Abstract
Bradyrhizobium japonicum Mur and Escherichia coli Fur are manganese- and iron-responsive transcriptional regulators, respectively, that belong to the same protein family. Here, we show that neither Mur nor Fur discriminate between Fe2+ and Mn2+ in vitro, nor is there a metal preference for conferral of DNA-binding activity on the purified proteins. When expressed in E. coli, B. japonicum Mur responded to iron, but not manganese, as determined by in vivo promoter occupancy and transcriptional repression activity. Moreover, E. coli Fur activity was manganese-dependent in B. japonicum. Total and chelatable iron levels were higher in E. coli than in B. japonicum under identical growth conditions, and Mur responded to iron in a B. japonicum iron export mutant that accumulated high levels of the metal. However, elevated manganese content in E. coli did not confer activity on Fur or Mur, suggesting a regulatory pool of manganese in B. japonicum that is absent in E. coli. We conclude that the metal selectivity of Mur and Fur depends on the cellular context in which they function, not on intrinsic properties of the proteins. Also, the novel iron sensing mechanism found in the rhizobia may be an evolutionary adaptation to the cellular manganese status.
INTRODUCTION
Approximately 40% of all proteins require a bound metal to function (Andreini et al., 2008). Because metals can be both scarce and toxic, cells must regulate gene expression to control metal-dependent processes and maintain homeostasis. Members of the Fur family of bacterial transcriptional regulators sense and respond to the cellular status of a specific divalent metal to regulate genes under their control (Bsat et al., 1998, Gaballa and Helmann, 1998, Hamza et al., 1998, Patzer and Hantke, 1998, Ahn et al., 2006, Diaz-Mireles et al., 2004, Chao et al., 2004, Platero et al., 2004).
Fur is the first and perhaps best studied member of this family. Fur binding to Fe2+ confers activity on the protein, resulting in the recognition and binding to cis-acting elements within the promoters of genes within its regulon. The Fur family regulators Zur, Nur and Mur sense and respond to Zn2+, Ni2+ and Mn2+, respectively, and appear to be mechanistically similar to Fur, except that they recognize different metals (Shin et al., 2011, Ahn et al., 2006, Diaz-Mireles et al., 2004, Platero et al., 2007, Hohle and O’Brian, 2010). The PerR protein found in Bacillus subtilis and some other gram-positive bacteria is an H2O2-responsive regulator in which bound Fe2+ catalyzes histidine oxidation to inactivate the regulator and derepress target genes (Lee and Helmann, 2006).
Fur is the global iron-dependent transcriptional regulator in E. coli, Bacillus subtilis and many other bacteria, and its wide phylogenetic distribution suggests that it is evolutionarily ancient. Fur has been described predominantly as a repressor, but examples of Fur-dependent activation have been reported (Yu and Genco, 2012). High affinity transport genes are among those repressed by Fur, which are expressed under low iron conditions to scavenge available iron from the environment.
Bradyrhizobium japonicum lives as a free-living organism or as the symbiont of soybean, where it converts atmospheric nitrogen to ammonia within plant cells of root nodules. B. japonicum belongs to the α-Proteobacteria, a diverse taxonomic group that that includes intracellular pathogens, symbionts, photosynthetic organisms, bacteria that degrade environmental pollutants, and the abundant marine bacterium Pelagibacter ubique (Rappe et al., 2002). B. japonicum and many related organisms do not use Fur for global iron-dependent transcriptional control. Instead, the Irr protein, also a Fur family member, positively and negatively regulates iron stimulon genes (O’Brian, 2015). Unlike other Fur family proteins, Irr does not directly bind its regulatory metal, but rather it senses and responds to an iron-dependent process, namely the synthesis of heme (Qi and O’Brian, 2002). Irr interacts directly with ferrochelatase, the enzyme that catalyzes the insertion of ferrous iron into protoporphyrin in the final step of heme biosynthesis. Irr is degraded (Qi and O’Brian, 2002) or inactivated (Singleton et al., 2010) by heme when iron is sufficient, but is active under iron limitation.
The Mur protein from B. japonicum and other rhizobia share about 50% similarity to E. coli Fur, and has been co-opted to respond to manganese rather than iron (Chao et al., 2004, Diaz-Mireles et al., 2004, Platero et al., 2004, Hohle and O’Brian, 2009, Menscher et al., 2012). Magnetospririllum gryphiswaldense and Caulobacter crescentus are α-Proteobacteria, but are more distantly related to the rhizobia, and their Fur homologs are iron-dependent regulators (da Silva Neto et al., 2009, Deng et al., 2015). The metal-binding residues of M. gryphiswaldense Fur have been elucidated by X-ray crystallography (Deng et al., 2015), and they are conserved in B. japonicum Mur as well as in Fur proteins from E. coli, Pseudomonas aeruginosa and other organisms (Fig. S1). Thus, the basis of metal specificities of Mur and Fur proteins in vivo are not readily apparent from amino acid sequences.
The function of Mur as a manganese-responsive regulator is consistent with observations indicating that B. japonicum is more reliant on manganese under non-stressed conditions than E. coli and perhaps other organisms as well. A B. japonicum mntH mutant is almost completely defective in high-affinity Mn2+-uptake, and has a severe growth defect under normal growth conditions (Hohle and O’Brian, 2009). Further, B. japonicum employs an outer-membrane channel specific for Mn2+ (Hohle et al., 2011). Lastly, B. japonicum has a single pyruvate kinase, PykM, that uses Mn2+ rather than Mg2+ (Hohle and O’Brian, 2012). The reliance on manganese is likely true for the rhizobia in general as judged by phenotypes of manganese transport mutants within that group (Anderson et al., 2009, Davies and Walker, 2007, Chao et al., 2004).
In the present work, we show that the metal selectivity of B. japonicum Mur and E. coli Fur are dependent on the cellular context in which they function, and has implications into the evolution of the unusual iron sensing mechanism in the rhizobia that is mediated by Irr.
RESULTS
B. japonicum Mur and E. coli Fur have similar metal-binding properties in vitro
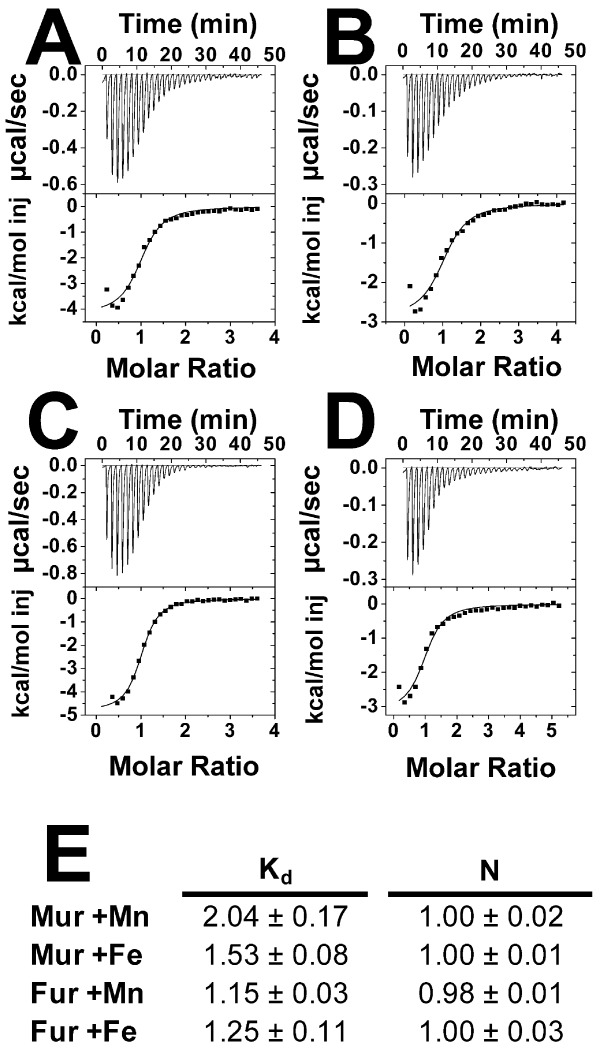
B. japonicum Mur and E. coli Fur share 49% similarity at the amino acid level, and respond to manganese and iron, respectively, in vivo. We wanted to determine if the metal responsiveness by B. japonicum Mur in vivo is due to differences in affinities between Mn2+ and Fe2+. Dissociation binding constants (Kd) of Mur for Mn2+ and mole ratios (N) were determined using isothermal titration calorimetry (ITC) (Fig 1). Each monomer of the Mur dimer bound one molecule of Mn2+ with a Kd value of 2.04 μM (Fig 1A,E). Furthermore, each Mur monomer bound one molecule of Fe2+, with a Kd value of 1.53 μM, similar to that seen with Mn2+ (Fig 1B,E). Rhizobium leguminosarum Mur was also reported to bind both metals similarly (Bellini and Hemmings, 2006). Because Mur bound both metals similarly, we wanted to determine if E. coli Fur showed differences in metal binding. Fur bound 1 Mn2+ or Fe2+ ion per monomer with similar Kd values of 1.15 μM and 1.25 μM, respectively (Fig 1C, D, and E). We conclude that Mur and Fur each bind Mn2+ and Fe2+ in vitro with similar affinities, which cannot explain metal selectivity in vivo.
Fig 1.
ITC analysis of B. japonicum Mur and E. coli Fur titrated with Mn2+ or Fe2+.
ITC raw data collection trace for titration of (A) Mn2+ to a sample of B. japonicum Mur, (B) Fe2+ to a sample of B. japonicum Mur, (C) Mn2+ to a sample of E. coli Fur, and (D) Fe2+ to a sample of E. coli Fur. (E) Dissociation binding constant (Kd) values and calculated number of sites per monomer (N) of B. japonicum Mur and E. coli Fur for Mn2+ and Fe2+ were determined using a one-binding-site model fitted to the titration data taken in triplicate.
