Abstract
A key aspect of cellular function is the proper assembly and utilization of ribonucleoproteins (RNPs). Recent studies have shown that hyper- or hypo-assembly of various RNPs can lead to human diseases. Defects in the formation of RNPs lead to "RNP hypo-assembly diseases", which can be caused by RNA degradation out-competing RNP assembly. By contrast, excess RNP assembly, either in higher-order RNP granules, or due to the expression of repeat-containing RNAs, can lead to "RNP hyper-assembly diseases”. Here, we discuss the most recent advances in understanding the cause of disease onset, as well as potential therapies from the aspect of modulating RNP assembly in the cell, which presents a novel route to the treatment of these diseases.
Keywords: RNP assembly, Stress granules, Spinal Muscular Atrophy, Dyskeratosis Congenita, RNA quality control, Therapy
The Expanding Spectrum of RNP Assembly Diseases
The proper interaction between RNA molecules and their protein partners is a ubiquitous event in the life cycle of every RNA. These interactions lead to the formation of ribonucleoprotein (RNP) particles (see Glossary) that allow RNA function and control [1].
The assembly of specific RNPs is generally in kinetic competition with RNA degradation (Figure 1). This competition both ensures that only fully assembled RNPs are produced, and it serves as a 'quality control' checkpoint for the fidelity of the RNP being formed. Given these competing RNA decay pathways, defects in RNP assembly can lead to degradation of key RNAs, and pathologies due to RNP loss in "RNP hypo-assembly diseases". Examples of these diseases include Spinal Muscular Atrophy (SMA) and Dyskeratosis Congenital (DC) for non-coding RNAs, and Constant Spring thalassemia for mRNAs.
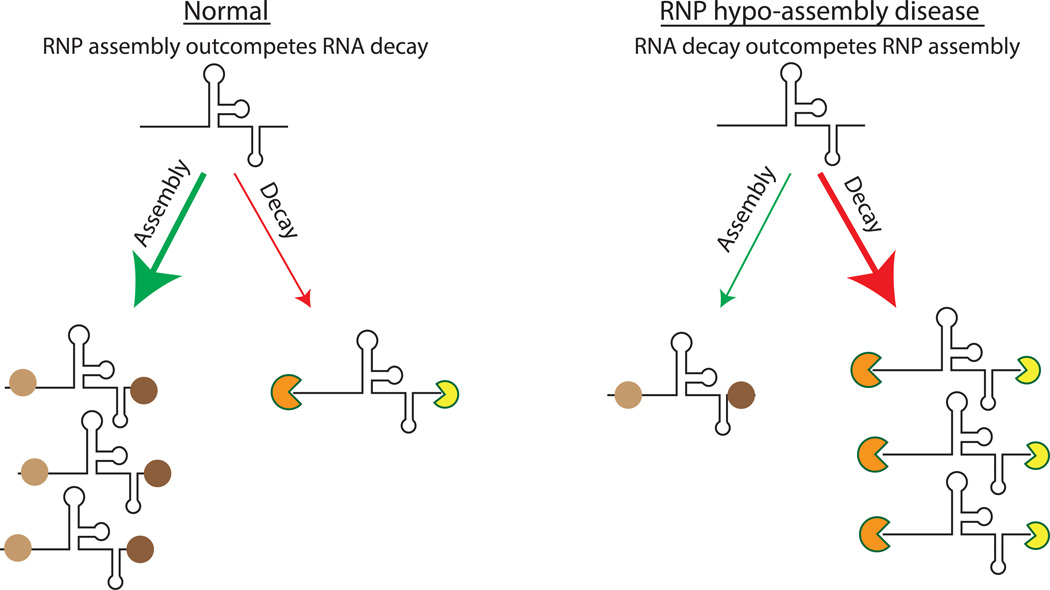
Figure 1. Mechanism of RNP Hypo-Assembly.
RNP hypo-assembly diseases are caused by RNA quality control pathways out-competing defective RNP assembly in the cell. Under normal physiological conditions, cellular RNAs interact with RNA binding proteins to form RNPs. In hypo-assembly diseases, RNP formation is reduced, leading to RNA degradation by competing RNA quality control mechanisms.
Individual RNPs also assemble into a variety of higher order structures, generally referred to as RNP granules [2]. For example, snRNPs, snoRNPs and telomerase RNPs traffic through a large RNP assembly in the nucleus referred to as Cajal bodies. Similarly, non-translating mRNPs in the cytosol can assemble into P-bodies, which contain a number of proteins involved in mRNA degradation, or stress granules, which contain a diverse proteome, including RNA binding proteins, and some translation factors. The formation of such mega-mRNPs is facilitated by protein-protein interactions of RNA binding proteins allowing multivalent interactions between individual RNPs, and leading to the assembly of these structures [3]. Hyper-assembly of such large RNP granules, either due to hyper-assembly of specific RNA binding proteins or to the expansion of nucleotide repeats in RNAs, can trigger several different pathologies, which we collectively refer to as "RNP hyper-assembly diseases". Examples of these diseases include Amyotrophic Lateral Sclerosis (mutations in TDP-43 and FUS, or C9orf72 expansions of G4C2 repeats) [4], Multisystem proteinopathy (mutations in hnRNPA1 and hnRNPA2B1) [5], and CUG or CCUG expansions in Myotonic Dystrophy Type 1 & 2 [6].
Despite recent discoveries of causative mutations associated with these diseases, there is a lack of clear understanding of disease etiology. Further, there are no treatments available for these diseases, which points to the urgency of finding ‘druggable’ targets in these pathologies. Here, we discuss this broad spectrum of human diseases from a RNP assembly perspective. We also suggest possible treatments based on the potential of altering the kinetics of RNP formation, which could provide novel therapeutic avenues for drug development.
