SUMMARY
Lipid droplets (LDs) are endoplasmic reticulum (ER)-derived lipid storage organelles uniquely encapsulated by phospholipid monolayers. LD membrane proteins are embedded into the monolayer in a monotopic hairpin-topology and therefore likely have requirements for their biogenesis distinct from those inserting as bitopic and polytopic proteins into phospholipid bilayers. UBXD8 belongs to a subfamily of hairpin-proteins that localize to both the ER and LDs, and are initially inserted into the cytoplasmic leaflet of the ER bilayer before partitioning to the LD monolayer. The molecular machinery responsible for inserting hairpin-proteins into membranes, however, is unknown. Here, we report that newly synthesized UBXD8 is posttranslationally inserted into discrete ER-subdomains by a mechanism requiring cytosolic PEX19 and membrane-integrated PEX3, proteins hitherto exclusively implicated in peroxisome biogenesis. Farnesylation of PEX19 uncouples ER/LD- and peroxisome targeting, expanding the function of this peroxin to an ER targeting pathway and suggesting a coordinated biogenesis of LDs and peroxisomes.
INTRODUCTION
Phospholipid bilayer membranes that ensheath cells and the organelles within them constitute a fundamental organizing principle of cellular life. Membrane-embedded proteins serve as conduits enabling selective permeability to solutes, as receptors transmitting signals between subcellular compartments, and as anchors segregating enzymes into functionally organized networks.
Not all organelles, however, are surrounded by lipid bilayers. Lipid droplets (LDs), cytoplasmic organelles that store metabolic energy as triglycerides (TG), are an exception to this principle of organelle architecture, as they are uniquely encapsulated by a phospholipid monolayer, which segregates their hydrophobic neutral lipid core from the aqueous cytosol1. As a consequence of their aliphatic interiors, LDs are unable to accommodate bi- or polytopic membrane proteins, and thus LD membrane proteins are integrated into the phospholipid monolayer monotopically through hydrophobic “hairpin” (HP) domains and expose all soluble domains to the cytosol2.
The prevailing model of LD biogenesis posits that local TG accumulation within the ER membrane bilayer triggers the budding of a LD from the cytoplasmic leaflet1. Several HP-proteins, including AUP13,4, GPAT45, AAM-B and UBXD86,7 exhibit a dual steady-state localization to LDs and the ER and are first integrated into the cytoplasmic leaflet of the ER membrane prior to localizing to LDs. Although the HP-domains of these proteins are necessary and sufficient for this dual localization, the molecular machinery by which HP-proteins are directed to and inserted into ER or LD membranes is unknown.
Most secreted and transmembrane proteins are directed to the ER by signal sequences that are engaged by signal recognition particle (SRP), which recruits ribosome-nascent chain complexes to ER-resident receptors, enabling cotranslational translocation of nascent polypeptides through the Sec61 translocon8,9. By contrast, C-terminal tail-anchored (TA) proteins are inserted into the ER membrane by a posttranslational pathway consisting of the cytosolic transmembrane-domain recognition complex (TRC)10,11 and the ER-resident receptors WRB and CAML12,13. Sequential transfer of TA-proteins from the initial recognition complex containing BAG6 to the membrane-embedded receptor complex is coordinated by cytosolic TRC4014.
In this study, we investigated the mechanism by which UBXD8, a HP-protein that partitions between the cytoplasmic leaflet of the ER and the LD monolayer6,7 and recruits the ATPase p97/VCP to membranes7, is targeted to and inserted into the ER. Our data reveal that UBXD8 is posttranslationally targeted to and inserted into the ER by a mechanism that is independent of the known SRP- or TRC-dependent pathways, but instead requires the peroxisome-biogenesis factors PEX19 and PEX3.
RESULTS
Posttranslational insertion of UBXD8 into ER membranes
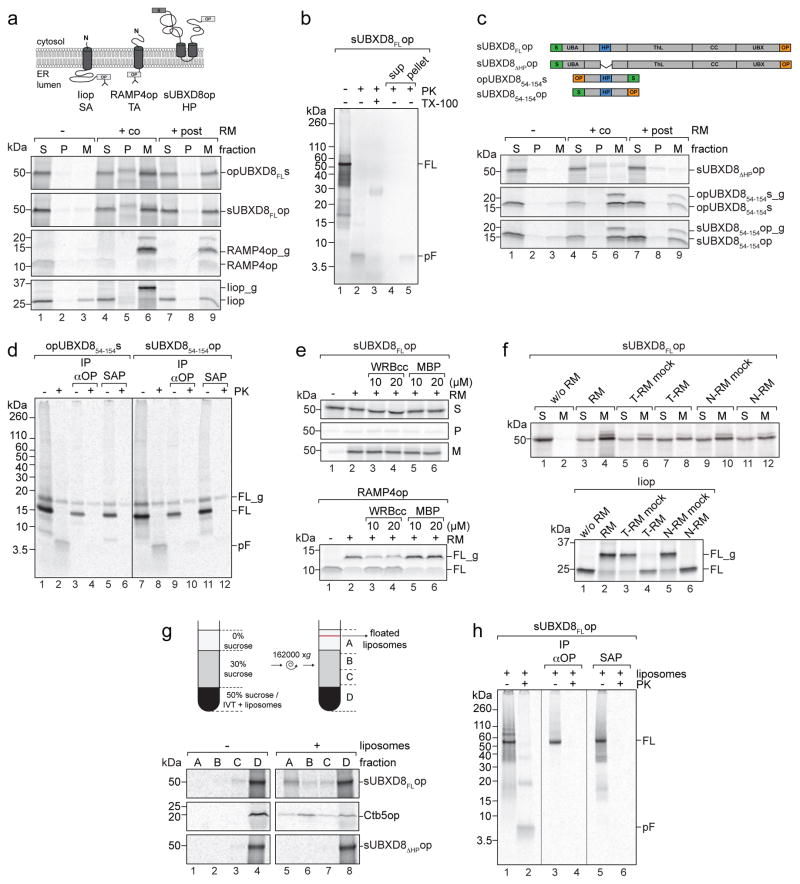
To study the mechanisms underlying membrane insertion of HP-proteins, we synthesized 35S-labeled, epitope-tagged UBXD8 in rabbit reticulocyte lysate (RRL) in the absence or presence of ER-derived rough microsomes (RMs) (Fig. 1a). Reactions were fractionated into soluble cytosolic proteins (S), peripheral membrane proteins released by extraction with sodium carbonate (P), and carbonate-resistant, membrane-integrated proteins (M). When translation was conducted in the presence of RMs, the majority of UBXD8 was detected in the membrane fraction (M) similar to the fractionation behaviour of an SRP-dependent signal anchor (SA) protein, invariant chain (Ii)15, and a tail-anchored (TA) protein, RAMP4op10. UBXD8 and RAMP4op were efficiently integrated into RMs when RMs were present during (cotranslational) or after termination (posttranslational) of protein synthesis. In contrast, Iiop was only cotranslationally integrated and glycosylated as expected for an SRP substrate (Fig. 1a). The opsin-tag contains a consensus N-glycosylation sequon that, upon ER membrane translocation, causes a ~2 kDa shift reflecting addition of an N-linked glycan. Iiop and RAMP4op were efficiently glycosylated when incubated with RMs (Fig. 1a). However, no UBXD8 glycosylation was detected, irrespective of whether the opsin-tag was at the N- or C-terminus, consistent with it being inserted into RMs in its native HP-topology where both termini face the cytosol. Protease treatment of RMs caused the 50 kDa band corresponding to full-length UBXD8 to collapse into a single fragment with the expected size (~5 kDa) of the protected HP-domain (Fig. 1b, lanes 1–2), which sedimented with membranes after re-fractionation and was digested upon detergent-solubilisation of RMs, consistent with it being membrane-integrated (Fig. 1b, lanes 3–5). Thus, in vitro synthesized UBXD8 can be posttranslationally integrated into RMs with the same HP-topology as in ER and LD membranes.
Figure 1. UBXD8 is posttranslationally inserted into membranes by a non-canonical ER-targeting pathway.
Indicated mRNAs were translated in RRL and membrane insertion monitored after incubation with RMs or liposomes by SDS-PAGE/autoradiography.
a) UBXD8 is posttranslationally inserted into RMs. Upper: Membrane topologies of opsin (op)-tagged signal-anchored (SA) invariant chain (Iiop), opsin-tagged tail-anchored (TA) RAMP4op, and opsin- and S-tagged UBXD8. Opsin-tag glycosylation sites are indicated. Lower: Proteins were synthesized in vitro without RMs (−), when RMs were present during translation (+co) or added after translation (+post). Membrane insertion was assessed by fractionation into soluble (S), carbonate-labile, peripheral membrane proteins (P) and carbonate-resistant, integral membrane proteins (M). _g, glycosylated.
b) Verification of UBXD8 topology. Proteinase K (PK) treatment of RMs with posttranslationally inserted full-length (FL) sUBXD8op in absence or presence of Triton X-100 (TX-100). Lanes 4+5: Refractionated samples. pF: protected fragment; sup: supernatant.
c) HP-domain is necessary and sufficient for posttranslational UBXD8 insertion into RMs. Upper: Constructs used. Lower: in vitro translation/translocation of radiolabeled proteins as in (a).
d) Verification of UBXD854-154 topology as in (b). Samples were precipitated with anti-opsin antibodies (IP αOP) or S-affinity purified (SAP).
e) TRC40 pathway-independent UBXD8 insertion. Posttranslational protein insertion into RMs in absence or presence of recombinant WRBcc or MBP. sUBXD8FLop (upper) was fractionated as in (a) and RAMP4op (lower) analysed directly. _g, glycosylated.
f) UBXD8 insertion persists after covalent modification of RMs. sUBXD8FLop (upper) and Iiop (lower) were cotranslationally incubated with RMs pre-treated with trypsin (T-RM) or N-ethylmaleimide (N-RM). sUBXD8FLop was fractionated as in (a) and Iiop analysed directly. _g, glycosylated.
g) UBXD8 associates with protein-free liposomes. Posttranslational liposome addition to in vitro translations followed by density gradient fractionation. Upper: Paradigm. Lower: Analysis of sUBXD8FLop, sUBXD8ΔHPop and opsin-tagged cytochrome b5 (Ctb5op).
h) UBXD8 HP-domain is inserted into liposomes. Posttranslational sUBXD8FLop insertion into liposomes as in (g) followed by protease digestion as in (d).
Representative autoradiograph from a single experiment are shown. The experiments in panels a, b, c, g were repeated twice and those in panels d, e, f, h were repeated once with similar results. Uncropped scans of all gels are available in Supplementary Figure 8.
