Abstract
Myelination is essential for nervous system function. Schwann cells interact with neurons and the basal lamina to myelinate axons, using known receptors, signals and transcription factors. In contrast, the transcriptional control of axonal sorting and the role of mechanotransduction in myelination are largely unknown. Yap and Taz are effectors of the Hippo pathway that integrate chemical and mechanical signals in cells. We describe a previously unknown role for the Hippo pathway in myelination. Using conditional mutagenesis in mice we show that Taz is required in Schwann cells for radial sorting and myelination, and that Yap is redundant with Taz. Yap/Taz are activated in Schwann cells by mechanical stimuli, and regulate Schwann cell proliferation and transcription of basal lamina receptor genes, both necessary for proper radial sorting of axons and subsequent myelination. These data link transcriptional effectors of the Hippo pathway and of mechanotransduction to myelin formation in Schwann cells.
Mechanical cues are important regulators of cell behavior, and are integrated with biochemical signals to control development, physiology and pathology. Yap and Taz, two related transcriptional co-activators downstream of the Hippo pathway, are also pivotal for mechanical signal transduction 1. Upon mechanical or chemical stimulation, Yap and Taz shuttle from the cytoplasm into the nucleus to associate with TEA domain (TEAD) transcription factors and regulate gene expression 2, 3. Whether the Hippo pathway and Yap/Taz are required for myelination is currently unknown. During development, peripheral nerves undergo significant morphogenetic changes that cause mechanical stimulation of Schwann cells as they interact with axons and the basal lamina. First, immature Schwann cells separate large axons from axon bundles in a process called radial sorting 4. After defasciculation, large axons acquire a 1:1 relationship with a Schwann cell, which then wraps the axon to form the myelin sheath. Schwann cells in nerves are also exposed to significant mechanical stimulation during limb growth and body movement throughout life. Finally, in response to injury, Schwann cells change their physical relationship with axons to undergo rapid demyelination and transition to a “repair” state that is required to clear cell debris, promote axonal regrowth and remyelinate regenerated axons 5. Thus, mechanotransduction should be critical for nerve development and response to injury, but the molecular mechanisms are poorly understood. In addition, while the network of transcription factors that control myelination has been explored in depth 6, the transcriptional control of radial sorting is largely unknown. Finally, interaction with the basal lamina during radial sorting is mediated by laminin receptors 7, but what controls their expression is also not known.
Here we ablated Yap and Taz in Schwann cells. We show that the absence of Yap and Taz causes a severe peripheral neuropathy due to a developmental impairment in axonal sorting, and that Yap/Taz-Tead1 are required for the transcriptional regulation of laminin receptors in Schwann cell. Thus, Yap/Taz downstream of mechanotransduction and the Hippo pathway are essential for Schwann cell development.
Results
Activation of Yap and Taz i Schwann cells
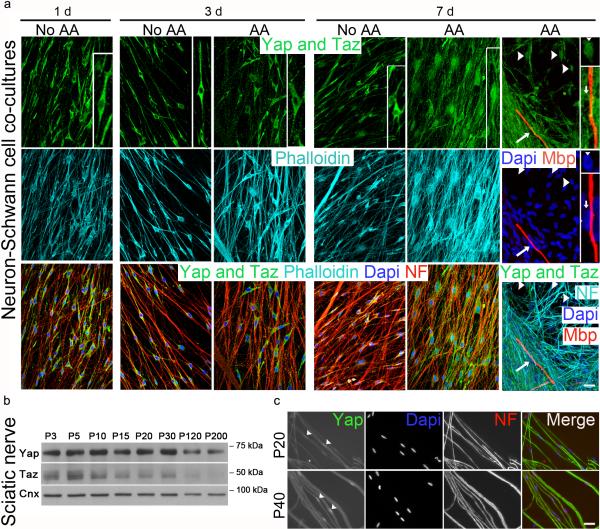
Yap and Taz are regulated by the Hippo pathway, but also by mechanotransduction independently of Hippo 1. Yap/Taz activation leads to their retention in the nucleus where they regulate gene expression that promotes proliferation or differentiation depending on the cell type 8. To ask how Yap/Taz are regulated in Schwann cells, we plated them on dorsal root ganglia (DRG) neurons and monitored their localization in different conditions. Contact with neurons or addition of ascorbic acid did not activate Yap and Taz, which were found in the cytoplasm of Schwann cells 1 and 3 days after plating (Fig. 1a). After 7 days in the presence of ascorbic acid, which causes proliferation, basal lamina deposition and myelination, Yap and Taz were found in the nuclei of many Schwann cells. However Yap/Taz activation did not correlate with myelination, because the nucleus of myelin-forming Schwann cells was devoid of Yap and Taz (Fig. 1a). In developing sciatic nerves Yap and Taz were expressed highly between postnatal day 3 (P3) and P15, when Schwann cells proliferate, sort axons and myelinate, but also between P15 and P30 during growth and maturation of myelin sheaths, nerves and limbs (Fig. 1b). Indeed Yap was in the nucleus of Schwann cells in sciatic nerves after myelination at P20 and P40 (Fig. 1c). Collectively, these data show that Yap and Taz are regulated in developing Schwann cells and suggest a role in myelination. Yap and Taz are activated early during proliferation and basal lamina deposition, and Yap is activated late during myelin maturation and nerve growth, but Yap/Taz are less activated during active myelin membrane wrapping. This suggests that it is not a specific molecular signal (e.g. axonally tethered neuregulin 9), rather varying physical stimulation that distinguishes these situations, and determines activation of Yap and Taz in Schwann cells.
Figure 1.
Yap/Taz expression and activation during Schwann cell development. (a) Yap and Taz staining (green) of Schwann cells plated on neurons for the time indicated, in myelinating (ascorbic acid, AA) or non-myelinating (no AA) conditions. Neurons are stained for neurofilaments (NF) in red in left panels and cyan in right panels; blue marks nuclei. cyan stains F-actin (phalloidin) on the left, and myelin basic protein (red) indicates myelin segments on the right. Yap/Taz remain cytoplasmic (i.e. inactive) after contact with neurons at 1 and 3 days. Only after 7 days and in the presence of AA can Yap/Taz be detected in the nucleus of Schwann cells at low density (arrowheads), but not in Schwann cells that have started myelination (arrows). One cell stained for Yap/Taz in each condition is enlarged in the inset on the right. Note the nucleus of a Schwann cell with nuclear Yap/Taz and no myelin, and the nucleus of another Schwann cell void of Yap/Taz and associated with myelin. The experiment was repeated twice. Scale bar, 20 μm. (b) Western blot analysis from sciatic nerve lysates shows that both Yap and Taz are highly expressed during development and myelination. The experiment was performed once. The western blots were cropped and the full-length blots are presented in Supplementary Fig. 4. (c) Teased fibers from P20 and P40 sciatic nerve stained for YAP (green), dapi (blue) and NF (red) show that Yap is nuclear after myelination. The experiment was repeated twice. Scale bar, 20 μm.
Yap and Taz respond to mechanic stimuli in Schwann cells
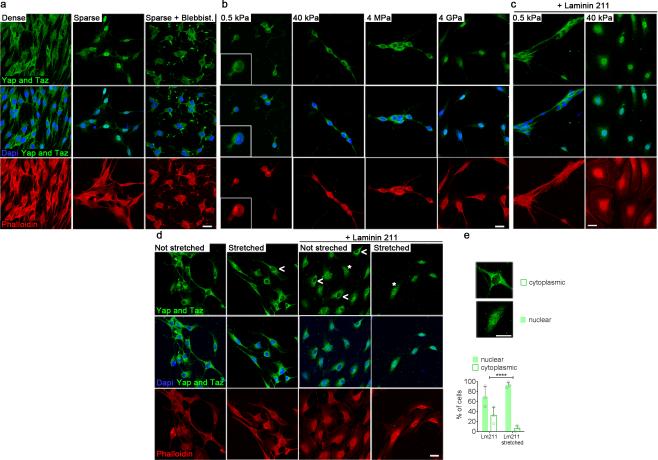
To determine directly if Yap and Taz respond to mechanostimulation in Schwann cells, we analyzed their subcellular distribution in response to various modifications of the physical environment. First we used cell density to modify cell geometry. Even when plated without axons, Yap/Taz remained nuclear in Schwann cells spreading at low density, but relocated to the cytoplasm in more confluent cultures (Fig. 2a). To exclude that the cytoplasmic localization of YAP/TAZ in Schwann cells at higher density was due Hippo signaling during contact inhibition, we inhibited non-muscle myosin with blebbistatin, which selectively blocks mechanical YAP/TAZ activation independently of the Hippo pathway1. In sparse Schwann cell cultures treated with blebbistatin Yap/Taz remained cytoplasmic (Fig. 2a), confirming that the actomyosin cytoskeleton is essential to transduce the mechanical signal that causes Yap/Taz relocalization. We next asked if Yap/Taz are regulated by substrates of increasing stiffness. We found that Yap/Taz remained cytoplasmic in Schwann cells plated on polyacrylamide hydrogels at elasticity moduli of 0.5kPa and 40kPa, and even on PDMS at 4MPa (Fig. 2b). Only on glass surfaces of extreme stiffness (4GPa) was Yap/Taz nuclear (Fig. 1b). However when laminin 211 was also coated on the substrates, Yap/Taz moved from cytoplasmic at 0.5kPa to nuclear at 40kPa (Fig. 2c), both rigidities being within the physiological range 10. Lastly, we analyzed the localization of Yap/Taz in response to direct mechanical stretching of the cells (Fig. 2d-e). We used a deformable silicone membrane coated with poly-L-lysine or poly-L-lysine plus laminin 211 and stretched Schwann cells for 30min at 150% static strain. Mechanical stretching with laminin 211 promoted the nuclear localization of Yap and Taz in Schwann cells (Fig. 2 d-e). Altogether, we show that Yap/Taz are modulated by laminin and mechanical stimuli in Schwann cells.