Mur and Fur bind DNA tightly irrespective of the regulatory metal bound
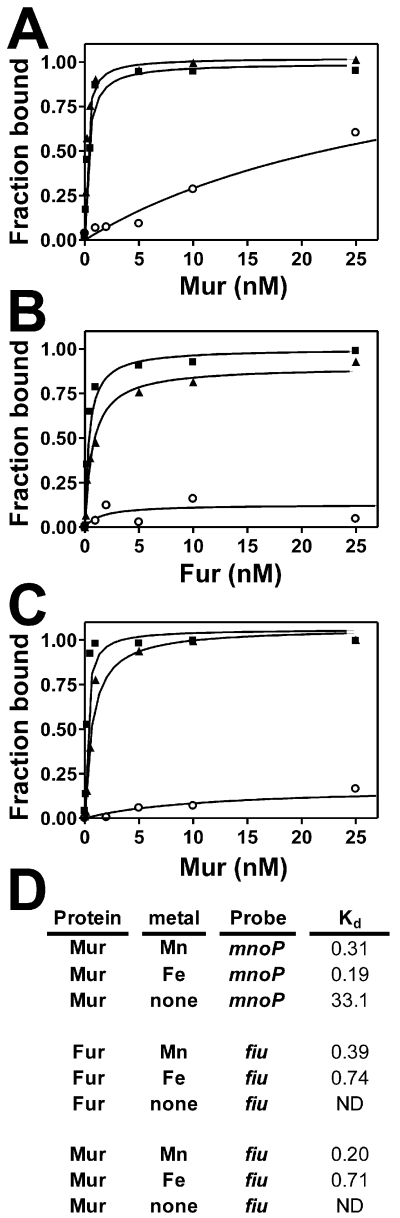
We sought to determine whether the affinity of Mn2+-bound Mur for DNA differed from that of Fe2+-bound Mur in vitro. Electrophoretic mobility shift assay (EMSA) reactions were carried out by titrating Mur in reactions with a 39 bp fragment containing the Mur binding site found within the mnoP gene promoter (Hohle et al., 2011). Mur bound DNA with a Kd of 33.1 nM in the absence of metal (Fig 2A,D). In the presence of Mn2+, Mur bound the mnoP DNA with about 100-fold greater affinity, with a Kd value of 0.31 nM (Fig 2A,D). A similar Kd value of 0.19 nM was observed in the presence of Fe2+ (Fig 2A,D), showing that Mn2+ does not confer greater DNA binding on Mur compared with Fe2+. Similar experiments were carried out with E. coli Fur, using a 39 bp fragment of DNA containing the Fur box located within the E. coli fiu gene promoter. Binding of Fur was undetectable in the absence of metal (Fig 2B,D). Fur bound the fiu DNA with a Kd value of 0.74 nM when bound by Fe2+ (Fig 2B,D). When Fe2+ was replaced with Mn2+, the affinity was modestly greater with a Kd value of 0.39 nM (Fig 2B,D). Thus, Fe2+ does not confer greater DNA-binding activity on Fur compared with Mn2+.
Fig 2.
Effects of Mn2+ and Fe2+ on binding of B. japonicum Mur or E. coli Fur to the mnoP or fiu promoter in vitro.
Electrophoretic mobility shift assay (EMSA) analysis was carried out using (A) B. japonicum Mur and 100 pmol mnoP containing the B. japonicum Mur binding site, (B) E. coli Fur and 100 pmol fiu containing the Fur binding site and (C) Mur and the fiu binding site. The binding reactions were carried out either with no metal (open circles), 100 μM Mn2+ (closed squares) or Fe2+ (closed triangles). Bound and unbound DNA were resolved on a 5% non-denaturing polyacrylamide gel and visualized by autoradiography. Autoradiographs were scanned, and bands were quantified. (D). Dissociation binding constants (Kd) were calculated from the binding data.
Mur is capable of binding the Fur box within the E. coli fiu gene promoter (Friedman and O’Brian, 2003), providing a control for the target DNA. Mur bound the fiu promoter with a subnanomolar Kd value in the presence of either metal (0.20 and 0.71 nM for Mn2+ and Fe2+ respectively). The data confirm that DNA-binding activities of Mur or Fur require a bound metal, but show that Mn2+and Fe2+ confer similar activities on each regulator. Apo-Mur bound mnoP promoter DNA with greater affinity than it bound the E. coli fiu DNA (Fig. 2A, C). However, the mnoP promoter is unbound by Mur in B. japonicum cells grown in low manganese medium (Hohle et al., 2011), and therefore the weak binding activity of apo-Mur is unlikely to be physiologically relevant.
Mur is Mn-responsive in B. japonicum, but Fe-dependent in E. coli
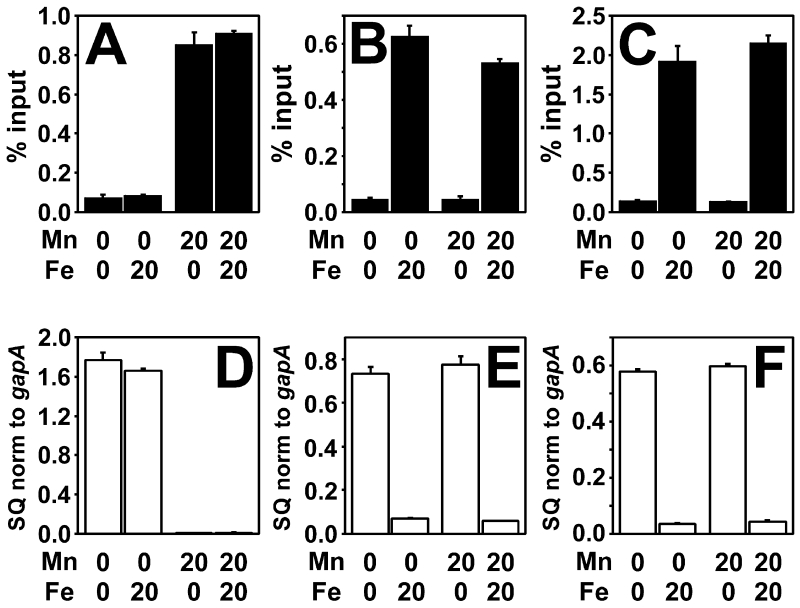
Mur occupies the promoter region of mnoP in a manganese-responsive manner in vivo, as determined by crosslinking/co-immunoprecipitation experiments using anti-Mur antibodies to precipitate bound DNA (Hohle et al., 2011) (Fig. 3A). Correspondingly, quantitative real time PCR (qPCR) showed that the mnoP gene transcript was low in B. japonicum cells when grown in 20 μM MnCl2 compared to when grown in low manganese medium (Fig. 3D). Mur occupancy and mnoP expression were independent of the iron status (Fig 3A,D).
Fig 3.
Effect of metals on in vivo promoter occupancy and transcriptional repression by Mur in B. japonicum and E. coli cells.
Cells were grown in media with the presence or absence of 20 μM MnCl2 or 20 μM FeCl3. (A) In vivo occupancy of the mnoP promoter by Mur in B. japonicum cells (B) In vivo occupancy of the fiu promoter by Mur in E. coli cells. (C) In vivo occupancy of the mnoP promoter by Mur in E. coli cells. In vivo occupancy data are expressed as the relative starting quantity (SQ) of DNA normalized to the input, and are presented as the average of triplicate samples with the error bars representing the standard deviation. (D) Analysis of mnoP mRNA by qPCR in B. japonicum cells. (E) Analysis of fiu mRNA in E. coli cells. (F) Analysis of mnoP mRNA in E. coli cells. The data are expressed as the relative starting quantity (SQ) of mntH mRNA normalized to the housekeeping gene gapA, and are presented as the average of triplicate samples with the error bars representing the standard deviation.
To assess whether the selectivity of Mur was dependent on cellular environment, we expressed B. japonicum mur in E. coli by replacing the fur ORF with that of mur. The E. coli cells were grown in the same media as was B. japonicum. We confirmed that the mur gene was expressed in E. coli by Western blot analysis (Fig. S2). In E. coli, Mur occupied the promoter region of the fiu gene when grown in the presence of iron (Fig. 3B), but not in its absence. Moreover, occupancy of the fiu promoter by Mur was independent of the manganese status even though the presence of protein was confirmed under that growth condition (Fig. S2). Consistent with the promoter occupancy, Mur-responsive expression of the fiu gene was dependent on iron, not manganese (Fig. 3B and E). The fiu gene was derepressed under all metal conditions in a fur mutant, confirming that the regulation observed in the mur+ E. coli strain was Mur dependent (Fig. S3). Thus, Mur is an iron-responsive transcription factor in E. coli cells
To determine if the iron-dependent activity of Mur observed in E. coli was influenced by the target gene, we introduced the B. japonicum mnoP gene in E. coli expressed from its own promoter containing the Mur binding site. The promoter region of mnoP in E. coli was occupied by Mur, and gene transcription was repressed, when iron was added to the growth medium, independent of the manganese concentration (Fig. 3C and F). Metal dependent gene expression was lost in an E. coli fur mutant (Fig. S3). Collectively, the findings suggest that the metal selectivity of Mur depends on the cellular context in which it functions, not on intrinsic properties of the protein.