The hydrophobic sequence in UBXD8 (amino acids 90-118) serves as a membrane HP-anchor6 that is necessary and sufficient for targeting UBXD8 to LDs in vivo16,17. To assess the role of the HP-domain in posttranslational insertion, we deleted the hydrophobic region (UBXD8ΔHP) or used a minimal UBXD8 version consisting of the HP-domain plus flanking residues (UBXD854-154) and monitored membrane insertion in vitro (Fig. 1c). No UBXD8ΔHP was detected in the membrane fraction after in vitro translation/translocation whereas UBXD854-154 was efficiently inserted into RMs under co- and posttranslational conditions. A minor fraction of UBXD854-154 became glycosylated (Fig. 1c), resisted protease treatment, and was efficiently affinity captured following protease treatment irrespective of the position of the tags (Fig. 1d). Therefore, a fraction of this minimal HP-construct was fully translocated across the ER membrane in vitro. The majority of membrane-associated UBXD854-154, however, gave rise to a protease-resistant ~5 kD fragment (Fig. 1d, lanes 2, 8) that failed to bind to N- or C- terminal affinity-capture reagents (Fig. 1d, lanes 4, 6, 10, 12), indicating correct UBXD854-154 insertion into RMs in a HP-topology. Thus, the HP-domain is necessary and sufficient for posttranslational UBXD8 insertion into the ER membrane.
UBXD8 membrane insertion is independent of SRP and TRC40
Posttranslational membrane insertion of UBXD8 could suggest employment of the TRC40-mediated ER-targeting pathway. To test this possibility, we used recombinant WRBcc, a soluble fragment of the TRC40 receptor WRB that binds to substrate-loaded TRC40, to block membrane insertion of TA-proteins by competing with endogenous WRB12. Inclusion of excess WRBcc in our in vitro translocation assays failed to alter UBXD8 insertion efficiency into RMs, despite substantially reducing the insertion of the TA-protein RAMP4op (Fig. 1e). Therefore, ER-insertion of UBXD8 is independent of the TRC40-WRB pathway.
To verify this conclusion and to assess the role of the canonical SRP pathway for UBXD8 insertion into RMs, we performed import assays using RMs that had been pre-treated with either N-ethylmaleimide or trypsin (Fig. 1f and Supplementary Fig. 1). Both conditions block SRP-18,19 and TRC40/WRB-dependent protein insertion10,20. While both treatments prevented insertion of the SRP substrate Iiop, neither interfered with UBXD8 insertion (Fig. 1f), establishing that UBXD8 integration is independent of the SRP and TRC40 pathways and, moreover, might not require ER-integrated proteins. Indeed, in vitro synthesized UBXD8 was present in buoyant fractions following incubation with protein-free liposomes, similar to the behaviour of cytochrome b5 (Fig. 1g), a protein known to insert into membranes independently of membrane-integrated proteins21. Association of UBXD8 with liposomes required the HP-domain as UBXD8ΔHP was retained in dense fractions upon sucrose gradient fractionation. Protease treatment of UBXD8-containing liposomes led to accumulation of a ~5 kDa protected fragment that bound to neither N- nor C-terminal affinity-capture reagents, indicating a correct HP-topology (Fig. 1h). Thus, UBXD8 can insert into membranes posttranslationally and independently of canonical SRP or TRC40 targeting pathways, protein-conducting channels or membrane protein receptors.
UBXD8 inserts into discrete ER-subdomains
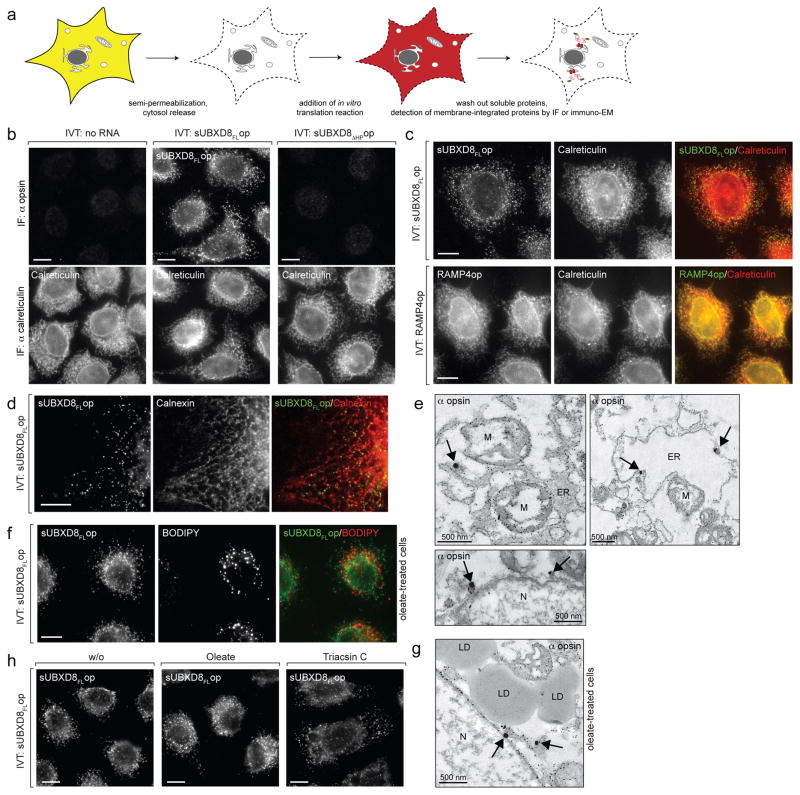
If UBXD8 membrane insertion is independent of membrane-integrated proteins, how then, is its strict localization in cells to the ER and LDs7 established and/or maintained? We used immunofluorescence microscopy to determine into which membranes in vitro synthesized UBXD8 inserts in semi-permeabilised cells (Fig. 2a). Full-length UBXD8 (sUBXD8FLop), but not sUBXD8ΔHPop, was recruited to discrete subcellular foci (Fig. 2b) that are strikingly different from the characteristic reticular distribution that endogenous UBXD8 exhibits in cells at steady-state. This punctate localization was not due to a general inability of proteins to insert into reticular ER in semi-permeabilised cells, since in vitro translated RAMP4op colocalised precisely with the ER marker calreticulin (Fig. 2c). By contrast, UBXD8 foci did not strictly colocalise with ER markers calreticulin (Fig. 2c) or calnexin (Fig. 2d) but exhibited a reticular pattern that closely followed the distribution of the ER (Fig. 2d and supplementary movie 1), suggesting UBXD8 insertion into distinct ER-subdomains. Indeed, immuno-electron microscopy (EM) of semi-permeabilised cells revealed labelling of sUBXD8FLop at ER membranes and on ~150 nm diameter electron-dense structures continuous with ER membranes (Fig. 2e, arrows). sUBXD8FLop failed to colocalise with LDs (Fig. 2f, g), indicating that newly synthesised UBXD8 preferentially inserts into ER-subdomains and not mature LDs. UBXD8 still inserted into foci after treatment with the long-chain fatty acyl CoA synthetase inhibitor, triacsin C (Fig. 2h), suggesting that neutral lipid synthesis is dispensable for recruitment of newly synthesized UBXD8 to ER-subdomains. These findings suggest that, while UBXD8 can spontaneously insert into protein-free membranes in vitro, it is specifically inserted into discrete ER-subdomains in semi-permeabilised cells before it distributes within the ER and eventually partitions to LDs. This implicates the existence of proteins specifying correct membrane targeting for nascent UBXD8 molecules.
Figure 2. UBXD8 is inserted into ER-subdomains in semi-permeabilised cells.
a) Experimental scheme: Cells grown on coverslips were semi-permeabilised to release cytosolic contents and incubated with in vitro translation reactions. membrane-integrated proteins were detected by immunofluorescence (IF) microscopy (b, c, d, f, h) or by immuno-electron microscopy (EM) (e and g).
b) The HP-region in UBXD8 is required for membrane insertion in semi-permeabilised cells. IF using anti-opsin and anti-calreticulin (ER marker) antibodies as indicated.
c) Full-length sUBXD8op inserts into discrete foci. IF of in vitro translated RAMP4op and sUBXD8FLop as in (b).
d) UBXD8 insertion sites align with the ER marker calnexin. IF of sUBXD8FLop co-stained with anti-calnexin antibodies. A single z-section is shown. Supplementary movie 1 shows a full z-stack.
e) UBXD8 insertion sites are associated with the ER. Electron micrographs showing immunogold labelled sUBXD8FLop using anti-opsin and fluoronanogold-coupled antibodies after insertion into semi-permeabilised cells. Arrows indicate specific gold-labelling at ER membranes. Small gold particles attached to all membrane structures represent non-specific background labelling due to gold enhancement, which is also found in the control specimen to which no in vitro translated protein was added (not shown). N: nucleus; M: mitochondria.
f – g) UBXD8 is not directly inserted into LDs. Before semi-permeabilisation and import of in vitro translated sUBXD8FLop, cells were treated with oleate to induce LD formation and analysed (f) by IF with anti-opsin antibodies (BODIPY labels LDs) or (g) by EM as in (e).
h) UBXD8 inserts into ER foci in LD-depleted cells. Cells were treated with triacsin C to inhibit neutral lipid synthesis and sUBXD8FLop was detected by IF using anti-opsin antibodies.
Representative images from a single experiment are shown. The experiment in panels a, b, c were repeated twice, in panels d, e were repeated thrice, and in panels f–h were repeated once, all with similar results.
Scale bars, 10 μm unless indicated.
