Abstract
Developmental nicotine exposure causes persistent changes in cortical neuron morphology and in behavior. We used microarray screening to identify master transcriptional or epigenetic regulators mediating these effects of nicotine and discovered increases in Ash2l, a component of a histone methyltransferase complex. We therefore examined genome-wide changes in H3K4 tri-methylation, a mark induced by the Ash2l complex associated with increased gene transcription. A significant number of regulated promoter sites were involved in synapse maintenance. We found that Mef2c interacts with Ash2l and mediates changes in H3K4 tri-methylation. Knockdown of Ash2l or Mef2c abolishes nicotine-mediated alterations of dendritic complexity in vitro and in vivo, and attenuates nicotine-dependent changes in passive avoidance behavior. In contrast, overexpression mimics nicotine-mediated alterations of neuronal structure and passive avoidance behavior. These studies identify Ash2l as a novel target induced by nicotinic stimulation that couples developmental nicotine exposure to changes in brain epigenetic marks, neuronal structure and behavior.
Keywords: Nicotine, histone modification, dendritic spines, Ash2l, Mef2c
High affinity nicotinic acetylcholine receptors (nAChRs) are involved in early brain development, nicotine-mediated alterations of neuronal structure, and persistent changes in behavior 1,2. Developmental nicotine exposure induces structural and synaptic changes in the developing brain 3 and these changes persist into later life 4. Accordingly, developmental exposure to nicotine is associated with many deleterious effects, including deficits in learning and later development of psychiatric disorders 5–7; however, the molecular mechanisms underlying these effects of cholinergic signaling on early brain development are largely unknown.
We used microarray screening to detect long-term changes in gene expression with the goal of identifying pleiotropic master transcriptional regulators responsible for persistent morphological alterations of cortical neurons. Among identified genes that could alter a transcriptional program, a component of a histone methylase complex, Ash2l, was significantly up-regulated by nicotine exposure during development. We therefore used next generation sequencing to determine whether nicotine could alter histone methylation associated with promoter regions of ensembles of genes involved in neuronal structure. Developmental nicotine exposure induced significant enrichment of trimethylated histone 3, lysine 4 (H3K4me3) at promoter regions of multiple genes involved in development and maintenance of excitatory synapses, including the transcription factor Mef2c, which assembles with Ash2l in a histone methyltransferase complex. Taken together, this study identifies a novel role for Ash2l in the regulation of neuronal morphology during early brain development and a new molecular mechanism underlying the long-term consequences of developmental nicotine exposure.
Results
Developmental nicotine treatment alters neuronal morphology
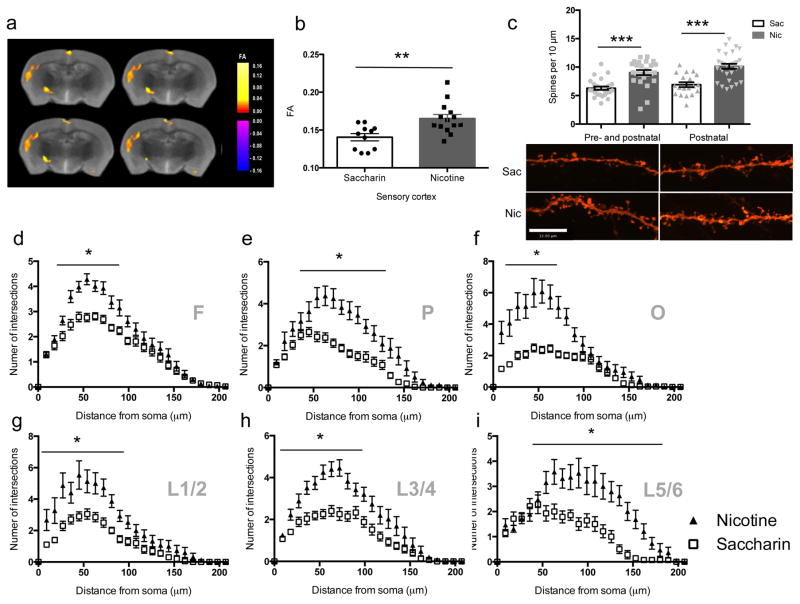
Exposure to tobacco smoke during human development can alter cortical structure as measured by diffusion tensor imaging (DTI) 8. Since nicotine can alter dendritic spine number and neuronal morphology 1, we determined whether nicotine in tobacco smoke could underlie changes in cortical fractional anisotropy (FA) measured by DTI. Among regimens used to deliver nicotine during development 7, drinking water administration results in significant blood levels in the dam and the offspring and has minimal stressful effects 9. We therefore exposed mice to saccharin or nicotine (200 μg/ml) from the time of conception through weaning (P21) and measured FA at 3 months of age. This regimen results in persistent neurochemical and behavioral changes in exposed pups, with no effects on maternal behavior 10. Developmental nicotine exposure induced significant increases in FA in a number of cortical areas, largely in gray matter (Fig. 1a, b). To determine whether changes in gray matter such as spine density and dendritic arborization underlie increased FA, we labeled neurons diolistically with DiI and performed Sholl analysis and spine counting in mice exposed to nicotine throughout pre- and postnatal development as in the DTI study, as well as in an additional group that was treated only from birth to weaning (postnatal exposure), a critical period for cortical development dependent on acetylcholine signaling 11. Nicotine significantly increased spine density in both the pre- and postnatal nicotine treated group and in the postnatal-only nicotine treated group (Fig. 1c). There was a significant effect of nicotine treatment on dendritic complexity across rostral, medial and caudal regions of cortex (Fig. 1d–f), and across all cortical layers, as demonstrated by a significant nicotine treatment by dendritic complexity interaction in each cortical layer compared to the saccharin-treated group (Fig. 1g–i). These results are consistent with previous studies showing that nicotine can induce persistent changes in spine density 3,4. Postnatal-only nicotine treated mice also showed a significant increase in dendritic complexity across all cortical regions and layers (supplementary Fig. 1).
Figure 1. Morphological changes in cortical neurons induced by developmental nicotine exposure.
Voxel-based analysis of FA changes induced by nicotine treatment during development measured by DTI (diagram from the Paxinos atlas 43 identifies affected regions). Two-tail t-tests of average FA values in nicotine and saccharin groups indicate a significant difference (n = 5 brains/condition; t(23) = 3.310, p = 0.00305573) in cortical regions investigated, including (a) somatosensory cortex and (b) sensory cortex. (c) Nicotine significantly increases spine density in nicotine treated groups compared to controls (pre- and postnatal: Sac (n = 26 slices from 6 mice), Nic (n = 25 slices from 5 mice), t49 = 5.151, p = 0.00000460; postnatal-only: Sac (n = 19 slices from 6 mice), Nic (n = 29 slices from 6 mice), t46 = 4.961, p = 0.00001002). (d–i) The number of intersections was assessed for the apical dendritic tree. Scale bar = 12 μm. (d) Frontal (F) cortical regions: (F(1,113) = 9.019, p = 0.00329026). The (e) parietal (P) and (f) occipital (O) regions of cortex showed a significantly higher number of intersections following nicotine exposure compared to the control group. (e) F(1,48) = 31.924; p = 0.00000085. (f) F(1,38) = 32.082, p = 0.00000165. (g–i) Layer-specific effects of developmental nicotine exposure on dendritic complexity. (g) Superficial layers of cortex showed the least change in complexity of dendritic arbors following early nicotine exposure (F(1,33) = 20.835, p = 0.0000661). Layers (h) 3/4 (F(1,85) = 25.927, p = 0.00000210) and (i) 5/6 (F(1, 30)=17.452, p = 0.00020270) show significant nicotine-induced increases in dendritic arborization compared to the control group. *, p < 0.05. Frontal (Sac, n = 41; Nic, n = 73), Parietal (Sac, n = 29; Nic, n = 23), Occipital (Sac, n = 23; Nic, n = 25), L1/2 (Sac, n = 20; Nic, n = 15), L3/4 (Sac: Sac, n = 36 ; Nic, n = 53), L5/6 (Sac, n = 14; Nic, n=18). “n” represents number of slices from 6 mice. Error bars represent s.e.m.
Nicotine induces a histone methyltransferase protein
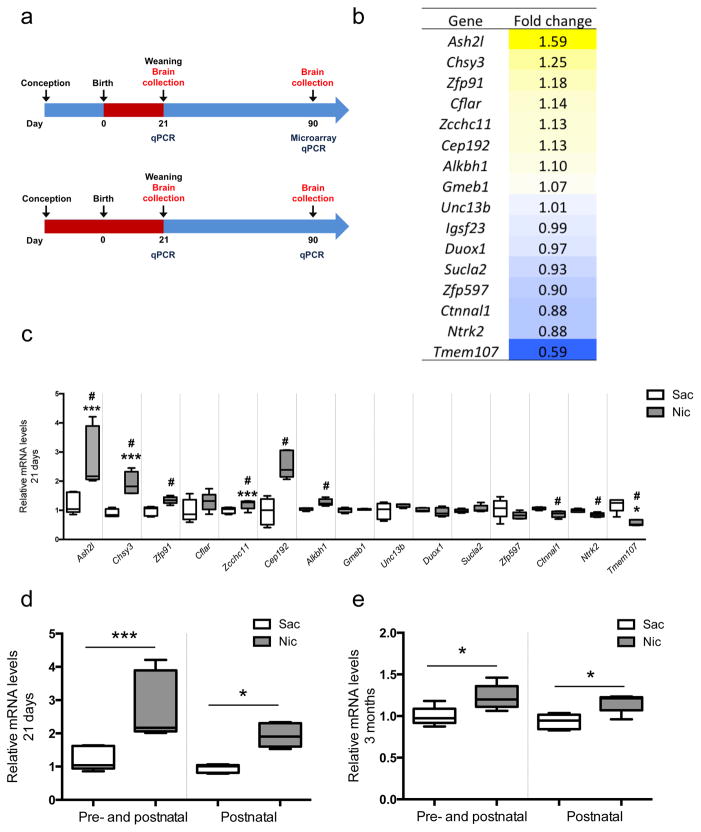
In order to identify persistent changes in transcriptional regulation that might be responsible for the effects of developmental nicotine treatment on cortical neuron morphology, we performed microarray analysis on mRNA prepared from dissected cortical tissue from mice administered saccharin or nicotine throughout the pre- and postnatal period until P21, that were then allowed to remain nicotine-free until 3 months of age (Fig. 2a; GEO accession #GSE80789). Our goal was to identify master transcriptional regulators that might maintain a program of gene expression responsible for persistent changes in neuronal morphology many weeks after developmental exposure to nicotine. We identified 18 probe sets that were significantly different between developmental nicotine-treated and control animals (Fig. 2b; supplementary Fig. 2a). To determine whether these changes in gene expression were an immediate or a later consequence of developmental nicotine exposure, we evaluated mRNA levels in independent cortical tissue samples by qRT-PCR immediately after nicotine exposure (21 days postnatal; Fig. 2c) or in adulthood following cessation of nicotine exposure (3 months postnatal; supplementary Fig 2b). At P21, 9 probes identified in the microarray study were significantly induced compared to the control group using qRT-PCR, including Ash2l, Chsy3, Ntrk2, Zfp91, Zcchc11, Cep192, Alkbh1, Ctnnal1, and Tmem107 (Fig. 2c). Five out of 15 probes initially identified by microarray were significantly different as measured by qRT-PCR compared to the control group at 3 months of age. These included Ash2l, Chsy3, Zcchc11, Cep192, and Tmem107. Among these, Ash2l was the most highly regulated by two regimens of developmental nicotine treatment (Fig. 2d, e) and met our criterion as a master regulator of transcription as measured in muscle development 12. Ash2l belongs to the trithorax (trx) gene family, a positive regulator of gene transcription particularly critical for development and cell differentiation through trimethylation of H3K4me3 13.