Fur is an iron-responsive regulator in E. coli, but responds to manganese in B. japonicum
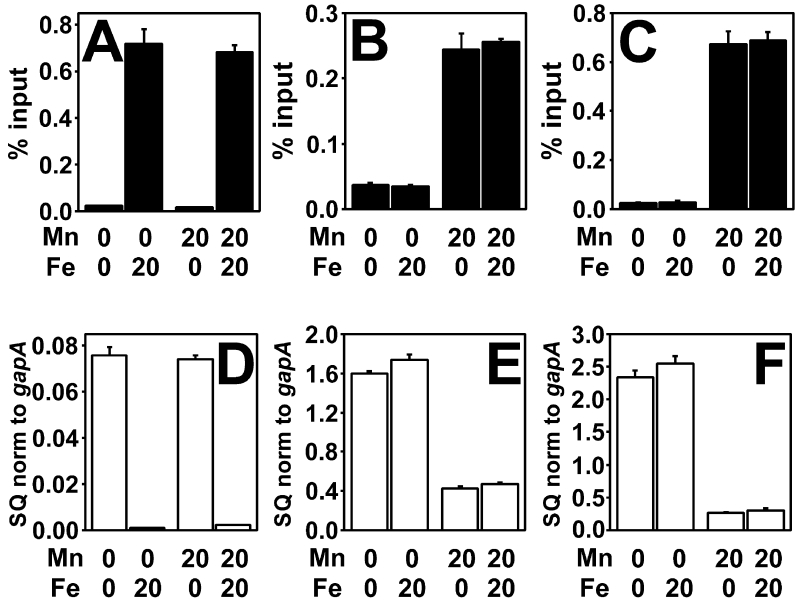
Because Mur responded to iron instead of manganese when expressed in E. coli, we sought to determine the metal specificity of E. coli Fur in B. japonicum. In E. coli, Fur occupied the fiu promoter when iron was present in the growth medium (Fig. 4A). Furthermore, the gene transcript of fiu was significantly reduced when grown in 20 μM FeCl3 compared to when it is grown in the absence of iron (Fig. 4D), corresponding to iron-dependent repression in E. coli. The E. coli fur gene was expressed in B. japonicum by replacing the ORF of the mur gene with that of fur so that transcription was initiated from the mur promoter. Expression of fur under all growth conditions examined was confirmed by Western blot analysis using anti-Fur antibodies (Fig. S2). Fur occupied the mnoP promoter in B. japonicum when manganese was added to the growth medium (Fig. 4B), resulting in low mnoP transcript under that growth condition (Fig. 4E). The addition of 20 μM FeCl3 to the growth medium did not confer activity on Fur in B. japonicum as determined by low occupancy of the mnoP promoter and derepression of the mnoP gene (Fig. 4B, E). This iron concentration was sufficient for Fur activity in E. coli.
Fig 4.
Effect of metals on in vivo promoter occupancy and transcriptional repression by Fur in B. japonicum and E. coli cells.
Cells were grown in media with the presence or absence of 20 μM MnCl2 or 20 μM FeCl3. (A) In vivo occupancy of the fiu promoter by Fur in E. coli cells. (B) In vivo occupancy of the mnoP promoter by Fur in B. japonicum cells. (C) In vivo occupancy of the fiu Fur box in B. japonicum cells. The Mur-binding site within the mntH promoter was replaced by the fiu Fur box, and the chimeric promoter placed upstream of the fiu open reading frame. In vivo occupancy data are expressed as the relative starting quantity (SQ) of DNA normalized to the input, and are presented as the average of triplicate samples with the error bars representing the standard deviation. (D) Analysis of fiu mRNA by qPCR in E. coli cells grown as described above. (E) Analysis of mnoP mRNA in B. japonicum cells. (F) Analysis of fiu mRNA in B. japonicum cells. The data are expressed as the relative starting quantity (SQ) of mntH mRNA normalized to the housekeeping gene gapA, and are presented as the average of triplicate samples with the error bars representing the standard deviation.
We wanted to determine whether the manganese-dependent activity of Fur in B. japonicum was due to the target DNA binding site. Because the fiu promoter was silent in B. japonicum, we constructed a Fur-responsive promoter in which the Mur binding site of the B. japonicum mntH promoter was replaced with the Fur box of the E. coli fiu gene. This chimeric promoter was placed upstream of the fiu open reading frame on a plasmid and introduced into B. japonicum cells. Fur occupied the mntH/fiu promoter in a manganese dependent manner (Fig. 4C). Moreover, fiu gene transcript expressed from the modified promoter was repressed in cells grown with 20 μM MnCl2 compared to when no exogenous manganese was added (Fig. 4F). Fur did not occupy the fiu promoter region in vivo in cells grown in low manganese medium in the presence of 20 μM FeCl3 (Fig. 4C). Metal-dependent repression of mnoP and fiu was lost in a B. japonicum mur mutant (Fig. S3) confirming that manganese-mediated expression of the fur+ B. japonicum strain depended on Fur. The findings suggest that metal selectivity of Fur depends on the cellular context in which it functions, not on intrinsic properties of the protein.
Mur responds to very high levels of iron in vivo in B. japonicum
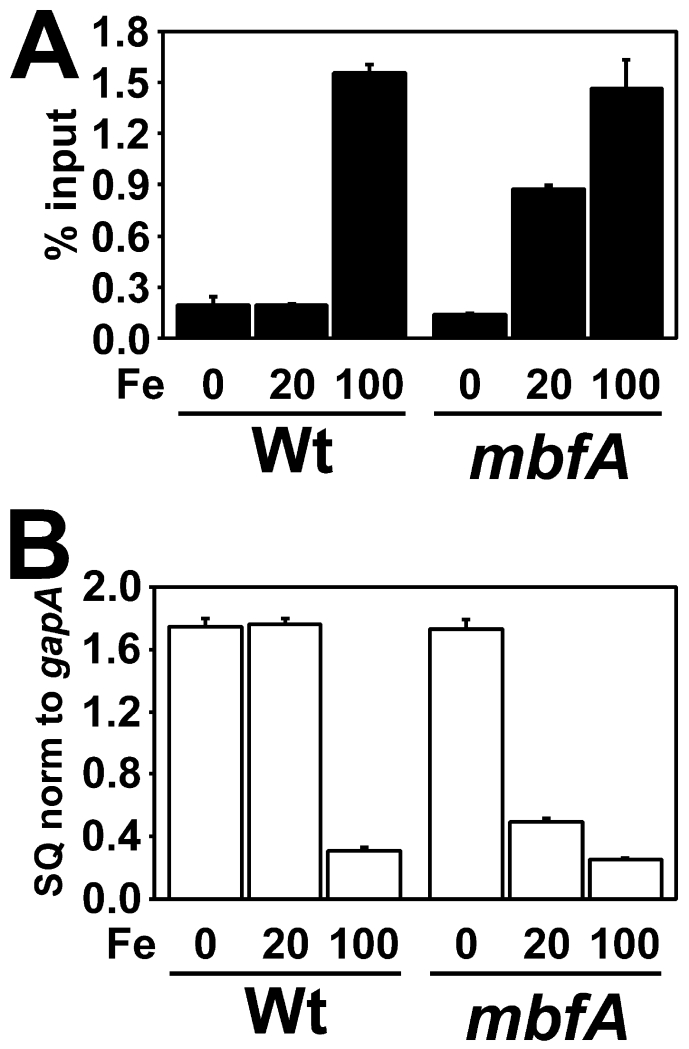
A B. japonicum mutant defective in the iron export gene mbfA was shown to accumulate higher intracellular levels than the wild type (Sankari and O’Brian, 2014). We wanted to determine if high concentrations of iron are capable of causing Mur to respond to iron in B. japonicum. Mur occupancy of the mnoP promoter was examined in B. japonicum wild type and mbfA cells grown under low manganese conditions with 0, 20, or 100 μM FeCl3 added to the growth medium. Mur occupancy of the mnoP promoter was low in both the wild type and mbfA strains grown under low iron (Fig 5A). Interestingly, Mur occupied the mnoP promoter in the mbfA mutant grown in 20 μM FeCl3 (Fig 5A), but not in the wild type. However, Mur occupied the mnoP promoter in the wild type strain when grown in 100 μM FeCl3 (Fig 5A). mnoP gene transcripts were low under conditions where Mur occupied the promoter region (Fig. 5B). This suggests that high intracellular concentrations of iron can promote Mur binding in vivo. Because Mur normally regulates the manganese transport genes mntH and mnoP, we assume that the high iron levels that confer Mur activity represent a dysregulated state.
Fig. 5.
Effects of high intracellular iron on in vivo promoter occupancy and transcriptional repression by Mur in B. japonicum cells
The intracellular iron concentration was elevated by growth in media containing 100 μM FeCl3 in the medium for B. japonicum, or by mutation of the iron exporter gene mbfA. Cells of the wild type and mutant were grown in media containing no added manganese and either 0, 20 μM or 100 μM FeCl3. (A) In vivo occupancy of the mnoP promoter by Mur in B. japonicum wild type (Wt) or mbfA strain. Data are expressed as the relative starting quantity (SQ) of DNA normalized to the input, and are presented as the average of triplicate samples with the error bars representing the standard deviation. (B) Analysis of mnoP mRNA by qPCR in B. japonicum wild type or mbfA strain. The data are expressed as the relative starting quantity (SQ) of mntH mRNA normalized to the housekeeping gene gapA, and are presented as the average of triplicate samples with the error bars representing the standard deviation.
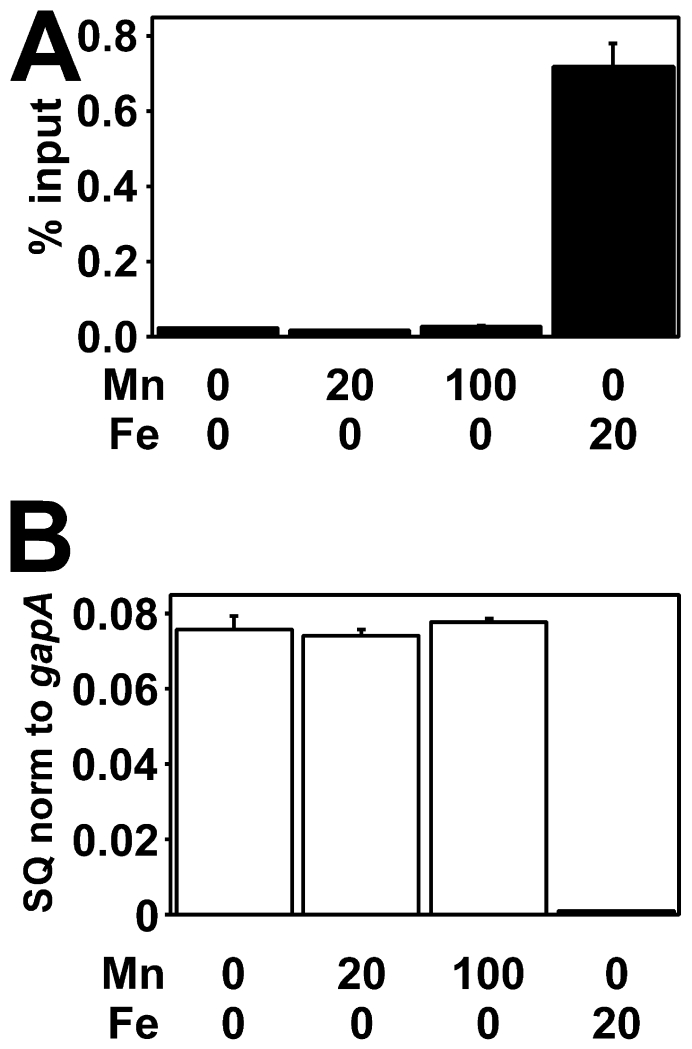
Because Mur responded to high iron in B. japonicum, we wanted to determine if Fur responds to high concentrations of manganese in E. coli. Cells were grown under low iron conditions with 0, 20, or 100 μM MnCl2 added to the growth medium. Fur did not occupy the fiu promoter in vivo or repress transcript levels at any manganese concentration tested (Fig. 6A), whereas high occupancy and fiu repression were observed with the 20 μM FeCl3 control (Fig. 6A, B). Thus, manganese-dependent Fur activity in E. coli was not achieved.
