome bd-II oxidase activity is approximately as resistant to sulfide as is the standard cytochrome bd oxidase (Fig. S5).
Of course, the fundamental difference between the bo and bd oxidases is that the bo oxidase contains copper, which may pull sulfide into the complex. Copper and sulfide have an antagonistic relationship with long roots in evolutionary history: in the anoxic primordial world, sulfur was present in the sulfide form and trapped copper into copper sulfide precipitates. The metal was unavailable to early organisms, as copper enzymes appeared only after the Great Oxygenation Event (Ridge et al., 2008, Ochiai, 1983). That pattern persists today, as obligate anaerobes do not employ copper as a cofactor for their enzymes. Indeed, copper enzymes are primarily involved in aerobic biochemistry that explicitly involves oxygen: in oxidases, superoxide dismutases, respiratory cytochrome oxidases, hemocyanins, and components of oxygenic photosystem II.
The biological niche of E. coli is as an oxygen-consuming microbe in a low-oxygen intestinal environment, although it has a secondary habitat in fully aerated surface waters during transit from one host to another. In the gut the oxygen levels are low, and local sulfur-reducing bacteria (SRB) raise the hydrogen sulfide content to millimolar levels (Macfarlane et al., 1992, Pitcher et al., 2000). At such an oxic-anoxic interface, the epithelial cells lining the intestine express high levels of sulfide oxidase activity in order to protect their cytochrome c oxidase from inhibition (Goubern et al., 2007, Blachier et al., 2010). Bacteria lack sufficient volume to try that strategy: sulfide instantly equilibrates across membranes, and clearance of sulfide from the entire habitat is implausible. Synthesis of a sulfide-resistant isozyme is a better tactic. Indeed, mutants of E. coli that lack cytochrome bo oxidase can competitively colonize the mouse intestine, but those lacking the cytochrome bd oxidase cannot (Jones et al., 2007).
Cytochrome bd oxidase is also more fit for hypoxic environments because of its high affinity for oxygen and the fact that its activity does not depend on acquisition of copper, which is likely to be sequestered by environmental thiols. Thus the sulfide resistance of the bd oxidase is probably a key attribute, but not the sole one, that suits it for the intestinal habitat. A similar case could be proposed for bacteria that live in the oxic/anoxic interfaces of sediments and still waters, where sulfide produced by sulfate-reducing bacteria diffuses from anoxic zones into microoxic ones. Indeed, cytochrome bd oxidase is common among facultative, microaerophilic, and even obligately anaerobic bacteria.
Sulfide production inside bacteria
Like reactive oxygen species or 2-aminoacrylate (Imlay, 2013, Ernst & Downs, 2015), sulfide is an unusual poison in that it is generated by the same bacterium that it can toxify. Three mechanisms stand out. The one revealed in this study is the inadvertent degradation of cysteine during cystine over-import (Fig. 9). As detailed here, this phenomenon arises from an unfortunate combination of enzyme attributes which otherwise serve important physiological purposes: the high flux capacity of TcyP, which enables growth on scant cystine but is excessive when cystine is more abundant; the rapid reduction of cystine by glutaredoxins, which avoids disulfide stress but precludes product inhibition of TcyP; the robust activity of tryptophanase, which is sufficient to employ tryptophan as a sole carbon source but has enough accidental cross-over activity to degrade cysteine; and the solvent exposure of the copper-heme binuclear site of cytochrome bo oxidase, which enables binding of dissolved oxygen but also of sulfide. Higher organisms also generate modest sulfide via cysteine degradation, and sulfide oxidase activity comprises a scavenging defense. Indeed, ethylmalonic encephalopathy is a human genetic disorder in which the oxidase activity is disabled and cytochrome c oxidase inhibition leads to fatal sequelae (Tiranti & Zeviani, 2013). Enzyme features that were selected for one purpose can lead to unfortunate off-target consequences for which organisms must develop coping mechanisms.