Figure 2. Nicotine exposure during development alters expression of a gene involved in histone methylation.
(a) Schematic of developmental nicotine exposure regimens. (b) List of the 18 probes that were significantly different between developmentally-treated and untreated animals. Primers (see supplementary Table 5) corresponding to 15 identified probes (excluding unannotated and predicted transcribed probe sets) were used to evaluate mRNA changes in cortical tissue from a separate set of animals using qRT-PCR. (c) 9 of 15 probe sets were significantly altered in independent samples immediately after the cessation of developmental nicotine treatment at 21 days of age (*p < 0.05; Ash2l: F(1,8) = 10.848, p = 0. 01096250; Chsy3: F(1,8) = 31.334, p = 0.00051163; Zfp91: F(1,8) = 14.28, p = 0.00539670; Cflar: F(1,8) = 1.83, p = 0.21311592; Zcchc11: F(1,8) = 5.49, p = 0.04718863; Cep192: F(1,8) = 30.084, p = 0.00058407; Gmeb1: F(1,8) = 0.416, p = 0.53699305; Alkbh1: F(1,8) = 16.474, p = 0.00363940; Unc13b: F(1,8) = 0.416, p = 0.53699305; Duox1: F(1,8) = 1.19, p = 0.30709203; Scula2: F(1,8) = 1.58, p = 0.24421686; Zfp597: F(1,8) = 2.15, p = 0.181; Ctnnal1: F(1,8) = 13.878, p = 0.00582675; Ntrk2: F(1,8) = 13.364, p = 0.00644127; Tmem107A : F(1,8) = 28.897, p = 0.00066513). Ash2l mRNA levels were significantly elevated following exposure to nicotine during development (d) immediately following nicotine exposure (21 days of age; F(3,16) = 6.898, p = 0.003411) and (e) in adulthood (3 months of age; F(3,16) = 10.736, p = 0.000413) in samples from mice treated throughout the pre- and postnatal period or during the postnatal period only. (*p <0.05; *** p <0.01 with multiple comparisons; # p <0.05). A total of 20 mice (Nic: n = 10; Sac, n = 10) were used for microarray studies. 5 animals were pooled for each experimental group and two replicate experiments were analyzed. 2 or 3 animals were pooled for each sample for qPCR analysis and 5 samples per each condition were analyzed (21 days: Nic: n = 13; Sac, n = 15. 3 months: Nic: n = 11; Sac, n = 15). Whiskers represent minimum to maximum value of data distribution.
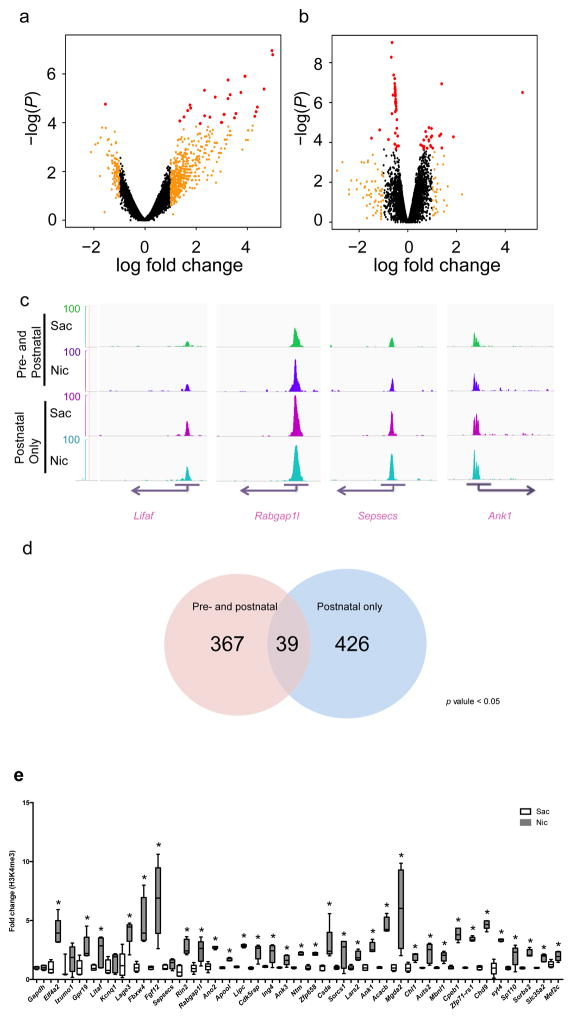
Histone methylation provides a potential mechanism for long-term regulation of gene expression that could maintain the effects of early nicotine exposure into adulthood. We therefore evaluated H3K4me3 across the genome in cortical tissue following pre- and postnatal (Fig. 3a) and postnatal-only (Fig. 3b) nicotine exposure and identified 14,881 gene loci (Fig. 3a, b; GEO accession # GSE80789). Short read sequences from the ELAND pipeline were performed for quality control, and those that did not meet criteria for fidelity were discarded. Identified peaks were viewed in Genome browser and their alignment was verified (examples shown in Fig. 3c). 367 differentially-enriched peaks with adjusted p values < 0.05 were identified in cortical samples from pre- and postnatal nicotine exposed mice, and 426 differentially enriched peaks were identified from postnatal-only exposed mice (Fig. 3d; supplementary Tables 1 and 2).
Figure 3. Differential enrichment of H3K4me3 at promoter sites associated with synapse function following developmental nicotine exposure. (a,b).
Scatterplots of relative enrichment level, degree of (fold) change, and P values (nominal p value cutoffs) of all 14,881 identified genomic sites in the pre- and postnatal, and postnatal-only, nicotine treated groups. Dots in red indicate genomic loci for which adjusted. p < 0.05 (c) Genome browser tracks of H3K4me3 epigenetic marks at four selected genomic sites (Litaf, Rabgab1l, Sepsecs, and Ank1). Images were acquired using igvtools integrated genome browser and confirmed with UCSC genome browser. The peak distribution pattern of each replicate of a representative locus is shown in supplementary Fig. 3b. (d) ChIP-seq analysis identified 367 differentially enriched peaks in cortical samples from mice exposed to nicotine during the pre- and postnatal periods, and 426 differentially enriched peaks in cortical samples from mice exposed to nicotine only in the postnatal period, with adjusted p values < 0.05. (e) Verification of identified gene loci that were enriched following both nicotine exposure regimens using ChIP-PCR showed that 36 of 39 genomic sites identified in the ChIP-seq analysis were differentially methylated in independent samples following pre- & postnatal nicotine exposure compared to the control group (*, p < 0.05; ***, p < 0.01 with multiple comparisons; #, p < 0.05; Gapdh: F(1,8) = 0.00, p = 1.000; Eif4a: F(1,8) = 31.364, p = 0.00051003; Izumo: F(1,6) = 0.900, p = 0.37940979; Gpr19: F(1,8) = 9.107, p = 0.01661552; Litaf: F(1,8) = 5.554, p = 0.04619611; kcnq1: F(1,8) = 0.792, p = 0.39946656; Lage3: F(1,8) = 16.418, p = 0.00367444; Fbxw4: F(1,8) = 17.9, p = 0.00287307; Fgf12: F(1,7) = 13.804, p = 0.00750159; Sepsecs: F(1,8) = 7.568, p = 0.02502024; Rin2: F(1,8) = 33.897, p = 0.00039509; Rabgap1l: F(1,8) = 11.344, p = 0.00981340; Ano2: F(1,8) = 107.443, p = 0.00000649; Apool: F(1,8) = 120.782, p = 0.00000418; Lipc: F(1,8) = 893.328, p = 0.00000001; Cdk5rap: F(1,8) = 23.198, p = 0.00132739; Ing4: F(1,8) = 10.019, p = 0.01328880; Ank3: F(1,8) = 11.424, p = 0.00964250; Ntm: F(1,7) = 330.474, p = 0.00000038; Zfp658: F(1,7) = 141.861, p = 0.00000669; csda: F(1,8) = 8.411, p = 0.01988645; Sorcs1: F(1,7) = 5.378, p = 0.05346909; Lars2: F(1,7) = 23.081, p = 0.00195662; Ank1: F(1,7) = 47.436, p = 0.00023397; Acacb: F(1,7) = 128.527, p = 0.00000930; Mgda2: F(1,7) = 10.811, p = 0.01333907; Chl1: F(1,7) = 18.662, p = 0.00348039; Auts2: F(1,8) = 11.318, p = 0.00986976; : F(1,8) = 29.295 Mbnl1, p = 0.00063647; : F(1,8) = 141.042 Cpeb1, p = 0.00000232; Zfp71-rs1: F(1,8) = 830.025, p = 0.0000001; Chd9: F(1,8) = 396.42, p = 0.00000004; Syt4: F(1,8) = 87.029, p = 0.00001422; Sp110: F(1,8) = 7.628, p = 0.02460310; Sorbs2: F(1,8) = 77.526, p = 0.00002176; Slc35a2: F(1,8) = 37.6, p = 0.00027950; Mef2c: F(1,8) = 10.881, p = 0.01088096). Each replicate was a pool of 2–4 brain samples and 5 replicates were used for each condition (Sac: n = 5 pools from 17 animals; Nic: n = 5 pools from 15 animals). Whiskers represent minimum to maximum value of data distribution.