Fig. 6.
Effects of high intracellular manganese on in vivo promoter occupancy and transcriptional repression by Fur in E. coli cells
E. coli cells were grown in media containing no added iron and either 0, 20 μM or 100 μM MnCl2, or 20 μM FeCl3 in the absence of manganese. (A) In vivo occupancy of the fiu promoter by Fur in E. coli cells. Data are expressed as the relative starting quantity (SQ) of DNA normalized to the input, and are presented as the average of triplicate samples with the error bars representing the standard deviation. (B) Analysis of fiu mRNA in E. coli cells. The data are expressed as the relative starting quantity (SQ) of mntH mRNA normalized to the housekeeping gene gapA, and are presented as the average of triplicate samples with the error bars representing the standard deviation.
Intracellular levels of iron correlate with iron-responsiveness of Fur and Mur
Both Fur and Mur are iron-dependent in E. coli, but respond to iron in B. japonicum only when grown in very high iron media or when the iron exporter gene mbfA is deleted. Therefore, we wanted to address whether E. coli accumulates more iron than B. japonicum under the same growth conditions. We measured the cellular iron content in B. japonicum and E. coli grown in medium containing 0, 20 or 100 μM FeCl3. When no manganese was added to the medium, E. coli accumulated more iron than was observed in B. japonicum at the same iron growth condition (Table 1). E. coli contained 214 nmol Fe/mg protein when grown in 20 μM Fe, which was over 4-fold more iron than found in B. japonicum. Under that condition, Fur and Mur are active in E. coli, but not B. japonicum. The B. japonicum iron content could be increased either by mutation of the mbfA gene (Table 2) (Sankari and O’Brian, 2014) or by growing in high iron media (100 μM Fe), both of which resulted in Mur activity in those cells (Fig 5A,B). Thus, activity correlated with the cellular iron content.
Table 1.
Determination of intracellular iron and manganese content of B. japonicum and E. coli. Data are expressed as average nmol metal per mg protein ±standard deviation of triplicate samples.
| Cellular iron content | |||
|---|---|---|---|
|
| |||
| Medium (μM) |
B. japonicum
|
E. coli
|
|
| Mn |
Fe |
nmol Fe/mg protein |
nmol Fe/mg protein |
| 0 | 0 | 0.96 ±0.01 | 2.0 ±0.02 |
| 0 | 20 | 46 ±3 | 214 ±3 |
| 0 | 100 | 307 ±5 | 1054 ±0 |
|
| |||
| 20 | 0 | 0.96 ±0.02 | 2.18 ±0.09 |
| 20 | 20 | 48 ±3 | 228 ±3 |
| 20 | 100 | 307 ±8 | 1268 ±52 |
|
| |||
| 100 | 0 | 0.95 ±0.00 | 2.09 ±0.01 |
| 100 | 20 | 47 ±1 | 274 ±2 |
| 100 | 100 | 307 ±7 | 1259 ±45 |
|
| |||
| Cellular manganese content | |||
|---|---|---|---|
|
| |||
| Medium (μM) |
B. japonicum
|
E. coli
|
|
| Mn |
Fe |
nmol Mn/mg protein |
nmol Mn/mg protein |
| 0 | 0 | 0.92 ±0.01 | 0.14 ±0.01 |
| 0 | 20 | 1.15 ±0.01 | 0.39 ±0.00 |
| 0 | 100 | 1.10 ±0.04 | 0.96 ±0.04 |
|
| |||
| 20 | 0 | 8.6 ±0.3 | 4.5 ±0.2 |
| 20 | 20 | 9.7 ±0.3 | 31 ±2 |
| 20 | 100 | 9.8 ±0.2 | 82 ±2 |
|
| |||
| 100 | 0 | 13.2 ±0.3 | 68 ±3 |
| 100 | 20 | 13.6 ±0.1 | 237 ±5 |
| 100 | 100 | 13.3 ±0.3 | 276 ±6 |
|
| |||
Table 2.
Determination of intracellular iron and manganese content of B. japonicum wild type and mbfA cells. Data are expressed as nmol metal per mg protein ±standard deviation of triplicate samples.
| Cellular Iron Content |
Cellular Manganese Content |
|||||
|---|---|---|---|---|---|---|
| Medium (μM) |
nmol Fe/mg protein |
nmol Mn/mg protein |
||||
| Mn |
Fe |
Wt |
mbfA
|
Wt |
mbfA
|
|
| 0 | 0 | 1.15 ±0.04 | 1.33 ±0.07 | 1.13 ±0.00 | 1.11 ±0.05 | |
| 0 | 20 | 43 ±3 | 93 ±4 | 1.16 ±0.04 | 1.11 ±0.03 | |
| 0 | 100 | 313 ±11 | 567 ±9 | 1.16 ±0.02 | 1.17 ±0.00 | |
|
| ||||||
| 20 | 0 | 0.91 ±0.06 | 1.12 ±0.03 | 10.5 ±0.7 | 10.2 ±0.3 | |
| 20 | 20 | 43 ±1 | 96 ±3 | 11.1 ±0.1 | 11.0 ±0.1 | |
| 20 | 100 | 308 ±8 | 574 ±27 | 12.7 ±0.4 | 11.5 ±0.5 | |
|
| ||||||
We also measured the cellular iron content as described above, except that 20 μM or 100 μM MnCl2 were also included in the growth medium (Table 1). We found that manganese did not affect the cellular iron content in B. japonicum or E. coli. Therefore, we can rule out that the observed manganese-responsiveness of Mur or Fur in B. japonicum is an indirect consequence of altering the iron content. In addition, we measured the iron content in mur+ E. coli cells and fur+ B. japonicum cells, and found them to be similar to their respective wild type cells under the same iron and manganese regimens (Table S1 and S2). Therefore, the observed metal-responsive activities of Mur or Fur in their heterologous hosts cannot be explained by changes in the iron content caused by their expression.
The chelatable iron pool correlates with Mur and Fur activity in B. japonicum and E. coli
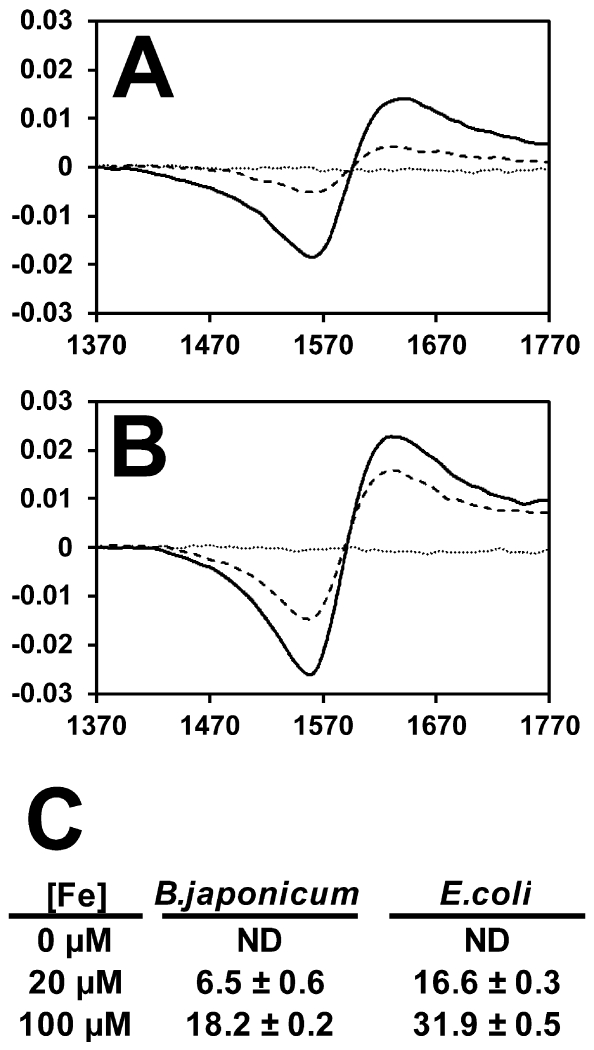
Most iron within cells is tightly associated with proteins, and therefore the regulatory iron that can be sensed by Fur (or Mur) is likely only a portion of the total iron content. We analyzed iron that could be chelated by the iron binding compound desferrioxamine as described previously (Woodmansee and Imlay, 2002). B. japonicum or E. coli cells were treated with the chelator, and bound iron was analyzed by EPR. No measureable signal was obtained for either bacterium when grown in medium with no added iron (Fig 7). When grown in 20 μM FeCl3, E. coli contained 16.6 nmol/mg protein of chelated iron, whereas B. japonicum contained only 6.5 nmol/mg protein (Fig 7). Under these conditions, Fur and Mur were active only in E. coli cells. Interestingly, the chelatable iron pool of B. japonicum grown in 100 μM FeCl3- was 18.2 nmol/mg protein (Fig. 7A,C), similar to the value obtained for E. coli when grown in 20 μM FeCl3- (Fig. 7B,C). This corresponds to iron-responsive Mur activity seen in B. japonicum under these growth conditions (Fig. 5A,B). The findings show that chelatable iron is higher in E. coli than in B. japonicum when grown under identical conditions, and which correlates with iron-responsive gene expression.