Authentic cysteine catabolism comprises a second potential source of endogenous sulfide. E. coli does not degrade cysteine quickly enough for cystine to serve as its sole carbon source (Imlay et al., 2015). Yet other bacteria deliberately degrade cysteine more robustly, and they generate sulfide when they do so. These bacteria include the related pathogen Salmonella, for which the rapid evolution of sulfide is used clinically as a diagnostic marker. The key enzyme is CdsH (Oguri et al., 2012), which E. coli lacks. How does Salmonella tolerate the sulfide that it produces?
Since sulfide freely equilibrates across membranes, it is possible that relatively dilute cells might be spared sulfide toxicity because the sulfide quickly clears the cell and the habitat. Our calculations show that infinitely dilute bacteria would contain only ~ 5 nM sulfide if it were generated at the rate observed during E. coli cystine overloading; accordingly, even a 100-fold increase in the rate of cysteine degradation would not boost the intracellular sulfide concentration to the micromolar level needed to poison cytochrome bo oxidase. Thus sulfide toxicity only occurs if sulfide accumulates in the local habitat, as when cells are in high density. Interestingly, cytochrome bd oxidase is indispensable even during intraperitoneal Salmonella infection (Craig et al., 2013), raising the possibility that CdsH action does elevate the local sulfide concentration in microhabitats. Other enzymes that enable anaerobic growth—pyruvate:formate lyase, anaerobic ribonucleotide reductase, and the Fnr transcription factor—are not needed for full virulence, so it is unlikely that this need for cytochrome bd oxidase arises from growth in host environments with very low oxygen concentrations.
The third endogenous source of sulfide is the sulfate/sulfite assimilation pathway itself. The conventional understanding is that sulfite reductase generates sulfide that is released to the bulk cytoplasm prior to capture by O-acetylserine sulfhydrase (CysK) (Mino et al., 2000). CysK condenses sulfide and O-acetylserine to form cysteine. At the outset the model seems problematic in two ways. First, the extremely high permeability of membranes suggests that this pathway intermediate could easily escape the cell. Such a situation is anathema to metabolism, which is configured to operate almost exclusively with charged metabolites that are membrane-impermeant. Second, the CysK binding constant for sulfide (KM = 6 μM, (Mino et al., 2000)) is high enough that concurrent inhibition of cytochrome bo oxidase (IC50 ~ 3 μM) seems plausible. Yet in this study we did not observe any sulfide release during sulfate assimilation (Fig. 1), and cytochrome bo oxidase remained active (Fig. 2C). One possible explanation is that sulfite reductase channels its sulfide product directly to CysK without release to the bulk solution. Another is that the CysK titers are calibrated to be so high that CysK quantitatively intercepts sulfide before it escapes the cell, while keeping steady-state sulfide levels too low to poison cytochrome bo oxidase. The second explanation is probably correct. CysK is by far the most abundant protein in the sulfate-assimilation pathway, and the total CysK activity exceeds by > 100-fold those of the other pathway enzymes (Li et al., 2014, Kredich, 1971). Indeed, our calculations (Experimental Procedures) indicate that the abundant CysK should keep sulfide at nanomolar levels, so that the bo oxidase can function under sulfate assimilatory conditions and little sulfide escapes the cell. In this view the high CysK activity can be construed as another device that circumvents sulfide stress.
In sum, it appears that sulfide can be produced at a substantial rate in E. coli by both deliberate and accidental intracellular processes. Cytochrome bo oxidase is the sole apparent target inside this bacterium, and the presence of the bd oxidase is sufficient compensation. It would be interesting to learn whether microbes that occupy highly aerobic habitats feature a greater number of sulfide-sensitive enzymes. Such aerobes might thereby be constrained from dwelling in hypoxic environments, much as the toxic actions of oxygen upon low-potential enzymes block anaerobes from growing in aerated habitats.