Hypo-assembly Diseases of RNPs
Dyskeratosis Congenita and Degradation of Telomerase RNA
Several mutations that cause dyskeratosis congentia (DC) do so by limiting assembly of the human telomerase RNP. DC is a telomere disorder belonging to a broader family of diseases characterized by shortening of telomeres and failure to renew stem cells [7]. The most common causes of DC are mutations in the RNA binding protein, dyskerin, or the human telomerase RNA (hTR), some of which disrupt the binding site for dyskerin in the 3' region of hTR [8]. Mutations in dyskerin and some mutations in the 3' region of hTR lead to low levels of hTR in the cell, which is directly related to reduced telomerase activity observed in DC models [9].
When dyskerin binding is defective, the hTR RNA is degraded by two distinct RNA decay mechanisms (Figure 2). First, hTR can be degraded in the nucleus by PAPD5-mediated oligoadenylation, which then provides a 3' single stranded region that allows efficient recruitment of a 3' to 5' exonuclease, EXOSC10. EXOSC10 can in turn function either on its own, or as a subunit of the larger nuclear exosome [10–13]. Alternatively, hTR can be degraded in the cytosol by DCP2-mediated decapping, which then exposes hTR to 5' to 3' degradation by the exonuclease XRN1 [11]. Importantly, it has been shown that inhibition of either decapping and/or nuclear EXOSC10-mediated degradation could rescue in dyskerin-depleted HeLa cells, both telomerase activity, as well as localization of the telomerase RNP (presumably lacking dyskerin) to Cajal bodies [11]. The rescue of activity and proper subcellular localization when RNA decay pathways are inhibited suggests that pharmaceutical approaches to block RNA nucleases might be a potential therapy for some alleles causing DC.
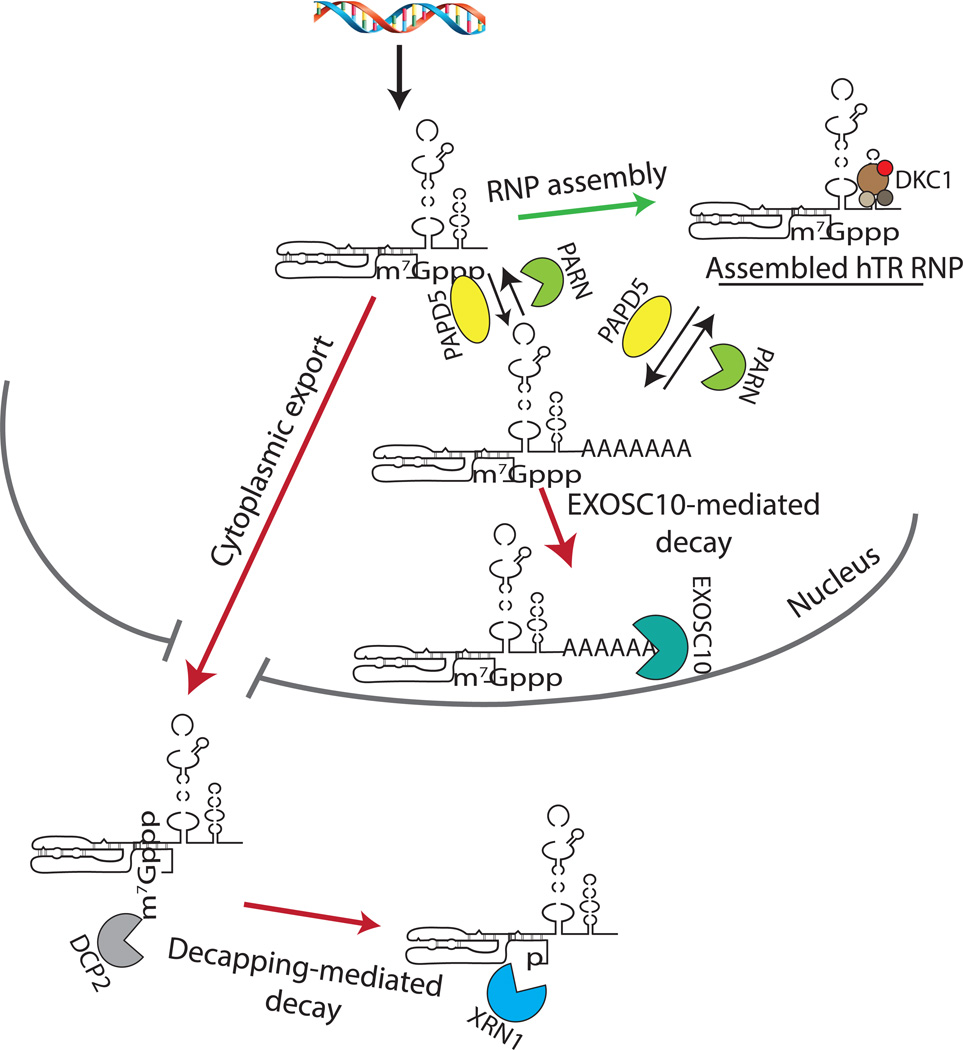
Figure 2. Competition Between hTR RNP Assembly and Quality Control.
Under normal conditions, hTR is bound by dyskerin and other H/ACA proteins to form the hTR RNP. In DC, unassembled hTR is degraded by EXOSC10 in the nucleus, and by DCP2/XRN1 in the cytoplasm. DCP2 removes the m7Gpp cap structure from the 5’ end of hTR, exposing the monophosphate to allow XRN1-mediated exonucleolytic digestion. Loss of PARN also leads to degradation of hTR by EXOSC10, which is aided by PAPD5-mediated oligoadenylation of hTR 3’ end, where it can destabilize both unbound and assembled hTR molecules.