Developmental nicotine exposure altered histone methylation at genomic sites responsible for the regulation of synapses, postsynaptic plasticity and cell junctions as identified by gene ontology (GO) analysis (using adjusted p values < 0.05; supplementary Tables 3 and 4). The genomic sites in these GO categories are highly overlapping (supplementary Fig. 3a). Most of the genomic sites identified in this screen are implicated in glutamatergic synaptic transmission (Grin1, Grin2c and Grin2d NMDA receptor subunits), and synapse formation (Dlg2, Camk2a, and Dlgap2).
Although there was substantial overlap in genomic sites associated with increased histone methylation in both nicotine-treated groups, developmental nicotine exposure resulted in different effects depending on time and duration of exposure. We identified common genomic sites from gene lists (adjusted p value < 0.05) of differentially enriched histone methylation following pre- and postnatal or postnatal-only nicotine exposure (example regulated locus; supplementary Fig. 3b). A total of 39 genes showed overlapping changes in both the pre- and postnatal and postnatal-only nicotine treated group (supplementary Tables 1 and 2).
In order to validate these changes, all 36 overlapping genomic sites (excluding 3 Riken genes with unknown function) were tested using chromosome immunoprecipitation (ChIP) followed by qPCR in independent samples. 34 of 36 tested genomic sites showed significant changes in associated H3K4me3 levels in the pre- and postnatal or postnatal-only groups compared to the control group (Fig. 3e, supplementary Fig. 3c).
Nicotine increases histone methylation at the Mef2c locus
Consistent with our search for master transcriptional regulators, we selected Mef2c for further functional characterization from among the 34 genomic sites that were verified by ChIP-PCR because the Mef2 family has been identified as part of the methylation complex along with Ash2l 12, and also participates in dendritic remodeling during neural differentiation 14,15. We first determined whether identified Ash2l and Mef2c binding sites in the genome 16 overlap with the sites of differentially enriched H3K4me3 sites we found following developmental nicotine exposure. A large proportion of Ash2l (mouse ES cell dataset) and Mef2c (early neuronal activity-induced cortical neuron datasets) binding sites overlapped with the H3K4me3 sites found here (supplementary Fig. 3d). GO analysis of these 6426 overlapping genomic sites (those sites differentially enriched as a result of nicotine exposure that overlapped with previously identified sites of Ash2l and Mef2c binding) identified synapse-related genes as a significant category (supplementary Fig. 3d). This suggested that Ash2l and Mef2c may normally interact at genomic sites related to synaptic function during neuronal development and induce histone tri-methylation and transcription of these loci, and that this complex is modulated as a result of developmental exposure to nicotine.
Mef2c expression is normally up-regulated during neuronal differentiation and maturation 14,15. Although it was not identified in our initial microarray screen, it is likely that the genome-wide evaluation missed some upregulated targets due to stringent statistical cut-offs. We therefore used targeted analyses to determine whether the mRNA encoding Mef2c is regulated following nicotine exposure during development. Using qPCR, we found that Mef2c mRNA levels were significantly elevated in the cortex of mice treated with nicotine throughout development (supplementary Fig. 4a). Mef2c genomic sites also showed significantly elevated H3 acetylation (supplementary Fig. 4b), an additional histone modification related to increased transcription.
At 3 months of age, H3K4me3 association with the Mef2c locus was elevated across all regions of cortex as measured by ChIP-PCR following developmental nicotine exposure, although changes were somewhat more modest in the caudal region (supplementary Fig. 4c). These data confirm that histone changes at the Mef2c locus are maintained for a long period following nicotine exposure, and that changes in histone methylation and gene expression occur broadly across cortical regions.
Ash2l and Mef2c expression levels are regulated by nAChRs
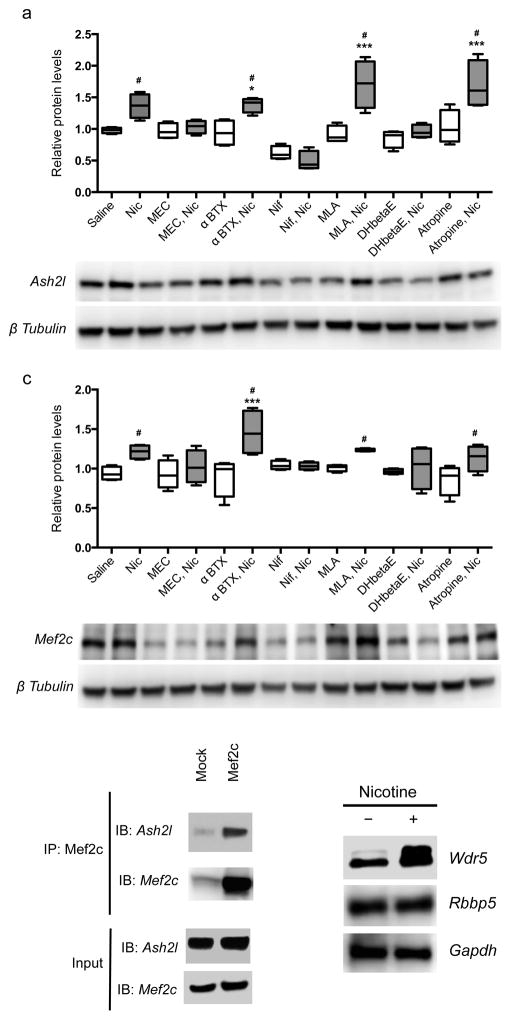
In vivo genome-wide screening can identify loci that may be relevant for long-term consequences of developmental nicotine exposure, but cannot distinguish between direct effects of nicotine and indirect changes of downstream pathways. To determine whether alterations of Ash2l and Mef2c levels are due to direct effects of nicotine mediated through nAChRs, Ash2l and Mef2c protein levels were evaluated in cultured neural progenitor cells. After 8–9 days in culture, neural progenitor cells had differentiated into neurons. Cells were treated with nicotine (1 μM) for 48 h and protein levels were evaluated by Western blotting. Nicotine exposure significantly increased Mef2c and Ash2l protein levels as compared to saline treatment (Fig. 4a, b). The involvement of nAChR activity in the effects of nicotine on Mef2c and Ash2l expression was verified by exposure to the nicotinic antagonists mecamylamine, α-bungarotoxin, dihydro-β-erythroidine (DHβE) and methyllycaconitine (MLA; Fig. 4a, b). Mecamylamine and DHβE significantly attenuated nicotine-induced increases in Ash2l and Mef2c protein levels, whereas α-bungarotoxin and MLA had little effect, suggesting that heteromeric (likely α4/β2 subunit containing) nAChRs are more likely to contribute to these effects than homomeric nAChRs containing the α7 subunit.
Figure 4. The Ash2l/Mef2c complex is regulated by nAChR activity.
(a, b) Nicotine treatment of neural progenitor cells in culture significantly elevated Ash2l (a) and Mef2c (b) protein levels (F(13,42) = 12.540, p = 0.000001 (Saline vs Nic: p = 0.009350; α-BTX vs α-BTX+Nic: p = 0.003186; MLA vs MLA+Nic: p = 0.000001; Atropine vs Atropine+Nic: p = 0.000030) and F(13,42) = 3.749, p = 0.000551 (Saline vs Nic: p = 0.023076; α-BTX vs α-BTX+Nic: p = 0.010348; MLA vs MLA+Nic: p = 0.05; Atropine vs Atropine+Nic: p = 0.023488), respectively, as measured by one-way ANOVA followed by LSD test for multiple comparisons). This was reversed by the broad spectrum nAChR antagonist mecamylamine (MEC) and the more selective heteromeric nAChR antagonist dihydro-β-erythroidine (DHβE). The selective α7 nAChR antagonists α-bungarotoxin (btx) and methyllyccaconitine (MLA) had no effect on their own and did not alter the effects of nicotine (Nic). The voltage-gated calcium channel blocker nifedipine (Nif) decreased levels of Ash2l and Mef2c at baseline and abolished the ability of nicotine to increase their expression. (c) Ash2l and Mef2c can be co-immunoprecipitated in a complex from neural progenitor cells (replicated twice). (d) Wdr 5 protein levels are increased by nicotine treatment. n = 4 replicates per condition for all neural progenitor experiments. Original Western blots presented in supplementary Fig. 7. Whiskers represent minimum to maximum value of data distribution.
Nicotine depolarizes neurons and increases levels of intracellular calcium, leading to induction of intracellular signaling cascades. To study the role of calcium signaling in nicotine-dependent alterations in Ash2l and Mef2c levels, we grew neural progenitor cells for 8–9 days, then treated with nifedipine (10 μg/ml) for 48 h with or without nicotine. Co-treatment with nifedipine blocked the effects of nicotine and significantly reduced basal levels of Ash2l and Mef2c (Fig. 4a, b), suggesting that expression of these proteins is calcium-dependent and could be induced by depolarization.
Direct interaction of Mef2c and Ash2l was next assessed by co-immunoprecipitation using a Mef2c antibody to pull down Ash2l (Fig. 4c). Ash2l was recognized in samples immunoprecipitated with the specific Mef2c antibody, but much less Ash2l was observed in the control condition. Thus, Ash2l and Mef2c form a complex in neural progenitor cells, as has been documented in muscle cells 12. We next determined whether additional methyltransferase complex subunits (Wdr5, Mll, Rbbp5) that form a complex with Ash2l in muscle were also present in neural progenitor cells. Wdr5 and Rbbp5 were detected following formalin cross-linking and immunoprecipitation of Ash2l at baseline, and levels of Wdr5 were significantly elevated following treatment of cells with nicotine (Fig. 4d). Thus, the methyltransferase complex is expressed in neural progenitors and multiple components are regulated by nicotine signaling.