BAG6 and PEX19 bind to newly synthesized UBXD8 in the cytosol
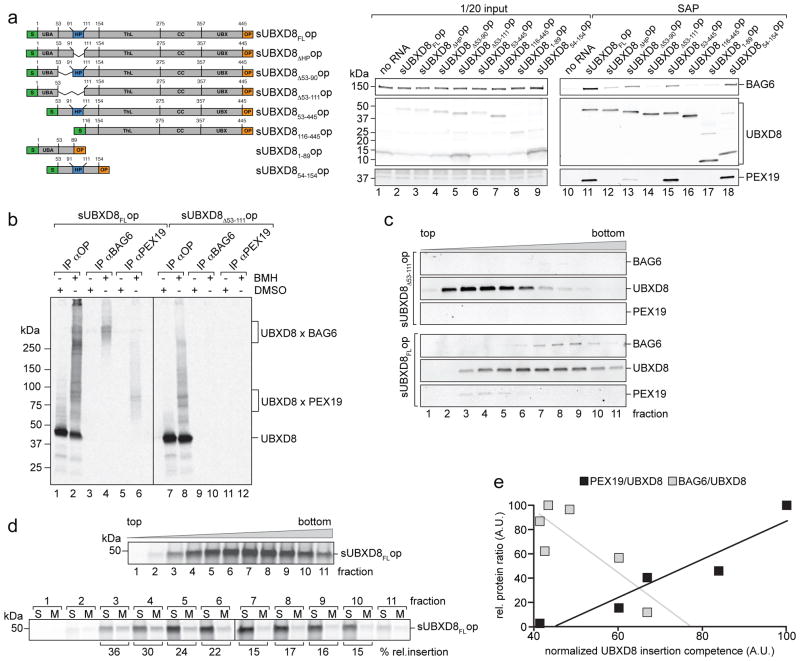
To identify potential targeting factors of newly synthesized UBXD8, we translated UBXD8 in RRL in the absence of RMs, which should favour prolonged engagement with cytoplasmic proteins maintaining its insertion-competent state, and used affinity capture followed by LC-MS/MS identification of proteins that specifically bound insertion-competent UBXD8FL but not UBXD8ΔHP. Two proteins, BAG6 and PEX19, were exclusively captured from UBXD8FL pull-downs (Supplementary Table 1). To verify these interactions and to map the interaction sites within UBXD8, we translated UBXD8 deletion mutants in vitro and assessed the amount of PEX19 and BAG6 present in complex with UBXD8 by affinity isolation and immunoblotting (Fig. 3a). The UBXD8 HP-region was both necessary and sufficient to engage PEX19 and BAG6. Because the deletion of the proline-rich sequence immediately preceding the annotated HP-region (UBXD8Δ53-90) reduced binding to PEX19 and BAG6 we extended the UBXD8 HP-deletion to include amino acids 53-111 for all subsequent experiments.
Figure 3. UBXD8 pre-insertion complexes contain PEX19 and BAG6.
a) HP-dependent UBXD8 binding to BAG6 and PEX19. Indicated UBXD8 constructs (left panel) were translated in RRL in the absence of membranes, S-affinity purified (SAP) and analysed by SDS-PAGE and immunoblotting with opsin- (UBXD8), BAG6- or PEX19-specific antibodies. Lanes 1–9 show 1/20 of input reaction, lanes 10–18 show corresponding elution fractions.
b) Direct interaction of UBXD8 with BAG6 and PEX19 assessed by chemical crosslinking. sUBXD8FLop or sUBXD8Δ53-111op were translated in RRL and treated with the crosslinker BMH, or DMSO, immunoprecipitated under denaturing conditions using the indicated antibodies and visualised by autoradiography. Mobilities of monomeric UBXD8 and crosslinked adducts for UBXD8-BAG6 and UBXD8-PEX19 are indicated.
c) Full-length UBXD8 forms distinct complexes with PEX19 and BAG6. sUBXD8FLop or sUBXD8Δ53-111op translated in RRL were fractionated on sucrose density gradients, UBXD8 complexes S-affinity purified from individual fractions and analysed by SDS-PAGE and immunoblotting using anti-opsin (UBXD8), -BAG6 and -PEX19 antibodies.
d – e) UBXD8 insertion competence correlates with PEX19 but not BAG6 association.
d) Sucrose gradient fractionation of radiolabeled sUBXD8FLop complexes as described in (c) but analysed by autoradiography (upper). RMs were added to individual fractions and UBXD8 insertion competence quantified by densitometry after fractionation into soluble (S) and membrane-integrated (M) proteins as in Fig. 1 (lower).
e) Relative UBXD8 insertion competence into RMs as quantified in (d) was plotted against the relative protein amounts of PEX19 or BAG6 associated with UBXD8 in the individual sucrose gradient fractions. Immunoblot analysis and protein quantification of a parallel sucrose gradient fractionation is shown in Supplementary Fig. 2. Linear least-squares regression analysis of association with PEX19 (R2 = 0.89) or BAG6 (R2 = 0.67) vs. UBXD8 insertion competence. A.U.: arbitrary units.
Representative autoradiographs from a single experiment are shown. The experiments in panels a–d were repeated once with similar results. Uncropped scans of all gels are available in Supplementary Figure 8.
To test whether BAG6 and PEX19 bind to UBXD8 directly, we used chemical crosslinking to generate radiolabeled, covalent pre-insertion complex adducts of in vitro synthesized UBXD8, which were immunoisolated following protein denaturation (Fig. 3b). The BAG6 antibody precipitated high molecular weight (>250 kDa) crosslinked adducts containing UBXD8FL (lane 4), while the PEX19 antibody captured adducts of ~80 kDa (lane 6). Neither antibody precipitated UBXD8Δ53-111 adducts. These findings confirm HP-domain-dependent interactions of UBXD8 with BAG6 and PEX19 and strongly suggest that they are direct.
To determine whether PEX19 and BAG6 bind UBXD8 in the same or in distinct complexes, we assessed their presence in affinity-isolated UBXD8 preinsertion-complexes fractionated on sucrose gradients (Fig. 3c). UBXD8FL forms higher molecular weight complexes compared to UBXD8Δ53-111 and cofractionated with BAG6 in fractions 6–10, whereas PEX19 was associated with UBXD8FL in fractions 3–6, indicating that UBXD8FL forms distinct complexes with PEX19 and BAG6.
UBXD8 in fractions containing PEX19 was 2-fold more efficiently integrated into RMs than was UBXD8 in BAG6-containing fractions (Fig. 3d). Insertion competence correlated positively with the ratio of PEX19 to UBXD8 in individual fractions (Fig. 3e and Supplementary Fig. 2), suggesting that PEX19-containing complexes facilitate insertion of newly synthesized UBXD8 into ER membranes.
PEX19 specifies the subcellular localization of UBXD8
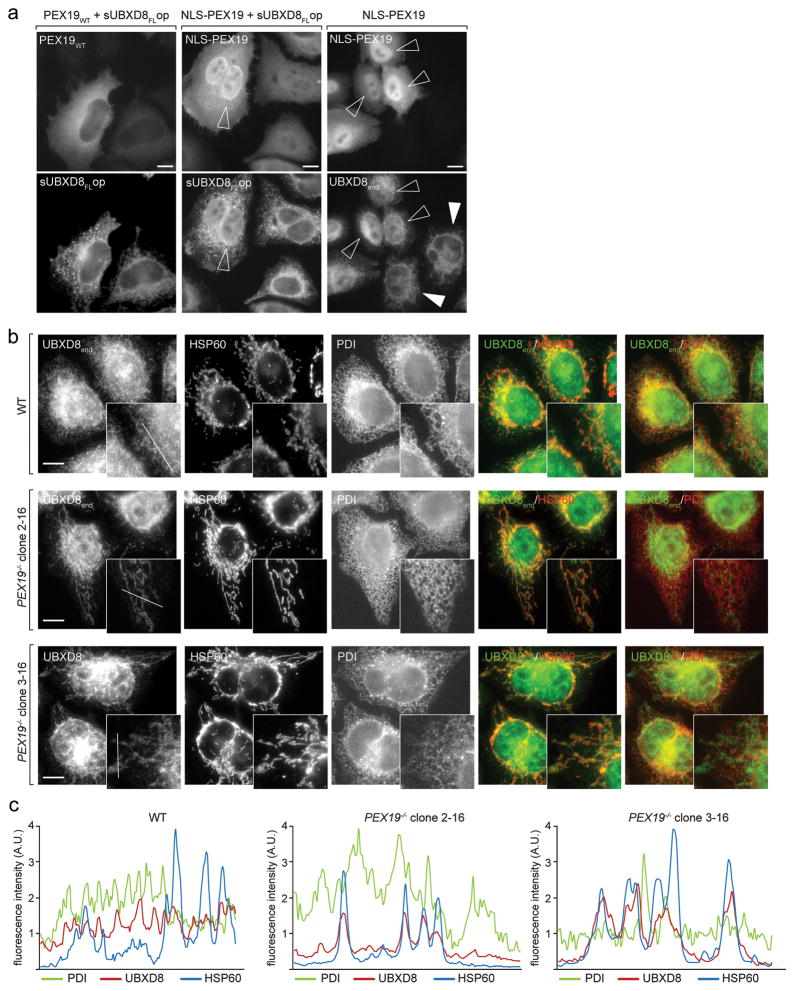
To test whether UBXD8 is a client for PEX19-mediated protein targeting in living cells, we overexpressed PEX19 appended with a nuclear localisation signal, NLS-PEX19, and monitored UBXD8 localisation (Fig. 4a). Re-direction of both, coexpressed sUBXD8op as well as endogenous UBXD8 to the nucleus demonstrates that PEX19 is sufficient to re-localize UBXD8 in cells.
Figure 4. PEX19 specifies the subcellular localization of UBXD8.
a) UBXD8 follows PEX19 redirected to the nucleus. Effect of wild-type (WT) or NLS-tagged PEX19 on co-expressed or endogenous UBXD8 localization assessed by IF microscopy. PEX19, sUBXD8FLop and endogenous UBXD8 were detected using anti-PEX19, anti-opsin or anti-UBXD8 antibodies, respectively. Open arrowheads indicate transfected cells with high nuclear NLS-PEX19 accumulation. White arrowheads indicate non-transfected cells with reticular staining for endogenous UBXD8. Representative images from a single experiment are shown. The experiment was repeated twice with similar results. All scale bars = 10 μm.
b) UBXD8 mislocalises to mitochondria in the absence of PEX19. Endogenous UBXD8 was detected by IF in WT cells or two different PEX19−/− clonal cell lines using anti-UBXD8 antibodies. Results are representative of nine individual PEX19−/− clonal cell lines derived from four different guide RNAs. Representative images from a single experiment are shown. The experiment was repeated twice with similar results. Anti-HSP60 and anti-PDI antibodies stain mitochondria and ER, respectively. All scale bars = 10 μm.
c) Fluorescence intensity line profiles as depicted in UBXD8 insets in (b) reveal increased correlation of UBXD8 localization with mitochondria in PEX19−/− cells. A.U.: arbitrary units. Representative line profiles from a single experiment are shown. The experiment was repeated twice with similar results.
To determine whether PEX19 is required for ER localization of UBXD8, we generated PEX19−/− cell lines by genome-editing (Supplementary Fig. 3a–c). Strikingly, the majority of endogenous UBXD8 in PEX19−/− cells was mislocalised to mitochondria and colocalised with HSP60 (Fig. 4b, c). UBXD8 localization to ER and LDs was restored when PEX19 was reintroduced into these cells, confirming that its mislocalisation was caused by the absence of PEX19 (Supplementary Fig. 3d). Thus, PEX19 specifies steady-state ER-localisation of endogenous UBXD8.