Fig 7.
Determination of intracellular chelatable iron in B. japonicum and E. coli cells by EPR.
EPR raw data traces for (A) B. japonicum or (B) E. coli cells treated with the iron chelator desferrioxamine after growth in media containing no added iron (dotted line), 20 μM FeCl3 (dashed line) or 100 μM FeCl3. (solid line) FeCl3. (C) Quantitation of intracellular chelatable iron as determined using EPR normalized to protein content. Data are expressed as average and standard deviation of triplicate trials. The data are expressed as nmol Fe per mg protein.
Manganese-dependent activities of Mur and Fur do not correlate with total cellular manganese content
Mur and Fur are manganese-dependent regulators in B. japonicum, but not in E. coli. We measured the cellular manganese content in each cell type grown in 0, 20 or 100 μM MnCl2, with no iron added to the medium (Table 1). The manganese content was higher in B. japonicum than in E. coli when cells were grown in manganese-limited media, but neither Fur nor Mur showed manganese-dependent activity in either cell type (Figs. 3 and 4). Previous work shows that the high affinity manganese transporter gene mntH is expressed in B. japonicum (Hohle and O’Brian, 2009), but not in E. coli (Anjem et al., 2009) when grown in manganese-limited media, which is consistent with greater manganese content in B. japonicum. The manganese content in B. japonicum was 8.6 nmol Mn/mg protein in cells grown in 20 μM MnCl2 with no iron added (Table 1), which was about twice as high as that found in E. coli. However, E. coli cells accumulated 68.9 nmol Mn/mg protein when grown in 100 μM MnCl2, much higher than the 13.3 nmol Mn/mg protein found in B. japonicum. However, neither Fur nor Mur were active in E. coli under that growth condition (Figs 3B,C,E,F and 4A,D) despite having over 8-fold greater manganese than B. japonicum grown in 20 μM MnCl2, where those regulators respond to manganese (Figs. 3A,D and 4B,C,E,F). These observations show that manganese-dependent activities of Mur and Fur do not correlate with the cellular manganese content. The findings implicate a regulatory pool of manganese in B. japonicum that is perceived by Mur or Fur that is absent in E. coli.
We measured the manganese content in mur+ E. coli cells and fur+ B. japonicum cells, and found them to be similar to the respective wild type under the same iron and manganese regimens (Table S1 and S2). Therefore, the observed metal-responsive activities of Mur or Fur in their heterologous hosts cannot be explained by changes in the manganese content caused by their expression.
Iron increases the manganese content in E. coli, but not in B. japonicum
We measured the cellular manganese content as described above, except that 20 μM or 100 μM FeCl3 were also included in the growth medium (Table 1). Iron in the medium did not affect the cellular manganese content in B. japonicum. However, we found that E. coli accumulated high levels of manganese in response to iron (Table 1). When E. coli cells were grown in 20 μM MnCl2, the manganese content increased about 40-fold when 100 μM FeCl3 was included in the medium compared to cells grown with no added iron. The manganese content reached 310 nmol/mg protein in E. coli cells grown with 100 μM each of MnCl2 and FeCl3.
Anjem et al (Anjem et al., 2009) found that E. coli cells accumulate manganese in response to H2O2 stress due to activation of the manganese transporter gene mntH by the transcriptional regulator OxyR. Because iron can promote oxidative stress due to the Fenton reaction, we addressed whether the observed iron-dependent accumulation of manganese was an oxidative stress response by examining an oxyR mutant. The cellular manganese content of an oxyR mutant increased with increasing manganese in the medium (Table 3), as was observed in the wild type. However, the manganese levels were independent of iron in the mutant. Moreover, iron levels in the oxyR strain were similar to the wild type under all metal regimens tested. Thus, accumulation of manganese in response to iron in E.coli appears to be an oxidative stress response.
Table 3.
Determination of intracellular manganese and iron content of E. coli and B. japonicum wild type and oxyR strains. Data are expressed as average nmol metal per mg protein ±standard deviation.
|
E. coli
|
B. japonicum
|
||||
|---|---|---|---|---|---|
| Medium (μM) |
nmol Mn/mg protein |
nmol Mn/mg protein |
|||
| Mn |
Fe |
Wt |
oxyR
|
Wt |
oxyR
|
| 0 | 0 | 0.33 ±0.01 | 0.38 ±0.02 | 1.57 ±0.03 | 1.38 ±0.04 |
| 0 | 20 | 0.62 ±0.01 | 0.53 ±0.01 | 1.47 ±0.17 | 1.53 ±0.08 |
| 0 | 100 | 0.91 ±0.04 | 0.70 ±0.01 | 1.53 ±0.05 | 1.51 ±0.04 |
|
| |||||
| 20 | 0 | 2.6 ±0.1 | 3.1 ±0.2 | 13.2 ±0.2 | 10.7 ±0.2 |
| 20 | 20 | 27 ±2 | 1.7 ±0.1 | 12.8 ±0.2 | 11.1 ±0.2 |
| 20 | 100 | 102 ±1 | 2.1 ±0.1 | 11.6 ±0.2 | 10.2 ±0.2 |
|
| |||||
| 100 | 0 | 67 ±1 | 64 ±5 | 16.0 ±0.3 | 16.7 ±0.8 |
| 100 | 20 | 220 ±4 | 73 ±1 | 17.6 ±0.6 | 16.8 ±0.3 |
| 100 | 100 | 311 ±2 | 56.3 ±0.1 | 16.9 ±0.4 | 16.2 ±0.3 |
|
| |||||
| Medium |
nmol Fe/mg protein |
nmol Fe/mg protein |
|||
|---|---|---|---|---|---|
| Mn |
Fe |
Wt |
oxyR
|
Wt |
oxyR
|
| 0 | 0 | 2.11 ±0.03 | 1.94 ±0.06 | 1.38 ±0.01 | 1.42 ±0.04 |
| 0 | 20 | 222 ±2 | 204 ±1 | 47 ±0.2 | 46 ±2 |
| 0 | 100 | 1132 ±8 | 1063 ±8 | 233 ±3 | 206 ±3 |
|
| |||||
| 20 | 0 | 2.12 ±0.01 | 2.02 ±0.01 | 1.10 ±0.00 | 1.17 ±0.05 |
| 20 | 20 | 212 ±2 | 208 ±2 | 53 ±1 | 47 ±1 |
| 20 | 100 | 1255 ±14 | 1192 ±38 | 239 ±16 | 229 ±16 |
|
| |||||
| 100 | 0 | 2.24 ±0.06 | 1.85 ±0.12 | 1.26 ±0.02 | 1.15 ±0.04 |
| 100 | 20 | 210 ±2 | 212 ±2 | 55 ±3 | 48 ±1 |
| 100 | 100 | 1236 ±20 | 1196 ±12 | 248 ±2 | 228 ±3 |
|
| |||||
Mutation of the oxyR gene in B. japonicum had no effect on metal accumulation under all growth conditions tested (Table 3). Importantly, wild type E. coli cells grown in 20 μM Fe accumulated about 210 nmol Fe/mg protein and showed elevated manganese levels, whereas B. japonicum cells grown in 100 μM Fe contained about 235 nmol Fe/mg protein, but did not show elevated manganese levels. Thus, under comparable cellular iron levels, E. coli exhibited a stress response that was not observed in B. japonicum. These observations suggest that B. japonicum and E. coli manage iron differently.
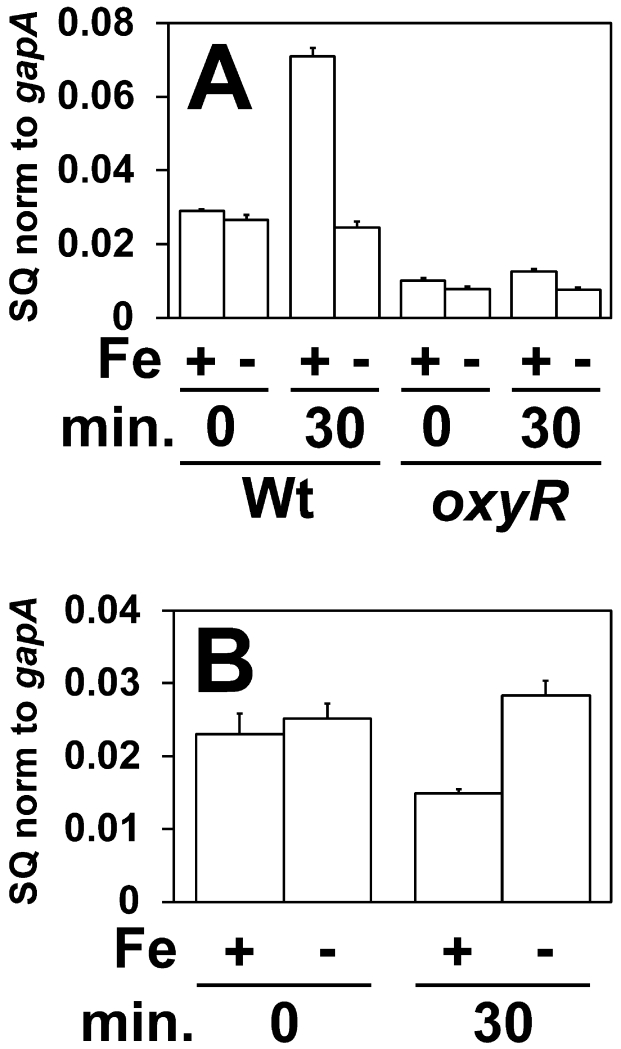
Iron exposure resulted in oxyR-dependent manganese accumulation in E. coli. Thus we addressed whether iron affected the expression of the Mn2+ transport gene. The mntH gene transcript level in E. coli wild type cells increased about 2-fold 30 minutes after 100 μM FeCl3 was added to the growth medium (Fig. 8A). mntH transcript levels remained constant when iron was not added to the growth medium. Iron-responsive induction was lost in an E. coli oxyR mutant (Fig 8A), showing that mntH induction depends on OxyR. No iron-responsive induction of mntH was seen in B. japonicum (Fig 8B).