The evidence that hTR can be subjected to oligoadenylation, which triggers 3' to 5' degradation, has allowed an understanding of why loss-of-function (LoF) mutations in the Poly(A) ribonuclease (PARN) can cause DC and idiopathic pulmonary fibrosis [14–16]. PARN removes the oligo(A) tails on the hTR 3' end which are added by PAPD5, thereby limiting its accessibility to EXOSC10 and/or the nuclear exosome (Figure 2) [11–13]. Further, analysis of hTR 3' ends, and sub cellular localization and function suggests that PARN is not required for hTR maturation, since a co-knockdown of EXOSC10 and PARN rescues hTR levels and telomerase activity in PARN-deficient HeLa cells [11]. Overall, these studies propose a model where hTR levels in the cell are regulated by multiple pathways. While a tighter regulation of hTR levels might be necessary to prevent excessive telomerase activity, which could otherwise lead to uncontrolled proliferation of human cells leading to tumor formation, these pathways can also rapidly degrade RNA when hTR cannot be assembled into stable RNP molecules.
Spinal Muscular Atrophy and snRNA Degradation
A second example of an RNP hypo-assembly disease is Spinal muscular atrophy (SMA). SMA is an autosomal recessive disease caused by mutations in the SMN1 gene, which is the principal coding gene for the Survival Motor Neuron (SMN) protein in humans [17]. SMN is an assembly factor for the binding of the Sm complex to the conserved Sm site on snRNAs in humans, which is an important step in the maturation and trafficking of snRNAs in the cell [18]. Humans also have a second copy of the SMN coding gene, SMN2, but a single point mutation in exon 7 leads to mis-splicing of the transcribed mRNA and production of an unstable truncated isoform of the SMN protein [19, 20]. SMA patients with LoF alleles in the SMN1 gene have reduced levels of SMN protein, and severity of the disease and survival are directly correlated to the number of copies of the compensatory SMN2 gene.
SMA mouse and Drosophila models, as well as tissue culture experiments have shown that reduction in the SMN protein leads to a non-uniform decrease in the levels of snRNAs [21–24]. Moreover, several observations suggest that snRNAs are degraded rapidly when they are unable to assemble into snRNPs. Mutations in the Sm site of budding yeast snRNAs, which do not express a SMN ortholog but assemble the snRNAs into snRNPs, lead to reduced steady-state levels and an increase in snRNA rate of decay [24]. Similarly, in human cells, knockdown of SMN or the SMN complex components reduces the levels of snRNAs [22, 24, 25].
Examination of mammalian snRNAs that are defective in the Sm protein binding, either due to mutations in the Sm binding site or in models of of SMN knockdown, have revealed that these mutant snRNAs are degraded in the cytoplasm by DCP2 and XRN1 (Figure 3)[24]. These findings are consistent with the cytoplasmic assembly of snRNPs in mammalian cells, followed by cap hypermethylation and nuclear import [26]. Importantly, inhibition of snRNA decapping through siRNA-mediated DCP2 knockdown in SMN knockdown mouse cells, can rescue splicing defects observed for U12-dependent introns in some mRNAs [23, 24]. This is consistent with a model where if SMN is limiting, sufficient numbers of snRNAs are degraded by Dcp2/Xrn1 to alter splicing, and inhibition of decapping by Dcp2 knockdown can alter the kinetic competition to at least partially restore snRNP assembly.
Figure 3. snRNP Biogenesis Competes with Programmed RNA Quality Control Pathways.
Under normal conditions, snRNAs are transcribed and exported to the cytoplasm, where the SMN protein as a part of the SMN complex promotes the binding of Sm complex to the Sm site on snRNAs. Following Sm complex binding, the 5’ cap of snRNAs is modified to m2,2,7Gpp, and snRNPs are re-imported to the nucleus for final maturation. When SMN levels are reduced, snRNAs are degraded in the cytoplasm by DCP2/XRN1 when Sm complex assembly is compromised, as observed in patients suffering from a severe form of SMA.
A recent study has suggested that quality control of snRNAs in the cytoplasm could also be triggered by bacterial infections such as Shigella sp., Listeria sp. and Salmonella sp. in human cell lines, causing a decrease in the levels of snRNAs and sequestering snRNAs in cytoplasmic foci defined as U-bodies [27]. Therefore, the snRNA quality control pathway in the cytoplasm seems to be a general tunable mechanism regulating snRNA availability in the cell in response to bacterial stress.
Two broader conclusions can be drawn from work on SMA and DC. First, in both diseases, specific RNA quality control pathways out-compete defective RNP assembly leading to RNA degradation, which could contribute to a pathological state when hTR RNP assembly is decreased or when SMN levels are reduced in the cell. Second, inhibition of RNA decay pathways rescues RNA levels, and more importantly, the function of the RNP, at least in model systems. How effective these strategies will be in treating animal models or possibly humans, rather than in cultured cells, remains to be seen. This work raises the possibility that there are other RNP or mRNP hypo-assembly diseases where RNA quality control pathways are responsible for the disease pathology.
RNP Hypo-assembly Diseases Include Pathologies of ncRNAs
Examination of the literature suggests other examples where defects in assembly of an RNP lead to degradation of the RNA thereby leading to a pathological condition (Table 1A). For example, mutations in the RMRP gene, which encodes the RNA component of the MRP ribonuclease (thought to function in rRNA biogenesis) [28–30], have been shown to lead to the human developmental disease Cartilage Hair Hypoplasia (CHH), which is characterized by sparse hair, skeletal abnormalities, immune deficiency and increased risk of cancer [31]. Interestingly, some of the RMRP mutations in the RNA itself and not the promoter region, lead to reduced levels and stability of the MRP RNA in cells derived from patients, as well as in cell culture models, suggesting that defects in RNP assembly can lead to degradation of the MRP RNA by as yet unknown pathways [32, 33]. Consequently, insufficient levels of MRP affect its normal function in yeast, mouse model and patient leukocyte cells [32–34]. Similarly, mutations in the U4atac snRNA coding gene have been shown to be the cause of the rare developmental disease Microcephalic Osteodysplastic Pulmonary Dwarfism Type 1 (MOPD 1) [35, 36]. One of the mutations adjacent to the Sm site has been shown to reduce the levels of the U4atac snRNA when expressed exogenously in a model rodent cell line, presumably due to a competing RNA decay mechanism, which could lead to defects in splicing dependent on the U12-spliceosome machinery, which the U4atac snRNP is a part of [37].