Experimental Procedures
Reagents
Potassium mono and dibasic phosphates, ammonium sulfate, sodium citrate, magnesium sulfate, glucose and glycerol were from Fisher Scientific. N,N-dimethyl-p-phenylenediamine was from Fluka, and Coomassie reagent was from Thermo Scientific. All other chemicals were from Sigma.
Cell cultures
Cells were grown in minimal A medium (Miller, 1972) supplemented with 0.5 mM each of leucine, isoleucine and valine. Branched-chain amino acids were provided because excess cysteine can impair Ile synthesis (Harris, 1981). Either 0.2% glucose, 0.7% glycerol or 50 mM sodium acetate were used as a carbon source. For induction of tryptophanase in the experiment of Fig. 1, 5 mM of tryptophan was added 2 hrs before cell harvesting. For typical experiments, cultures were grown overnight in the specified medium, resuspended to 0.005 OD600 in 25 ml of the same medium, and then grown to 0.15–0.2 OD600 in 125- ml flasks at 37° C with vigorous shaking. Eleven ml of cell culture was placed into each of two 50-ml flasks, and 0.6 mM of cystine were added into one of them, leaving the other flask as a control. Since nonfermentable carbon sources provide slow growth rates, the subsequent increase in optical density was monitored at 420 nm in 6 to 10 min increments depending on the experiment. The lower wavelength increased the sensitivity about two-fold.
Strain constructions
Strains are all congenic derivatives of the wild-type K12 strain MG1655 and are listed in Table S1. P1 transduction was performed by standard methods (Miller, 1972) with antibiotic selection on LB plates.
Total thiol measurements
A 1-ml aliquot of cell culture (0.15 OD600) was filtered using 220 nm syringe filter, and then 0.5 ml of the filtrate was mixed with 0.5 ml of 0.4 mM 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) in 100 mM KPi buffer, pH 7.0 at room temperature. The mixture was incubated in the dark at room temperature for 5 min, and then the absorbance at 412 nm was measured using an extinction coefficient of 14 mM−1 cm−1. Authentic reduced glutathione and sodium sulfide nonahydrate were used as standards.
Sulfide measurements
Hydrogen sulfide release was quantified using the methylene blue method (Siegel, 1965). A 20 mM stock of N,N-dimethyl-p-phenylenediamine (DPMDA) in 50% HCl was freshly prepared for each experiment. A 30 mM stock solution of ferric chloride was dissolved in 1 M HCl. Cells were grown to proper OD, collected by centrifugation, washed with 100 mM KPi buffer (pH 7.0, room temperature) and resuspended in fresh medium in 50 ml flasks at 37° C. After the addition of cystine, 1 ml aliquot of cells suspension was added to a 2 ml plastic tube, 0.1 ml of ferric chloride and 0.1 ml of DMPDA were added as quickly as possible, and the tube was sealed with parafilm and kept in the dark at room temperature for 15 min. The suspension was then centrifuged for 4 min on a table-top centrifuge, and the absorbance of the supernatant was determined at 650 nm. Calibration was performed for each medium using sodium sulfide nonahydrate.
Vesicle preparation
For most work, cells were grown to 0.2 OD600 in glycerol medium. Two tubes of 40 ml of cell culture were centrifuged, resuspended in 4 ml of 100 mM KPi buffer, pH 7.0, and lysed by French press (Messner and Imlay, 1999). The lysates were centrifuged for 10 min at 10,000 × g at 4° C. Vesicles were sedimented from the lysates by centrifugation for 1 hr at 100,000 × g, and they were then resuspended in 1 ml of cold 100 mM KPi buffer, pH 7.0, and stored on ice. The total protein concentration was determined using Coomassie blue reagent. In respiration experiments a final vesicle concentration of 0.01–0.02 mg ml−1 of protein was used. To obtain cells with substantial content of cytochrome bd-2 oxidase, SSK12 was grown in anoxic medium containing 0.2% glucose and 0.2% casamino acids to 0.5 OD600 in an anaerobic chamber. This enzyme is induced either anaerobically or in stationary phase (Atlung and Brondsted, 1994); under these growth conditions, this cell density comprises early stationary phase.