Ash2l/Mef2c mediate nicotine effects on dendrite morphology
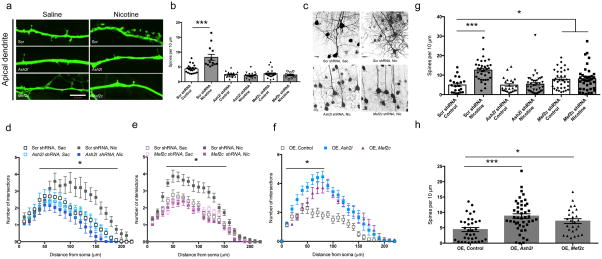
If the Ash2l/Mef2c complex is involved in transcriptional regulation of genes involved in synapse and spine formation, these proteins may be necessary for the ability of nicotine to induce changes in dendritic structure. To determine whether Ash2l and Mef2c are involved in dendritic spine formation, we generated shRNAs specific for mouse Ash2l or Mef2c and transfected these into neural progenitor cells. Down-regulation of Ash2l or Mef2c protein was confirmed by Western blotting (supplementary Fig. 5). Neural progenitor cells were transfected with Ash2l shRNA, Mef2c shRNA or scrambled control shRNA (Fig. 5a–f) and exposed for 4 days to saline or nicotine. Four days of nicotine exposure significantly increased the number of spines on dendritic shafts of cultured neurons (Fig. 5a, b). Conversely, knockdown of Ash2l or Mef2c (Fig. 5a, b) significantly reduced the number of dendritic spines on apical dendrites of cultured neural progenitor cells and abolished the increase in spines induced by 4 days of nicotine exposure.
Figure 5. The Ash2l/Mef2c complex mediates dendritic remodeling by nicotine in neural progenitor cells and in vivo.
(a, b) shRNAs targeting Ash2l or Mef2c were transfected into neural progenitor cells and spine density was measured on the apical dendrite in control cells (Saline) or following 4 days of exposure to nicotine in the bath (Nicotine). Nicotine-induced increases in spine density are significantly attenuated by Ash2l shRNA and Mef2c shRNA delivery (F(5,108)=28.45, p=0.000001 with LSD correction for multiple comparisons). Scrambled Saline vs. Scrambled Nicotine: p = 0.000001; Scrambled shRNA: Saline, n = 20, Nicotine, n=13; Ash2l shRNA: Saline, n = 26, Nicotine = 11; Mef2c shRNA: Saline, n = 17, Nic = 11. “n” sizes for neural progenitor studies represent total cells counted across 3 independent cultures. Scale bar = 18 μm. (c – f) shRNAs or overexpression constructs for Ash2l and Mef2c were introduced into the developing cortex at E14 by electroporation, with or without nicotine treatment. (c) Representative images of cortical neurons following nicotine treatment or electroporation of shRNA constructs (supplementary Fig. 6 shows extent of construct spread). (d) Nicotine exposure during the first 21 days of postnatal development increased the complexity of cortical neuron dendrites as measured by Sholl analysis (F(1, 174) = 14.570, p = 0.0002) and this was blocked by electroporation of shRNAs targeting Ash2l into the developing cortex at E14. Gene x treatment interaction: F(1, 174) = 16.633, p = 0.00006893 by two-way ANOVA with repeated measures followed by Tukey’s test for multiple comparisons (Scrambled Saccharin (n = 32) vs. Scrambled Nicotine (n = 57): p = 0.000581, Ash2l Saccharin (n = 22) vs. Ash2l Nicotine (n = 67): p = 0.229149. “n” sizes for in vivo studies represent number of slices from 5 mice. (e) Nicotine-mediated changes in dendritic complexity are abolished by electroporation of Mef2c shRNA into developing cortex. Gene x treatment interaction: F(1,110) = 95.076, p = 0.0000001 by two-way repeated measures ANOVA with repeated measures followed by Tukey’s test for multiple comparisons (Scrambled Saccharin (n = 26) vs. Scrambled Nicotine (n = 31): p = 0.000581, Ash2l Saccharin (n = 27) vs. Ash2l Nicotine (n = 30): p = 0.229149). (f) Overexpression of Ash2l or Mef2c increases dendritic complexity (F(2,71) = 100.450, p = 0.0000001 by one-way ANOVA with repeated measures). OE control (n = 28) vs. OE Ash2l (n = 22): p = 0.0000001; OE control (n = 28) vs. OE Mef2c (n = 24): p = 0.0000001 with Tukey’s test for multiple comparisons. (g) Nicotine-induced increases in spine density are significantly attenuated by electroporation of Ash2l or Mef2c shRNAs into developing cortex (F(5,183) = 10.748, p = 0.0000001 with LSD test for multiple comparisons). Scrambled Saccharin vs. Scrambled Nicotine: p = 0.0000001; Scrambled Saccharin vs. Mef2c Saccharin: p = 0.042553; Scrambled Saccharin vs. Mef2c Nicotine: p = 0.023182; Scrambled Nicotine vs. Mef2c Saccharin: p = 0.000158; and Scrambled Nicotine vs. Mef2c Nicotine: p = 0.000295. Scrambled shRNA: (Sac, n = 21; Nic, n = 36). Ash2l shRNA: (Sac, n = 34; Nic, n = 37). Mef2c shRNA: (Sac, n = 23; Nic, n = 38). (h) Overexpression of Ash2l or Mef2c significantly increases spine density increases (F(2,102) = 19.692, p = 0.00000006 with LSD test for multiple comparisons). OE control (n = 34) vs. OE Ash2l (n = 45): p = 0.000033; OE control (n = 34) vs. OE Mef2c (n = 26): p = 0.019220. Differences measured by one-way ANOVA followed by LSD test for multiple comparisons. Whiskers represent minimum to maximum value of data distribution. Error bars represent s.e.m.
To determine whether the Ash2l/Mef2c complex could also be critical for effects of nicotine on cortical neuron structure in vivo, in utero electroporation was used to introduce the shRNAs targeting Ash2l or Mef2c into a subset of neurons in the cortex of mice at embryonic day 14 (E14). Control (scrambled) shRNA was electroporated into the right hemisphere of a subset of the developing embryos and Ash2l or Mef2c shRNA was electroporated into the left hemisphere of remaining littermates (supplementary Fig. 6). Dams were administered nicotine or saccharin in the drinking water starting from the day of surgery and pups were euthanized on postnatal day 21 for analysis of cortical neuron dendritic structure. Exposure to nicotine increased dendritic complexity in neurons expressing the scrambled construct, whereas knockdown of Ash2l or Mef2c abolished the ability of nicotine exposure to alter dendritic branching (Fig. 5c–e). Consistent with the possibility that the Ash2l/Mef2c complex mediates the effects of nicotine on dendritic structure, overexpression of Ash2l or Mef2c mimicked the ability of nicotine to increase dendritic complexity in the absence of drug exposure (Fig. 5f).
Similarly, electroporation of the Ash2l shRNA abolished the ability of nicotine to increase density of dendritic spines (Fig. 5g). Unexpectedly, knockdown of Mef2c resulted in significantly increased basal spine density compared to control animals; however, there was no further increase in spine density by nicotine exposure following Mef2c knockdown (Fig. 5g). These results suggest that Mef2c signaling likely has multiple roles in spine formation and stabilization 14,17–22, but among those roles, it contributes to the ability of nicotine to increase spine number. In contrast, overexpression of Ash2l or Mef2c increased spine density (Fig. 5h).
Ash2l/Mef2c mediate nicotine effects on passive avoidance
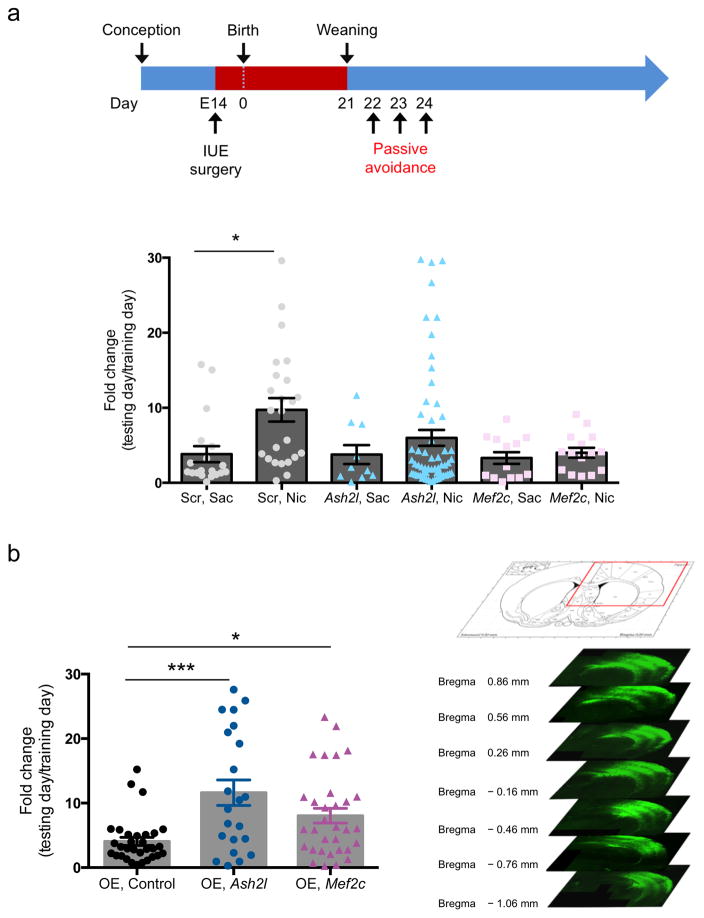
Knockdown of Ash2l or Mef2c in vivo prevents developmental nicotine-induced remodeling of dendritic morphology and spine density. In previous studies, we showed that nicotine exposure during the first 3 postnatal weeks induces hypersensitive passive avoidance learning in response to a subthreshold foot shock that is normally ignored 23,24. Furthermore, this effect is mediated through β2 subunit-containing nAChRs on corticothalamic neurons. We hypothesized that this behavioral consequence of developmental nicotine exposure could result from morphological changes in sensory cortex as a result of Ash2l and Mef2c induction. We tested passive avoidance behavior in mice following shRNA-mediated knockdown or cDNA overexpression of Ash2l or Mef2c in sensory cortex via in utero electroporation. While we targeted sensory cortex based on the DTI findings and the role of sensory stimuli in passive avoidance behavior, all animals showed broad cortical expression of the shRNA constructs, and some expression was seen bilaterally. Mice were exposed to saccharin or nicotine from the time of surgery until weaning on postnatal day 21 (Fig. 6a). As predicted, nicotine-exposed mice demonstrated hypersensitive passive avoidance performance, as shown by increased latency to enter a dark chamber paired with a mild foot shock, compared to saccharin-exposed mice (Fig. 6a). Cortical delivery of Ash2l or Mef2c shRNA by in utero electroporation significantly attenuated the hypersensitive passive avoidance induced by nicotine (Fig. 6a). Despite this significant attenuation, Ash2l knockdown did not completely abolish the effects of nicotine, likely due to incomplete knockdown or variability in cortical targeting. Finally, Ash2l or Mef2c overexpression via electroporation mimicked the effect of nicotine exposure, resulting in hypersensitive passive avoidance behavior as compared to controls (Fig. 6b), demonstrating bidirectional modulation of passive avoidance by Ash2l and Mef2c.