Figure 2.
Laminins and mechanical stimulation regulate Yap and Taz in primary Schwann cells. Confocal immunofluorescence images of Yap/Taz (green), Phalloidin (red) and dapi (blue) in Schwann cells are shown. (a) Schwann cells plated at different densities and treated or not with the non-muscle myosin inhibitor blebbistatin (Blebbist. 25 μM). Yap/Taz are enriched in the nucleus when Schwann cells are sparse. When cells are dense or treated with blebbistatin, Yap/Taz remain mainly cytoplasmic. The experiment was repeated two times. Scale bar, 20 μm. (b-c) Schwann cells plated sparsely on substrates with different rigidity: Polyacrylamide (0.5kPa, 40kPa), PDMS (4MPa) or glass (4GPa). Yap/Taz are enriched in the nuclei on the most rigid support (4GPa). When laminin 211 is also added, Yap/Taz are enriched in the nucleus at 40kPa, but not 0.5 kPa. The experiment was repeated three times. Scale bar, 20 μm. (d-e) Schwann cells plated on a silicone substrate with or without laminin 211 were stretched for 30min. Without laminin 211, Yap/Taz are in the cytosol (open arrowhead). With laminin 211, Yap/Taz are found in the nucleus of 68% of cells (asterisk). After mechanical stretching most cells (93%) have nuclear Yap/Taz. The experiment was repeated three times. Quantification of Yap/Taz localization scored in over 500 cells (777 in not stretched, 531 in stretched). Fischer exact test **** P < 0.0001. Scale bar, 20 μm.
Taz is required for radial sorting of axons
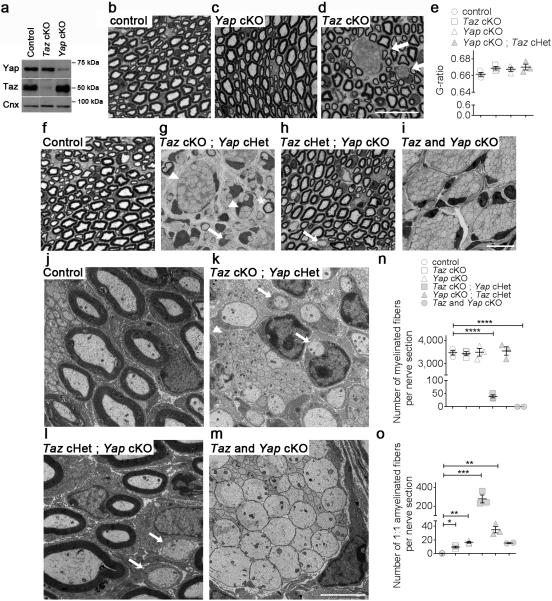
To determine the function of Yap and Taz in Schwann cells, we generated mice in which either was specifically ablated. Mice with the Yap (Yap1) or Taz (Wwtr1) gene flanked by LoxP sites 11 were crossed with P0-Cre 12 that drives Cre expression in the Schwann cell lineage from E13.5 13. As a result, Yap and Taz expression were drastically reduced in sciatic nerves at P20 (Fig. 3a). Either of Yap or Taz conditional knock-out (cKO) mice were grossly normal. At P20, cross sections of sciatic nerves from control littermates showed numerous properly myelinated axons (Fig. 3b, f, j). Yap cKO nerves did not show abnormalities of radial sorting or myelination (Fig. 3c). In contrast, in Taz cKO mice many large caliber axons were not myelinated, but grouped in immature bundles (Fig. 3d), the hallmark of a partial defect in axonal sorting (reviewed in 7). Myelin thickness was not significantly affected (Fig. 3e). These results indicate that Taz is required for the proper sorting of axons by Schwann cells in vivo.
Figure 3.
Ablation of Taz in Schwann cells impairs radial sorting of axons. (a) Western blot analysis shows that Taz and Yap protein levels are drastically reduced in sciatic nerves of Taz fl/fl ; P0-Cre (cKO) and Yap cKO, respectively. Also, Taz is up-regulated in Yap conditional null nerves. The experiment was repeated twice. Western blots were cropped and the entire blots are presented in Supplementary Fig. 4. (b-d) Toluidine blue stained semithin cross-sections of sciatic nerves from control (b) Yap cKO (c) and Taz cKO (d) at P20. Bundles of unsorted axons can be observed in Taz cKO (arrows). Bar, 20 μm. Three animals per genotype were analyzed. (e) Myelin thickness as measured by g-ratio is not decreased in mutant nerves. Each circle indicates the average value from one nerve from a different animal. Error bars indicate s.e.m. n=3 (animal). One way ANOVA. (f-o) Depletion of Yap from Schwann cells worsens the radial sorting defects of Taz cKO. (f-i) Toluidine blue stained semithin cross-sections and (j-m) electron micrographs of sciatic nerves from control, Taz and Yap double cKO at P20. The radial sorting defects of Taz cKO worsen with conditional ablation of one (g) or two (i) Yap alleles in a dose dependent manner. Bars, 10 μm (d), 20 μm (i) and 2 μm (m). Myelinating Schwann cell (asterisk), promyelinating Schwann cells (arrows) and immature Schwann cells (arrowhead) are indicated in g, h, k and l. (n, o) Quantification of myelinated (n) and amyelinated (o) fibers at P20. All mutants manifest an increase in the number of amyelinated fibers, indicating a delay in myelination. The data are presented as mean ± s.e.m. Each circle indicates the average value from one nerve from a different animal. n = 3 mice per genotype, except for Taz and Yap double cKO (in grey) where only two animals were analyzed. Taz cKO Yap cHet myelinated fibers, P < 0.0001, t = 15.08, df=4; Taz cKO Yap cKO myelinated fibers, P < 0.0001, t = 26.55, df=3; Taz cKO amyelinated fibers P = 0.013, t = 6.261, df=4; Yap cKO amyelinated fibers P = 0.0009, t = 13.1, df=4; Taz cKO Yap cHet myelinated fibers P = 0.011, t = 6.572, df=4; Yap cKO Taz cHet myelinated fibers P = 0.0068, t = 7.545, df=4; two-tailed unpaired Student's t test with Bonferroni correction. *P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.001.
Ablation of both Taz and Yap prevents radial sorting
Taz and Yap physically interact and have redundant roles. Taz was upregulated in Yap cKO nerves at P20 (Fig. 3a), which prompted us to ablate both Yap and Taz. By P20, double cKO mice showed a severe neuromuscular phenotype with weight loss, muscular atrophy and wide base walking. By P40, mutant mice manifested nearly paralyzed hind limbs and severe atrophy and had to be euthanized. Morphological analysis of Yap/Taz cKO sciatic nerves at P20 revealed a complete arrest in radial sorting; only large bundles of naked axons were present (Fig. 3i, m) similar to embryonic nerves. Interestingly, removing one allele of Yap in Taz cKO mice at P20 created a similar severe external phenotype, with an intermediate morphological phenotype (Fig. 3g, k, n, o), with bundles of unsorted axons (Fig. 3g, k arrowheads), promyelinating Schwann cells (Fig. 3g, k arrows) and few myelinated fibers (Fig. 3g asterisk). In contrast, removing one allele of Taz in Yap cKO mice did not produce any clinical phenotype, and sciatic nerves at P20 showed no radial sorting impairment, but minor defects with some Schwann cells blocked at the promyelinating stage (arrows in Fig. 3h, l). Thus, only one allele of Taz is sufficient to compensate for Yap cKO. Collectively the data indicate that Taz has a more prominent role during Schwann cell development, and they indicate a functional redundancy between Taz and Yap in axonal sorting and myelination.