PEX19 and PEX3 cooperate in UBXD8 insertion into ER-subdomains
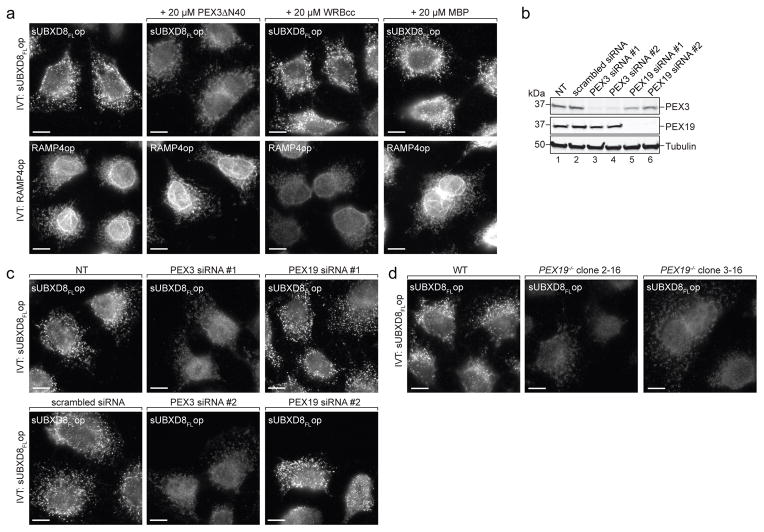
To investigate how PEX19 targets newly synthesized UBXD8 to ER-subdomains, we tested the effect of recombinant PEX3ΔN40, a soluble variant of the membrane-resident PEX19-receptor PEX322, which is required for membrane insertion of peroxisomal membrane proteins (PMPs)23, on the insertion of in vitro synthesized UBXD8 in semi-permeabilised cells. In the presence of excess PEX3ΔN40, but not WRBcc or MBP, sUBXD8FLop was not inserted into ER foci but instead distributed more diffusely consistent with mitochondrial mislocalisation (Fig. 5a, upper). In contrast, RAMP4op insertion was unaffected by PEX3ΔN40 addition but sensitive to WRBcc (Fig. 5a, lower). These data establish an essential role for PEX19 in specifying insertion of newly synthesized UBXD8 into ER-subdomains, which we propose to be ER entry-sites for newly synthesised UBXD8.
Figure 5. PEX19 and PEX3 are essential for UBXD8 insertion into ER-subdomains.
a) UBXD8 insertion into ER foci is inhibited by competition with PEX3ΔN40. sUBXD8FLop or RAMP4op were translated in RRL, incubated with semi-permeabilised wild-type (WT) cells in the presence of indicated purified proteins and detected by immunofluorescence (IF) as described in Fig. 2. Representative images from a single experiment are shown. The experiment was repeated twice with similar results. All scale bars = 10 μm.
b–c) UBXD8 insertion into ER foci requires PEX3.
b) WT cells were transfected with indicated siRNAs and analysed by SDS-PAGE and immunoblotting with indicated antibodies. NT: non-transfected. Uncropped scans are available in supplementary Figure 8. Representative immunoblots from a single experiment are shown. The experiment was repeated once with similar results.
c) In vitro translated sUBXD8FLop was added to semi-permeabilised WT cells transfected with siRNA constructs as in (b) and analysed by IF. NT: non-transfected. Representative images from a single experiment are shown. The experiment was repeated once with similar results. All scale bars = 10 μm.
d) UBXD8 insertion into ER foci requires PEX19/PEX3. In vitro translated sUBXD8FLop was incubated with semi-permeabilised WT or two individual PEX19−/− clonal cell lines and detected by IF as in Fig. 2. Representative images from a single experiment are shown. The experiment was repeated once with similar results. All scale bars = 10 μm.
Scale bars = 10 μm.
PEX3 knock-down in wild-type cells (Fig. 5b) abolished the insertion of in vitro translated sUBXD8FLop into foci after semi-permeabilisation (Fig. 5c), indicating that PEX3 is essential for correct UBXD8 insertion. In contrast, PEX19 knockdown did not affect sUBXD8FLop insertion (Fig. 5b, c), likely because of the presence of RRL-derived PEX19 bound to in vitro translated sUBXD8FLop (Fig. 3). Semi-permeabilised PEX19−/− cells, however, were not competent for inserting in vitro translated sUBXD8FLop into ER-subdomains (Fig. 5d), most likely because these cells are also depleted of PEX3 (Supplementary Fig. 3b), consistent with a PEX19 role in stabilizing PEX324. Thus, both cytosolic PEX19 and membrane-integrated PEX3 are required for correct targeting and insertion of UBXD8 into ER-subdomains.
UBXD8 ER insertion-sites colocalise with endogenous PEX3 but are distinct from mature peroxisomes
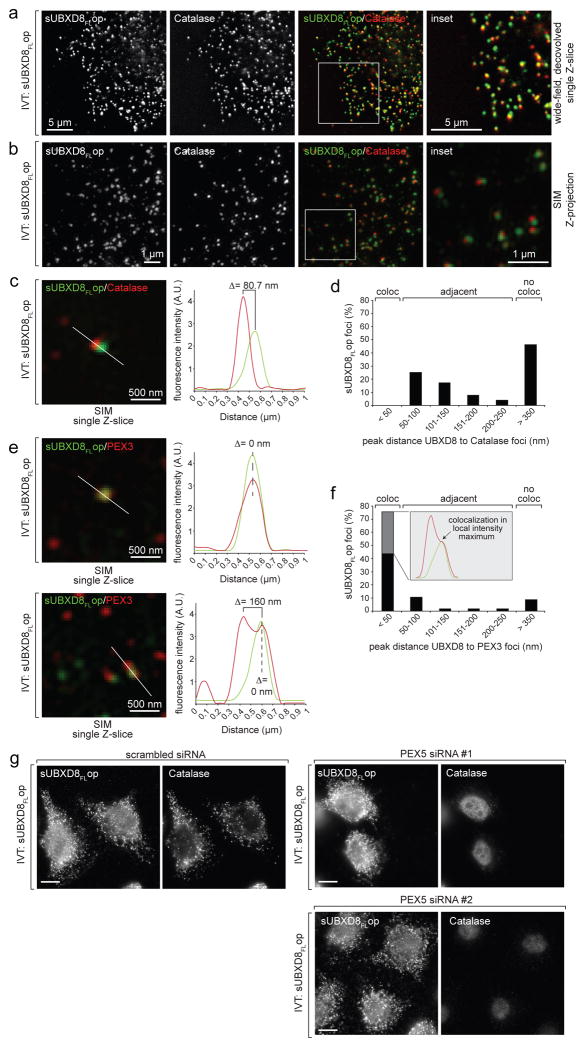
Since PEX19 and PEX3 are known to insert peroxisomal membrane proteins (PMPs) into peroxisomes, we investigated the spatial relationship of UBXD8 insertion-sites with peroxisomes in semi-permeabilised cells. Catalase-positive peroxisomes did not colocalise with UBXD8 foci but were found in close apposition (50–250 nm) in about half the cases (Fig. 6a–d). This relationship between UBXD8 insertion-sites and peroxisomes was confirmed with the peroxisomal membrane protein PEX14 (Supplementary Fig. 4).
Figure 6. UBXD8 insertion sites co-localize with endogenous PEX3 but not with mature peroxisomes.
a–d) UBXD8 insertion sites are in close proximity to but distinct from peroxisomes. In vitro translated sUBXD8FLop was imported into semi-permeabilised wild-type (WT) cells and visualized by IF microscopy as in Fig. 2. Anti-catalase antibodies mark mature peroxisomes. or SIM (b, c, e). Positions of insets are indicated by white boxes. A.U.: arbitrary units.
a) Representative single Z-slice from deconvolution wide-field microscopy.
b) Representative SIM Z-stack projection.
c) Representative SIM z-slice inset (left) and fluorescence intensity profile from indicated line scan (right) illustrates how colocalisation of sUBXD8FLop foci with catalase-positive foci was quantified. Data are from a single cell image representative of >50 cells and at least 3 SIM images.
d) Quantification of the distances between sUBXD8FLop foci and catalase-positive foci from SIM micrographs analysed as illustrated in (c).
e–f) Two distinct colocalisation phenotypes for sUBXD8FLop foci with endogenous PEX3.
e) Left, two representative fields of single SIM z-slices after co-staining for sUBXD8FLop and endogenous PEX3 are shown to illustrate either complete colocalisation (upper) or double foci with one colocalised spot adjacent to a PEX3-positive but sUBXD8FLop-negative spot (lower). Right, fluorescence intensity profiles from indicated line scans as they were used to quantify colocalisation of sUBXD8FLop foci with PEX3-positive foci in (f).
f) Quantification of the distances between sUBXD8FLop foci and PEX3-positive foci obtained from line profiles of fluorescence intensity as in (e). Grey-shaded bar indicates percentage of sUBXD8FLop foci that colocalised with PEX3 foci and were also found to be adjacent to sUBXD8FLop-negative PEX3 foci within a distance of 50–250 nm.
g) Insertion of UBXD8 into ER foci is independent of mature peroxisomes. WT cells were transfected with indicated siRNAs for 120h, used for semi-permeabilisation and import of sUBXD8FLop, and stained with anti-opsin or anti-catalase antibodies as indicated. Scale bars = 10 μm.
Representative images from a single experiment are shown. The experiment in panel a was repeated 4 times, in panels b, d, f, once, and in panel g twice, with similar results. In panels d, f, 57 foci were analysed.
In contrast, the majority (75%) of UBXD8 foci colocalised with endogenous PEX3 (Fig. 6e, f). However, we observed two distinct colocalisation phenotypes: While 44% of total UBXD8 foci colocalised with a single PEX3 focus, an additional 31% of UBXD8 foci colocalised with PEX3 and were also adjacent (50–250 nm) to an additional PEX3-positive but UBXD8-negative focus. PEX3 resides in peroxisomes and the ER25–28. Since UBXD8 insertion-sites are positive for PEX3 but negative for catalase, our data suggest that UBXD8 is specifically inserted into PEX3-containing sites that are not peroxisomes, potentially corresponding to pre-peroxisomal ER29.
To assess the role of mature peroxisomes in UBXD8 insertion into ER-subdomains, we depleted cells of PEX5, an essential peroxin for import of peroxisomal matrix proteins30 (Supplementary Fig. 5a). Although these cells lacked mature peroxisomes (Supplementary Fig. 5b), no defect in the import of in vitro translated sUBXD8op after semi-permeabilisation was observed, indicating that mature peroxisomes are dispensable for UBXD8 insertion (Fig. 6g).