Figure 6. Developmental nicotine-induced changes in passive avoidance behavior require expression of Ash2l and Mef2c.
(a) Nicotine treatment from E14 through P21 induces hypersensitive passive avoidance behavior that is attenuated by knockdown of Ash2l or Mef2c (F(5,135) = 2.759; p = 0.02090053, LSD correction for multiple comparisons). Scrambled Saccharin vs. Scrambled Nicotine: p = 0.015343; Ash2l Saline vs. Ash2l Nicotine: p = 0.009136 with LSD multiple comparison test. Scrambled shRNA: (Sac, n = 19; Nic, n = 24). Ash2l shRNA: (Sac, n = 10; Nic, n = 60). Mef2c shRNA: (Sac, n = 13; Nic, n = 15). F(5,130) = 2.713; p = 0.023 (b) Overexpression of Ash2l or Mef2c induces hypersensitive passive avoidance behavior (F(2,82) = 8.858; p = 0.00031264, LSD correction for multiple comparisons). OE-control vs. OE-Ash2l: p = 0.000076; OE-control vs. OE-Mef2c: p = 017160 with LSD multiple comparison test). OE, control: (n = 31), OE, Ash2l: (n = 22), OE, Mef2c: (n = 32). (c) Representative images of an in utero electroporated mouse brain. Efficiency of electroporation was variable but expression was consistently spread broadly across the parietal cortex. Error bars represent s.e.m. Competing financial interests
Discussion
The current results demonstrate that Ash2l and Mef2c are critical for the ability of nAChR activity to alter cortical neuron architecture and behavior during development. We propose that nicotine-dependent induction of the Ash2l/Mef2c complex during cortical development induces alterations of histone methylation associated with promoter regions of genes involved in structure and function of glutamatergic synapses, and increases spine number, dendritic branching and hypersensitive passive avoidance behavior, a task modulated by nAChRs expressed in cortical neurons. Together, these results identify a molecular mechanism underlying the ability of nAChRs to influence synaptic and dendritic structure and provide a potential cellular mechanism underlying a persistent behavioral consequence of developmental nicotine exposure.
Among the small number of genes that are persistently regulated in adulthood following early nicotine exposure, we focused on Ash2l because it fit the profile of a protein that could maintain a broad program of gene expression changes into adulthood. Ash2l is the mammalian homolog of ash2, a gene that belongs to the trithorax (trx) gene family and is a positive regulator of homeotic genes through DNA binding 25 and maintenance of chromatin in a transcriptionally active conformation 26. The Ash2l gene is highly conserved across evolution and functions as a transcription factor 27. It is a subunit of several Trx complexes that associate with the Set1 family of histone H3K4 methyltransferases 28. Decreased expression of Ash2l impairs histone H3K4 trimethylation (H3K4me3) and expression of SET family target genes 29.
In mammals, Ash2l is expressed in fetal human and mouse brain and its expression decreases in adult tissue 30. A study of alcohol-induced changes in brain gene expression identified changes in trx-related genes, including Ash2l 31, suggesting that modulation of Ash2l may also be important for effects of alcohol and other drugs on brain development. Based on the human proteome atlas database, Ash2l protein in brain is detected only in neuronal and glial cells of the cerebral cortex and cerebellum 32. We cannot exclude the possibility that Ash2l signaling in glial cells is involved in nicotine effects on cortical morphology or behavior; however, the in utero electroporation studies may indicate that this is unlikely. Electroporation into the cortex at E14 largely targets pyramidal neurons because GABA interneurons have not yet migrated from the ganglionic eminence at E14, whereas astroglia develop later and electroporated DNA is diluted by the time glia start to differentiate 33. In addition, the genes most significantly regulated by nicotine exposure during development fall into neuronally-relevant GO groups, including those involved in glutamatergic synaptic formation and maintenance. Taken together, these observations support a primary role for Ash2l and Mef2c in neuronal signaling following nicotine exposure during development.
This study also identifies a novel mechanism of gene regulation during brain development that could be more generally involved in activity-dependent structural changes of cortical neurons. Ash2l-mediated changes in H3K4me3, specifically at promoters of genes involved in synapse maintenance, likely reflect transcriptional alterations at these loci during a critical period of development that become fixed in the genome 34. H3K4me3 has been identified as an active promoter mark 35 and may regulate formation of the pre-initiation complex and gene transcription by interacting with other transcription factors 36.
Of the 34 promoter regions associated with H3K4me3 changes following nicotine exposure, the Mef2c locus is of particular interest because interaction between Ash2l and Mef2d selectively induces trimethylation of H3K4, which subsequently induces gene expression changes required for muscle differentiation 12. While Mef2c mRNA levels increase in parallel with the increase in H3K4me3 associated with its promoter, it is not yet known whether these histone methylation changes alter Mef2c expression directly. In myoblasts undergoing terminal differentiation, p38α-dependent phosphorylation of Mef2d recruits the Ash2l complex to myogenic gene promoters, enhancing histone H3K4 methylation 12, promoting formation of the transcription initiation complex and recruiting associated transcription factors that maintain the expression of myogenic genes 37. Thus, the enrichment of H3K4me3 and persistence of elevated Ash2l expression many weeks after developmental nicotine exposure may maintain transcription of loci associated with H3K4me3 enrichment into adulthood.
Although differential H3K4me3 enrichment is positively correlated with transcription of specific subsets of genes, it is weakly correlated with genome wide transcription changes. The involvement of H3K4me3 in specific gene expression changes is therefore still somewhat controversial. The current study showing differential enrichment of histone methylation at genomic sites following developmental nicotine exposure may suggest that this subset of promoter regions is among those activated by the H3K4me3 mark. While the mechanisms that distinguish genomic loci transcribed as a result of this histone methylation event are not yet clear, the ability of nicotinic signaling to stimulate synaptically-related loci may help identify components of the complex that confer specific transcriptional activation. We note that although a small subset of loci were significantly enriched by both pre- and postnatal and postnatal-only nicotine exposure compared to saccharin treatment, 99% of loci identified with the H3K4me3 mark were identified in both nicotine treatment groups. The two regimens of nicotine exposure resulted in differential enrichment of H3K4me3 patterns across this set of gene loci, likely because histone methylation changes are highly dynamic across this period of neural precursor differentiation resulting in a cumulative representation of changes at each stage of development. Thus, the subset of gene loci showing differential H3K4me3 enrichment common to the two regimens of nicotine exposure are likely to be those loci most critical for the functional effects of nicotine on adult neuronal morphology and behavior.
Surprisingly, most of the genes implicated in developmental response or neuroadaptation to nicotine are implicated in glutamatergic neurotransmission, synaptic structure or spine formation. In addition to its role in the Ash2l complex, the Mef2 family is involved in synaptic maturation and dendritic structure 14,17–22. Members of the Mef2 family are expressed throughout the developing central nervous system 38 and can regulate activity-dependent synaptic morphogenesis in hippocampal neurons 39,40. Mef2c regulates gene expression 18,19 and facilitates neuronal plasticity by regulating synaptic number and function in adult rodents 14. The identification of mutations in genes regulated by the Mef2 family in human neurological diseases, including autism spectrum disorders, suggests that Mef2 activity is critical for normal brain development. Conditional knockout of the Mef2c gene in mouse brain results in changes in neuronal density, disorganized cortical structure and immature electrophysiological properties, along with abnormalities in anxiety-like behaviors and cognitive tasks 21. The current results support the idea that Mef2c plays multiple roles in neuronal development, some of which are dependent on Ash2l and others that are likely to be independent.
Although loci with synapse-related functions were most clearly identified by gene ontology analysis following developmental nicotine exposure, genomic sites in the channel binding protein and activity modulator groups were also identified, including a number of calcium and potassium channel subunits (see supplementary Tables 3 and 4). In addition, developmental nicotine administration induced changes in histone methylation of acetylcholine receptor loci, including the α3 subunit of the nAChR (Chrna3) and the muscarinic subunit M1 (Chrm1). Thus, developmental nicotine exposure has persistent effects on histone structure of loci associated with glutamatergic synaptic transmission, neuronal excitability and cholinergic signaling, and common molecular and cellular changes can be induced by different durations of nicotine exposure.
The early postnatal weeks appear to be a critical period for nicotine-induced molecular, cellular and behavioral alterations. The first 2–3 weeks of postnatal development in the mouse corresponds to the third trimester of human pregnancy with respect to cortical synaptic development 41,42. Previous studies suggest that stimulation of nAChRs and the resulting calcium influx contributes to maturation of glutamatergic synapses 11 and stimulation of β2* nAChRs induces spine formation in hippocampal and cortical neurons in a cell-autonomous manner 1. In previous studies we have shown that developmental nicotine exposure during the first three weeks of postnatal development induces persistent hypersensitive passive avoidance behavior that lasts into adulthood 23,24. Increased passive avoidance learning in response to a mild footshock that is normally ignored could represent an inability to screen out irrelevant sensory stimuli, consistent with the increased incidence of attentional disorders in children exposed in utero to maternal smoking 7. Maternal smoking is also associated with increased smoking in adult offspring, although this is not modeled with the passive avoidance paradigm. The hypersensitive passive avoidance phenotype requires expression of β2* nAChRs on layer 6 corticothalamic neurons, consistent with the finding that manipulation of Ash2l or Mef2c levels in cortex can modulate passive avoidance learning.