Schwann cell proliferation is reduced in mutant mice
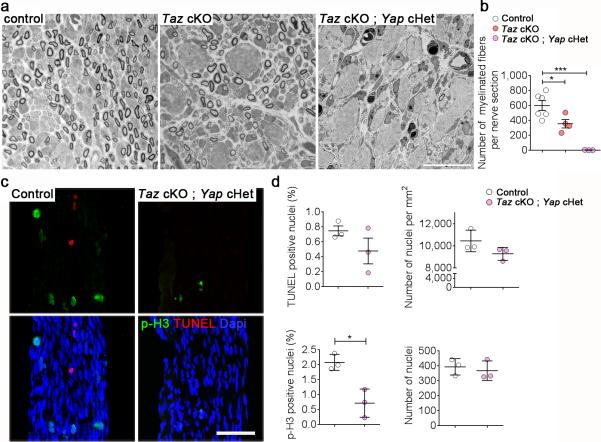
Reduction of Schwann cells available to engage axon can impair radial sorting 14. Such a failure in generating Schwann cells could be due to defects in proliferation or survival. To verify whether Yap or Taz control Schwann cell proliferation or apoptosis, we measured the fraction of anti-phospho-Histone 3 and TUNEL positive cells in P3 sciatic nerves, a time when matching of the number of Schwann cells and axons by apoptosis, proliferation and radial sorting is ongoing 15. We focused our analysis on Taz cKO; Yap cHet animals as their sciatic nerves show a severe arrest of radial sorting at P20. We first confirmed that the same defect was already present at P3 by semithin section analysis. Indeed, while in control nerves radial sorting was ongoing and myelination had started, Schwann cells of Taz cKO and Taz cKO; Yap cHet animals were already blocked at an immature stage (Fig. 4a-b). Only 0.72 % of Schwann cells were proliferating in Taz cKO; Yap cHet sciatic nerves, versus 2.08 % of Schwann cells in control sciatic nerves (Fig. 4c-d), while the rate of Schwann cell apoptosis was not significantly different. The density and total number of Schwann cell nuclei did not differ significantly between Taz cKO; Yap cHet and control sciatic nerves, even after taking into account that mutant nerves were smaller and amyelinated, which could increase the density of all cells and mask an actual reduction of Schwann cell numbers in the mutant (Fig. 4c-d). No significant differences in Schwann cell proliferation and apoptosis could be detected among all genotypes at P20, after completion of radial sorting (Supplementary Fig. 1). However, by P20 the total number of Schwann cells was decreased in double cKO, which were not used for subsequent studies. Thus, the reduction of proliferation in Taz cKO; Yap cHet Schwann cells might contribute to the severe radial sorting defects.
Figure 4.
Radial sorting defects in Taz cKO; Yap cHet are associated with a reduction in Schwann cell proliferation at P3. (a) Semithin cross-sections of sciatic nerves stained with Toluidine blue from control, Taz cKO and Taz cKO; Yap cHet at P3. Bar, 10 μm. At least three animals per genotype were analyzed. (b) Quantification of the number of myelinated fibers at P3 shows that they are absent in the double mutant. n = 3 mice per genotype. One way ANOVA P = 0.0003, F (2, 10) = 20.88 with Bonferroni post hoc test Taz cKO P = 0.0325, Taz cKO Yap cHet myelinated fibers P = 0.0001. (c) TUNEL staining (red), pH3 staining (green) and DAPI (blue) analysis on longitudinal section of sciatic nerves from control and Taz cKO; Yap cHet at P3. Bar, 50 μm. Three animals per genotype were analyzed. (d) Relative number of TUNEL and pH3 positive nuclei, density of nuclei per mm2 of sciatic nerve and total number of nuclei in sciatic nerve (length of sciatic nerve measured: 400 μm). n = 3 mice per genotype. TUNEL P = 0.021, t = 1.461, df=4; p-H3 P = 0.012, t = 4.341, df=4; two-tailed unpaired Student's t test.
In b and d each circle indicates the average value from one nerve from a different animal. Error bars indicate s.e.m. * P < 0.05, *** P < 0.001.
Taz and Yap control the expression of laminin receptors
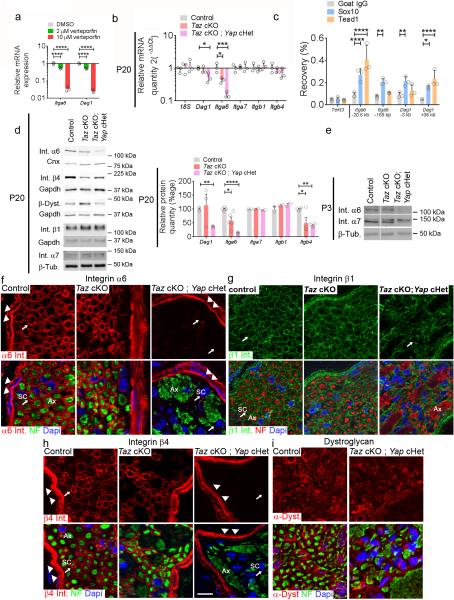
Members of the Tead family are the main transcription factors that interact with Yap and Taz 3, 16. Tead binding sites are enriched in enhancers of active genes in peripheral nerves during myelination 17 and Teads might cooperate with the myelin gene transcription factor Sox10 18. We hypothesized that the developmental defects observed in Taz cKOs were caused by a misregulation of Tead regulated genes. Using previously published ChIP-Seq data of sciatic nerves, we first identified active enhancers marked by acetylation of histone H3 lysine 27 (H3K27ac) that contained Tead motifs and were associated with genes controlling Schwann cell development and axonal sorting 18. Potential Tead-regulated enhancers were identified for ErbB2, Cdc42, Egr2 and Sox10 genes, which are involved in the transduction of the axonal signal that guides axonal sorting and myelination 14, 19-22 (Supplementary Fig. 2a). Treatment of a Schwann cell line with Verteporfin, a drug that interferes with Yap/Tead interactions 23, decreased the expression of ErbB2, Cdc42, Egr2 and Sox10 mRNA (Supplementary Fig. 2b). However in vivo we found no evidence of decreased ErbB2, Cdc42, or Sox10 mRNA or protein levels in mutant sciatic nerves, at least at P20 (Supplementary Fig. 2c-d). Egr2 protein was also not decreased at P20, but the expression of Egr2 mRNA was decreased by RNA profiling in Taz cKO; Yap cHet P3 nerves (see below). We then tested regulation of the Itga6 gene, that codes for an integrin required for Schwann cells to sort axons 24 and also possessed potential Tead-regulated enhancers (Supplementary Fig. 3a). Treatment of primary rat Schwann cells with Verteporfin decreased the expression of Itga6 mRNA (Fig. 5a). Importantly the mRNA and protein levels for integrin α6 subunit were reduced in mutant sciatic nerves (Fig. 5b-d), and correlated with the severity of the radial sorting phenotype observed in Taz cKO (protein decreased by 59%) and Taz cKO; Yap cHet (protein decreased by 85%).
Figure 5.
Taz and Yap control expression of integrin α6 and dystroglycan in Schwann cells. (a) Relative mRNA levels of Dag1 and Itga6 are decreased upon Verteporfin treatment in Schwann cells. n = 3 independent experiments. One way ANOVA P < 0.0001, F (2, 6) = 228.9 with Bonferroni post hoc test Dag1 2μM P < 0.0001, Dag1 10μM P < 0.0001, Itga6 2μM P < 0.0001, Itga6 10μM P < 0.0001. A logarithmic scale was used for the y-axis and the origin was set to 1. (b) Expression of Itga6 mRNA is decreased in Taz cKO, while both Itga6 and Dag1 mRNA are decreased in Taz cKO; Yap cHet at P20. n = 3 mice; One way ANOVA Dag1 P = 0.0263, Dag1 F (2, 6) = 7.082 with Bonferroni post hoc test Taz cKO Yap cHet P = 0.0242; One way ANOVA Itga6 P = 0.0003, F (2, 6) = 42.47 with Bonferroni post hoc test Taz cKO P = 0.0114, Taz cKO Yap cHet P = 0.0002. A logarithmic scale was used the y-axis and the origin was set to 1. (c) ChIP-qPCR on P15 rat sciatic nerves. Sox10 and Tead1 binding are enriched, relative to a negative control IP with goat IgG, on Itga6 and Dag1 enhancer regions. The negative control is a region 17.8 kb from the Tekt3 gene, not expressed in Schwann cells. n = 3 independent experiments; Two way ANOVA P < 0.0001, F (2, 30) = 28.79 with Bonferroni post hoc test. Sox10 Itga6 −20.6 kb P < 0.0001, Sox10 Itga6 −165 bp P = 0.0027, Sox10 Dag1 −3 kb P = 0.0063, Sox10 Dag1 +36 kb P = 0.0156 0.05, Tead1 Itga6 −20.6 kb P < 0.0001, Tead1 Dag1 +36 kb P < 0.0001. (d-e) Western blot at P20 (d) and P3 (e) shows a reduction of α6 integrin protein in Taz cKO and Taz cKO; Yap cHet sciatic nerves with no compensation by α7 integrin. Integrin β4 in Taz cKO, and both integrin β4 and β-dystroglycan in Taz cKO; Yap cHet mutants are reduced. At least three animals per genotypes were analyzed. For Dag1 n = 3 control mice, 3 Taz cKO, 3 Taz cKO Yap cHet; for Itga6 n = 5 control animal, 5 Taz cKO, 4 Taz cKO Yap cHet; for Itgb4 n = 4 control animal, 4 Taz cKO, 4 Taz cKO Yap cHet. Two way ANOVA P = 0.0004, F (4, 32) = 9.947 with Bonferroni post hoc test. Dag1 Taz cKO Yap cHet P = 0.006, Itga6 Taz cKO P = 0.0206, Itga6 Taz cKO Yap cHet P < 0.0001, Itgb4 Taz cKO P = 0.011, Itgb4 Taz cKO Yap cHet P = 0.0029. Western blots were cropped and the entire blots are presented in Supplementary Fig. 4. (f-i) Localization of integrin α6 (f), β1 (g) and β4 subunits (h) and α-dystroglycan (i) in cross-sections of sciatic nerves at P20. Integrin α6 and β4 subunits (red) are observed around the perineurium (arrowheads) and Schwann cells (arrows, SC) in control and Taz cKO nerves but not in Schwann cells (arrows, SC) surrounding bundles of amyelinated axons (Ax) in Taz cKO; Yap cHet nerves. Integrin β1 (green) is maintained in Schwann cells and dystroglycan is variably decreased in Schwann cells. Error bars indicate s.d. in (a, c) and s.e.m in (b, d) P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001. Bar, 10 μm.