Table 1.
| A List of Possible hypo-ncRNP Assembly Diseases | ||
|---|---|---|
| Gene | Disease association | Effect on ncRNA |
| DKC1/TERC/PARN | Dykseratosis Congenita (DC) | Reduction in hTR levels [16, 117, 118] |
| SMN1 | Spinal Muscular Atrophy (SMA) | Non-uniform reduction in snRNA levels [17] |
| RMRP | Cartilage hair Hypoplasia (CHH) | Reduction in MRP RNA levels [31, 33] |
| RNU4atac | Microcephalic Osteodysplastic Primordial Dwarfism 1 (MOPD1) | Reduction in U4atac RNA levels [37] |
| B List of Possible hypo-mRNP Assembly Diseases | ||
| Gene | Disease association | mRNA mutation |
| HBA2 | Constant Spring α thalassemia | UAA-CAA mutation of the stop codon [38] |
| FKTN | Fukuyama congenital muscular dystrophy | Retrotransposon insertion in 3’ UTR of FKTN mRNA [41] |
| SEPN1 | Rigid spine muscular dystrophy | T-C or G-A mutation in the 3’ UTR [40, 119] |
| LRRK2 | Parkinson’s disease | SNP in 3’ UTR [42] |
Hypo-assembly Diseases of mRNPs
mRNA destabilization due to a loss of RNP formation has similarly been implicated in human disease (Table 1B). One example of this is Constant Spring thalassemia, where a mutation in the stop codon substituting it for a Gln codon (UAA to CAA) leads to the ribosome moving into the 3' UTR of the α-globin mRNA [38]. This leads to the removal of the α-globin mRNA stability complex from the 3' UTR of the mRNA, which normally stabilizes the mRNA during erythrocyte development [39]. As a result, patients suffering from Constant Spring α thalassemia exhibit reduced levels and stability of the α globin mRNA in red blood cells, resulting in hemoglobin deficiency and thalassemia. Similarly, a mutation in the SECIS element in the 3' UTR of SEPN1 mRNA, which codes for Selenoprotein N, leads to loss of SBP2 protein binding to the mRNA, and in the absence of this bound protein, SEPN1 mRNA is rapidly degraded, giving rise to a mild form of rigid spine muscular dystrophy [40]. Another example of disease caused by hypo-assembly of an mRNP is the retrotransposon insertion in the 3’ UTR of the FKTN mRNA, which codes for the Fukutin protein whose function is not well understood, in Fukuyama congenital muscular dystrophy. The retrotransposon insertion leads to a strong reduction in the levels of the FKTN mRNA by an unknown mechanism [41]. Similarly, a SNP in the 3’ UTR of the LRRK2 mRNA leads to a strong reduction in the levels of the LRRK2 mRNA in a specific region of the brain in patients suffering from Parkinson’s disease, increasing the risk of disease onset [42]. One anticipates that a growing number of disease-related mutations in non-coding regions of mRNAs will lead to some form or other of pathology, due to alterations in RNP assembly, which in some cases (e.g. Constant Spring α thalassemia), may even trigger competing RNA decay pathways. The effect of mutation(s) on mRNA or protein levels need not be large, as small reductions in the levels of a critical RNP component could be sufficient to reduce the rate of RNP formation and elevate RNA decay, also considering that RNP assembly in the cell is highly stochastic and varies for individual RNAs.
Hyper-assembly Diseases of RNPs
There are two general classes of known scenarios where excessive RNP assembly can lead to a variety of human diseases. First, mutations that lead to either hyper-assembly, or persistence, of stress granules can contribute to a spectrum of multisystem neurodegenerative diseases, including ALS, FTLD, some muscle myopathies, as well as specific cancers. Second, expression of mRNAs containing aberrantly long repeat expansions can lead to the formation of large RNP assemblies that sequester RNA binding proteins thereby altering neuronal and muscular cellular metabolism, contributing to the pathogenesis of myotonic dystrophy type 1 and 2, and potentially ALS. The common theme of "RNP hyper-assembly diseases" is the aberrant formation, or persistence, of a large RNP particle, which then causes toxic consequences for various cells by numerous mechanisms.
Hyper-assembly of Stress Granules and Disease
Mutations that increase stress granule formation, or limit their clearance, have been implicated in several diseases, including ALS, FTLD, and some cancers. Stress granules are concentrated large RNA-protein assemblies containing non-translating mRNAs, some RNA binding proteins and translation initiation factors, as well as a number of non-RNA binding proteins, including proteins involved in signaling pathways [43]. Although stress granules are often observed during stress responses, when pools of non-translating mRNAs increase, related assemblies exist in neurons and during early development [44, 45]. Stress granules assemble through a variety of protein-protein interactions, including intrinsically disordered regions of proteins, some of which are "prion-like domains", predicted to have a high probability of forming β-amyloid like structures [46]. The high concentration prion-like domains in stress granules has been proposed to lead to increased rates of β-amyloid initiation within stress granules, and therefore excess stress granule formation or persistence might trigger disease. Consistent with this hypothesis, patients with ALS or FTLD often accumulate RNP aggregates in affected tissues that share components of stress granules [46].