The correlation between the ability of the Ash2l/Mef2c complex to regulate nicotine-dependent remodeling of dendritic morphology and changes in passive avoidance behavior suggest that the alterations in neuronal structure may mediate a persistent behavioral consequence of early nicotine exposure. The current study provides a novel molecular mechanism that underlies these cellular and behavioral consequences of nicotine administration. Moreover, the fact that DTI has the sensitivity to detect these morphological changes of dendritic spine density provides clinical opportunities to evaluate these changes in behaving human subjects. In addition to mediating the effects of developmental exposure to the nicotine in tobacco, these molecular mechanisms likely contribute to normal brain development, suggesting that dysregulation of cholinergic induction of Ash2l and Mef2c as a result of genetic or environmental factors could increase the risk for neurodevelopmental disorders such as attention deficit hyperactivity disorder 5–7.
Online Methods
Animals
All procedures were approved by the Yale University Institutional Animal Care and Use Committee and conformed to the standards of the NIH Guide for Care and Use of Laboratory Animals. C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) used in the developmental nicotine exposure studies acclimated for 1 week, had ad libitum food and water and a 12-h light–dark cycle (lights on at 7AM).
Reagents
Nicotine hydrogen tartrate (Sigma, St Louis, MO, USA) was dissolved in phosphate-buffered saline (PBS) pH 7.4 and placed at 20°C until use. Inhibitors were incubated with the cultures for the following durations before nicotine stimulation: mecamylamine (20 min; Sigma), α-bungarotoxin (α-BTX; 20 min; Sigma and Calbiochem) and nifedipine (15 min; Sigma). Most inhibitors were used at a stock concentration of 1000X of the desired stimulating concentration. Anti-Ash2l antibody (abcam, ab176334) and anti-Mef2c antibody (Cell Signaling, 5030) were validated using cells overexpressing Ash2l and Mef2c. Anti-Wdr5 antibody (Abcam, ab56919) and anti-Rbbp5 antibody (Abcam, ab52084) have been validated in published studies (supplementary methods checklist). Anti-β tubulin antibody (Abcam, ab6046) and anti-Gapdh antibody (Sigma, G8795-200UL) were used as loading controls for Western blots and have been validated in published studies (supplementary methods checklist).
Developmental Nicotine Exposure
After vaginal plug identification, gestating mice were singly housed with ad libitum access to food and water containing 200 μg/ml nicotine hydrogen tartrate (calculated as free base) in 2% (w/v) saccharin or pH-matched 2% saccharin with 0.2% (v/v) tartaric acid (Sigma-Aldrich, St Louis, MO). Solutions were prepared twice a week. At 21 days, offspring were weaned and left undisturbed until brain tissue harvesting except for routine husbandry. Female mice were used for genome wide screening, structural analyses and biochemical studies described below and animals were randomly assigned to treatment groups for all studies.
Diffusion tensor imaging (DTI) and analysis
The DTI datasets were obtained using a custom-made 43 H radio frequency volume coil (40mm diameter) on a 9.4T horizontal bore magnet (Bruker, Billerica, MA, USA) The DTI experiments were performed using the Stejskal-Tanner spin-echo diffusion-weighted sequence with a diffusion gradient of 5 ms and a delay between the two diffusion gradients of 15 ms as previously described 44. Briefly, twenty contiguous coronal slices of 0.5 mm thickness were acquired with a repetition time of 2 s and an echo time of 25.1 ms. Two 1ms Shinnar–Le Roux (SLR) pulses were used for excitation and inversion, respectively. Eighteen averages were acquired and the 128 × 64 images were zero-filled to 256 × 256, resulting in an in-plane resolution of 100 μm × 100 μm. Sixteen different images were acquired for each slice, fifteen corresponding to noncollinear diffusion weighting directions (b=1000 s/mm2) and one with no diffusion weighting. In addition, a 256 x 256 fast spin-echo anatomical image was acquired for each brain with the same slice geometry and position as the DTI images. The anatomical image was used for registration of each individual brain to a common reference image.
The diffusion tensor was calculated from the intensities of the 16 diffusion-weighted images and the tensor eigenvalues (λ1, λ2 and λ3) were obtained by matrix diagonalization. The fractional anisotropy (FA) was calculated using the following equation:
where λavg = (λ1 + λ2 + λ3)/3. The fast spin-echo anatomical images were non-rigidly registered to a reference isotropic anatomical image (100 μm) using a non-linear intensity-based warping parameterized in terms of a tensor b-spline grid with uniform control point spacing of 1.5 mm. These transformations were used to warp the smoothed FA (Gausian smoothing sigma=0.2 mm) maps to the same isotropic reference anatomical image at 100μm resolution. The mean and the standard deviation FA maps for both nicotine and saccharin groups were then calculated. To measure the differences between the two groups, two tail t-test maps were generated from the mean and standard deviation FA maps. The t-test maps were used to define brain regions (Fig. 1a), for which the mean FA difference becomes significant (p<0.05). An average FA value was calculated first for each individual brain in each of these regions. Then, an average FA value and the corresponding standard deviation in these regions were calculated for nicotine and saccharin groups separately.
Diolistic labeling of cortical neurons in mouse brain slices
Tungsten particles (1.3 μm diameter, Bio-Rad, Berkeley, CA) were coated with the lipophilic carbocyanine dye DiI (Invitrogen, Calrsbad, CA). DiI-coated particles were delivered diolistically into tissue at 120 psi using a Helios Gene Gun system (Bio-Rad, Berkeley, CA) fitted with a polycarbonate filter (3.0 μm pore size; BD Biosciences, Franklin Lakes, NJ). DiI was allowed to diffuse along neuronal dendrites and axons in PBS for 16 – 24 h at 4°C, and then labeled sections were fixed again in 4% paraformaldehyde for 1 h. After brief washing in PBS, sections were mounted with Fluoromount G mounting media (Electron Microscopy Sciences, Hatfield, PA) onto glass slides (Thermo Scientific, Waltham, MA).
Confocal imaging and three-dimensional reconstructions
Cortical pyramidal neurons were imaged using an UltraVIEW VoX spinning disc confocal microscope with a 60x CFI Plan Apo objective and equipped with a Hamamatsu C9100-50 camera, with excitation at 561 nm. Z-stacks were collapsed into projection images and 2- 3 neurons/brain were manually analyzed per section.
Neuronal tracing and data analysis
Cortical regions were identified using stereotactic coordinates according to the Paxinos mouse brain map 1: Frontal cortex: Bregma +1 mm; Parietal cortex: Bregma -2.06 mm; Occipital cortex: Bregma -2.46 mm (from rostral to caudal). Pyramidal neurons from layers 1/2, layer 3/4, and layer 5/6 of entire cortex with complete cellular and dendritic staining were selected for analysis. For apical spine density quantitative analysis, collected data were processed for reconstruction with NeuronStudio software as described in the manufacturer’s manual (CNIC, Mt. Sinai School of Medicine, New York, NY; http://research.mssm.edu/cnic/help/ns/index.html). A 200 μM segment of the apical dendritic branch was analyzed. Maximum and minimum spine length was set at < 3.5 μm and > 0.5 μm respectively. After automatic detection of spines by the NeuronStudio software, spots that were not on the quantified dendrite were removed manually. For Sholl analysis of apical dendritic arborization, Simple Neurite Tracer (SNT) software was used. SNT is open source software, licensed under the GNU General Public License and based on the public domain image processing software ImageJ. To build a simple path, successive points were selected along the midline of a neural process, and the software would automatically find a path between them. Connected neural processes were confirmed by generating a 3D reconstruction image.
The morphological parameters measured for each apical dendrite and complexity of dendritic arborization were measured using Sholl analysis with NeuronStudio software (MicroBrightField) by calculating the number of crossings that intersected defined concentric circles spaced 10 μm-apart starting from the center of cell body as a function of distance from the soma and is based on a total of 214 neurons from 6 animals in the pre- & postnatal nicotine treated group and 181 neurons from 5 animals in the postnatal-only nicotine treated group. All data were collected by an investigator blinded to condition.
Microarray analysis
Mouse tissue samples were cut into slices less than 1 mm in thickness, transferred to Trizol reagent (Invitrogen, Carlsbad, CA) and disrupted with a Dounce tissue homogenizer (Wheaton, Millville, NJ). An equal volume of chloroform was added and samples were centrifuged for 5 min (4°C, 12,000g). The aqueous phase was transferred to a column from an RNeasy tissue kit (Qiagen, Redwood City, CA) and RNA was isolated according to the manufacturer's recommendations. Subsequently, RNAs from 4 animals were pooled to obtain 5 pools per experimental group (20 subjects total). Generation of double-stranded cDNA, preparation and labeling of cRNA, hybridization to GeneChip® Mouse Genome 430 2.0 Arrays (Affymetrix, Santa Clara, CA) and washing was performed according to the standard Affymetrix protocol. Data analysis was performed using different R packages from the Bioconductor project (www.bioconductor.org). Microarray experiments were done on sample sizes based on successful experiments in the literature. All confirmation experiments were performed on multiple pools of brain samples to reduce biological variability and provide adequate statistical power.
Chromatin Immunoprecipitation (ChIP)
Mouse tissue samples (50 mg) were cut into slices less than 1 mm in thickness, and fixed in 3 ml of 1% formaldehyde/PBS solution for 10 min at room temperature to cross-link chromatin DNA and proteins. After being washed with PBS, the tissue samples were homogenized in a glass-Teflon homogenizer. Following homogenization, chromatin was isolated using the Magna ChIP G kit (Millipore, Temecula, CA) according to the Millipore protocol. Briefly, cells were lysed in Cell Lysis Buffer in the presence of protease inhibitor cocktail. Nuclei were isolated from the lysed cells by centrifugation, and resuspended in Nuclear Lysis Buffer. The chromatin DNA was then fragmented into 150 – 500 base-pair range by sonication using a Branson Sonifer (Danbury, Connecticut).