PEX19 farnesylation is essential for UBXD8 localisation to the ER and LDs
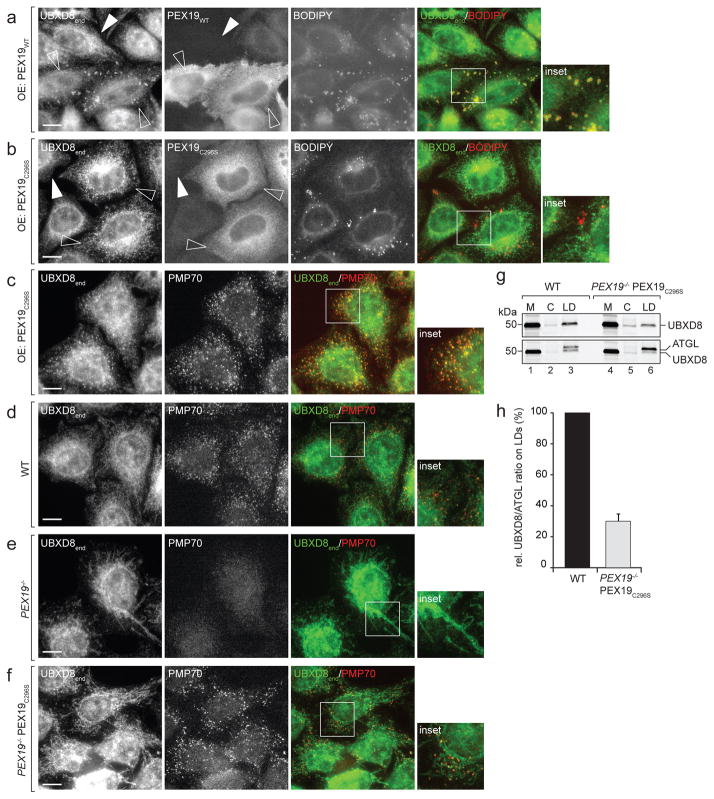
PEX19 is known to be farnesylated in cells but this posttranslational modification is dispensable for peroxisome biogenesis31. To test whether PEX19 farnesylation affects UBXD8 localisation, we overexpressed either PEX19WT or the farnesylation-deficient mutant PEX19C296S in wild-type cells and monitored the steady-state localization of endogenous UBXD8 by immunofluorescence (Fig. 7a–c). Overexpression of either PEX19WT (Fig. 7a) or PEX19C296 (Fig. 7b) did not disrupt the ER distribution of endogenous UBXD8, presumably because endogenous wild-type PEX19 is still present in these cells. Interestingly, however, overexpression of PEX19WT led to pronounced accumulation of UBXD8 on LDs as revealed by BODIPY co-staining (Fig. 7a). We previously reported that UBXD8 accumulation on LDs results either from overexpression of UBXD8 or from induction of LD biogenesis by oleate treatment7. The observation that PEX19WT overexpression induces endogenous UBXD8 accumulation on LDs in cells not loaded with oleate suggests that PEX19 is limiting for UBXD8 trafficking to LDs.
Figure 7. PEX19 farnesylation determines UBXD8 localization in cells.
a–c) Dominant-negative effect of PEX19 farnesylation mutant on ER and LD targeting of endogenous UBXD8. Wild-type (WT) cells were transfected with either WT PEX19 (a) or with the farnesylation mutant PEX19C296 (b and c). PEX19, UBXD8 and PMP70 were detected by IF microscopy using antibodies and LDs were visualized using BODIPY staining. Insets show boxed regions; Open and white arrowheads indicate transfected and non-transfected cells, respectively. Scale bars = 10 μm..
a) Overexpression of PEX19WT in WT cells promotes trafficking of endogenous UBXD8 to LDs. Representative images from a single experiment are shown, the experiment was repeated twice with similar results.
b–c) Overexpression of PEX19C296 in WT does not promote trafficking of endogenous UBXD8 to LDs but instead causes mislocalisation to peroxisomes. Representative images from single experiments are shown, the experiments were repeated once with similar results.
d–f) Stable expression of PEX19C296 in PEX19−/− cells rescues peroxisome biogenesis but not mitochondrial mislocalization of endogenous UBXD8.
WT cells, PEX19−/− cells or PEX19−/− cells stably expressing PEX19C296 (PEX19−/− PEX19C296) as indicated were analysed by IF microscopy using UBXD8- and PMP70-specific antibodies. Insets show boxed regions; Scale bars = 10 μm. Representative images from single experiments are shown, the experiments were repeated once with similar results. Images in (f) are representatives of three individual clonal PEX19−/− PEX19C296 cell lines.
g–h) UBXD8 levels on LDs are severely reduced in PEX19−/− PEX19C296 cells.
g) Oleate-treated WT and PEX19−/− PEX19C296 cells were fractionated into membranes (M), cytosol (C) and LDs. A representative immunoblot from a single experiment is shown, the experiment was repeated twice with similar results. Membranes were first decorated with UBXD8-specific antibodies (upper) and then redecorated with anti-ATGL antibodies (lower). Uncropped scans are available in supplementary Figure 8.
h) Relative UBXD8 amounts in LD fractions compared to the LD-resident membrane protein ATGL were quantified by densitometry from immunoblots as shown in (g). n=3 independent experiments normalised to values obtained from WT cells. Mean ± s.e.m. (=4.9%)..
In striking contrast, PEX19C296S overexpression in wild-type cells led to enrichment of UBXD8 in punctate structures that did not correlate with LDs (Fig. 7b), but colocalised with the peroxisome marker PMP70 (Fig. 7c). This dominant-negative effect of overexpressed PEX19C296S on UBXD8 distribution in wild-type cells suggests that PEX19 farnesylation is essential to prevent delivery of UBXD8 to peroxisomes and to promote partitioning to ER sites from where it can be mobilized to LDs. Indeed, while stable expression of PEX19C296S rescued peroxisome biogenesis in PEX19−/− cells (Fig. 7d–f, Supplementary Fig. 6), it failed to complement PEX19 function on the localisation of endogenous UBXD8, which remained mislocalised to mitochondria as in PEX19−/− cells (Fig. 7e, f). Moreover, a fraction of endogenous UBXD8 in PEX19−/− PEX19C296S cells was also present on peroxisomes (Fig. 7f), reflecting the profound mistargeting of UBXD8 to peroxisomes upon PEX19C296S overexpression in wild-type cells (Fig. 7b, c). Additionally, we found that the amount of endogenous UBXD8 on LDs isolated from oleate-treated PEX19−/− PEX19C296S cells was strongly (~70%) reduced compared to wild-type cells (Fig. 7g, h). Together, these findings demonstrate an essential role for PEX19 farnesylation in directing UBXD8 to ER and LD membranes.
DISCUSSION
Posttranslational protein integration into membranes implies that biosynthesis on cytosolic ribosomes is mechanistically uncoupled from membrane insertion and raises the question of how organelle-specific UBXD8 targeting within a cellular context is achieved. Selective targeting requires favoured delivery to the correct target membrane and prevention of promiscuous integration into inappropriate membranes. In this study we identified PEX19 and BAG6 as direct HP-domain-specific binding partners of newly synthesized UBXD8 and provide three lines of evidence supporting the conclusion that PEX19 specifies correct targeting of UBXD8 to ER membranes in cells. First, misdirecting PEX19 to the nucleus leads to nuclear accumulation of UBXD8. Second, in the absence of PEX19 endogenous UBXD8 is mislocalized to mitochondria, consistent with our observation that UBXD8 can insert into protein-free membranes and with studies showing that other posttranslationally inserted membrane proteins can accumulate in mitochondrial membranes when their respective organelle targeting pathways are disrupted32–34. Third, blocking PEX19 function with a soluble fragment of PEX3 in semi-intact cell import-assays prevents insertion of in vitro synthesized UBXD8 into ER-subdomains and also causes mitochondrial mislocalisation.
The role for BAG6 in UBXD8 biogenesis is less clear. This multifunctional chaperone has been implicated in a variety of cellular processes including ER-membrane targeting, protein quality control, and ERAD14,35–37. BAG6 could contribute to UBXD8 targeting independently of or in collaboration with PEX19, or alternatively, could participate in UBXD8 turnover (Supplementary figure 7). Further investigation is required to assess these possibilities.
PEX19 and PEX3 are essential for de novo peroxisome biogenesis at the ER and for posttranslational insertion of PMPs into peroxisomal membranes30,38. PEX3 can be biosynthetically inserted into the ER25–27, where it concentrates in a discrete subdomain termed the preperoxisomal ER (pER)28,29. Preperoxisomal vesicles bud from the pER in a process dependent on PEX3’s interaction with PEX1924,39,40 and a PEX3 function in intra-ER sorting and packaging of PMPs has been suggested40. Our finding that newly synthesized UBXD8 inserts into PEX3-containing ER-subdomains, leads us to speculate that the pER may have a more general role; perhaps as an ER-domain specialized for sorting of membrane proteins that are targeted to the ER by non-canonical insertion pathways. Our data establish that PEX19 farnesylation, which is dispensable for peroxisome biogenesis31, is essential for UBXD8 insertion into the ER and LD partitioning. It may therefore serve to segregate HP-anchored proteins destined for LDs from bilayer-spanning peroxisomal proteins. Further studies will reveal whether and how this covalent lipid modification influences PEX19’s association with PEX3-containing ER-subdomains.
LDs and peroxisomes both originate from the ER membrane and have complementary roles in lipid metabolism41. LDs store neutral lipids and hydrolyse them into fatty acids, which are further metabolised in peroxisomes. Conversely, peroxisomes uniquely synthesize ether lipids, which account for up to 20% of the neutral lipid content of LDs and are absent from LDs in cells lacking peroxisomes42,43. LDs and peroxisomes are spatially associated and their juxtaposition with the ER44 suggests that all three organelles are intimately coupled to balance lipid storage and consumption. Our finding that peroxisomal proteins and LD-destined HP-proteins share targeting machinery raises the hypothesis that LD and peroxisome biogenesis may be mechanistically coordinated in the ER. We previously reported that UBXD8 positively regulates LD abundance by controlling the activity of the major lipase on LDs7. Thus, coordinating the biogenesis of such a protein with peroxisome biogenesis could allow mutual control of metabolic functions fulfilled by these organelles that have to act in concert during metabolic change.
METHODS
Reagents
Canine pancreas rough microsomes were a gift from Bernhard Dobberstein and stored at 2 eq/μl in RM buffer (250 mM sucrose, 50 mM Hepes/KOH pH 7.6, 50 mM KOAc, 2 mM Mg(OAc)2, 1 mM DTT). Purified WRBcc and MBP was a kind gift from Fabio Vilardi and have been described earlier45.