Oxygen consumption
The rate of oxygen consumption by whole cells was monitored at 37° C using a Clark cell–based Micrometrix model 10 oxygen system at 0.2 OD600 cell density. Consumption of oxygen by vesicles was observed using 0.01–0.02 mg ml−1 total protein. The oxygen concentration was deliberately varied in the chamber by incubating the cell suspension until the oxygen concentration had fallen to the desired level. Respiration was temporally linear in every experiment until 95–98% of oxygen was consumed. To demonstrate the reversibility of sulfide inhibition, 4 ml of 0.2 OD cells were incubated in a 15 ml plastic tube with sodium sulfide for 10 min. Respiration was then measured. Cells were bubbled with nitrogen gas for 1 min, which removed 99% of the sulfide, and oxygen was reintroduced by vigorously shaking the capped tube. Respiration was then measured again.
Assays of respiratory enzymes
Total NADH oxidase activity of membranes represented electron flow from NADH through the NADH dehydrogenases, quinone pool, and cytochrome oxidases to molecular oxygen. These assays were performed using 0.1 mM NADH in 200 mM KPi buffer, pH 7.1, and the activity was determined by monitoring the disappearance of NADH at 340 nm (ε = 6.22 mM−1 cm−1).
The NADH dehydrogenase enzyme activity was determined by performing the same reaction in the presence of 3 mM KCN and 0.4 mM of ethanol-dissolved plumbagin. In this situation plumbagin abstracts the electrons directly from the NADH dehydrogenase enzymes. The final ethanol concentration did not exceed 1%.
The activities of the terminal cytochrome oxidases (cytochrome bo plus cytochrome bd oxidases) was monitored by oxygen consumption after the addition of reduced coenzyme Q1 (Thomas et al., 1993). Cell suspensions were incubated with 0.4 mM of Q1 (from 40 mM ethanol solution stock) and 5 mM of dithiothreitol (as a Q1 reductant).
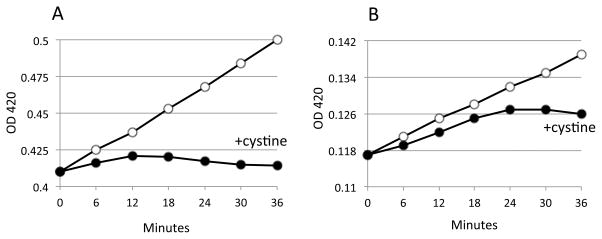
To test the reversibility of sulfide inhibition, membranes were prepared from the cytochrome bo oxidase-overproducing strain SSK151 pRG110. Vesicles were suspended in 20 ml of 70 mM KPi buffer (pH 7.0, room temperature) to a final protein concentration of 0.02 mg ml−1. The suspension was then split into two suspensions of 10 ml apiece, and 25μM sodium sulfide was added to one. After one min of incubation, both control and sulfide-treated samples were tested for their ability to oxidize 100 μM NADH. Then 4 ml of each suspension was placed in a 10 ml beaker and bubbled with nitrogen gas for 1 min. Air was then bubbled briefly (about 3 sec) for re-oxygenation. DTNB analysis confirmed the removal of 99% of sulfide from the sulfide-treated samples. Aliquots were then re-tested for their ability to oxidize NADH. The gassing protocol modestly diminished the activities of the untreated membranes (ca. 30%), and this correction was made in analyzing the data.