One class of mutations that increase stress granule formation are mutations in RNA binding proteins such as TDP-43, FUS, Atx2, and hnRNPA1 in a spectrum of multisystem neurodegenerative diseases (Figure 4). Moreover, a wide range of abundant RNA binding proteins with prion-like domains have been linked to neurodegenerative diseases [47]. In several cases, when pathogenic mutations have been examined in model systems or in vitro, the mutations are seen to increase the formation of stress granules, or some related aggregates, and promote self-assembly into β-amyloid like fibers [5, 48]. For example, pathogenic mutations in TDP-43 are found in its Gly-rich domain or the unstructured C-terminus domain, and have been shown to lead to aggregate formation in human cell culture models, and to increase β-amyloid formation in vitro [49, 50]. Similarly, hnRNPA1 and hnRNPA2B1 mutations in their prion-like domains lead to the formation of cytoplasmic aggregates that also contain TDP-43, and in vitro form β-amyloids more efficiently than the wild-type protein [5]. Disease causing mutations in Atx2 are typically polyQ expansions, which when short in length, increase the risk of ALS [51], possibly due to their interaction with TDP-43, and when further expanded can lead to spinocerebellar ataxia type 2 (SCA2) [52, 53]. Since Atx2 is a key component of stress granule and neuronal granule assembly in mammals, Drosophila and yeast [54–56], a reasonable hypothesis is that expansion of the polyQ domain increases the aggregation of Atx2 and other interacting components of stress granules. In contrast, some pathogenic mutations in FUS are often found in a nuclear localization sequence, and these mutations may increase stress granule assembly by leading to an increased concentration of FUS in the cytosol [57].
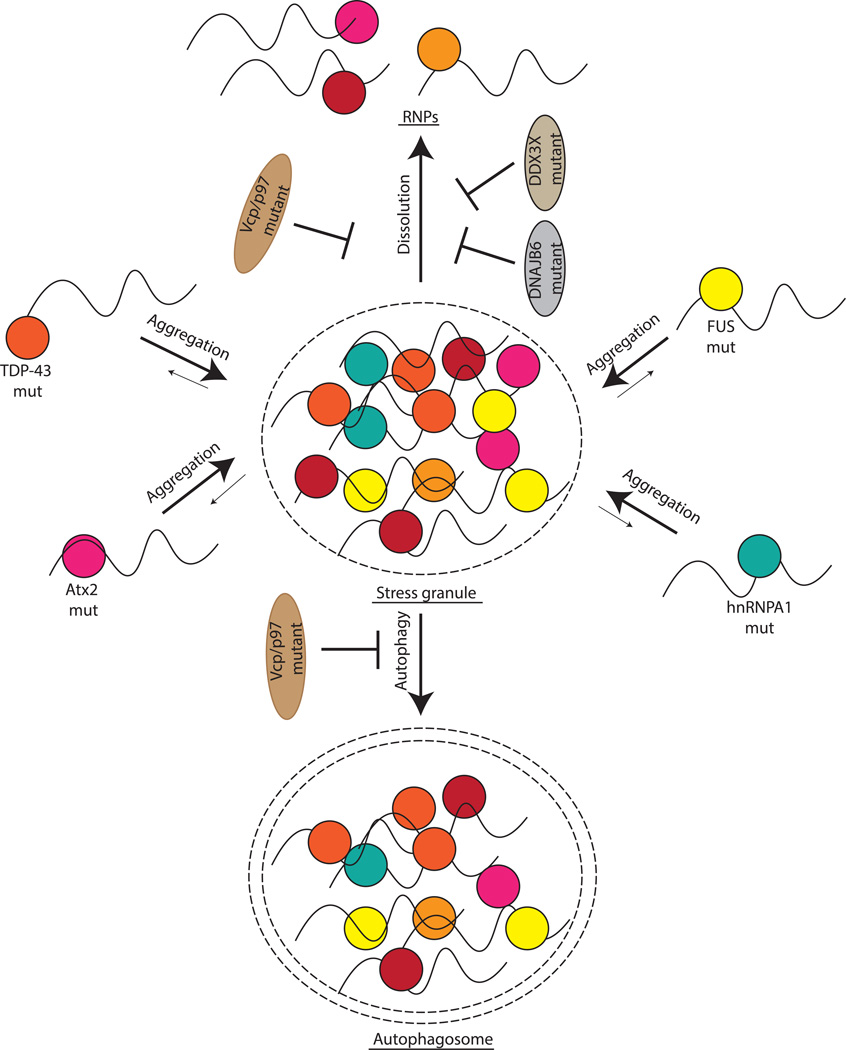
Figure 4. Formation and Persistence of Stress Granules Potentially Cause RNP Hyper-Assembly Diseases.
Disease-causing mutations in RNA binding proteins, such as TDP-43, FUS and hnRNPA1/A2B1, can lead to formation of aberrant RNP granules, which share components of stress granules. Similarly, mutations that lead to increased persistence of stress granules, such as VCP or DNAJB6, can also lead to a variety of human diseases.
Defects in cellular machinery for disassembly or clearance of stress granules have also been reported to contribute to similar diseases (Figure 4). For example, specific mutations in VCP/p97, a AAA-ATPase ubiquitin segregase that remodels protein complexes containing ubiquitinated proteins [58], have been found to lead to ALS, similar to hyper-assembly mutations in TDP-43, hnRNPA1, or hnRNPA2B1. These pathogenic mutations in VCP/p-have been shown to lead to constitutive stress granule formation in human tissue culture cells, and furthermore, the yeast ortholog of VCP, Cdc48, can affect clearance of stress granules by autophagy [59]. Hence, VCP/p97 appears to affect stress granule disassembly or clearance. Similarly, mutations in the heat shock HSP40 protein, DNAJB6, have been reported to lead to the human disease limb-girdle muscular dystrophy [60], and pathogenic mutations in DNAJB6 lead to the formation of stress granule-like assemblies in model systems [61, 62]. These findings suggest that DNAJB6 also plays a role in the disassembly/clearance of stress granules. Consistent with this model, the yeast ortholog of DNAJB6, Sis1, targets yeast stress granules for autophagy [63].