Plasmids and antibodies
Expression constructs for RAMP4op and Ctb5op were a gift from Bernhard Dobberstein and are described elsewhere46. UBXD8 constructs used in this study are derived from previously published UBXD8 expression plasmids 47 that were used as templates for PCR-based cloning using primers either encoding an s-tag (MKETAAAKFERQHMDS) or an opsin-tag (GPNFYVPFSNKTG) as well as either an XbaI or a NotI restriction site. PCR products were digested with the indicated enzymes and ligated into an empty pCDNA3.1(−) vector cut with the same enzymes. UBXD8 constructs lacking internal amino acid sequences (ΔHP, Δ53-90, Δ53-111) were generated by primer-extension overlap PCR. Following digestion of the parental plasmid DNA by DpnI the PCR reaction was transformed into E. coli DH5alpha, positive clones identified by restriction digest/sequencing and then subcloned into an empty pCDNA3.1(−) vector using XbaI and NotI. Similarly, to introduce a PreScission protease cleavage site preceding the C-terminal S-tag in UBXD8 constructs, primers encoding the amino acid sequence LEVLFQGP were used for primer extension overlap PCR. pRK5rs-Iiop48 was used for cloning Iiop into pCDNA3.1(−) by PCR and ligation of the XbaI/NotI fragment. PEX19 expression constructs are derived from a cDNA clone (Thermo clone ID: 2820701) and cloned into pCDNA3.1(−) by generating PCR products with XbaI/NotI restriction sites. The reverse primer used to generate PEX19C296S encoded the G887C mutation. To generate N-terminally NLS-tagged PEX19, a forward primer encoding the amino acid sequence MAPKKKRKVGDGS was used. To generate N-terminally hexa-histidine-tagged PEX3ΔN40 for bacterial expression and purification we used a PEX3 cDNA (true clone origene SC117821), a forward primer encoding an NcoI restriction site followed by caccaccaccaccaccac encoding six consecutive histidines and complementary to the authentic PEX3 sequence lacking the fist 40 amino acids as well as a reverse primer encoding an NotI restriction site. The PCR product was cloned into a pET21d vector using the indicated enzymes.
All constructs were verified by sequencing and detailed sequence information will be made available upon request.
The mouse monoclonal anti-opsin (R2-15; 1:1000 IB, 1:300 IF, 3 μl IP), rabbit anti-PEX349 (1:1000 IB, 1:100 IF) and rabbit anti-BAG650 (1:1000 IB; 5 μl IP) antibodies were kindly provided by Bernhard Dobberstein, Gabriele Dodt and Stephen High, respectively. Antibodies against calnexin (ADI-SPA-865, Enzo Life Sciences; 1:2000 IB, 1:300 IF), calreticulin (ADI-SPA-680, Enzo Life Sciences, 1:1000 IB, 1:100 IF), PEX19 (ab137072, Abcam; 1:1000 IB, 1:100 IF, 5 μl IP), PDI (ADI-SPA-891, Enzo Life Sciences; 1:1000 IB, 1:100 IF), UBXD8 (16251-1-AP, PTG; 1:1000 IB, 1:100 IF), PMP70 (NBP2-36770, Novus Biologicals; 1:200 IF), Catalase (D4P7B, Cell Signaling; 1:1000 IB, 1:500 IF), tubulin (T6199, Sigma-Aldrich; 1:10000 IB), PEX14 (10594-1-AP, PTG; 1:100 IF), PEX5 (12545-1-AP, PTG; 1:1000 IB), ATGL (21385, Cell signalling; 1:2000 IB) and HSP60 (sc-1052; Santa Cruz Biotechnology, 1:50 IF) are commercially available. IRDye secondary antibodies (926-68020, 926-68021, 926-32211, 926-32214, Licor) were used for immuno blotting (1:15000) and donkey-derived, Cy3-, Alexa488- or Cy5- conjugated secondary antibodies (715-165-151, 711-165-152, 715-545-151, 711-545-152, 705-485-147, 715-175-151, Jackson Immunoresearch) for immunofluorescence (1:1000).
In vitro transcription/in vitro translation
All mRNAs were generated from PCR products using the RiboMax large-scale RNA production system T7 supplemented with m7G cap analogue (Promega), DNase I digested, and purified using Microspin G-25 columns (GE Healthcare) according to manufacturer’s instructions. Proteins were translated in rabbit reticulocyte lysate (RRL; Promega) either supplemented with complete amino-acid mix or, to synthesize radiolabeled proteins, with amino-acid mix lacking methionine and (35S)-protein labelling mix from Perkin Elmer (11 μCi/μl) for 45 min at 30 °C.
Protein insertion assays into RMs, liposomes and semi-permeabilised cells
For cotranslational protein insertion, 0.2 eq/μl RMs were present during the in vitro translation reaction, whereas for posttranslational insertion RMs were added for 30 min after the translation reaction was stopped by addition of 2.5 mM puromycin. Soluble proteins (fraction S) were separated from membranes by centrifugation (100000 ×g, 5 min) through a sucrose cushion. Peripheral proteins were released from these membranes with 0.1 M carbonate pH 11.0 (fraction P) and membrane-integral proteins (fraction M) re-isolated by centrifugation (130000 ×g, 10 min). Proteins from all fractions were precipitated by adding 2 volumes saturated ammonium sulfate and analysed by SDS-PAGE and autoradiography.
Liposomes were generated from egg PC (L-α-phosphatidylcholine; Avanti Polar Lipids) in liposome buffer (50 mM Hepes/KOH pH 7.6, 50 mM KOAc, 2 mM Mg(OAc)2 pH 7.5) to a final concentration of 8 mg/ml and an average diameter of 100 nm by extrusion. 0.5% Texas-Red DHPE (Invitrogen) was incorporated to visualize the liposome-containing fractions. Liposomes were posttranslationally added to the in vitro translation reaction to a final concentration of 0.8 mg/ml PC and incubated for 30 min. For liposome flotation, a 12 μl reaction was mixed with 236 μl liposome buffer containing 50% sucrose, overlaid with 500 μl liposome buffer containing 30% sucrose and 250 μl liposome buffer, centrifuged in a TLS-55 for 3h at 162000 ×g and 4 °C. 4 fractions (300, 200, 200, 300 μl) were collected from to top to bottom, supplemented with 50 μg insulin as a carrier and proteins precipitated with ammonium sulfate followed by SDS-PAGE and autoradiography.
For protein insertion into semi-permeabilised cells, cells were seeded onto glass cover slips and semi-permeabilised by 0.003% digitonin in S-buffer (250 mM sucrose, 20 mM Hepes/KOH pH 7.4, 2.5 mM Mg(OAc)2, 25 mM KCl, 2.5 mM EGTA, 1 mM DTT, protease inhibitors) for 5 min. After washing out the cytosol, in vitro translation reactions were added to the cells posttranslationally for 30 min, non-inserted proteins removed by washing in S-buffer and cells fixed with 4% formaldehyde in PBS. Proteins of interest were detected by standard immunofluorescence protocols or samples processed for immuno-electron microscopy.
Pre-treatment of RMs
For trypsin-treatment, pelleted RMs were resuspended in 20 μg/ml trypsin (Promega, sequencing grade) freshly dissolved in PSB (50 mM Hepes/KOH pH7.4, 100 mM KOAc, 2 mM Mg(OAc)2) and incubated for 1h on ice. The reaction was stopped by adding 20 μg/ml aprotinin (SIGMA) and 2 mM PMSF and incubation on ice for 15 min. RMs were collected by centrifugation through a 500 mM sucrose cushion in PSB, washed in aprotinin- and PMSF-containing PSB and re-collected by centrifugation through a sucrose cushion. For NEM-treatment, pelleted RMs were resuspended in PSB containing 2 mM NEM and incubated for 30 min at 25 °C. The reaction was quenched with 20 mM DTT and RMs collected by centrifugation through a 500 mM sucrose cushion in PSB.
In both cases the final RM pellet was resuspended in RM buffer to a final volume equal to the starting material and stored in aliquots at −80°C after flash freezing. As controls, RMs were treated as outlined above without adding trypsin or NEM, respectively.
Protease-protection assays
After protein insertion into RMs, membranes were resuspended in 50 mM Hepes/KOH pH 7.6, 50 mM KOAc, 2 mM Mg(OAc)2 and incubated with 2 mg/ml Proteinase K (Invitrogen) for 45 min at 30 °C. Digestion was stopped by adding 5 mM PMSF and samples were either directly added to boiling SDS-sample buffer, further fractionated by centrifugation through a sucrose cushion containing 5 mM PMSF (130000 ×g, 10 min), or subjected to affinity purification after protein denaturation in boiling SDS (1% in Tris/HCl pH 8.0) and 1:10 dilution with 1% Triton X-100, 100 mM NaCl, 50 mM Hepes pH 7.4.
For digestion of proteins inserted into liposomes, proteinase K was added to the in vitro insertion reaction and liposomes isolated by flotation as described. Proteins in the liposome-containing top fraction were either TCA precipitated in presence of 0.5% Triton X-100 as a carrier or affinity isolated after solubilisation with 1% Triton X-100 in 10 mM Tris/HCl pH 7.5, 150 mM NaCl, 2 mM EDTA. Proteins were analysed by autoradiography after separation on 12% BisTris NuPAGE precast gels with MES buffer (Invitrogen).
Analyses of UBXD8 pre-insertion complexes by S-affinity isolation, chemical crosslinking and sucrose gradient fractionation
S-tagged UBXD8 variants were translated in RRL (40 μl reaction volume) in the absence of membranes, the reaction stopped by addition of 2.5 mM puromycin and diluted with 900 μl PBS. UBXD8 complexes were affinity isolated using S-agarose beads (Novagen). After washing in cleavage buffer (20 mM Tris/HCl pH 7.5, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA) proteins were eluted from the beads by addition of SDS-sample buffer and heating at 65 °C for 10 min. Presence of UBXD8, PEX19 and BAG6 in the purified complexes was assessed by immunoblotting following SDS-PAGE.