The rate of thiol efflux per cell
In closed tubes we determined that upon cystine addition to glycerol-grown cells, they released 35 μM total thiol min−1 OD−1 into the medium. The cytoplasmic volume is 0.5 ml per 1 L of 1 OD bacteria (Imlay & Fridovich, 1991), and so the thiols released represented 70 mM thiol min−1 lost from cells relative to the cytoplasmic volume. Cellular demand for sulfur from the medium is 64 μM to achieve 1 OD of biomass (Imlay et al., 2015), indicating a total sulfur content of 130 mM. At a doubling time of 60 min in glycerol medium (k = 0.0116 min−1), the intracellular use of cysteine is 130 mM × 0.0116 min−1 = 1.5 mM min−1. Therefore, the rate of thiol efflux from cystine-treated cells occurs at a rate 50 times the cellular sulfur demand, due to the over-import of cystine by TcyP.
Calculation of the competition for sulfide between CysK and efflux
Extract measurements indicate 15.5 U mg−1 of O-acetylserine sulfhydrylase (OAS) activity, primarily due to CysK, where 1 U generates 1 μmol product/min (Kredich, 1971). The total protein concentration in E. coli is ~ 300 mg ml−1 (Seaver & Imlay, 2001), so that one cell [3.2 × 10−15 L cytoplasm (Seaver & Imlay, 2001)] contains 1.5 × 10−8 units of OAS, or a Vmax of 2.5 × 10−16 mol sec−1 cell−1. Since exponentially growing E. coli under the studied growth condition synthesizes 1.5 mM min−1 cysteine (above section), the flux through OAS is 8 × 10−20 mole sec−1 cell−1. OAS behaves with Michaelis-Menten kinetics with an apparent KM of 6μM for sulfide (Mino et al., 2000). Solution of the Michaelis-Menten equation indicates that the observed flux occurs when the sulfide concentration is 1.9 nM.
Efflux by diffusion through the membranes can be described using the permeability coefficient (3 cm/sec, (Cuevasanta et al., 2012)) and area of cytoplasmic membrane (1.4 × 10−7 cm, (Seaver & Imlay, 2001)):
| (4) |
At pH 7.4, when total sulfide = 1.9 nM, the concentration of H2S = 0.6 nM (Li et al., 2014). The predicted rate of efflux is 2.5 × 10−19 mole sec−1 cell−1. This value is slightly higher than the measured rate of cysteine synthesis (0.8 × 10−19 mole sec−1 cell−1) and at face value suggests that 24% the endogenous sulfide is incorporated by OAS before it escapes the cell. Considering that OAS was assayed in vitro at room temperature, it is likely that this value is a considerable underestimate. It appears that CysK activity is plausibly high enough to trap most endogenous sulfide and that the internal steady-state sulfide concentration would be ~1000-fold below the OAS KM. That concentration is also several orders of magnitude too low to interfere with cytochrome bo oxidase activity. Approximations of enzyme kinetics and imprecision in cell measurements lead to imprecision in the final values; but if neither oxidase poisoning nor efflux were a possibility, the CysK titer would provide an adequate flux even if it were diminished by three orders of magnitude. It seems reasonable to conclude that the high titer of CysK comprises an adaptation to solve the poisoning and efflux problems.
Supplementary Material
Acknowledgments
This study was funded by grant GM101012 from the National Institutes of Health.