Fig 8.
Effects of iron on expression of the mntH gene in E. coli and B. japonicum
Cells were grown to mid log phase, then 0 or 100 µM FeCl3 was added to the medium, and cells were harvested after 30 minutes. (A) Analysis of mntH mRNA by qPCR in E. coli wild type (Wt) or oxyR strain. (B) Analysis of mntH mRNA in B. japonicum cells. The data are expressed as the relative starting quantity (SQ) of mntH mRNA normalized to the housekeeping gene gapA, and are presented as the average of triplicate samples with the error bars representing the standard deviation.
DISCUSSION
In the present study, we found that the metal specificities of B. japonicum Mur and E. coli Fur are strongly dependent on the cellular environment in which they function rather than on the physical properties of the protein. In addition, B. japonicum and E. coli manage manganese and iron differently to affect homeostasis and gene expression. Finally, the inability of Mur or Fur to function as iron metalloregulators in B. japonicum may explain why it, and related organisms, sense iron differently than has been described in many other bacterial phyla.
Mur behaves as Fe2+-responsive regulator in E. coli cells, and Fur responds to Mn2+ rather than Fe2+ in B. japonicum cells. Thus, B. japonicum confers manganese selectivity on these proteins. When grown identically in media containing up to 20 μM manganese, B. japonicum accumulates more manganese as long as iron is not also added (Table 1). Indeed, E. coli appears to need very little manganese unless challenged with hydrogen peroxide (Anjem et al., 2009), whereas B. japonicum requires the metal under unstressed conditions (Hohle and O’Brian, 2012, Hohle and O’Brian, 2014). However, the total cellular manganese content is not sufficient to explain the behavior of Mur and Fur. Neither regulator was active in iron-limited E. coli cells containing 8-fold more cellular manganese than was found in B. japonicum cells where Mur was active (Table 1, Fig. 3 and 4). This suggests that the population of manganese perceived by Mur in B. japonicum differs from E. coli and does not correlate with total content of the metal.
Selectivity of metal sensing has been described in terms of affinity, access and allostery (Waldron et al., 2009). We have ruled out affinity as the basis for selectivity by showing that Mur and Fur each bind Fe2+ and Mn2+ with similar affinities (Fig. 1). In addition, Mn2+ and Fe2+ confer similar DNA binding affinities on Fur and Mur (Fig. 2), thereby ruling out allostery as a basis of selectivity. The observed dependence on cellular milieu for metal selectivity by Fur and Mur strongly argues that access to metal is a major factor. The NmtR repressor from Mycobacterium tuberculosis responds to Ni2+ in its natural host, but only to Co2+ when expressed in Synechococcus PCC7942, which corresponds to a higher cellular nickel content in the natural host (Cavet et al., 2002). The iron-specific transcription factor DtxR from Corynebacterium diphtheria mediates both iron-and manganese-responsive gene expression when expressed in Bacillus subtilis, and can be made highly manganese-specific upon mutation of its metal binding site (Guedon and Helmann, 2003). Thus, DtxR metal selectivity is based on both affinity and access.
Whereas affinity and allostery can be readily addressed in vitro, access to metal in cells is more difficult to characterize mechanistically. Metals are predominantly tightly associated with proteins, and only a portion of the total cellular content is perceived by a sensor such as Fur or Mur to act as so-called regulatory metal or a metal pool. Strategies associated with protein folding during biogenesis can ensure correct metallation of some enzymes (Tottey et al., 2008, Leach and Zamble, 2007, Lee et al., 1993), but Mur and Fur must bind metals reversibly to function as sensors. There may be a chaperone or some other trafficking mechanism that ensures that Mur is bound by the correct metal as has been described, for example, for copper trafficking in Enterococcus hirae (Cobine et al., 1999). Further studies needs to be done to fully elucidate the mechanism in which Mur and Fur obtain their metal.
E. coli and B. japonicum manage iron differently as observed by the higher level of total and chelatable iron in E. coli when cells were grown under identical conditions (Table 1, Fig. 7). Although chelatable iron cannot be assumed to be a quantitative measure of the regulatory population perceived by Fur or Mur, it gives an assessment of iron that is not tightly bound in macromolecules. Our data suggests that the lower level of iron in B. japonicum cells allows Mur to function as a manganese-dependent regulator. Raising the cellular iron level by loss of the iron exporter MbfA or by growing cells in very high iron medium resulted in active Mur in B. japonicum even under low manganese conditions (Fig. 5A and B). Thus, high iron would be expected to interfere with manganese responsiveness. However, observations in E. coli indicate that low iron may be necessary but insufficient to render Fur or Mur manganese-responsive. The regulators are not active in E. coli grown in low iron media in the presence of manganese (Figure 3B, C, E, and F, Fig 4A and D). This reinforces the conclusion that there is a mechanism of manganese perception by Mur or Fur in B. japonicum apart from low iron levels that is missing in E. coli. We note that growth of an E.coli mutant that accumulates high levels of manganese was able to activate Fur (Martin et al., 2015) when grown with 500 μM manganese, showing that Fur can be manganese-responsive under extreme conditions.
Increasing the iron level in cells by iron supplementation in the growth medium resulted in a concomitant increase in manganese levels as well in E. coli, but not B. japonicum (Table 1). A previous study reported that exposure of E. coli to H2O2 elevates manganese levels by oxyR-dependent activation of the manganese transporter gene mntH (Anjem et al., 2009). Indeed, we found here that manganese accumulation by E. coli in response to iron also depended on oxyR expression, showing that iron elicits an oxidative stress response. When the cellular iron content was elevated in B. japonicum to levels found in E. coli, it did not elicit a manganese accumulation response (Table 1). It is somewhat counterintuitive that E. coli, which maintains iron at a higher level than B. japonicum, appears to be more stressed by the metal.
B. japonicum and related bacteria sense iron through an unusual mechanism via Irr not found in E. coli or other model systems (O’Brian, 2015), and the current work offers a plausible explanation for this. B. japonicum is very reliant on manganese (Hohle et al., 2011, Hohle and O’Brian, 2012, Hohle and O’Brian, 2014), and the cellular accommodations to this are incompatible with Fur functioning as an iron-dependent regulator. The Irr protein does not respond to iron directly, but rather responds to an iron-dependent process, namely the synthesis of heme (Qi and O’Brian, 2002). Irr interacts directly with the heme biosynthetic enzyme ferrochelatase, which uses iron as a substrate, and heme triggers Irr inactivation under iron replete conditions. Fe2+ and Mn2+ share similar sizes and coordination geometries, potentially allowing mismetallation of proteins. However, because Irr binds heme rather than Fe2+, it is able to discriminate iron in the form of heme from manganese, which is likely to be crucial in a cell containing a high level of bioavailable manganese. As a result, Irr can function as an iron-responsive regulator in a cellular environment where Fur cannot.
It is likely that this conclusion is generally applicable to the α-Proteobacteria that contain both Irr and Mur. Irr is a Fur family protein, and so it is likely that it arose from a gene duplication event. As a rhizobial ancestor adapted to a greater manganese metabolism, the fur gene product became manganese responsive, and the duplicated gene became irr. The latter event would have presumably required additional mutations that allow Irr to bind heme and recognize ferrochelatase. A bioinformatic and phylogenetic analysis of the a-Proteobacteria indicate that most species that contain Irr also have Mur (Rodionov et al., 2006), which agrees with studies in which Irr and Mur have been described experimentally.
Some α-Proteobacteria within the Family Rhizobiaceae contain RirA as well as Irr to mediate iron-responsive gene expression (Ngok-Ngam et al., 2009, Todd et al., 2002, Viguier et al., 2005, Ojeda et al., 2012). Although the mode of iron sensing by RirA has not been elucidated, it is proposed to be active as an iron-sulfur protein (Rodionov et al., 2006). This idea is consistent with the conclusion that these bacteria have evolved iron-sensing mechanisms that do not rely on binding to the free metal.
MATERIALS AND METHODS
Strains and media
Bacterial strains used in this study are listed in Table 4. B. japonicum strains were routinely grown at 29°C in glycerol-salts-yeast (GSY) medium as described elsewhere (Frustaci et al., 1991). E. coli strains were routinely grown at 37°C in Luria-Bertani (LB) broth. For all experiments, B. japonicum and E. coli were grown in modified GSY medium, containing 0.5 g l-1 yeast extract instead of 1 g l-1 supplemented with MnCl2 or FeCl3 at the concentrations indicated for each experiment. The concentrations of manganese and iron present in the unsupplemented media were 0.2 μM and 0.3 μM, respectively, as determined using a Perkin Elmer model 1100B atomic absorption spectrometer.
Table 4.
Strains used in this study
| B. japonicum | ||
| Strains | Relevant Characteristics | Reference |
| USDA110 | Parent Strain | |
| GEM4 | mur::Ω-cassette | (Hamza et al., 1999) |
| Bjfur+ | B. japonicum mur ORF replaced by E. coli fur ORF | |
| This study | ||
| 110mbfAΔΩ | mbfA:: Ω-cassette | (Sankari and O’Brian, 2014) |
| oxyR Δ Ω | oxyR:: Ω-cassette | (Panek and O’Brian, 2004) |
| Plasmids | ||
| pVK102FurBox-mntHprom-fiu | pVK102 with fiu fused to mntH | |
| promoter containing fiu Fur Box | This study | |
| E.coli | ||
| Strains | Relevant Characteristics | Reference |
| BW25113 | E. coli parent strain | (Datsenko and Wanner, 2000) |
| JW0669 | fur::kan | (Datsenko and Wanner, 2000) |
| JW0669:kan::camR/sacB | fur::camR/sacB | This study |
| Ecmur+ | E. coli fur ORF replaced by | This study |
| B. japonicum mur ORF | ||
| JW3933-3 | oxyR::kan | (Datsenko and Wanner, 2000) |
| Plasmids | ||
| pVK102 mnoP | pVK102 with mnoP and promoter | This study |
Construction of B. japonicum mutant strains
To construct Bjfur+, a B. japonicum mutant expressing E. coli fur, the open reading frame of B. japonicum mur and 500-bp flanking DNA on each side of it was amplified by PCR using USDA110 genomic DNA as the template, and ligated into pBluescriptSK+. The open reading frame was deleted using inverse PCR as previously described (Panek and O’Brian, 2004). The open reading frame of E. coli fur was amplified by PCR using BW25113 genomic DNA as template, and ligated into the inverse PCR product to create an in-frame fur construct run off of the native B. japonicum mur promoter. The construct was introduced into pLO1 (Lenz et al., 1994), mobilized into the mur strain GEM4, and selected for single recombinant mutants on GSY medium containing 75 μg ml-1 kanamycin, 100 μg ml-1 spectinomycin, and 100 μg ml-1 streptomycin. Double recombinants were then selected on GSY medium containing 5% sucrose. Double recombinants were confirmed using antibiotic sensitivity, PCR, and Western blot.