For chemical crosslinking, radiolabeled UBXD8 variants were translated in RRL as described above. After diluting the reactions with 10 volumes PBS, they were incubated with 250 mM BMH crosslinker (Pierce), or DMSO as a negative control, for 30 min at 25°C. The reactions were quenched with 20 mM DTT, supplemented with 10 mM Tris/HCl pH7.5 and proteins denatured by adding 1% SDS and heating for 10 min at 55 °C. After diluting the reaction 10fold with IP buffer A (10 mM Tris/HCl pH7.5, 150 mM NaCl, 2 mM EDTA, 0.4% Triton X-100) proteins were immuno-precipitated. After washing the beads with IP buffer A, IP buffer B (10 mM Tris/HCl pH7.5, 500 mM NaCl, 2 mM EDTA, 0.2% NP-40) and 10 mM Tris/HCl pH7.5 proteins were eluted with SDS-sample buffer and incubation at 65 °C for 10 min followed by SDS-PAGE and visualization by autogradiography.
To separate UBXD8 pre-insertion complexes, 100 μl in vitro translation reactions were layered onto 2 ml 5–20% sucrose gradients (in 50 mM Hepes/KOH pH7.4, 100 mM KOAc, 2 mM MgCl2) and centrifuged for 5 h at 55000 rpm at 4 °C in a TLS-55 rotor. Ten fractions (200 μl each) were collected from the top, diluted with 900 μl PBS and subjected to S-affinity purification, SDS-PAGE and immunoblotting as described above. For membrane insertion assays, radiolabeled UBXD8 complexes were separated on sucrose gradients, 100 μl of each fraction supplemented with 8 eq RMs for 30 min at 30 °C, and insertion reactions fractionated into soluble (S), peripheral (P) and membrane-integrated proteins (M). UBXD8 amounts in the individual S and M fractions were quantified by densitometry and relative amounts in the M fraction compared to the sum of S and M calculated. For regression analyses, protein amounts of affinity-isolated UBXD8 and associated PEX19 and BAG6 in the individual sucrose gradient fractions were quantified from a parallel immunoblotting experiment by densitometry and plotted against UBXD8 membrane insertion efficiency.
Cell culture and transfection
HeLa Kyoto cells51 were cultivated at exponential growth rates in DMEM containing 4.5 g/L glucose and glutamine (Corning) supplemented with 10% FCS (Gemini) at 37°C and 5% CO2 and regularly tested for the absence of mycoplasma. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. We did not attempt to authenticate our cell lines. Fugene 6 (Promega) was used for transient plasmid- and Lipofectamine 2000 (Life Technologies) for transient siRNA-transfections according to manufacturer’s instructions. Two individual Silencer Select pre-designed siRNAs specific for PEX3, PEX19 and PEX5, respectively, and a scrambled siRNA control were used at a final concentration of 3.3 nM (Life Technologies; IDs: s16154, s16156, s11612, s11613, s11630, s11632, 4390843). After 4h of siRNA transfection cells were supplemented with fresh medium. Cells were then either grown for additional 72h before further processing (PEX3 and PEX19 knock-down) or 48h later seeded for a second round of siRNA transfection and processed 120h after the first transfection (PEX5 knock-down).
For LD induction, cells were treated with 200 μM oleic acid in complex with 0.2% BSA in standard medium for 16h. To deplete LDs from cells 10 μM triacsin C was added to cells for 16h.
For generating clonal PEX19−/− cell lines stably expressing PEX19C296S, selection medium (DMEM / 10% FBS / 500 μg/ml geneticin) was added to cells 48h post-transfection. Pools stably expressing PEX19C296S were seeded for clonal selection after 2 weeks. Single cell-derived clones were isolated, individually expanded under selection pressure and analysed by immuno-fluorescence and -blotting.
Immunoblotting
For quantitative immunoblotting, cells were harvested in PBS, lysed in 1% Triton X-100, 50 mM Hepes pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM PMSF, Complete EDTA-free protease inhibitors (Roche) for 15 min on ice and samples cleared by centrifugation (16000 ×g, 10 min, 4°C). Protein concentration was determined by BCA assay (Pierce), equal protein amounts separated by SDS-PAGE, followed by wet-transfer onto nitrocellulose membranes. 5% skim milk in TBS-T was used to block unspecific binding and to dilute antibodies. IRDye secondary antibodies (LiCor) were used for signal detection by Odyssey imaging (LiCor). Band intensities were quantified by densitometry using either ImageJ or Image Studio Lite software (LiCor).
Immunofluorescence
For immunofluorescence, cells were seeded onto glass coverslips (#1.5 high precision), fixed with 4% formaldehyde in PBS for 20 min and permeabilised with 0.1% Triton X-100 for 10 min. Unspecific binding sites were blocked with either 1% BSA or 3% BSA/ 5% FBS in PBS followed by antibody incubation in the same solution. BODIPY 493/503 (Invitrogen) was used at 5 μg/ml to stain LDs. Specimen were mounted on glass slides using Fluoromount G (EMS) or slow fade gold (Invitrogen) and analysed using a Zeiss AxioImager.M1 microscope with PlanApochromat oil objectives (63x or 100x/ 1.4 N.A.) and appropriate filter sets. Usually 8 individual z-sections in 300 nm intervals were collected using a CoolSNAP HQ camera (Photometrics). Where indicated, images were deconvolved using the Slidebook software and the nearest neighbour setting. Individual datasets were normalized for brightness/contrast and merged, and pseudo-colored pictures generated using ImageJ software. Micrographs were cropped and assembled using Adobe Photoshop and Illustrator.
For structured illumination microscopy, specimens were examined with the Deltavision OMX Blaze 3D-SIM microscope equipped with 3 emCCD (Photometrics, Evolve 512) and an U-PLANAPO SIM objective (100x/1.42 N.A.). Individual z-sections of 125 nm were taken, reconstructed and aligned using the SoftWoRx software. Parameters for channel alignment were verified on the same day by calibration with fluorescent beads. For assessment of colocalisation, fluorescence intensity profiles were plotted and intensity peak distances measured using Image J. All SIM micrographs have a pixel size of 40.35 nm.
Electron Microscopy
After protein insertion into semi-permeabilised cells, cells were fixed with 4% formaldehyde/PBS for 20 min, washed in PBS, and quenched with 50 mM glycine/PBS for 5 min. After washing in PBS, unspecific binding sites were blocked with 3%BSA/ 5% FBS/ PBS for 20 min followed by incubation with anti-opsin antibodies for 1h. Fluoro-nanogold, which is a 1.4 nm nanogold particle and Alexa Flour 488 fluorophore coupled to an affinity-purified Fab′ fragment (Nanoprobes, Cat. 7202) was used as a secondary probe in a 1:300 dilution. Specific labelling of sUBXD8op insertion sites as seen with conventional immunofluorescence was verified by examining a parallel sample by fluorescence microscopy. For ultrastructural characterization specimens were fixed in 4% PFA/ 2% glutaraldehyde/ 0.1M Cacodylate. Gold-particles were enhanced for 6 min using GoldEnhance EM Plus (Nanoprobes) followed by post-fixation in 1% OsO4 for 1h and en bloc staining with 1% uranyl acetate over night. After dehydration samples were embedded into Epon resin and 80 nm ultra-thin sections collected on carbon-coated copper grids. Contrasting was performed with a 1:1 mix of 3% uranyl acetate and acetone for 30 sec followed by lead citrate staining (0.2%) for 3 min. Specimens were examined with a Joel, JEM1400 transmission electron microscope equipped with a Gatan Orius 10.7 megapixel CCD camera at 120kV.
Large-scale affinity-purification and mass spectrometry of UBXD8 preinsertion complexes
Large-scale (1 ml) in vitro translation reactions without mRNA or supplemented with mRNAs of opUBXD8FLPPs or opUBXD8ΔHPPPs encoding a PreScission protease cleavage site (LEVLFQGP) preceding the C-terminal S-tag were carried out for 45 min at 30°C and terminated by adding 2.5 mM puromycin. Potential aggregated protein species and ribosomes were removed by centrifugation (260000 ×g, 30 min) and opUBXD8-PPs-complexes isolated on S-agarose beads. Beads were extensively washed in a spin column format with cleavage buffer (20 mM Tris/HCl pH7.5, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA) and incubated in 2 bed volumes cleavage buffer supplemented with 8 units PreScission protease (GE Healthcare) for 4h at 8 °C with shaking. Proteins were eluted from the column by centrifugation (2 min at 200 ×g) and incubated with glutathione sepharose beads to retain GST-tagged PreScission protease during a second elution. Proteins in the final eluates were precipitated by TCA and separated by SDS-PAGE. Whole lanes were excised from the gel, cut into pieces and subjected to in-gel tryptic digestion, peptide extraction and subsequent peptide identification by LC-MS/MS.
Expression and purification of recombinant HIS-tagged PEX3ΔN40
Expression of N-terminally hexa-histidine-tagged PEX3ΔN40 was induced in E. coli Rosetta (DE3) cells (Novagen) with 1 mM IPTG at 0.5 OD600 for 16h at 18°C. Cells were pelleted and resuspended in buffer A (20 mM Tris/HCl pH8.0, 150 mM NaCl, 5% glycerol, 10 mM imidazole) and cells lysed by adding 0.5 mg/ml lysozyme, 1 mM EDTA, 1 mM PMSF, and Complete protease inhibitors (Roche) for 30 min. After addition of 10 mM MgCl2 and 10 μg/ml DNase I, the lysate was cleared by centrifugation (90000 ×g for 30 min) and loaded onto a HisTrap HP column (GE Healthcare). After washing with 30 column volumes (CV) buffer A containing 500 mM NaCl and with 20 CV of buffer A, proteins were eluted with a linear imidazole gradient over 10 CV using buffer A and buffer B (20 mM Tris/HCl pH8.0, 150 mM NaCl, 5% glycerol, 500 mM imidazole). Fractions containing PEX3ΔN40 were subjected to ultra-filtration and buffer exchange to 20 mM Tris/HCl pH8.0, 150 mM NaCl, 5% glycerol, 1 mM DTT and protein purity of greater than 95% was assessed by SDS-PAGE and coomassie staining.
Generation of PEX19 knock-out cell lines by CRISPR/Cas9-based genome-editing
HeLa Kyoto cells lacking endogenous PEX19 were generated by CRISPR/Cas9-mediated genome editing according to Ran et al.52 and protocols published by the Zhang lab (rev20130212 at http://www.genome-engineering.org/crispr/). In brief, we used an online CRISPR design tool (http://crispr.mit.edu/) to select four individual guide RNA sequences targeting exon1 or exon 2 of genomic PEX19 and designed the following oligos accordingly:
PEX19-Exon1-guide1: caccGTGTCGGGGCCGAAGCGGAC and aaacGTCCGCTTCGGCCCCGACAC;
PEX19-Exon1-guide2: caccgTGTCGGGGCCGAAGCGGACA and aaacTGTCCGCTTCGGCCCCGACAc;
PEX19-Exon1-guide3: caccgTGAGGAAGGCTGTAGTGTCG and aaacCGACACTACAGCCTTCCTCAc;
PEX19-Exon2-guide1: caccGGGCCCCAGAAGAGATCGCC and aaacGGCGATCTCTTCTGGGGCCC;
Oligos were phosphorylated, annealed and cloned into pX330 (Addgene) using BbsI. CRISPR constructs were verified by sequencing and transfected into cells using Fugene 6 (Roche). After 48h, transfected cells were diluted to select clonal cell lines, which were screened for the absence of PEX19 by immunoblotting using anti-PEX19 antibodies (Abcam) and by immunofluorescence using anti-catalase antibodies to verify the absence of peroxisomes. At least two individual clonal cell lines derived from each gRNA were used to verify PEX19-mediated effects on UBXD8-insertion into semi-permeabilised cells and on endogenous UBXD8 localization.