References
- Abramson J, Riistama S, Larsson G, Jasaitis A, Svensson-Ek M, Laakkonen L, Puustinen A, Iwata S, Wikstrom M. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat Struct Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- Atlung T, Brondsted L. Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation, and growth phase. J Bacteriol. 1994;176:5414–5422. doi: 10.1128/jb.176.17.5414-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awano N, Wada M, Mori H, Nakamori S, Takagi H. Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl Environ Microbiol. 2005;71:4149–4152. doi: 10.1128/AEM.71.7.4149-4152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachier F, Davila AM, Mimoun S, Benetti PH, Atanasiu C, Andriamihaja M, Benamouzig R, Bouillaud F, Tome D. Luminal sulfide and large intestine mucosa: friend or foe? Amino Acids. 2010;39:335–347. doi: 10.1007/s00726-009-0445-2. [DOI] [PubMed] [Google Scholar]
- Bolic B, Popovic-Bijelic AJ, Nikolic-Kokic A, Spasic S, Blagojevic D, Spasic MB, Spasojevic I. Reactions of superoxide dismutases with HS−/H2S and superoxide radical anion: An in vitro EPR study. Nitric Oxide. 2015;51:19–23. doi: 10.1016/j.niox.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Cooper CE, Brown GC. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J Bioenerg Biomembr. 2008;40:533–539. doi: 10.1007/s10863-008-9166-6. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Melville SB, Albrecht JA, Gunsalus RP. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol Microbiol. 1997;25:605–615. doi: 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- Craig M, Sadik AY, Golubeva YA, Tidhar A, Slauch JM. Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol Microbiol. 2013;89:887–902. doi: 10.1111/mmi.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Vera LR, Yanofsky C. Conserved residues Asp16 and Pro24 of TnaC-tRNAPro participate in tryptophan induction of tna operon expression. J Bacteriol. 2008;190:4791–4797. doi: 10.1128/JB.00290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevasanta E, Denicola A, Alvarez B, Moller MN. Solubility and permeation of hydrogen sulfide in lipid membranes. PLoS One. 2012;7:e34562. doi: 10.1371/journal.pone.0034562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’mello R, Hill S, Poole RK. The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology. 1996;142:755–763. doi: 10.1099/00221287-142-4-755. [DOI] [PubMed] [Google Scholar]
- Ernst DC, Downs DM. 2-aminoacrylate stress induces a context-dependent glycine requirement in ridA strains of Salmonella enterica. J Bacteriol. 2015 doi: 10.1128/JB.00804-15. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettiplace R. The influence of the lipid on the water permeability of artificial membranes. Biochim Biophys Acta. 1978;513:1–10. doi: 10.1016/0005-2736(78)90106-2. [DOI] [PubMed] [Google Scholar]
- Fu H, Chen H, Wang J, Zhou G, Zhang H, Zhang L, Gao H. Crp-dependent cytochrome bd oxidase confers nitrite resistance to Shewanella oneidensis. Environ Microbiol. 2013;15:2198–2212. doi: 10.1111/1462-2920.12091. [DOI] [PubMed] [Google Scholar]
- Goubern M, Andriamihaja M, Nubel T, Blachier F, Bouillaud F. Sulfide, the first inorganic substrate for human cells. FASEB J. 2007;21:1699–1706. doi: 10.1096/fj.06-7407com. [DOI] [PubMed] [Google Scholar]
- Gu M, Imlay JA. Superoxide poisons mononuclear iron enzymes by causing mismetallation. Mol Microbiol. 2013;89:123–134. doi: 10.1111/mmi.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CL. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as the target enzyme. J Bacteriol. 1981;145:1031–1035. doi: 10.1128/jb.145.2.1031-1035.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill BC, Woon TC, Nicholls P, Peterson J, Greenwood C, Thomson AJ. Interactions of sulphide and other ligands with cytochrome c oxidase. Biochem J. 1984;224:591–600. doi: 10.1042/bj2240591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266:6957–6965. [PubMed] [Google Scholar]