Aberrant formation or persistence of stress granules may also contribute to the etiology of a subtype of pediatric medulloblastoma. The key observation is that specific mutations in the DDX3X gene contribute to the Wnt-dependent subclass of pediatric medulloblastoma [64–66]. This may be connected to stress granule persistence since these cancer promoting mutations in DDX3X inhibit its ATPase activity [67]. Moreover, similar mutations in the yeast ortholog of DDX3X, Ded1, have been reported to lead to constitutive and larger stress granule formation [68].
Excessive stress granule formation and/or persistence is likely to contribute to disease by at least three overlapping mechanisms. First, the accumulation of the mutated protein in these large assemblies is likely to lead to a loss-of-function state for that protein, leading to alteration in that protein’s function. For example, mutations in TDP-43 leading to its accumulation in aggregates result in splicing alterations [69]. Second, the formation of aberrant stress granules is likely to sequester other RNA binding proteins, as well as mRNAs and microRNAs (miRNAs), which could lead to a wide range of aberrant aspects of RNA regulation. Finally, since many stress granule components are involved in signaling pathways, the sequestration of such components may very likely inhibit or enhance various signaling pathways [70], with deleterious downstream consequences for cell viability or replication.
Repeat-containing Toxic RNAs and RNP Hyper-assembly
In some cases, the expression of a repeat-containing mRNA can lead to hyper-assembly of RNPs with toxic consequences (Figure 5A and B). The best understood example of this phenomenon occurs in the expansion of CUG repeats in the DMPK mRNA and CCUG repeat in the CNBP mRNA, which are the genetic causes of Myotonic Dystrophy Type 1 & 2, respectively [71–74]. The expansion of the CUG repeat in patients leads to the sequestration of a number of RNA binding proteins in nuclear RNP foci, including Muscleblind-like proteins (MBNL), and hnRNP H (Figure 5A) [75–77]. These nuclear RNA foci are believed to cause disease by a gain-of-function (GoF) mechanism; sequestration of proteins, most prominently MBNL proteins, has been found to lead to defects in alternative splicing in mouse models and human cell lines [78, 79]. Similarly, misregulation of CUGBP1-associated alternative splicing due to hyperphosphorylation of CUGBP1 has been observed in human cell lines, patient fibroblasts and mouse model expressing the CUG repeat expansion [80].
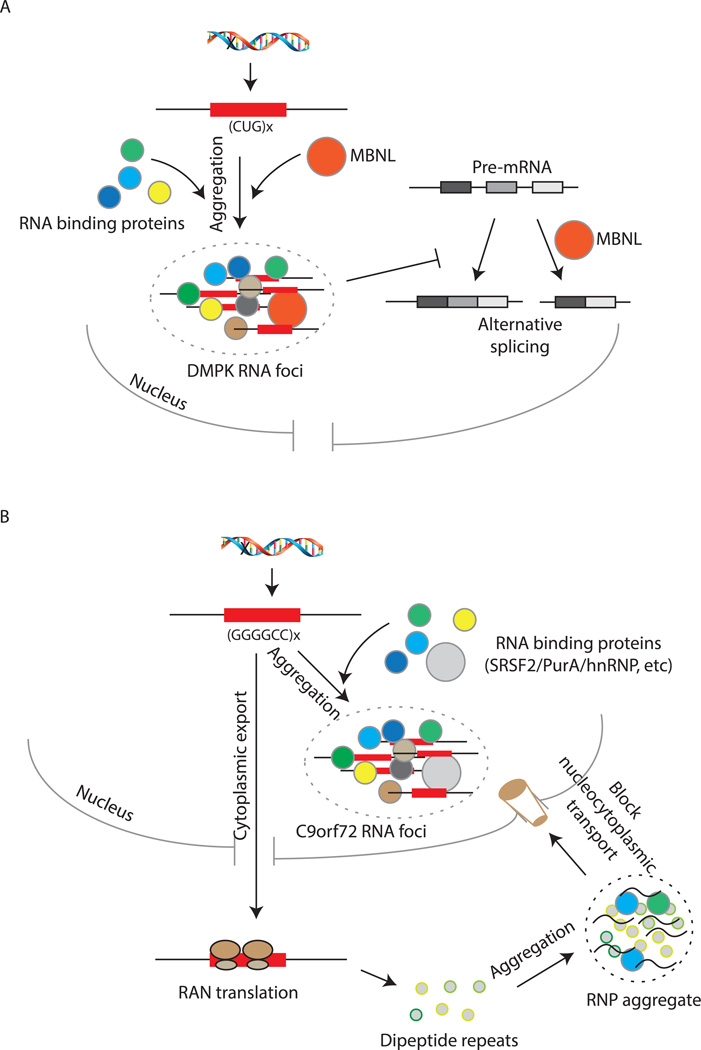
Figure 5. Repeat Expansion in mRNAs as a Cause of RNP Hyper-Assembly Diseases.
A) CUG repeat expansion in the DMPK mRNA can sequester other RNA-binding proteins like MBNL in RNA foci, and inhibit alternative splicing of pre-mRNAs leading to disease pathology. B) G4C2 repeat expansion in the C9orf72 mRNA could sequester RNA binding proteins in nuclear foci, or can be translated into dipeptide repeats that can also aggregate with other RNA-protein complexes in the cytoplasm. These cytoplasmic aggregates could possibly inhibit nucleo-cytoplasmic transport of cellular components.