To ask if Teads bind directly to Itga6 regulatory regions, we performed ChIP-qPCR using anti-Tead1 antibodies, since profiling studies indicate that Tead1 is most highly expressed among Tead family members in Schwann cells (data not shown). We first performed Tead1 ChIP in the S16 Schwann cell line, in which Itga6 expression was previously shown to be Sox10-dependent 25. After testing multiple sites, we found binding of Tead1 only at the enhancer at −20 kb upstream of Itga6 (Supplementary Fig. 3b). Although the Tead motif (GGAATG) was not found at this enhancer by HOMER analysis (Supplementary Fig. 3a), we found a conserved AGAATG variant, which also binds Tead1 26. By performing ChIP-qPCR in P15 sciatic nerves we confirmed binding of Tead1 and Sox10 to the same enhancer at −20.6 kb in vivo, but not to the promoter region that is bound by Sox10 (Fig. 5c).
We previously reported that deletion of Itga6 in Schwann cells in mice causes an early and compensatory up-regulation of integrin α7β1, requiring ablation of both Integrin α6β1 and α7β1 to reveal radial sorting defects 24. In contrast in our mutant animals the reduction of integrin α6 was not compensated by an increase of integrin α7 at P3 or P20 (Fig. 5d-e). By staining sciatic nerve sections we confirmed that integrin α6 was undetectable in Taz cKO; Yap cHet Schwann cells, while its expression was preserved in perineurial cells, in which P0-Cre is not expressed (Fig. 5f). The integrin α6 subunit pairs with integrin β4 or β1 subunits to form laminin receptors 27, and previous studies revealed that integrin α6β1 is the relevant receptor in Schwann cells for radial sorting 13, 24. Integrin β1, which can interact with several other integrins α subunits in Schwann cells (α1, α3, α5, α7) 28, was still present adjacent to the Schwann cell basal lamina in Taz cKO; Yap cHet mutant nerves (Fig. 5g). In contrast integrin β4 can only dimerize with the integrin α6 subunit, and it was postulated that failed hetero-dimerization of integrin subunits in the synthetic pathway leads to degradation 27. In agreement, we detected no integrin β4 protein in mutant Schwann cells (Fig. 5h), despite normal levels of Itgb4 mRNA (Fig. 5b).
Because deletion of Taz and 50% Yap caused an even more severe phenotype than that seen in the absence of both integrins α6β1 and α7β1 24, we asked if Taz and Yap also control the expression of a third redundant laminin receptor required for radial sorting, dystroglycan 29. Indeed, we examined two prominent H3K27ac-marked enhancers near the Dag1 gene, one of which contained a predicted Tead binding site (Supplementary Fig. 3a). Binding of Tead1 to the −36 kb site was detected by ChIP-qPCR in vivo using anti-Tead1 antibodies (Fig. 5c), while the other enhancer bound Sox10 but lacked significant Tead1 binding. Verteporfin treatment decreased Dag1 mRNA (Fig. 5a), and mRNA and protein levels for β-dystroglycan were reduced in Taz cKO; Yap cHet, but not Taz cKO Schwann cells (Fig. 5b, d, i).
Yap and Taz regulate expression of lipid synthetic enzymes
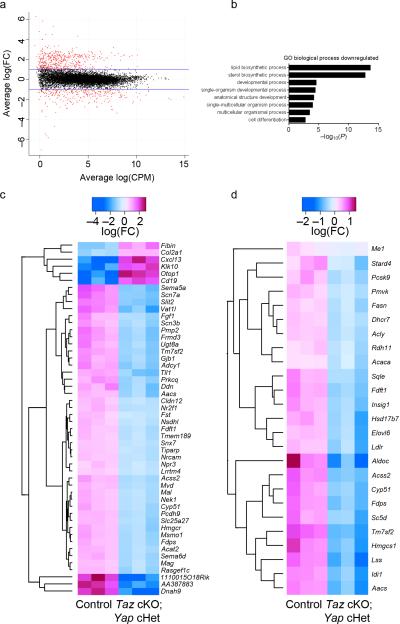
To examine the function of Yap/Taz in Schwann cells at a genome-wide level, we performed RNAseq transcriptome profiling of P3 developing peripheral nerves in Taz cKO; Yap cHet and control mice. We identified 2076 misregulated transcripts in mutant nerves, 982 were increased and 1093 were decreased at a 5% false discovery rate (Fig. 6a and Supplementary Table 1). We confirmed that the mRNA for Itga6 and Dag, as well as the known Yap/Taz/Tead targets genes Nov and Wisp, were decreased in mutant nerves. Among the top disregulated genes were signaling molecules predicted to be important in nerve development such as protein kinase C (Prkcq) and adenylate cyclase (Adcy1); and novel proteins potentially important such as the up-regulated P2Y receptor inhibitor Otopetrin (Otop1) and the chemokine Cxcl13 or the down-regulated Protocadherin 9 (Pcdh9) and the secreted glycoprotein Slit2 (Fig. 6c). Differentiation genes such as Mag, Pmp22 and Mbp were downregulated while Sox2 and Pou3f1 that are expressed in immature and promyelinating Schwann cells were upregulated. Interestingly, we found that the most misregulated genes coded for lipid and cholesterol biosynthetic and regulatory enzymes (Fig. 6b, d). We performed a gene set enrichment analysis using the MSigDB C2 database of ~5000 gene sets 30. Among genes down-regulated in Taz cKO; Yap cHet mice, we found a significant enrichment of sterol regulatory element-binding protein (Srebf) target genes (gene set enrichment P = 5 × 10−4) 31 with all 25 genes down-regulated in Taz cKO; Yap cHet mice (Fig. 6d). Srebf2 mRNA itself was significantly reduced by 48.7% in Taz cKO; Yap cHet cKO mice compared to controls. A similar decrease of lipid biosynthetic genes and Srebf2 was also observed in Egr2-deficient mice at P7 32, 33, therefore the modest decrease in Egr2 expression may partially account for these observations. Together these data suggest that Yap/Taz are required for normal expression of Srebf2, and their deletion substantially impairs the transcription of genes involved in lipid/sterol biosynthesis in Schwann cells.
Figure 6.
RNAseq analysis of Taz cKO; Yap cHet sciatic, brachial and peripheral trigeminal nerves at P3. (a) Log2 fold-change in Taz cKO; Yap cHet versus control mice was plotted against the average count size (log-counts-per-million) for every gene. Genes highlighted in red were significantly differentially expressed at 5% false discovery rate (FDR). Blue lines indicate 2-fold change. (b) Gene ontology analysis of biological processes downregulated significantly in Taz cKO; Yap cHet mice. (c) Heatmap of the top 50 most significant differentially regulated genes. Scale indicates expression relative to median of six samples. (d) Heatmap of Srebf target genes. Target genes were obtained from 31. Scale indicates expression relative to median of six samples. All genes are found downregulated in Taz cKO; Yap cHet mice. The data can be accessed at NCBI GEO accession number GSE79115.
The basal lamina is preserved in Taz mutant nerves
Taz regulates the transcription of laminin 511 and the organization of the extracellular matrix in breast cancer cells, and this in turn engages integrins in a positive regulatory loop 34. Our transcriptomic revealed a reduction of Lama2 transcripts, required to synthesize laminin 211. To determine if Yap/Taz had a major effect on laminins and the extracellular matrix in our system, we analyzed the expression and localization of laminins and the organization of the Schwann cell basal lamina in control and mutant nerves at P20. Fig. 7 shows that, while laminin 211 protein level was slightly reduced in Taz cKO; Yap cHet mice by Western blot (Fig. 7b), we did not find major alterations of laminins or morphological evidence of basal lamina disorganization in our mutants at this age.
Figure 7.