- Imlay KRC, Korshunov S, Imlay JA. Physiological roles and adverse effects of the two cystine importers of Escherichia coli. J Bacteriol. 2015;197:3629–3644. doi: 10.1128/JB.00277-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Chowdhury FZ, Fabich AJ, Anderson A, Schreiner DM, House AL, Autieri SM, Leatham MP, Lins JJ, Jorgensen M, Cohen PS, Conway T. Respiration of Escherichia coli in the mouse intestine. Infect Immun. 2007;75:4891–4899. doi: 10.1128/IAI.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junemann S. Cytochrome bd terminal oxidase. Biochim Biophys Acta. 1997;1321:107–127. doi: 10.1016/s0005-2728(97)00046-7. [DOI] [PubMed] [Google Scholar]
- Kredich NM. Regulation of L-cysteine biosynthesis in Salmonella typhimurium I Effects of growth on varying sulfur sources and O-acetyl-L-serine on gene expression. J Biol Chem. 1971;246:3471–3484. [PubMed] [Google Scholar]
- Kredich NM. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Molecular Microbiology. 1992;6:2747–2753. doi: 10.1111/j.1365-2958.1992.tb01453.x. [DOI] [PubMed] [Google Scholar]
- Kredich NM. Biosynthesis of cysteine. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Washington, D.C: ASM Press; 1996. pp. 514–527. [Google Scholar]
- Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, Gibson GR, Cummings JH. Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol. 1992;72:57–64. doi: 10.1111/j.1365-2672.1992.tb04882.x. [DOI] [PubMed] [Google Scholar]
- Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci USA. 2004;101:13318–13323. doi: 10.1073/pnas.0403064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini S, Imlay JA. The induction of two biosynthetic enzymes helps Escherichia coli sustain heme synthesis and activate catalase during hydrogen peroxide stress. Mol Microbiol. 2015;96:744–763. doi: 10.1111/mmi.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NM, Maricle BR. Species-specific enzymatic tolerance of sulfide toxicity in plant roots. Plant Phyiol Biochem. 2015;88:36–41. doi: 10.1016/j.plaphy.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, Poole RK, Cooper CE. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat Chem Biol. 2009;5:92–96. doi: 10.1038/nchembio.135. [DOI] [PubMed] [Google Scholar]
- Mathai JC, Missner A, Kugler P, Saparov SM, Zeidel ML, Lee JK, Pohl P. No facilitator required for membrane transport of hydrogen sulfide. Proc Natl Acad Sci USA. 2009;39:16633–16638. doi: 10.1073/pnas.0902952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner KR, Imlay JA. The identification of primary sites of superoxide and hydrogen peroxide in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. 1999;274:10119–10128. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1972. [Google Scholar]
- Mino K, Yamanoue T, Sakiyama T, Eisaki N, Matsuyama A, Nakanishi K. Effects of bienzyme complex formation of cysteine synthetase from Escherichia coli on some properties and kinetics. Biosci Biotechnol Biochem. 2000;64:1628–1640. doi: 10.1271/bbb.64.1628. [DOI] [PubMed] [Google Scholar]
- Nagy P. Mechanistic chemical perspective of hydrogen sulfide signaling. Meth Enzymol. 2015;554:3–29. doi: 10.1016/bs.mie.2014.11.036. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC. Chemical composition of Escherichia coli. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella. ASM Press; Washington, D.C: 1996. [Google Scholar]
- Newton WA, MOrino Y, Snell EE. Properties of crystalline tryptophanase. J Biol Chem. 1965;240:1211–1218. [PubMed] [Google Scholar]
- Nicholls P, Marshall DC, Cooper CE, Wilson MT. Sulfide inhibition of and metabolism by cytochrome c oxidase. Biochem Soc Trans. 2013;41:1312–1316. doi: 10.1042/BST20130070. [DOI] [PubMed] [Google Scholar]
- Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- Ochiai EI. Copper and the biological evolution. BioSystems. 1983;16:81–86. doi: 10.1016/0303-2647(83)90029-1. [DOI] [PubMed] [Google Scholar]