The open reading frame of E. coli fiu was amplified by PCR using BW25113 genomic DNA as template, and ligated into pBluescriptSK+. A B. japonicum mntH-promoter-fiu fusion was created so that the Mur binding site of mntH was replaced by the fiu Fur Box. The construct was introduced into a very low copy vector, pVK102, to create pVK102FurBox-mntHprom-fiu. The pVK102 construct was mobilized into USDA110, GEM4, and Bjfur+, and selected on GSY medium containing 75 μg ml-1 kanamycin and 75 μg ml-1 tetracycline. Mutants were confirmed using PCR.
Construction of E. coli mutant strains
To construct Ecmur+, an E. coli mutant expressing B. japonicum mur, the open reading frame of E. coli fur and 500-bp flanking DNA on each side of it was amplified by PCR using BW25113 genomic DNA as the template, and ligated into pBluescriptSK+. The open reading frame of fur was deleted using inverse PCR. A cassette expressing the camR and sacB genes, conferring chloramphenicol resistance and sucrose sensitivity, respectively, was ligated into the inverse PCR product. The camR/sacB cassette containing the fur flanking DNA was isolated using PCR, purified using a QIAGEN PCR purification kit, and transformed into JW0669 using lambda Red recombineering (Datsenko and Wanner, 2000, Yu et al., 2000). Constructs were verified using antibiotic resistance and sucrose sensitivity. The open reading frame of B. japonicum mur was amplified by PCR using USDA110 genomic DNA as template, and ligated into the inverse PCR product containing the fur genomic flanks. The mur gene containing the fur flanking DNA was isolated using PCR, purified using a QIAGEN PCR purification kit, and transformed into JW0669:fur::camR/sacB using lambda Red recombineering. Constructs were selected on LB containing 5% sucrose. The construct was confirmed using antibiotics sensitivity, colony PCR, and Western blot.
DNA including the open reading frame and promoter region of the B. japonicum mnoP gene was amplified by PCR using USDA110 genomic DNA as template, and ligated into pBluescriptSK+. The construct was introduced into pVK102, to create pVK102mnoP and transformed into BW25113, JW0669, and Ecmur+. Constructs were confirmed by PCR and analysis of plasmid preparations.
Overexpression and purification of B. japonicum Mur and E. coli Fur
B. japonicum Mur and E. coli Fur proteins were overexpressed and purified as described elsewhere (Friedman and O’Brian, 2003). Purified protein was dialyzed one time against ITC dialysis buffer (20 mM Tris, pH 7.5 and 150 mM NaCl) containing 0.5 mM EDTA to chelate any metals bound to the proteins. The proteins were dialyzed 3× against ITC dialysis buffer without EDTA. The final dialysate was saved for protein dilutions and metal preparations for ITC analysis.
Determination of binding affinities for manganese and iron for Mur and Fur using isothermal titration calorimetry (ITC)
All proteins, buffers, and metal solutions were degassed using a ThermoVac (ThermoScientific) at 24°C prior to running on ITC. Protein dilutions and metal salt preparations were carried out using the dialysate from the final dialysis step. Proteins were diluted to a monomer concentration of 30 μM before being loaded into the sample cell of a MicroCal VP-ITC. A 5 mM MnSO4 solution was prepared by diluting a 1 M MnSO4 stock solution. Titrations of manganese were carried out at a cell temperature of 25°C by injecting 1 μl of 5 mM MnSO4 at a rate of 0.5 μl s-1 every 90 seconds for 45 minutes, with reference power set to 20 μcal s-1 and a stirring speed set to 242. Working stocks of FeSO4 used for titrations were made fresh in degassed buffer to avoid oxidation of Fe2+ to Fe3+ and were 5.8 mM for Mur, and 7.0 mM for Fur. Titrations of iron were carried out in an anaerobic chamber at a cell temperature of 28°C by injecting 1 μl of FeSO4 at a rate of 0.5 μl s-1 every 90 seconds for 45 minutes, with reference power set to 20 μcal s-1 and a stirring speed set to 242. Data were analyzed by a version of Origin modified for ITC data analysis as supplied by MicroCal. All experiments were carried out in triplicate.
Determination of metal dependent binding affinities for Mur and Fur to DNA
Binding affinities of Mur and Fur were determined using electrophoretic gel mobility shift assays modified from that previously described (Hohle and O’Brian, 2009). Briefly, binding reactions containing 39-bp DNA fragments containing either the B. japonicum mnoP Mur binding site or the E. coli fiu Fur box were titrated with varying amounts of Mur or Fur in the presence of 0.1 mM MnCl2, 0.1 mM FeSO4, or no metal. 1 mM ascorbate was added to reactions containing FeSO4 as a reducing agent. No metal was added to the non-denaturing polyacrylamide gel.
In vivo binding of Fur or Mur to promoter DNA by cross-linking and immunoprecipitation
This technique was used to analyze the occupancy of the mnoP or fiu promoters by Mur or Fur in vivo. 50-ml cultures of USDA110, GEM4, and Bjmur+ expressing fiu were grown under low or high manganese and low or high iron conditions to mid-log phase (OD540 0.35 - 0.4). 10-ml cultures of BW25113, JW0669, and Ecmur+ expressing mnoP were grown under low or high manganese and low or high iron conditions to mid-log phase (OD600 0.35 - 0.4). In vivo cross-linking of DNA to protein and subsequent immunoprecipitation with antibodies specific to Mur or Fur were carried out as described elsewhere (Small et al., 2009). Immunoprecipitated DNA (1 μl) was analyzed by qPCR using primers that amplify the promoter regions of interest. The data are expressed as the SQ of immunoprecipitated DNA normalized to the input.
Analysis of RNA by quantitative real time PCR
Expression levels of selected genes were determined by qPCR with iQ SYBR supermix (BIO-RAD) using CFX-96 Touch Real Time PCR thermal cycler (BIO-RAD). RNA was isolated from B. japonicum and E. coli using a hot phenol-extraction method as previously described (Yang et al., 2006). cDNA was synthesized from 1 μg total RNA using iScript cDNA synthesis kit (BIO-RAD). qPCR reactions were carried out as previously described (Hohle and O’Brian, 2009). Data are expressed as average of three triplicates and the standard deviation is represented by the error bars.
Determination of intracellular iron and manganese using atomic absorption spectroscopy
Fifty milliliters of B. japonicum cultures were grown in 0, 20, or 100 μM MnCl2 and 0, 20, or 100 μM FeCl3 and harvested by centrifugation at 13,000 × g at 4°C for 5 min. The cell pellet was washed once with PBS containing 0.5 mM EDTA, twice with PBS, and twice with metal-free water. Cell pellets were digested in 69% HNO3 (JT Baker, AAS grade) at 98°C for 2 hours, followed by an additional digestion in 5 mM H2O2 at 70°C for 2 hours. The intracellular iron content was determined on a Perkin Elmer Atomic Absorption Spectrometer model 1100B equipped with a model HGA 700 graphite furnace as previously described (Yang et al., 2006). The same samples were used to measured intracellular manganese content as previously described (Hohle and O’Brian, 2009). Determination of intracellular iron and manganese in E. coli was done similarly, starting with 5 ml cultures. Protein concentrations were determined using the Bio-Rad protein assay using BSA as a standard.
Determination of intracellular chelatable iron by electron paramagnetic resonance spectroscopy
The quantification of intracellular chelatable iron was determined using EPR spectroscopy modified from previously described (Woodmansee and Imlay, 2002). B. japonicum or E. coli cells (1 μl) were grown in low-metal medium supplemented with 0, 20, or 100 μM FeCl3. Cell pellets were harvested and resuspended in low metal medium containing 20 mM desferrioxamine. Cultures were incubated, with shaking, at their respective growth temperatures (29°C for B. japonicum; 37°C for E. coli). After the incubation, cells were harvested, washed 3× in 20 mM Tris-HCl, pH 7.4, and resuspended in 0.5 ml Tris-HCl, pH 7.4 containing 10% glycerol. Final volume of cell suspension was approximately 1.2 ml. 0.4 ml of the cell suspension was loaded into 5-mm quartz EPR tube (Wilmad), immediately frozen in liquid nitrogen and stored at −80°C. Remaining cell suspension was used for protein quantification using the BIO-RAD protein assay. An iron standard curve was generated using 0.1 – 2.0 mM FeSO4 solutions containing 1 mM desferrioxamine. Samples were run on a Bruker Elexsys E 500 EPR spectrometer. Each run was 100 scans consisting of 200 points and was acquired and processed using Bruker Xenon software. Final data was analyzed in Microsoft Excel. EPR settings were as follows: field center, 1570 G; field sweep, 400 G, sweep time, 4 sec. Samples were run in a dewar to maintain liquid nitrogen temperatures during the course of the run. Each sample was run in triplicate, with the average and standard deviations being reported.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 GM099667 to M.R.O’B.