Cellular fractionation of lipid droplets
Four sub-confluent 10 cm dishes of oleate-loaded cells were harvested in ice-cold PBS, resuspended and incubated for 10 min in hypotonic lysis medium (HLM; 20 mM Tris/HCl pH7.4, 1 mM EDTA) containing 250 mM sucrose and Complete EDTA-free protease inhibitors (Roche), and lysed by passaging ten times through a 25G1/2 needle. The post-nuclear supernatant (5 min, 500 ×g) was adjusted to a final concentration of 20% sucrose, overlaid with HLM and centrifuged for 1h at 172000 ×g and 4°C in a TLS-55 rotor. The buoyant LD fraction was collected using a tube slicer (Beckman), the cytosolic fraction by pipetting and the membrane-containing pellet washed three times with HLM. Proteins in the LD fraction were solubilized for 20 min in 2% Triton X-100 at 65 °C, precipitated with 10% TCA and washed twice with acetone. Equivalent percentages of cytosolic and membrane fractions were analysed next to TCA-precipitated LD proteins by SDS-PAGE and quantitative immunoblotting.
Data availability
All data supporting the findings of this study are available without undue qualification from the corresponding author on request.
Statistics and reproducibility
Uncropped scans of all gels are available in supplementary Figure 8.
Experiments used for statistical quantification were repeated independently three times (n=3) and normalised to values obtained from WT cells. The mean and the standard error of the mean are displayed as bar graph with error bars.
Micrographs, autoradiographs and immunoblots shown are representative for at least two independent experiments as depicted in the individual figure legends. For assessment of sUBXD8op foci colocalisation with endogenous proteins at least 50 foci, as depicted in the individual figure legends, were analysed from a representative single experiment. The experiment itself was repeated at least once. For genome-edited PEX19−/− cells nine independent clonal cell lines were characterized with similar results. For PEX19−/− cell lines stably expressing PEX19C296S three independent clonal cell lines were analysed with similar results.
Supplementary Material
Acknowledgments
We thank Josh Elias and Fiona McAllister for protein identification by mass spectrometry, John Perrino for gold-enhancement, embedding and sectioning of the EM-samples, and Jon Mulholland for help with structured illumination microscopy. We are grateful to Bernhard Dobberstein, Fabio Vilardi, Vincenzo Favaloro, Stephen High and Gabriele Dodt for generously sharing valuable reagents, to Shizuka Bridget Yamada for technical help, to Maxence Nachury for access to his fluorescence microscope, and to David Mick and Maggie Pearce for critical reading of the manuscript. This work was supported by a National Institutes of Health grant GM074874 to R.R.K. and, in part, by the Shared Instrumentation Grants (SIG) 1S10OD01227601 and 1S10RR02678001.
Footnotes
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
B.S. performed and analysed all experiments, prepared the figures and wrote the first draft of the manuscript. B.S. and R.R.K jointly conceived the experimental design, interpreted the results and wrote subsequent drafts of the manuscript.
References
- 1.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiele C, Spandl J. Cell biology of lipid droplets. Curr Opin Cell Biol. 2008;20:378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Stevanovic A, Thiele C. Monotopic topology is required for lipid droplet targeting of ancient ubiquitous protein 1. J Lipid Res. 2013;54:503–513. doi: 10.1194/jlr.M033852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klemm EJ, Spooner E, Ploegh HL. Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J Biol Chem. 2011;286:37602–37614. doi: 10.1074/jbc.M111.284794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilfling F, et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zehmer JK, et al. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J Cell Sci. 2009;122:3694–3702. doi: 10.1242/jcs.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olzmann JA, Richter CM, Kopito RR. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 9.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 10.Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J Cell Sci. 2008;121:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 12.Vilardi F, Lorenz H, Dobberstein B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J Cell Sci. 2011;124:1301–1307. doi: 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto Y, Sakisaka T. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol Cell. 2012;48:387–397. doi: 10.1016/j.molcel.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 14.Mariappan M, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipp J, Dobberstein B. Signal recognition particle-dependent membrane insertion of mouse invariant chain: a membrane-spanning protein with a cytoplasmically exposed amino terminus. The Journal of cell biology. 1986;102:2169–2175. doi: 10.1083/jcb.102.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki M, et al. Derlin-1 and UBXD8 are engaged in dislocation and degradation of lipidated ApoB-100 at lipid droplets. Mol Biol Cell. 2012;23:800–810. doi: 10.1091/mbc.E11-11-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JN, et al. Identification of Ubxd8 protein as a sensor for unsaturated fatty acids and regulator of triglyceride synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21424–21429. doi: 10.1073/pnas.1011859107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer DI, Dobberstein B. Identification and characterization of a membrane component essential for the translocation of nascent proteins across the membrane of the endoplasmic reticulum. The Journal of cell biology. 1980;87:503–508. doi: 10.1083/jcb.87.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. The Journal of cell biology. 1982;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favaloro V, Vilardi F, Schlecht R, Mayer MP, Dobberstein B. Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J Cell Sci. 2010;123:1522–1530. doi: 10.1242/jcs.055970. [DOI] [PubMed] [Google Scholar]
- 21.Brambillasca S, et al. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 2005;24:2533–2542. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt F, et al. Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19. J Biol Chem. 2010;285:25410–25417. doi: 10.1074/jbc.M110.138503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang Y, Morrell JC, Jones JM, Gould SJ. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. The Journal of cell biology. 2004;164:863–875. doi: 10.1083/jcb.200311131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt F, et al. The role of conserved PEX3 regions in PEX19-binding and peroxisome biogenesis. Traffic. 2012;13:1244–1260. doi: 10.1111/j.1600-0854.2012.01380.x. [DOI] [PubMed] [Google Scholar]
- 25.Aranovich A, Hua R, Rutenberg AD, Kim PK. PEX16 contributes to peroxisome maintenance by constantly trafficking PEX3 via the ER. J Cell Sci. 2014;127:3675–3686. doi: 10.1242/jcs.146282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toro A, et al. Evaluation of the role of the endoplasmic reticulum-Golgi transit in the biogenesis of peroxisomal membrane proteins in wild type and peroxisome biogenesis mutant CHO cells. Biol Res. 2007;40:231–249. doi: 10.4067/s0716-97602007000200014. [DOI] [PubMed] [Google Scholar]
- 27.Mayerhofer PU, Bano-Polo M, Mingarro I, Johnson AE. Human Peroxin PEX3 Is Co-translationally Integrated into the ER and Exits the ER in Budding Vesicles. Traffic. 2016;17:117–130. doi: 10.1111/tra.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. The Journal of cell biology. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geuze HJ, et al. Involvement of the endoplasmic reticulum in peroxisome formation. Mol Biol Cell. 2003;14:2900–2907. doi: 10.1091/mbc.E02-11-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim PK, Hettema EH. Multiple Pathways for Protein Transport to Peroxisomes. J Mol Biol. 2015;427:1176–1190. doi: 10.1016/j.jmb.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vastiau IM, et al. Farnesylation of Pex19p is not essential for peroxisome biogenesis in yeast and mammalian cells. Cell Mol Life Sci. 2006;63:1686–1699. doi: 10.1007/s00018-006-6110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halbach A, et al. Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J Cell Sci. 2006;119:2508–2517. doi: 10.1242/jcs.02979. [DOI] [PubMed] [Google Scholar]
- 33.Sacksteder KA, et al. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. The Journal of cell biology. 2000;148:931–944. doi: 10.1083/jcb.148.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JG, Ye Y. Bag6/Bat3/Scythe: a novel chaperone activity with diverse regulatory functions in protein biogenesis and degradation. Bioessays. 2013;35:377–385. doi: 10.1002/bies.201200159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hessa T, et al. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodrigo-Brenni MC, Gutierrez E, Hegde RS. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol Cell. 2014;55:227–237. doi: 10.1016/j.molcel.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agrawal G, Subramani S. Emerging role of the endoplasmic reticulum in peroxisome biogenesis. Front Physiol. 2013;4:286. doi: 10.3389/fphys.2013.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 40.Agrawal G, Fassas SN, Xia ZJ, Subramani S. Distinct requirements for intra-ER sorting and budding of peroxisomal membrane proteins from the ER. The Journal of cell biology. 2016;212:335–348. doi: 10.1083/jcb.201506141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19:380–392. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hajra AK, Das AK. Lipid biosynthesis in peroxisomes. Ann N Y Acad Sci. 1996;804:129–141. doi: 10.1111/j.1749-6632.1996.tb18613.x. [DOI] [PubMed] [Google Scholar]
- 43.Bartz R, et al. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 44.Tabak HF, Murk JL, Braakman I, Geuze HJ. Peroxisomes start their life in the endoplasmic reticulum. Traffic. 2003;4:512–518. doi: 10.1034/j.1600-0854.2003.00110.x. [DOI] [PubMed] [Google Scholar]
- 45.Vilardi F, Lorenz H, Dobberstein B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J Cell Sci. 2011;124:1301–1307. doi: 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J Cell Sci. 2008;121:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olzmann JA, Richter CM, Kopito RR. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schrul B, Kapp K, Sinning I, Dobberstein B. Signal peptide peptidase (SPP) assembles with substrates and misfolded membrane proteins into distinct oligomeric complexes. Biochem J. 2010;427:523–534. doi: 10.1042/BJ20091005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt F, et al. The role of conserved PEX3 regions in PEX19-binding and peroxisome biogenesis. Traffic. 2012;13:1244–1260. doi: 10.1111/j.1600-0854.2012.01380.x. [DOI] [PubMed] [Google Scholar]
- 50.Leznicki P, High S. SGTA antagonizes BAG6-mediated protein triage. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19214–19219. doi: 10.1073/pnas.1209997109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landry JJ, et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 2013;3:1213–1224. doi: 10.1534/g3.113.005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available without undue qualification from the corresponding author on request.