The G4C2 expansion in the C9orf72 mRNA observed in ALS/FTD may represent another RNP-hyper assembly disease, although the toxic consequences of this expansion are complex. The C9orf72 mRNA expansion is the most common genetic cause of sporadic and familial ALS in the world, with patients presenting 100–2000 repeats in the first intron of the C9orf72 gene [81, 82]. The expanded repeat sequence also accumulates in nuclear RNA foci, at least in patient brain samples, and may sequester key RNA binding proteins through a hyper-assembly mechanism (Figure 5B) [83, 84]. Consistent with that possibility, the C9orf72 expansion binds to a subset of RNA binding proteins including Nucleolin, hnRNPH1, hnRNPF, SRSF2 and hnRNPA1 in brain and spinal cord samples from ALS patients, as well as human cell lines [85, 86]. However, whether such sequestration of RNA binding proteins in nuclear foci contributes to disease remains to be determined. This is an unresolved issue and the toxicity of C9orf72 may be complex since this expansion can cause multiple perturbations, ranging from LoF of the C9orf72 protein, to repeat-associated non-ATG (RAN) translation of the repeat, the latter producing dipeptide repeats in both sense and antisense directions [87–92]. Several studies have also attempted to converge the different cellular effects of hexanucleotide expansion to a defined molecular cascade contributing to disease. As such, it has been suggested that the formation of RNA foci and protein aggregates are able to affect regular cellular functions in neuronal cells, manifesting in defects such as nucleolar stress and aberrant nucleo-cytoplasmic transport of cellular components (Figure 5B) [93–96].
CUG expansions have also been reported in the ATXN8OS mRNA, which causes Spinocerebellar Ataxia Type 8 [97, 98]. This repeat also sequesters RNA binding proteins, and can undergo RAN translation in human cell lines [99–101]. Therefore, it may also lead to toxicity by multiple mechanisms including a hyper-assembly of RNPs. Taken together, repeat expansions in RNAs can sequester other RNA binding proteins and lead to RNP hyper-assembly, or can undergo RAN translation which can cause peptide aggregation and cellular toxicity. Which of these mechanisms can be targeted for therapy needs to be further investigated.
Discovery of Possible Treatments for RNP Assembly Diseases
One possible way to treat RNP hypo-assembly diseases will be to inhibit the competing RNA decay pathways. Although many RNA decay enzymes are important for organismal viability, two reasons suggest this might be a viable option. First, RNA decay pathways are often redundant, and inhibiting one pathway may not impact general viability [102]. For example, hypomorphic mice where DCP2 protein levels have been reduced to ~10% of wild-type levels are still viable [103]. This viability could be due to redundancy in decapping enzymes, since mammalian cells express multiple other enzymes with decapping activity, including the enzyme NUDT16 [104–106]. Second, for many nucleases, there are specific co-factors for various substrates; thus targeting a key co-factor for a general nuclease may result in a more specific mode of RNA decay inhibition. Interestingly, the general splicing inhibitor, Isoginkgetin, has shown a similar effect as the knockdown of the exosome on hTR processing, leading to longer oligo(A) tails and more oligoadenylated hTR precursor species in HeLa cells, suggesting that it might also potentially block exosome-mediated degradation [12]. Importantly, although warranting further investigation, the identification of compounds capable of targeting specific RNA decay mechanisms may provide useful therapeutic leads to treat some of these diseases.
Treatment of RNP hyper-assembly diseases may have more options. Since many of these diseases are due to dominant expression of a mutated protein or toxic RNA, antisense oligonucleotides (ASOs) or siRNA based therapies to reduce expression of the pathogenic RNA molecule are reasonable. For example, ASOs have also been developed as a means to target RNAs involved in a number of diseases, most prominently myotonic dystrophy (DM1), amyotrophic lateral sclerosis (ALS) and Huntington's disease, and have shown promise in mouse models and human cell lines [107–111]. Alternatively, in cases where the consequence of a pathogenic mutation is aberrant stress granule formation, small molecules that disrupt stress granule formation or decrease their persistence may be effective treatments.
One way to decrease stress granule persistence is to identify compounds that promote stress granule dissolution or clearance from patient cells. For example, Aquinnah Pharmaceuticals is trying to develop proprietary compounds that clear stress granules from cells (http://aquinnahpharma.com). Similarly, upregulation of autophagy, which has been shown to promote stress granule clearance from cells, could represent a viable therapeutic strategy. Interestingly, compounds such as rapamycin or carbamazepine are known promoters of autophagy in human cells and have been FDA-approved for the treatment of a number of different diseases such as prevention of kidney transplant rejection and epilepsy. However, of note, there have also been some contrasting results regarding the benefit of rapamycin treatment on animal survival in mouse models of ALS [112, 113], and the effect of rapamycin on disease outcome seems to depend on the type of model used. Consequently, there is a pressing need for further preclinical exploration of rapamycin in other mouse models as well as in patient cells. A key issue in these cases will be to achieve a better understanding of how stress granule persistence contributes to pathology in specific diseases. One appealing possibility is that aberrant stress granules exert toxicity by hijacking signaling pathways, and given the number of drugs that currently target cell signaling, some of these might putatively rescue the effect of aberrant stress granules.
Finally, for cases where a specific protein-RNA interaction drives aberrant RNP formation, compounds that directly target given interactions may be of promising therapeutic outlook. For example, rational design of small molecules that are capable of binding to RNA secondary structures with high affinity have so far been shown to target the CUG-repeat in DM1 and the CCUG-repeat in DM2 in a mouse cell line, and to disrupt the binding of MBNL proteins to the CUG repeat, thereby correcting splicing defects and reduce RNA toxicity [114–116].
Concluding Remarks
Based on current evidence of misregulated RNP assembly in several human diseases, including SMA, DC and ALS, it will be important to understand not only the molecular mechanisms behind disease onset, but also the changes in the cellular function that occur during disease progression. This will facilitate the characterization of the most optimal therapeutic approaches targeting RNP assembly, as well as the temporal windows in which they might be used to treat some of the pathologies discussed here (see Outstanding Questions). An important step in this process will undoubtedly be the identification of appropriate model systems that best recapitulate/mimic the appearance and progression of given diseases in patients. Complementary in vitro, in vivo, and ex vivo (e.g. patient cells) model systems constitute important components allowing a proper validation of experimental hypotheses regarding aberrant RNP assembly and disease. As such, placing the most recent data on RNP assembly in this context will be helpful in explaining the often conflicting results obtained from different studies on one given pathology, as in the case of ALS. Future research should also focus on translating ASO and small molecule treatments discovered in cell lines to clinical trials as quickly as possible, as there are few to none treatment options available for most of these devastating diseases.