REFERENCES
- Ahn BE, Cha J, Lee EJ, Han AR, Thompson CJ, Roe JH. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol Microbiol. 2006;59:1848–1858. doi: 10.1111/j.1365-2958.2006.05065.x. [DOI] [PubMed] [Google Scholar]
- Anderson ES, Paulley JT, Gaines JM, Valderas MW, Martin DW, Menscher E, Brown TD, Burns CS, Roop RM., 2nd The manganese transporter MntH is a critical virulence determinant for Brucella abortus 2308 in experimentally infected mice. Infect Immun. 2009;77:3466–3474. doi: 10.1128/IAI.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorgan. chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- Anjem A, Varghese S, Imlay JA. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol. 2009;72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini P, Hemmings AM. In vitro characterization of a bacterial manganese uptake regulator of the Fur superfamily. Biochemistry. 2006;45:2686–2698. doi: 10.1021/bi052081n. [DOI] [PubMed] [Google Scholar]
- Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. Bacillus subtilis contains multiple Fur homologs: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- Cavet JS, Meng W, Pennella MA, Appelhoff RJ, Giedroc DP, Robinson NJ. A nickel-cobalt-sensing ArsR-SmtB family repressor. J. Biol. Chem. 2002;277:38441–38448. doi: 10.1074/jbc.M207677200. [DOI] [PubMed] [Google Scholar]
- Chao TC, Becker A, Buhrmester J, Puhler A, Weidner S. The Sinorhizobium meliloti fur gene regulates, with dependence on Mn(II), transcription of the sitABCD operon, encoding a metal-type transporter. J Bacteriol. 2004;186:3609–3620. doi: 10.1128/JB.186.11.3609-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobine P, Wickramasinghe WA, Harrison MD, Weber T, Solioz M, Dameron CT. The Enterococcus hirae copper chaperone CopZ delivers copper(I) to the CopY repressor. FEBS Lett. 1999;445:27–30. doi: 10.1016/s0014-5793(99)00091-5. [DOI] [PubMed] [Google Scholar]
- da Silva Neto JF, Braz VS, Italiani VCS, Marques MV. Fur controls iron homeostasis and oxidative stress defense in the oligotrophic alpha-proteobacterium Caulobacter crescentus. Nucleic Acids Research. 2009;37:4812–4825. doi: 10.1093/nar/gkp509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Walker GC. Disruption of sitA compromises Sinorhizobium meliloti for manganese uptake required for protection against oxidative stress. J Bacteriol. 2007;189:2101–2109. doi: 10.1128/JB.01377-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Wang Q, Liu Z, Zhang M, Machado AC, Chiu TP, Feng C, Zhang Q, Yu L, Qi L, Zheng J, Wang X, Huo X, Qi X, Li X, Wu W, Rohs R, Li Y, Chen Z. Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat Commun. 2015;6:7642. doi: 10.1038/ncomms8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Mireles E, Wexler M, Sawers G, Bellini D, Todd JD, Johnston AW. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology. 2004;150:1447–1456. doi: 10.1099/mic.0.26961-0. [DOI] [PubMed] [Google Scholar]
- Friedman YE, O’Brian MR. A novel DNA-binding site for the ferric uptake regulator (Fur) protein from Bradyrhizobium japonicum. J Biol Chem. 2003;278:38395–38401. doi: 10.1074/jbc.M306710200. [DOI] [PubMed] [Google Scholar]
- Frustaci JM, Sangwan I, O’Brian MR. Aerobic growth and respiration of a δ-aminolevulinic acid synthase (hemA) mutant of Bradyrhizobium japonicum. J. Bacteriol. 1991;173:1145–1150. doi: 10.1128/jb.173.3.1145-1150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedon E, Helmann JD. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol Microbiol. 2003;48:495–506. doi: 10.1046/j.1365-2958.2003.03445.x. [DOI] [PubMed] [Google Scholar]
- Hamza I, Chauhan S, Hassett R, O’Brian MR. The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 1998;273:21669–21674. doi: 10.1074/jbc.273.34.21669. [DOI] [PubMed] [Google Scholar]
- Hamza I, Hassett R, O’Brian MR. Identification of a functional fur gene in Bradyrhizobium japonicum. J. Bacteriol. 1999;181:5843–5846. doi: 10.1128/jb.181.18.5843-5846.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohle TH, Franck WL, Stacey G, O’Brian MR. Bacterial outer membrane channel for divalent metal ion acquisition. Proc Natl Acad Sci U S A. 2011;108:15390–15395. doi: 10.1073/pnas.1110137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohle TH, O’Brian MR. The mntH gene encodes the major Mn2+ transporter in Bradyrhizobium japonicum and is regulated by manganese via the Fur protein. Mol Microbiol. 2009;72:399–409. doi: 10.1111/j.1365-2958.2009.06650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohle TH, O’Brian MR. Transcriptional control of the Bradyrhizobium japonicum irr gene requires repression by Fur and antirepression by Irr. J Biol Chem. 2010;285:26074–26080. doi: 10.1074/jbc.M110.145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohle TH, O’Brian MR. Manganese is required for oxidative metabolism in unstressed Bradyrhizobium japonicum cells. Mol. Microbiol. 2012;84:766–777. doi: 10.1111/j.1365-2958.2012.08057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohle TH, O’Brian MR. Magnesium-dependent processes are targets of bacterial manganese toxicity. Mol Microbiol. 2014;93:736–747. doi: 10.1111/mmi.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach MR, Zamble DB. Metallocenter assembly of the hydrogenase enzymes. Curr Opin Chem Biol. 2007;11:159–165. doi: 10.1016/j.cbpa.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- Lee MH, Pankratz HS, Wang S, Scott RA, Finnegan MG, Johnson MK, Ippolito JA, Christianson DW, Hausinger RP. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993;2:1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J. Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JE, Waters LS, Storz G, Imlay JA. The Escherichia coli small protein MntS and exporter MntP optimize the intracellular concentration of manganese. PLoS Genet. 2015;11:e1004977. doi: 10.1371/journal.pgen.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menscher EA, Caswell CC, Anderson ES, Roop RM. Mur regulates the gene encoding the manganese transporter MntH in Brucella abortus 2308. J. Bacteriol. 2012;194:561–566. doi: 10.1128/JB.05296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngok-Ngam P, Ruangkiattikul N, Mahavihakanont A, Virgem SS, Sukchawalit R, Mongkolsuk S. Roles of Agrobacterium tumefaciens RirA in iron regulation, oxidative stress response, and virulence. J Bacteriol. 2009;191:2083–2090. doi: 10.1128/JB.01380-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brian MR. Perception and homeostatic control of iron in the Rhizobia and related bacteria. Annu Rev Microbiol. 2015;69:229–245. doi: 10.1146/annurev-micro-091014-104432. [DOI] [PubMed] [Google Scholar]
- Ojeda JF, Martinson DA, Menscher EA, Roop RM., 2nd The bhuQ gene encodes a heme oxygenase that contributes to the ability of Brucella abortus 2308 to use heme as an iron source and is regulated by Irr. J Bacteriol. 2012;194:4052–4058. doi: 10.1128/JB.00367-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek HR, O’Brian MR. KatG is the primary detoxifier of hydrogen peroxide produced by aerobic metabolism in Bradyrhizobium japonicum. J Bacteriol. 2004;186:7874–7880. doi: 10.1128/JB.186.23.7874-7880.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- Platero R, de Lorenzo V, Garat B, Fabiano E. Sinorhizobium meliloti Fur-like (Mur) protein binds a Fur box-like sequence present in the mntA promoter in a manganese-responsive manner. Appl Environ Microbiol. 2007;73:4832–4838. doi: 10.1128/AEM.00686-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero R, Peixoto L, O’Brian MR, Fabiano E. Fur is involved in manganese-dependent regulation of mntA (sitA) expression in Sinorhizobium meliloti. Appl Environ Microbiol. 2004;70:4349–4355. doi: 10.1128/AEM.70.7.4349-4355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, O’Brian MR. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol. Cell. 2002;9:155–162. doi: 10.1016/s1097-2765(01)00431-2. [DOI] [PubMed] [Google Scholar]
- Rappe MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-Proteobacteria. PLoS Comput Biol. 2006;2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankari S, O’Brian MR. A bacterial iron exporter for maintenance of iron homeostasis. J. Biol. Chem. 2014;289:16498–16507. doi: 10.1074/jbc.M114.571562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Jung HJ, An YJ, Cho YB, Cha SS, Roe JH. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc Natl Acad Sci USA. 2011;108:5045–5050. doi: 10.1073/pnas.1017744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C, White GF, Todd JD, Marritt SJ, Cheesman MR, Johnston AW, Le Brun NE. Heme-responsive DNA binding by the global iron regulator Irr from Rhizobium leguminosarum. J Biol Chem. 2010;285:16023–16031. doi: 10.1074/jbc.M109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SK, Puri S, Sangwan I, O’Brian MR. Positive control of ferric siderophore receptor gene expression by the Irr protein in Bradyrhizobium japonicum. J Bacteriol. 2009;191:1361–1368. doi: 10.1128/JB.01571-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JD, Wexler M, Sawers G, Yeoman KH, Poole PS, Johnston AW. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology. 2002;148:4059–4071. doi: 10.1099/00221287-148-12-4059. [DOI] [PubMed] [Google Scholar]
- Tottey S, Waldron KJ, Firbank SJ, Reale B, Bessant C, Sato K, Cheek TR, Gray J, Banfield MJ, Dennison C, Robinson NJ. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455:1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- Viguier C, Cuiv PO, Clarke P, O’Connell M. RirA is the iron response regulator of the rhizobactin 1021 biosynthesis and transport genes in Sinorhizobium meliloti 2011. FEMS Microbiol Lett. 2005;246:235–242. doi: 10.1016/j.femsle.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- Woodmansee AN, Imlay JA. Quantitation of intracellular free iron by electron paramagnetic resonance spectroscopy. Methods Enzymol. 2002;349:3–9. doi: 10.1016/s0076-6879(02)49316-0. [DOI] [PubMed] [Google Scholar]
- Yang J, Sangwan I, Lindemann A, Hauser F, Hennecke H, Fischer HM, O’Brian MR. Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol Microbiol. 2006;60:427–437. doi: 10.1111/j.1365-2958.2006.05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Genco CA. Fur-mediated activation of gene transcription in the human pathogen Neisseria gonorrhoeae. J. Bacteriol. 2012;194:1730–1742. doi: 10.1128/JB.06176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.