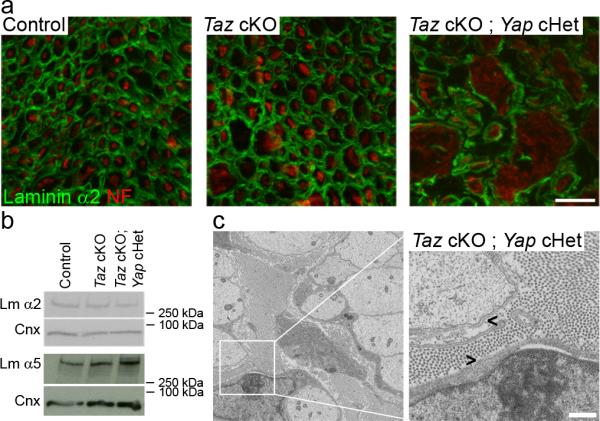
Laminins are expressed and the basal lamina is properly organized in Taz cKO and Taz cKO; Yap cHet nerves. (a) Immunolocalization of laminin α2 chain (green) and neurofilament (red) in cross sections of control, Taz cKO and Taz cKO; Yap cHet sciatic nerves at P20. Laminin staining can be observed around myelinated fibers (control, Taz cKO) or around bundles of amyelinated axons (Taz cKO; Yap cHet). Scale bar, 10μm. The experiment was repeated twice. (b) Western blot analysis for laminin α2 and α5 chains at P20 shows only a slight decrease in laminin α2 levels, with no other major alteration in the mutants. The experiment was repeated twice. Western blots were cropped and the entire blots are presented in Supplementary Fig. 4. (c) Electron micrographs of Taz cKO; Yap cHet sciatic nerves at P20. Basal lamina is properly organized around bundles of amyelinated axons (arrowhead). Scale, 500 nm.
Collectively, our data indicate that Yap, Taz and Teads regulate the expression of crucial laminin receptors in Schwann cells, namely integrin α6β1, α6β4 and dystroglycan. These defects correlate with the increasing severity in Taz cKO and Yap/Taz cKO mice, and paired with a decrease in Schwann cell proliferation and a defect in lipid biosynthesis, can account for the radial sorting phenotype.
Discussion
We demonstrate that Yap and Taz are required for normal peripheral nerve development. Yap and Taz control radial sorting and subsequent myelination by regulating Schwann cell proliferation, and via their role as Tead transcriptional coactivators. Yap and Taz are redundant in Schwann cells, and mice deficient for both genes have a complete arrest at the developmental step of axonal sorting. Also, Yap is not able to compensate for Taz ablation, while one Taz allele prevents radial sorting defects in Yap cKO; Taz cHet mutants.
We identify Yap, Taz and Tead1 as transcriptional regulators of radial sorting in Schwann cells. Radial sorting is a pre-requisite for peripheral myelination because it allows Schwann cells to engage in a 1:1 relationship with a large axon (promyelinating fiber). Although the transcriptional network that controls the transition from pro-myelination to myelination is well established (reviewed in 6), the transcriptional control of radial sorting is poorly understood. Our work places Yap/Taz-Tead1 at the core of the transcriptional regulation of axonal sorting. Differently from Schwann cells, oligodendrocytes in the central nervous system myelinate multiple axons, and as such they don't need to engage in a 1:1 relationship in order to myelinate. Also oligodendrocytes do not deposit a basal lamina and the mechanical stimuli placed upon them are likely to be different from Schwann cells 35, 36. Of note, Tead motifs were identified in conjunction with Sox10 binding sites in Schwann cells, but not oligodendrocytes 18. Based on these considerations, it is tempting to speculate that Yap, Taz and Teads fulfill different roles in oligodendrocytes and in Schwann cells, although this remains to be determined.
We show that laminin-binding integrins and dystroglycan are important downstream targets of Yap/Taz-Tead transcription. Previous work showed that Integrin α6β1 and dystroglycan, but not integrin α6β4, are required for radial sorting (reviewed in 7), but the factors that control their expression were unknown. Here we show that Yap/Taz are required for the expression of integrin α6 and β-dystroglycan in Schwann cells. Absence of Taz and one Yap allele in Schwann cells results in profound decrease of major receptors (integrin α6β1, α6β4 and dystroglycan) required to interact with laminins in the basal lamina. Previously we could not assess the independent function of integrin α6β1 as it was compensated by integrin α7β1 24 and deletion of integrin β1 subunit in Schwann cells removes 12 different integrin receptors 13. The selective decrease of integrin α6 subunit in Yap/Taz deficient nerves reinforces the idea that integrin α6β1 is the crucial integrin receptor for radial axonal sorting in Schwann cells.
Matching of Schwann cell and axon number is also critical for radial sorting, and mice deficient for Yap and Taz show a decrease of Schwann cell proliferation at P3, that could also contribute to the phenotype that we have observed. This is consistent with the notion that Yap and Taz control cell number and proliferation in a variety of other cell types (reviewed in 37), that they regulate many cell cycle genes with Tead and AP1 factors 38 and that they constitute important check-point through the Hippo pathway to neoplastic growth 39. For ChIP studies we used Tead binding motifs because they are highly enriched in Schwann cell-specific enhancers18 and previous ChIP-seq studies showed that the majority of Yap/Taz sites colocalize with Tead binding sites. However, Yap/Taz also utilize other partners such as Runx 1/2 to bind DNA 38, and they can also repress gene expression 40. Thus, we expect that many genes downstream of Yap/Taz contribute to orchestrate axonal sorting and myelination. To begin address this issue we performed genome-wide transcriptomic profiling, and revealed that Yap/Taz are required for proper expression of many genes, including the network that controls Schwann cell differentiation and myelination. Among these, lipid and sterol synthetic and regulatory genes were particularly affected, including all the targets of Srebf2. Whether Yap/Taz directly regulate these genes remains an important future question to be determined. Overall, we propose that the combined reduction of dystroglycan and integrin α6β1 without integrin α7β1 compensation, together with the reduction of Schwann cell proliferation and of the cholesterol biosynthesis pathways represent possible mechanisms by which the phenotype might be caused.
An important question is what controls Yap/Taz activation in Schwann cells. Yap/Taz lie downstream of mechano-transduction and of the Hippo pathway, which can be activated by multiple upstream inputs, such Wnt, G-coupled protein receptors, EGF and TGFβ signaling 41. Some of these pathways are involved in Schwann cell development and could conceivably regulate Yap/Taz in Schwann cells 42-44. Also, mechanical stimuli such as compression and traction forces exerted by neighboring cells or by the extracellular matrix participates in the activation of Yap/Taz in several cell types 41. We find that a combination of laminin 211 and mechanical stimuli activate Yap and Taz in Schwann cells, it will be interesting in the future to determine which receptors are involved. Reverse genetic and in vitro experiments suggested that Schwann cells respond to basal lamina composition and its mechanical properties during radial sorting and myelination 13, 44-47, but the molecular mechanisms are not known. Indeed we expected that integrins, potential mechanotransducers 48 would lie upstream of Yap/Taz, yet we were surprised to find that Yap/Taz instead regulated them. Because laminin211 synergizes with mechanical stimulation to activate Yap/Taz, it is possible that basal lamina components activate Yap/Taz via integrins or Gpr126 in a positive feedback loop44, 49. Diseases such as Hereditary Neuropathy with Liability to Pressure Palsy also underscore the importance of physical forces in peripheral myelination, where ablation of one copy of the Peripheral Myelin Protein 22 gene in Schwann cells causes demyelination after minor mechanical stimuli by largely unknown mechanisms 50. Thus, mechanotransduction is likely critical for nerve development and disease, and our work begins to shed light on its molecular mechanisms.
Online Methods
Animal models and morphology
All experiments involving animals followed experimental protocols approved by the Roswell Park Cancer Institute, University at Buffalo, and the University of Wisconsin School of Veterinary Medicine Institutional Animal Care and Use Committees. Taz and Yap floxed mice in C57BL6/129 mixed background, and P0-Cre transgenic in the congenic C57BL6/N background were described previously 11, 12. The resulting mutant mice contained a mixed background and only littermates were compared. Genotyping of mutant mice was performed by PCR on tail genomic DNA, as described previously 11, 12. Mutant and control littermates were sacrificed at the indicated ages, and sciatic nerves were dissected. Males and females were included in the study. No animals were excluded from the study. Animals were housed in cages of 5 animals in 12 h light / dark cycles. 3 animals per age and per genotype were analyzed, which is the minimum number required to obtain statistically significant results. Semithin section and electron microscopic analyses of sciatic nerves were performed as described 51. For g-ratio (axon diameter/fiber diameter) and axonal distribution, 4 semithin images per sciatic nerve were acquired at the 100x objective. Axon and fiber diameters were quantified using the Leica QWin software (Leica Microsystem). Blinding was not possible because of the severity of the phenotype. Fibers were quantified using ImageJ (http://imagej.nih.gov). Data were analyzed using GraphPad Prism 6.01.
Cell culture
Primary rat Schwann cells were produced as described 52 and cultured in DMEM supplemented with 4.5 g l−1 glucose, L-glutamine, sodium pyruvate, 5% bovine growth serum, penicillin, streptomycin, 0.2% bovine pituitary extract, and 2 μM forskolin. Schwann cells were not used beyond the fourth passage. Rat DRG neurons were isolated from embryonic day 14.5 embryos and established on collagen-coated glass coverslips as described 53. Explants were cycled with fluoroxidine (FUDR, Sigma-Aldrich) to eliminate all non-neuronal cells. Neuronal medium was supplemented with 50 ng ml−1 NGF (Harlan, Bioproducts for Science). Rat Schwann cells were added (50000 or 200000 cells per coverslip) to establish myelinating cocultures of DRG neurons, and myelination was initiated by supplementing the medium with 50 μg ml−1 ascorbic acid (Sigma-Aldrich).