To immuno-precipitate specific genomic regions of chromatin DNA, the isolated chromatin fragments were incubated with antibodies against histone H3 trimethylated at Lysine 4 (H3K4-me3, Abcam, Cambridge, MA) and magnetic protein G beads (Millipore) at 4° C for 2.5 hr. Following incubation, the beads were washed with low salt, high salt, LiCl salt, and TE buffers, and chromatin fragments were reverse cross-linked by proteinase K digestion at 62° C for 2 hr.
The eluted DNA was purified after reverse cross-linking by column purification. H3k4me3-specific enrichment was validated by PCR amplification of select positive and negative genomic regions and amplification signals were normalized to the input DNA samples which were extracted from the same pools of chromatin fragments without enrichment by immunoprecipitation. To control for any differences in DNA across conditions, all ChIP-PCR data were normalized to GAPDH.
High-throughput, massively parallel sequencing
Sample preparation and sequencing on the Genome Analyzer (Illumina, San Diego, CA) were carried out according to the Illumina protocols with some modifications. Briefly, the double stranded cDNA and ChIP-enriched genomic DNA were treated with T4 DNA polymerase and the Klenow fragment for end repair. The 5’ ends of the DNA fragments were then phosphorylated by T4 polynucleotide kinase, and an adenosine base was added to the 3’ end of the fragments by Klenow (3’-5’ exo). A universal adaptor was then added to both ends of the DNA fragments by A-T ligation. Following 18 cycles of PCR with the Phusion DNA polymerase, the DNA library was then purified on a 2% agarose gel, and fragments of 170 – 300 base-pair in size were recovered. Around 15 ng of the DNA library was then used for cluster generation on a grafted GAII Flow Cell, and then sequenced on the Genome Analyzer for 36 cycles using the “Sequencing-by-synthesis” method.
Sequence base-calling, mapping to genome, data normalization and statistical analysis
Sequences were called from image files with the Illumina Genome Analyzer Pipeline (GApipeline) and aligned to the reference genome (UCSC mm9) using Extended Eland in the GApipeline. Uniquely mapped H3K4-me3 reads for each sample were retrieved from export.txt files (output of Extended Eland). Based on their mapping locations, these selected reads were parsed with in-house Perl scripts to generate base coverage in WIG file format. After moving average smoothing, the chromosome locations of enrichment peaks were identified from pooled WIG files using in-house Perl scripts.
The uniquely aligned reads by ELAND pipeline were considered in calling peaks. To eliminate noise and account for unequal total numbers, we used a defined analysis model (Model-based Analysis of ChIP-Seq, MACS 45) with default parameters to find peaks, which were called “peaks” of H3K4me3. For comparison analysis, a customized genomic interval file was created for EdgeR 46 analysis.
Gene Ontology analysis was performed by using the Functional Annotation Tool of the DAVID Bioinformatics Resources 6.7, National Institute of Allergy and Infectious Diseases (NIAID), NIH (http://david.abcc.ncifcrf.gov/). Statistical methods were not used to predetermine sample sizes, but ‘n’ sizes were based on other published studies. All confirmation experiments were performed on multiple pools of brain samples to reduce biological variability and provide adequate statistical power.
Cell culture
Neural progenitor cell cultures were generated from C57BL/6J mice at embryonic day 15 as described in the Cold Spring Harbor protocol for neural progenitor cell culture 5. Each embryonic mouse brain was placed in a clean dish containing ice-cold 1X PBS. Brains were separated into two hemispheres, followed by separation of the dorsal telencephalon. Isolated brain regions were carefully transferred to a non-treated polystyrene dish containing cold 1X PBS and tissue was disrupted with a Pasteur pipette. Papaine Enzyme solution prepared as described in protocol 47 was added and incubated for 20 min at 37°C. Neural progenitor cell media (NEP) was added after adding enzyme inhibitor solution and centrifugation. Cells were counted using a hemacytometer and 4x106 cells were added to a non-treated culture dish containing NEP medium with EGF and bFGF (NEP complete medium). Once spheres formed, neural spheres were transferred to a PDL and laminin coated culture dish. The next day, after the neural spheres settled and attached to the culture dish, NEP complete medium was replaced with NEP medium containing 1% horse serum. For histological analysis, shRNA constructs were introduced into cells at DIV 8/9 when progenitor cells had already differentiated into neurons, and cultures then underwent a further 4 days of exposure to nicotine or saline. Histological analyses were performed at DIV 12/13.
Western blotting
Approximately 40 μg of each sample was separated by 10% SDS–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Blots were blocked with 5% milk in Tris-buffered saline with 0.05% (v/v) Tween-20 (TBS-T) for 60 min at 25°C, washed three times in TBS-T for 15 min, and then incubated at 4°C overnight in primary antibody diluted in TBS-T. Blots were washed three times in TBS-T and incubated for 60 min at 25°C in secondary antibody in TBS with 0.1% Tween. Blots were washed three times in TBS-T and two times in water before scanning with an Odyssey Infrared Scanner (Li-Cor Biosciences, Lincoln, NE, USA). The scans were performed with the following scan parameters: intensity = 5.0, for each wavelength (700 and 800 nm), resolution = 169, quality = medium, and focus offset = 0 mm. The scanned intensity of the two secondary antibodies’ fluorophores was then used by the Odyssey Software v1.2 (Li-Cor Biosciences) to produce individual images for each wavelength, and a pseudocolor overlay of the 700 (red) and 800 nm (green) scans. The area of each band was selected for quantification and the integrated intensity of the 700 and 800 nm wavelengths was measured using Odyssey Software without investigator manipulation. Since Li-Cor uses the absolute intensities for quantitation, the pseudocolor images are reflections of the absolute value measured by the program. The integrated intensity values were then exported to Excel (Microsoft, Redmond, WA, USA) for further analysis. For Ash2l expression detection, Chemiluminescence (ECL detection kit from GE health care, Pittsburgh, PA, USA) was used.
Immunoprecipitation assay
Approximately 4 x 106 neural progenitor cells (NPCs) were treated with nicotine or PBS. Following treatment, NPCs were lysed in Ripa buffer (0.7 % Na-Deoxycholate, 500 mM LiCl, 50mM HEPES-KOH, pH 7.6, 1% NP-40, 1mM EDTA). Samples were sonicated and protein was measured using a D/C protein assay kit (Bio-rad). 500–1000 ug of protein was used for each immunoprecipitation assay. The anti-H3K4me3 antibody (Millipore) was coupled to Protein-A beads (Novex Lifetechnologies) and immunoprecipitation and elution were performed according to the manufacturer’s recommendation.
Design and construction of shRNAs
shRNAs targeting Ash2l and Mef2c were constructed by selecting unique 24 base or 19 base sequences in the consensus coding region of the mRNA encoding the different isoforms of Ash2l and Mef2c (Ash2l; GenBank accession NM_011791.3, Mef2c; GenBank accession NM_025282.3) as previously described 48. A random sequence of 24 bases without similarities to any known mRNA was selected for the scrambled shRNA. Synthetic oligonucleotide duplexes were designed as previously described. Annealed oligonucleotides were ligated into a pAAV-EGFP-shRNA vector as described previously and positive clones were verified by sequencing 48–50. Sequences of shRNAs were as follows:
Ash2l shRNA (5’ to 3’: 1. GTGCCATCACAGTGGGAATACCTA; 2. GCCATCACAGTGGGAATACCTACT; 3. CCGGAAGCAAGCAAATTTA).
Mef2c shRNA (5’ to 3’: 1. GAATAGTATGTCTCCTGGT; 2. GGTAACCTGAACAAGAATA; 3. GGACAAACTCAGACATTGT)
In utero electroporation
DNA plasmids encoding either scrambled or Ash2l-targeting shRNA were diluted to a final concentration of 3 μg/μl in PBS containing 0.1% fast green dye tracer. DNA solution (1–2 μl) was unilaterally injected into a lateral ventricle of E14 embryos using a glass pipette and positive pneumatic pressure. PBS soaked Tweezertrodes with 5 mm electrode diameter (45-0489; BTX) were then positioned across the heads of the embryos followed by five 44 volt square pulses lasting 50 ms each at 950 ms interval applied using a pulse generator (ECM830; BTX).
Statistical analysis and computational analysis
All statistical analyses were accomplished using SPSS (IBM, Inc.). Shapiro-Wilks test of normality and the Kolmogorov-Smirnov Test were conducted to test for normal distribution. Levene’s test was conducted for homogeneity variance. Differences in fractional anisotrophy (FA) were analyzed by two-tailed t tests. One-way ANOVA with repeated mesures or two-way ANOVA were conducted on the test sholl analysis of nicotine-mediated or IUE-induced morphological changes. One-way ANOVA, followed by Tukey or LSD test for multiple comparisons, was used to analyze experiments confirming changes in mRNA and protein levels in independent samples. Fold-change in ChIP (H3K4m3) experiments was analyzed by one-way ANOVA. A supplementary methods checklist is available with complete statistical details of all experiments.
Passive avoidance behavior
Passive avoidance testing was conducted at 21 days of age in a step-through, two chamber apparatus (Ugo Basile, Comerio, Italy) using a 3-day paradigm 24. Day 1: mice were allowed to explore the apparatus for 5 min. Day 2: mice were placed in the light chamber and received a mild, inescapable footshock (0.2 mA, 2s) after entry into the dark chamber. Day 3: mice were placed in the light chamber and time to cross into the dark chamber was measured. Latency to cross was measured on days 2 (training day) and 3 (testing day), with 5 min maximum. Passive avoidance studies were performed by an investigator blinded to condition. Both male and female mice were used for passive avoidance behavior and no sex differences were observed, as has been published previously. No animals were excluded from analysis in studies of passive avoidance behavior. No statistical methods were used to pre-determine sample sizes, but sample sizes are similar to those reported in previous publications 24.
Supplementary Material
Acknowledgments
This work was supported by grants DA14241 (MRP), DA10455 (MRP), NS052519 (FH) and NS086329 (FH) from the National Institutes of Health, the State of Connecticut, Department of Mental Health and Addiction Services and the Kavli Institute for Neuroscience at Yale.