- Oguri T, Schneider B, Reitzer L. Cysteine catabolism and cysteine desulfhydrase (CdsH/STM0458) in Salmonella enterica serovar typhimurium. J Bacteriol. 2012;194:4366–4376. doi: 10.1128/JB.00729-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J Bacteriol. 2003;185:1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenninger M, Lerp H, Tobler M, Passow C, Kelley JL, Funke E, Greshake B, Erkoc UK, Berberich T, Plath M. Parallel evolution of cox genes in H2S-tolerant fish as key adaptation to a toxic environment. Nature Commun. 2013;5:3873–3880. doi: 10.1038/ncomms4873. [DOI] [PubMed] [Google Scholar]
- Pietri R, Lewis A, Leon RG, Casabona G, Kiger L, Yeh SR, Fernandez-Alberti S, Marden MC, Cadilla CL, Lopez-Garriga J. Factors controlling the reactivity of hydrogen sulfide with hemeproteins. Biochemistry. 2009;48:4881–4894. doi: 10.1021/bi801738j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietri R, Roman-Morales E, Lopez-Garriga J. Hydrogen sulfide and hemeproteins: knowledge and mysteries. Antiox & Redox Signal. 2011;15:393–404. doi: 10.1089/ars.2010.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher MCL, Beatty ER, Cummings JH. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut. 2000;46:64–72. doi: 10.1136/gut.46.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB. The catalytic mechanism of peroxiredoxins. Subcell Biochem. 2007;44:61–81. doi: 10.1007/978-1-4020-6051-9_4. [DOI] [PubMed] [Google Scholar]
- Reiffenstein RJ, Hulbert WC, Roth SH. Toxicology of hydrogen sulfide. Annu Rev Pharmacol Toxicol. 1992;32:109–134. doi: 10.1146/annurev.pa.32.040192.000545. [DOI] [PubMed] [Google Scholar]
- Ridge PG, Zhang Y, Gladyshev VN. Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS One. 2008;3:e1378. doi: 10.1371/journal.pone.0001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver LC, Imlay JA. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J Bacteriol. 2001;183:7182–7189. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel LM. A direct microdetermination for sulfide. Anal Biochem. 1965;11:126–132. doi: 10.1016/0003-2697(65)90051-5. [DOI] [PubMed] [Google Scholar]
- Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK. Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. J Am Chem Soc. 2001;123:11103–11104. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- Thomas JW, Puustinen A, Alben JO, Gennis RB, Wikstrom M. Substitution of asparagine for aspartate-135 in subunit I of the cytochrome bo ubiquinol oxidase of Escherichia coli eliminates proton-pumping activity. Biochemistry. 1993;32:10923–10928. doi: 10.1021/bi00091a048. [DOI] [PubMed] [Google Scholar]
- Tiranti V, Zeviani M. Altered sulfide (H2S) metabolism in ethylmalonic encephalopathy. Cold Spring Harb Persp Biol. 2013;5:a011437. doi: 10.1101/cshperspect.a011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler M, Henpita C, Bassett B, Kelley JL, Shaw JH. H2S exposure elicits differential expression of candidate genes in fish adapted to sulfidic and non-sulfidic environments. Comp Biochem Physiol Pt A. 2014;175:7–14. doi: 10.1016/j.cbpa.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Tseng CP, Albrecht J, Gunsalus RP. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol. 1996;178:1094–1098. doi: 10.1128/jb.178.4.1094-1098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MS, Woodberry T, Hafner LM, Giffard PM. The bspA locus of Lactobacillus fermentum BR11 encodes an L-cystine uptake system. J Bacteriol. 1999;181:2192–2198. doi: 10.1128/jb.181.7.2192-2198.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitvitsky V, Yadav PK, Kurthen A, Banerjee R. Sulfide oxidation by a noncanonical pathway in red blood cells generates thiosulfate and polysulfides. J Biol Chem. 2015;290:8310–8320. doi: 10.1074/jbc.M115.639831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Wan F, Fu H, Li N, Gao H. A matter of timing: contrasting effects of hydrogen sulfide on oxidative stress response in Shewanella oneidensis. J Bacteriol. 2015;197:3563–3572. doi: 10.1128/JB.00603-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang TX, Anderson B. The relationship between permeant size and permeability in lipid bilayer membranes. J Mem Biol. 1994;140:111–122. doi: 10.1007/BF00232899. [DOI] [PubMed] [Google Scholar]
- Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.