Western blot
Sciatic nerves were dissected, stripped of the epineurium, frozen in liquid nitrogen, pulverized and resuspended in lysis buffer (95mM NaCl, 25mM Tris-HCl pH 7.4, 10mM EDTA, 2% SDS, 1mM Na3VO4, 1mM NaF and 1:100 Protease Inhibitor Cocktail (Roche)). Protein lysates were incubated at 4°C for 30 min then centrifuged at 16,000 rpm for 30min at 4°C. Supernatant protein concentrations were determined by BCA protein assay (Thermo Scientific) according to the manufacturer's instructions. Equal amounts of homogenates were diluted 3:1 in 4X Laemmli (250mM Tris-HCl pH 6.8, 8% SDS, 8% β-Mercaptoethanol, 40% Glycerol, 0.02% Bromophenol Blue), denatured 5min at 100°C, resolved on SDS-polyacrylamide gel, and electroblotted onto PVDF membrane. Blots were then blocked with 5% BSA in 1X PBS, 0.05% Tween-20 and incubated over night with the appropriate antibody. Abcam anti-ErbB2 1/250 (ab2428), BD Pharminogen anti-integrin β1 subunit 1/200 (553715), Cell Signaling anti-Yap 1/1000 (4912), Enzo anti-laminin α2 chain 1/200 (ALX-804-190), Novocastra anti-β-dystroglycan 1/50 (NCL-b-DG), Proteintech anti-Taz 1/1000 (23306-1-AP), Santa Cruz anti-Cdc42 1/200 (sc-87), anti-integrin α6 subunit 1/1000 (sc-6597), anti-Yap 1/1000 (sc-101199), Sigma anti-Calnexin 1/3000 (C4731), anti-Gapdh 1/2000 (G9545), anti-β-Tubulin 1/2000 (T4026), anti-Egr2 1/200 was gifted by Dr. Meijer, Centre for Neuroregeneration, Edinburg, anti-integrin α7 subunit 1/1000 was gifted by Dr. Tarone, Univ. of Turin, anti-integrin β4 subunit 1/1000 was gifted by Dr. Brophy, Centre for Neuroregeneration, Edinburgh, anti-laminin α5 chain 1/200 was gifted by Dr. Sorokin, University of Muenster, anti-Sox10 1/200 was gifted by Dr. Wegner, FAU, Germany. Membranes were then rinsed in 1X PBS and incubated for 1h with secondary antibodies. Blots were developed using ECL, ECL plus (GE Healthcare) or Odyssey CLx infrared imaging system (Li-Cor). Western blots were quantified using Image J software (http://imagej.nih.gov/ij). Each experiment was performed at least two times on nerves from at least 3 animals.
TUNEL and proliferation assays
TUNEL and proliferation assays were performed on longitudinal sections of sciatic nerves. Slices were fixed with 4% PFA, permeabilized 1 min in methanol, blocked for 1h in 20% FBS, 2% BSA, 0.1% Triton-X in PBS, and stained overnight with anti-rabbit PH3 antibody 1/400 (Millipore: #06-576), followed by Jackson DyLight 488-conjugated secondary antibody. Slices were then incubated 15 min in TDT buffer (30 mM Tris, 140 mM sodium cacodylate trihydrate, 1 mM cobalt chloride) followed by enzyme mixture (60 U ml−1 TdT (Sigma), 6.25 mM Biotin-16-dUTP (Roche)) in TDT buffer for 1 h at 37 °C. The reaction was terminated with TB buffer (300 mM NaCl, 40 mM sodium citrate). After blocking in 2% BSA, coverslips were incubated with Streptavidin-Rhodamine secondary antibody. Nuclei were counterstained with DAPI and analyzed with fluorescence Leica DM6000B microscope. For longitudinal sections, nerves from 3 different animals per genotype were analyzed. The proportion of p-H3 or TUNEL-positive nuclei were determined blindly from 5 random fields for each nerve. Stained nuclei were counted manually on images using ImageJ software (http://imagej.nih.gov/ij). The total number of nuclei in sciatic nerves was obtained by measuring the total area of sciatic nerve over 400 μm of length and multiplying the total area by the nuclei density. Data were analyzed using Graph pad Prism 6.01.
Immunohistochemistry
Longitudinal sections and teased fibers from sciatic nerves were fixed in 4% PFA, permeabilized with acetone, washed in PBS, blocked for 1 h in 5% fish skin gelatin, 0.5% Triton X-100 in 1X PBS. Primary Schwann cells were fixed in 1% PFA, 0.5% DOTMAC, washed in PBS, blocked for 1 h in 5% normal goat serum, 0.1% Triton in 1X PBS. Co-cultures were fixed in 1% PFA, 0.5% DOTMAC, permeabilized with methanol, washed in PBS, blocked for 1 h in 20% fetal calf serum, 1% bovine serum albumin, 0.05% Triton in 1X PBS. The following primary antibodies were incubated overnight: Abcam anti-integrin β4 subunit 1/150 (ab25254), BD Pharminogen anti-integrin β1 subunit 1/150 (553837), Cell Signalling anti-Yap 1/100 (4912), Covance anti-NFH 1/700 (PCK-593P), anti-MBP 1/1000 (SMI-99P), Enzo anti-laminin α2 chain 1/250 (ALX-804-190), Millipore ani-NFH 1/500 (AB1989), anti-α-dystroglycan IIH6 (05-593); Proteintech anti-Yap/Taz 1/100 (23306-1-AP), anti-integrin α6 subunit 1/10, was gifted by Dr. Sonnenberg, Netherlands Cancer Institute. Sections were rinsed in PBS, incubated 1 h with phalloidin-TRITC 1/100 (Sigma, P1951), Jackson DyLight 488 1/1000, 549-conjugated 1/1000 or 649-conjugated 1/1000 secondary antibodies, stained with DAPI, and mounted with Vectashield (Vector Laboratories). Images were acquired with a Zeiss ApoTome or a confocal microscope Leica SP5II. Microscopy equipment and settings have been detailed in Supplementary Fig. 5. Maximum intensity projection from Z-stacks were created with Fiji software (fiji.sc). The localization of Yap/Taz on various substrates and in the stretched experiment was assessed blindly.
Preparation of substrates of differing stiffness
Polyacrylamide (PA) gel and Polydimethylsiloxane (PDMS) were used to create substrates with different elastic modulus (E) ranging from softer PA gel with E = 0.5 kPa and 40kPa to stiff PDMS with E = 4Mpa. The PA gels with two elastic moduli of 0.5 and 40 kPa were fabricated according to the protocol described by Tse et al. 54. Briefly, 18mm round coverslips were cleaned with 70% ethanol and treated with 0.1M NaOH on hotplate to create a uniform thin NaOH film on the surface. The surface was then coated with 3-Aminopropyltriethoxysilane (TCI Cat. No. A0439) for 5 minutes followed by treatment with 0.5% glutaraldehyde (ACROS organics, Cat. No. 233280250) solution in PBS for 30 minutes. Functionalized coverslips were dried for gel coating. Acrylamide to Bis-acrylamide (BIO-RAD) ratio (v/v) of 3%:0.1% and 8%:0.48% were used to create 0.5 and 40 kPa PA gel substrates, respectively. After gel formation the gel surface was functionalized using sulfo-SANPAH (CovaChem, Cat. No. DF3363) by exposing to UV light. Finally, PA gels were coated with Poly-L-lysine (0.01 mg/ml) with or without laminin 211 (10 μg ml−1, produced as described 53. PDMS elastomer to curing agent (Sylgard 184) ratio of 4:1 was used to create PDMS substrate with 4 MPa elastic modulus as previously described by Zhao et al. 55. Briefly, the elastomer and curing agent were mixed thoroughly and degassed, and small amount of mixture was added to coverslips and spun to create a thin layer. The coverslips were then cured at 60°C followed by 120°C in the oven. Surface of the PDMS was coated using Poly-L-Lysine with or without laminin at the concentrations previously mentioned. Schwann cells were trypsinized, counted and plated on the different substrates (glass, PA gel and PDMS) at 200,000 cells per coverslip. Cells were allowed to spread for 24 hours before fixation.
Stretching experiment
Silicone sheets of 0.01 inch thickness (SMI silicone sheeting, 0.01” NRV G/G 40D, Saginaw, MI) were used for static stretching experiment. The membrane was cut into 2.5 cm wide stripes and sterilized with 70% ethanol and under UV lamp. The surface was coated with PLL and laminin at aforementioned concentrations. The membranes were kept in PBS prior to use. Schwann cells were seeded on silicone sheet at 200,000 cells in a patterned area comparable to an 18 mm coverslip and were allowed to grow for 24 hours. The membrane was then mounted on a custom-made uniaxial stretching device and stretched to 150% strain. The samples were kept under static strain for 30 minutes and then relaxed and fixed immediately. Non-stretched membrane with plated cells was used as a negative control.