Footnotes
Accession Codes: All microarray and DNA sequencing data have been submitted to the GEO database at NCBI. The accession number for all datasets in this study is GSE80789.
The authors declare no competing financial interests.
Author Contributions: Y.J. designed the study, performed the experiments and data analyses, wrote and edited the manuscript. Z.Z and Q.Y. prepared samples and analyzed data for the ChIP-seq experiments. D.G provided critical resources, experimental design and advice on the ChIP-seq experiments. D.C. performed the imaging studies and data analyses for the diffusion tensor imaging experiments. F.H. provided valuable resources and critical advice on interpretation of the diffusion tensor imaging experiments. Y.S.M designed shRNA constructs, helped troubleshoot knockdown studies and provided critical advice on behavioral design. A.B. provided critical resources, experimental design and advice on the in utero electroporation studies. L.S.H performed in utero electroporation studies. A.M.L performed in utero electroporation surgeries and contributed to data collection. C.J.H. designed and prepared samples for the diffusion tensor imaging experiment and contributed to the microarray experiment. M.R.P. designed the project, assisted in interpretation of all studies, wrote and edited the manuscript. All authors contributed to editing of the manuscript.
Data Availability
H3K4me3 ChIP-seq and microarray datasets have been deposited in the GEO database at NCBI. All datasets for this study are available under a single accession number: GSE80789. Original Western blots are available in supplementary Figure 7. Gene Ontology data are available in supplementary Tables 1–4. Statistical analyses for all experiments are detailed in the Figure Legends and in the supplementary Data Checklist. All data that support the findings of this study are available from the corresponding author upon request.
Literature cited
- 1.Lozada AF, et al. Induction of dendritic spines by beta2-containing nicotinic receptors. The Journal of Neuroscience. 2012;32:8391–8400. doi: 10.1523/JNEUROSCI.6247-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballesteros-Yanez I, Benavides-Piccione R, Bourgeois JP, Changeux JP, DeFelipe J. Alterations of cortical pyramidal neurons in mice lacking high-affinity nicotinic receptors. PNAS. 2010;107:11567–11572. doi: 10.1073/pnas.1006269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muhammad A, et al. Prenatal nicotine exposure alters neuroanatomical organization of the developing brain. Synapse. 2012;66:950–954. doi: 10.1002/syn.21589. [DOI] [PubMed] [Google Scholar]
- 4.Mychasiuk R, Muhammad A, Gibb R, Kolb B. Long-term alterations to dendritic morphology and spine density associated with prenatal exposure to nicotine. Brain Research. 2013;1499:53–60. doi: 10.1016/j.brainres.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 5.Sorenson CA, Raskin LA, Suh Y. The effects of prenatal nicotine on radial-arm maze performance in rats. Pharmacology, Biochemistry & Behavior. 1991;40:991–993. doi: 10.1016/0091-3057(91)90117-k. [DOI] [PubMed] [Google Scholar]
- 6.Sobrian SK, Marr L, Ressman K. Prenatal cocaine and/or nicotine exposure produces depression and anxiety in aging rats. Progress in Neuro-psychopharmacology & Biological Psychiatry. 2003;27:501–518. doi: 10.1016/S0278-5846(03)00042-3. [DOI] [PubMed] [Google Scholar]
- 7.Heath CJ, Picciotto MR. Nicotine-induced plasticity during development: modulation of the cholinergic system and long-term consequences for circuits involved in attention and sensory processing. Neuropharmacology. 2009;56(Suppl 1):254–262. doi: 10.1016/j.neuropharm.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobsen LK, Picciotto MR, Heath CJ, Mencl WE, Gelernter J. Allelic variation of calsyntenin 2 (CLSTN2) modulates the impact of developmental tobacco smoke exposure on mnemonic processing in adolescents. Biological Psychiatry. 2009;65:671–679. doi: 10.1016/j.biopsych.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pauly JR, Sparks JA, Hauser KF, Pauly TH. In utero nicotine exposure causes persistent, gender-dependant changes in locomotor activity and sensitivity to nicotine in C57Bl/6 mice. Int J Dev Neurosci. 2004;22:329–337. doi: 10.1016/j.ijdevneu.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Heath CJ, Horst NK, Picciotto MR. Oral nicotine consumption does not affect maternal care or early development in mice but results in modest hyperactivity in adolescence. Physiology & Behavior. 2010;101:764–769. doi: 10.1016/j.physbeh.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dwyer JB, McQuown SC, Leslie FM. The dynamic effects of nicotine on the developing brain. Pharmacology & Therapeutics. 2009;122:125–139. doi: 10.1016/j.pharmthera.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rampalli S, et al. p38 MAPK signaling regulates recruitment of Ash2l-containing methyltransferase complexes to specific genes during differentiation. Nature Structural & Molecular Biology. 2007;14:1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wysocka J, Allis CD, Coonrod S. Histone arginine methylation and its dynamic regulation. Frontiers in Bioscience. 2006;11:344–355. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 14.Barbosa AC, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. PNAS. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho EG, et al. MEF2C enhances dopaminergic neuron differentiation of human embryonic stem cells in a parkinsonian rat model. PloS One. 2011;6:e24027. doi: 10.1371/journal.pone.0024027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan M, et al. The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J Biol Chem. 2013;288:5039–5048. doi: 10.1074/jbc.M112.424515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gossett LA, Kelvin DJ, Sternberg EA, Olson EN. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Molecular and Cellular Biology. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 20.Zweier M, et al. Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Human Mutation. 2010;31:722–733. doi: 10.1002/humu.21253. [DOI] [PubMed] [Google Scholar]
- 21.Li H, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. PNAS. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakai Y, et al. Neuroendocrine phenotypes in a boy with 5q14 deletion syndrome implicate the regulatory roles of myocyte-specific enhancer factor 2C in the postnatal hypothalamus. European Journal of Medical Genetics. 2013;56:475–483. doi: 10.1016/j.ejmg.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 23.King SL, et al. Conditional expression in corticothalamic efferents reveals a developmental role for nicotinic acetylcholine receptors in modulation of passive avoidance behavior. The Journal of Neuroscience. 2003;23:3837–3843. doi: 10.1523/JNEUROSCI.23-09-03837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heath CJ, King SL, Gotti C, Marks MJ, Picciotto MR. Cortico-thalamic connectivity is vulnerable to nicotine exposure during early postnatal development through alpha4/beta2/alpha5 nicotinic acetylcholine receptors. Neuropsychopharmacology. 2010;35:2324–2338. doi: 10.1038/npp.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazo AM, Huang DH, Mozer BA, Dawid IB. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. PNAS. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends in Genetics. 1990;6:416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- 27.Ikegawa S, Isomura M, Koshizuka Y, Nakamura Y. Cloning and characterization of ASH2L and Ash2l, human and mouse homologs of the Drosophila ash2 gene. Cytogenetics and Cell Genetics. 1999;84:167–172. doi: 10.1159/000015248. 15248. [DOI] [PubMed] [Google Scholar]
- 28.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature Structural & Molecular Biology. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 29.Steward MM, et al. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nature Structural & Molecular Biology. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 30.Adamson AL, Shearn A. Molecular genetic analysis of Drosophila ash2, a member of the trithorax group required for imaginal disc pattern formation. Genetics. 1996;144:621–633. doi: 10.1093/genetics/144.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng MK, Shearn A. The direct interaction between ASH2, a Drosophila trithorax group protein, and SKTL, a nuclear phosphatidylinositol 4-phosphate 5-kinase, implies a role for phosphatidylinositol 4,5-bisphosphate in maintaining transcriptionally active chromatin. Genetics. 2004;167:1213–1223. doi: 10.1534/genetics.103.018721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhlen M, et al. Proteomics Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 33.Nadarajah B, Parnavelas JG. Modes of neuronal migration in the developing cerebral cortex. Nat Rev Neurosci. 2002;3:423–432. doi: 10.1038/nrn845. [DOI] [PubMed] [Google Scholar]
- 34.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 36.Lauberth SM, et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan CC, et al. Transcription factor Ap2delta associates with Ash2l and ALR, a trithorax family histone methyltransferase, to activate Hoxc8 transcription. PNAS. 2008;105:7472–7477. doi: 10.1073/pnas.0711896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. The Journal of Neuroscience. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flavell SW, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 40.Shalizi A, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 41.Krmpotic-Nemanic J, Kostovic I, Kelovic Z, Nemanic D. Development of acetylcholinesterase (AChE) staining in human fetal auditory cortex. Acta Oto-laryngologica. 1980;89:388–392. doi: 10.3109/00016488009127153. [DOI] [PubMed] [Google Scholar]
- 42.Krmpotic-Nemanic J, Kostovic I, Kelovic Z, Nemanic D, Mrzljak L. Development of the human fetal auditory cortex: growth of afferent fibres. Acta Anatomica. 1983;116:69–73. [PubMed] [Google Scholar]
- 43.Franklin KBJ, Paxinos G. The Mouse Brain in Stereotatic Coordinates. 1997 [Google Scholar]
- 44.Duque A, et al. Neuroanatomical changes in a mouse model of early life neglect. Brain Structure & Function. 2012;217:459–472. doi: 10.1007/s00429-011-0350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hutton SR, Pevny LH. Isolation, culture, and differentiation of progenitor cells from the central nervous system. CSH Protocols. 2008;2008 doi: 10.1101/pdb.prot5077. pdb prot5077. [DOI] [PubMed] [Google Scholar]
- 48.Hommel JD, Sears RM, Georgescu D, Simmons DL, DiLeone RJ. Local gene knockdown in the brain using viral-mediated RNA interference. Nature Medicine. 2003;9:1539–1544. doi: 10.1038/nm964. [DOI] [PubMed] [Google Scholar]
- 49.Mineur YS, et al. Nicotine decreases food intake through activation of POMC neurons. Science. 2011;332:1330–1332. doi: 10.1126/science.1201889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mineur YS, et al. Cholinergic signaling in the hippocampus regulates social stress resilience and anxiety- and depression-like behavior. PNAS. 2013;110:3573–3578. doi: 10.1073/pnas.1219731110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.