Verteporfin treatment
Primary cells were treated with 2 and 10 μM Verteporfin (Sigma SML0534). At 4 h after treatment, cells were stimulated with 20 ng μl−1 neuregulin-1 β isoform (heregulin-β 1, R&D Systems). RNA was harvested at 24 h after treatment, and cDNA was analyzed by RT-qPCR using the following primers:
18S (Forward: cgccgctagaggtgaaattct, Reverse: cgaacctccgactttcgttct),
ErbB2 (Forward: aggtctggagggaacatcct, Reverse: tgggatgcatgtgtctcagt),
Cdc42 (Forward: gctctggagatgcgttcatag, Reverse: gaagcaatattggctgccttg),
Egr2 (Forward: gcactctgtggccctagaaca, Reverse: ggctgagatggctcgagaaa),
Sox10 (Forward: cgaattgggcaaggtcaaga, Reverse: caccgggaacttgtcatcgt),
Itga6 (Forward: cgagagatcaacgacgagaaac, Reverse: tctttcctacaccctcctctatg),
Dag1 (Forward: ctcccagggtgtttcagact, Reverse: tcagagcaaccaaggtgaca).
The S16 rat Schwann cell line 56 was obtained from Richard Quarles, cultured as described 52, and expresses relatively high levels of myelin genes 25.
RNA preparation and RT-qPCR
Sciatic nerves were dissected, stripped of epineurium, frozen in liquid nitrogen and pulverized. Total RNA was prepared from sciatic nerve or Schwann cells with TRIzol (Roche Diagnostic). 1 μg of RNA was reverse transcribed using Superscript III (Invitrogen). For each reaction, 5 μM of oligo(dT)20 and 5 ng μl−1 random hexamers were used. Quantitative PCR were performed using the 20ng of cDNA combined with 1X FastStart Universal Probe Master (Roche Diagnostic). Data were analyzed using the threshold cycle (Ct) and 2(−ΔΔCt) method. Actb gene was used as endogenous gene of reference and the average expression of control animals was set as the reference for normalizing the data. For data representation in Figure 5b and Supplementary Fig 2c, only one control animal was used for normalizing the data. The primers and probe used are the following:
18S (Forward: ctcaacacgggaaacctcac, Reverse: cgctccaccaactaagaacg, Probe:#77),
Actb (Forward: aaggccaaccgtgaaaagat, Reverse: gtggtacgaccagaggcatac, Probe:#56),
Cdc42 (Forward: tgtttccgaaatgcagacaa, Reverse: tgactgcatagttgtcaaaaacag, Probe:#9),
Dag1 (Forward: ctgctgctgctccctttc, Reverse: gcagtgttgaaaaccttatcttcc, Probe:#99),
ErbB2 (Forward: tgtgtggacctggacgaac, Reverse: tgcaatgatgaatgtcactgg, Probe:#3),
Itga6(Forward: cctgaaagaaaataccagactctca, Reverse: ggaacgaagaacgagagagg, Probe:#99),
Itga7 (Forward: agaaggtggagcctagcaca, Reverse: gctgaacaccacacacttgg, Probe:#49),
Itgb1 (Forward: caaccacaacagctgcttctaa, Reverse: tcagccctcttgaattttaatgt, Probe:#2),
Itgb4 (Forward: cttggtcgccgtctggta, Reverse: tcgaaggacactaccccact, Probe:#9).
Chromatin Immunoprecipitation (ChIP)
ChIP was carried out as described previously 25, 57 with the following antibodies: goat IgG (Santa Cruz Biotechnology, sc-2028), Sox10 (R&D, AF2864), and Tead1 (BD Biosciences, 610923). Primers used included the following:
Tekt3 +7.8kb (Forward: aataccagcagatccggaagac, Reverse: ttgactcgttctcccaggtttt),
Itga6 −22.8kb (Forward: tgcttctgataagccccaac, Reverse: aagtcccagacacgtccttg),
Itga6 −20.6kb (Forward: tggaggcagaaggagaaaaa, Reverse: aggggcctgttaggaacact),
Itga6 −16.2kb (Forward: gacttgagctgtctgcatgg, Reverse: tggactgactaggcttccaca),
Itga6 −8.5kb (Forward: aaaagccaaacaaacccaga, Reverse: ggctagggcaagctaaggat),
Itga6 −7.8kb (Forward: tgcttttggtcatgtggttg, Reverse: accactggctagctcagcat),
Itga6 −165bp (Forward: tcgataaaacgccggagagt, Reverse: gtagctagcagccgctcaat),
Dag1 +3kb (Forward: atgaacccctcttcctgacc, Reverse: ccttgctgagactgtgctca),
Dag1 −36kb (Forward: ggatggaagactgaaaggcc, Reverse: ccctttccctctggtgtgag).
Bioinformatics
H3K27ac enrichment at select loci was obtained from previously published ChIP-Seq data 18, and Tead motifs were found using Homer 58.
RNA seq analysis
Sciatic, trigeminal and brachial nerves at P3 were dissected, frozen in liquid nitrogen and pulverized. Total RNA was prepared from pools of nerves (8 trigeminal, 8 brachial and 8 sciatic nerves per pool) with TRIzol (Roche Diagnostic), then purified with RNeasy column (Qiagen). Samples were quantified using Ribogreen Assay (Invitrogen) and the quality of samples were checked using Agilent Bioanalyzer 2100 RNA nano 6000 chip (Agilent). Illumina TruSeq RNA sample preparation kit (Illumina) was used to prepare cDNA libraries from RNA samples. Samples were poly A selected to isolate mRNA, the mRNA was cleaved into fragments, the first strand reverse transcribed to cDNA using SuperScript II reverse Transcriptase (Invitrogen) and random primers, followed by second strand cDNA synthesis using Second Strand Master Mix supplied with the kit. After end repair, the addition of a single ‘A’ base, and ligation with adapters, the products were enriched and purified with PCR to create the final cDNA library as per manufacturer's protocol. cDNA libraries were quantified using Picogreen Assay (Invitrogen) and Library Quantification kit (Kapa Biosystems). Agilent Bioanalyzer 2100 DNA 7500 chip was used to confirm the quality and size of the cDNA libraries. The cDNA libraries were then normalized, pooled and paired-end sequenced (100 standard cycles) using the Illumina HiSeq2500 following the manufacturer's instructions at the UB Genomics and Bioinformatics Core Facility (Buffalo, NY). Sequences were aligned to the UCSC Mm10 mouse genome using tophat (v2.0.13) and counts per gene determined using htseq (v0.6.1). R/Bioconductor was used for subsequent analysis. Following loading of read counts using DESeq, edgeR was used for differential expression analysis of read counts 59. Genes with low counts (less than one count-per-million) in at least three libraries were removed from the analysis, and TMM normalized applied to account for differences between libraries. For differential expression analysis, common NB dispersion was estimated and a generalized linear model likelihood ratio test was used to determine differential expression between cKO and wild-type mice. P-values were controlled for multiple testing by determining the false discovery rate (FDR). Pathway analysis was performed in edgeR using gene ontology (GO) enrichment analysis, KEGG pathway enrichment analysis, and gene set enrichment analysis of Broad C2 gene sets (v5) using roast. Heatmaps were plotted using log2-counts-per-million, with a prior count of 1. Data are available at http://www.ncbi.nlm.nih.gov/geo/ accession number: GSE79115.
Statistical analyses
Experiments were not randomized, but data collection and analysis were performed blind to the conditions of the experiments. No data were excluded from the analyses. Data obtained were presented as mean ± s.e.m or s.d. Two tailed unpaired Student t-test, two tailed unpaired Student t-test Bonferroni corrected, Fischer exact test, One way ANOVA and Two way ANOVA were used for statistical analysis of the differences among multiple groups according to the number of samples. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar those generlly employed in the field. Data distribution was assumed to be normal but this was not formally tested. Values of P < 0.05 were considered to represent a significant difference.
Supplementary Material
Acknowledgments
We thank E. Hurley for superb technical assistance; A. Sonnenberg, Netherlands Cancer Institute, D. Meijer and P. Brophy, Centre for Neuroregeneration, Edinburgh, the late G. Tarone, Univ. of Turin, Dr. Sorokin, University of Muenster, and M. Wegner, FAU, Erlanger-Nurberg for antibodies and the late R. Quarles for the S16 cells. This work was funded by grants NS045630 (to M.L.F), NS096104 (to L.W.) and NS075269 (to J. S.).
Footnotes
The authors declare no competing financial interests.
Author contributions
Y.P, K.C., C.B., M.P. and M.L.F designed research and interpreted data; Y.P performed experiments with assistance from C.L-A, K.C., C.B., M.P., C.W., D.A., K.A. Y.H; C.L.A. and J.S. designed and performed ChIP sequencing and promoter analysis. M.A. and R.Z. designed and helped to perform biomechanical experiments; A.G. and J.W and L.W. contributed analytical tools; F.J.S. analyzed RNAseq data; Y.P. and M.L.F wrote the manuscript; Y.P., C.L.A, R.Z., F.J.S., J.S., L.W. and M.L.F. analyzed data and critically reviewed the manuscript.
Accession codes
RNAseq data are available at http://www.ncbi.nlm.nih.gov/geo/ accession number: GSE79115.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its supplementary information files. All original data are available from the corresponding author upon reasonable request.
References
- 1.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 2.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webster HD, Martin R, O'Connell MF. The relationships between interphase Schwann cells and axons before myelination: a quantitative electron microscopic study. Dev Biol. 1973;32:401–416. doi: 10.1016/0012-1606(73)90250-9. [DOI] [PubMed] [Google Scholar]
- 5.Jessen KR, Mirsky R, Lloyd AC. Schwann Cells: Development and Role in Nerve Repair. Cold Spring Harbor perspectives in biology. 2015;7 doi: 10.1101/cshperspect.a020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svaren J, Meijer D. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 2008;56:1541–1551. doi: 10.1002/glia.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feltri ML, Poitelon Y, Previtali SC. How Schwann Cells Sort Axons: New Concepts. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2015 doi: 10.1177/1073858415572361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 9.Taveggia C, et al. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. doi: 10.1016/j.neuron.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 11.Reginensi A, et al. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet. 2013;9:e1003380. doi: 10.1371/journal.pgen.1003380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feltri ML, et al. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Annals of the New York Academy of Sciences. 1999;883:116–123. [PubMed] [Google Scholar]
- 13.Feltri ML, et al. Conditional disruption of beta 1 integrin in Schwann cells impedes interactions with axons. The Journal of cell biology. 2002;156:199–209. doi: 10.1083/jcb.200109021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benninger Y, et al. Essential and distinct roles for cdc42 and rac1 in the regulation of Schwann cell biology during peripheral nervous system development. The Journal of cell biology. 2007;177:1051–1061. doi: 10.1083/jcb.200610108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grinspan JB, Marchionni MA, Reeves M, Coulaloglou M, Scherer SS. Axonal interactions regulate Schwann cell apoptosis in developing peripheral nerve: neuregulin receptors and the role of neuregulins. J Neurosci. 1996;16:6107–6118. doi: 10.1523/JNEUROSCI.16-19-06107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung HA, Sun G, Keles S, Svaren J. Dynamic regulation of Schwann cell enhancers after peripheral nerve injury. J Biol Chem. 2015;290:6937–6950. doi: 10.1074/jbc.M114.622878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Anido C, et al. Differential Sox10 genomic occupancy in myelinating glia. Glia. 2015 doi: 10.1002/glia.22855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garratt AN, Voiculescu O, Topilko P, Charnay P, Birchmeier C. A dual role of erbB2 in myelination and in expansion of the schwann cell precursor pool. The Journal of cell biology. 2000;148:1035–1046. doi: 10.1083/jcb.148.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo L, Moon C, Zheng Y, Ratner N. Cdc42 regulates Schwann cell radial sorting and myelin sheath folding through NF2/merlin-dependent and independent signaling. Glia. 2013;61:1906–1921. doi: 10.1002/glia.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topilko P, et al. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 22.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellegatta M, et al. alpha6beta1 and alpha7beta1 integrins are required in Schwann cells to sort axons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:17995–18007. doi: 10.1523/JNEUROSCI.3179-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srinivasan R, et al. Genome-wide analysis of EGR2/SOX10 binding in myelinating peripheral nerve. Nucleic acids research. 2012;40:6449–6460. doi: 10.1093/nar/gks313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anbanandam A, et al. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17225–17230. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Flier A, Sonnenberg A. Function and interactions of integrins. Cell Tissue Res. 2001;305:285–298. doi: 10.1007/s004410100417. [DOI] [PubMed] [Google Scholar]
- 28.Previtali SC, et al. Expression of laminin receptors in schwann cell differentiation: evidence for distinct roles. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:5520–5530. doi: 10.1523/JNEUROSCI.23-13-05520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berti C, et al. Non-redundant function of dystroglycan and beta1 integrins in radial sorting of axons. Development. 2011;138:4025–4037. doi: 10.1242/dev.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horton JD, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leblanc SE, et al. Regulation of cholesterol/lipid biosynthetic genes by Egr2/Krox20 during peripheral nerve myelination. Journal of neurochemistry. 2005;93:737–748. doi: 10.1111/j.1471-4159.2005.03056.x. [DOI] [PubMed] [Google Scholar]
- 33.Le N, et al. Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2596–2601. doi: 10.1073/pnas.0407836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang C, et al. A laminin 511 matrix is regulated by TAZ and functions as the ligand for the alpha6Bbeta1 integrin to sustain breast cancer stem cells. Genes & development. 2015;29:1–6. doi: 10.1101/gad.253682.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Fagundo C, Bar-Kochba E, Livi LL, Hoffman-Kim D, Franck C. Three-dimensional traction forces of Schwann cells on compliant substrates. Journal of the Royal Society, Interface / the Royal Society. 2014;11:20140247. doi: 10.1098/rsif.2014.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jagielska A, et al. Mechanical environment modulates biological properties of oligodendrocyte progenitor cells. Stem Cells Dev. 2012;21:2905–2914. doi: 10.1089/scd.2012.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irvine KD, Harvey KF. Control of organ growth by patterning and hippo signaling in Drosophila. Cold Spring Harbor perspectives in biology. 2015;7 doi: 10.1101/cshperspect.a019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zanconato F, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nature cell biology. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo L, Teng L. YAP/TAZ for cancer therapy: opportunities and challenges (review). International journal of oncology. 2015;46:1444–1452. doi: 10.3892/ijo.2015.2877. [DOI] [PubMed] [Google Scholar]
- 40.Kim M, Kim T, Johnson RL, Lim DS. Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell reports. 2015;11:270–282. doi: 10.1016/j.celrep.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 41.Aragona M, et al. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 42.Anliker B, et al. Lysophosphatidic acid (LPA) and its receptor, LPA1 , influence embryonic schwann cell migration, myelination, and cell-to-axon segregation. Glia. 2013;61:2009–2022. doi: 10.1002/glia.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grigoryan T, et al. Wnt/Rspondin/beta-catenin signals control axonal sorting and lineage progression in Schwann cell development. Proc Natl Acad Sci U S A. 2013;110:18174–18179. doi: 10.1073/pnas.1310490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen SC, et al. The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron. 2015;85:755–769. doi: 10.1016/j.neuron.2014.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen ZL, Strickland S. Laminin gamma1 is critical for Schwann cell differentiation, axon myelination, and regeneration in the peripheral nerve. J Cell Biol. 2003;163:889–899. doi: 10.1083/jcb.200307068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKee KK, et al. Schwann cell myelination requires integration of laminin activities. Journal of cell science. 2012;125:4609–4619. doi: 10.1242/jcs.107995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grove M, Brophy PJ. FAK is required for Schwann cell spreading on immature basal lamina to coordinate the radial sorting of peripheral axons with myelination. J Neurosci. 2014;34:13422–13434. doi: 10.1523/JNEUROSCI.1764-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz MA. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harbor perspectives in biology. 2010;2:a005066. doi: 10.1101/cshperspect.a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paavola KJ, Sidik H, Zuchero JB, Eckart M, Talbot WS. Type IV collagen is an activating ligand for the adhesion G protein-coupled receptor GPR126. Science signaling. 2014;7:ra76. doi: 10.1126/scisignal.2005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chance PF, et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72:143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 51.Quattrini A, et al. Beta 4 integrin and other Schwann cell markers in axonal neuropathy. Glia. 1996;17:294–306. doi: 10.1002/(SICI)1098-1136(199608)17:4<294::AID-GLIA4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 52.Gokey NG, Srinivasan R, Lopez-Anido C, Krueger C, Svaren J. Developmental regulation of microRNA expression in Schwann cells. Molecular and cellular biology. 2012;32:558–568. doi: 10.1128/MCB.06270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colombelli C, et al. Perlecan is recruited by dystroglycan to nodes of Ranvier and binds the clustering molecule gliomedin. The Journal of cell biology. 2015;208:313–329. doi: 10.1083/jcb.201403111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Current protocols in cell biology / editorial board, Juan S. Bonifacino ... [et al.] 2010:16. doi: 10.1002/0471143030.cb1016s47. Chapter 10, Unit 10. [DOI] [PubMed] [Google Scholar]
- 55.Zhao R, Chen CS, Reich DH. Force-driven evolution of mesoscale structure in engineered 3D microtissues and the modulation of tissue stiffening. Biomaterials. 2014;35:5056–5064. doi: 10.1016/j.biomaterials.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toda K, Small JA, Goda S, Quarles RH. Biochemical and cellular properties of three immortalized Schwann cell lines expressing different levels of the myelin-associated glycoprotein. Journal of neurochemistry. 1994;63:1646–1657. doi: 10.1046/j.1471-4159.1994.63051646.x. [DOI] [PubMed] [Google Scholar]
- 57.Jones EA, et al. Distal enhancers upstream of the Charcot-Marie-Tooth type 1A disease gene PMP22. Human molecular genetics. 2012;21:1581–1591. doi: 10.1093/hmg/ddr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.