Outstanding questions.
SMN is expressed ubiquitously and is an abundant protein in most cells of the human body. SMN has been proposed to play an integral role in pre-mRNA splicing, mRNA transcription and transport of RNAs in neurons. What exactly is the mechanism behind the onset of Spinal Muscular Atrophy, and why are motor neurons specifically affected by SMN deficiency?
In multisystem neurodegenerative spectrum disorders such as ALS and FTLD, mutated RNA binding proteins are often ubiquitously expressed in the human body. What makes neurons particularly vulnerable to the formation of persistent RNP granules? What are the consequences of persistent RNP granule formation and how can this persistence be alleviated?
C9orf72 repeat expansion has been proposed to cause disease onset by various possible mechanisms including gain of function of the repeat containing RNA or loss of function of the encoded protein. How do cellular changes originate in the presence of the expansion, and are one or all of these putative changes important for the disease onset?
Mutations preventing RNP granule clearance can also cause a wide variety of human diseases. Since RNP granules are typically cleared by a number of redundant mechanisms in human cells, why do RNP granules continue to persist in a disease state, and more importantly, can other RNP granule clearance pathways be therapeutically modulated to clear these aggregates in affected individuals?
Trends.
Mutations in RNP assembly components can lead to RNP hypo-assembly diseases by triggering RNA decay.
Prevention of RNA decay could be a viable therapeutic strategy for RNP hypo-assembly diseases.
Mutation in RNA binding proteins lead to RNP hyper-assembly and formation of persistent stress granules.
Persistence of stress granules due to loss of disassembly pathways could also lead to the same family of RNP hyper-assembly diseases.
Repeat expansions in RNAs could lead to toxicity through multiple potential mechanisms.
Therapeutic strategies for RNP hyper-assembly diseases include prevention of stress granule persistence and modulation of cellular signaling pathways, as well as targeted disruption of constituent RNA-protein interactions.
Acknowledgments
We would like to thank Anne Webb for her help with illustrations used in this manuscript. This work was supported by funds from NIH R01 GM45443 to R.P. R.P. is an investigator of the Howard Hughes Medical Institute.
Glossary
- Ribonucleoproteins (RNPs)
Particles consisting of RNAs bound to their binding partner proteins; interactions which are often mediated by other protein chaperones. A single mRNA can form many different RNPs depending on the different permutations of RNA binding proteins bound to it, and can form bigger RNPs when those bound proteins in turn, interact with other protein partners.
- Spinal Muscular Atrophy
Genetic disease characterized by failure of lower motor neurons, leading to loss of muscle control and eventual respiratory problems. Severity ranges from fatal to mild depending on the number of SMN2 copies in the patient’s genome.
- Dyskeratosis Congenita
Rare human disease characterized by loss of self-renewal capacity of stem cells, eventually leading to bone marrow failure.
- Non-coding RNAs
RNAs that are not translated and do not produce protein.
- Constant Spring α thalassemia
Rare form of hemoglobin deficiency leading to moderate to severe anemia.
- RNP granules
Higher order assemblies of RNPs in the cell, characterized by microscopically visible non-membranous foci.
- snRNPs
Small nuclear ribonucleoproteins consisting of U-rich snRNAs and Sm proteins, along with other proteins specific to each individual snRNP. Important for pre-mRNA splicing.
- snoRNPs
Small nucleolar ribonucleoproteins consisting of box C/D or box H/ACA snoRNAs and a specific set of binding proteins. They carry out post-transcriptional pseudouridylation or 2’-methylation of tRNA, rRNAs and snRNAs.
- Telomerase RNP
Particle consisting of telomerase RNA bound to H/ACA snoRNP proteins and TCAB1, which associates with the telomerase reverse transcriptase enzyme TERT in a cell cycle-dependent manner, and extends telomeres. Only active in stem cells and germline cells in healthy controls, and activated in most forms of cancers.
- Cajal bodies
Non-membranous nuclear foci consisting of scaRNAs, snRNAs and telomerase RNA, characterized by the concentration of the Coilin protein.
- P-bodies
Non-membranous cytoplasmic foci consisting of RNA decay enzymes. Believed to be the site of mRNA storage and turnover in eukaryotic cells.
- Amyotrophic Lateral Sclerosis
Human disease of upper and lower motor neurons characterized by loss of muscle function, rapid progression from onset, leading to death.
- Multisystem proteinopathy
Inherited pleiotropic disease, symptoms of which can be ALS and inclusion body myopathy, among others. Affects motor neurons as well as bone function in patients.
- Myotonic Dystrophy Type 1 and 2
Disease characterized by progressive muscle wasting and weakness in patients, eventually causing eye and hormonal defects. Type 1 is generally more severe than type 2.
- PAPD5-mediated oligoadenylation
Non-templated post-transcriptional addition of 1–20 A’s at the 3’ end of RNAs by the PAPD5 polymerase.
- Nuclear exosome
Protein complex comprising 10 core proteins including the exo/endonuclease DIS3, along with the exonuclease EXOSC10. Involved in 3’ to 5’ degradation of a number of nuclear and cytoplasmic RNAs in the cell.
- DCP2-mediated decapping
Removal of the 5’ m7Gpp cap structure from RNA polymerase II-transcribed mRNAs and ncRNAs by the hydrolase DCP2, leaving a 5’ p on the RNA. Necessary for the eventual degradation of the RNA in a 5’ to 3’ manner by the exonuclease XRN1.
- Idiopathic pulmonary fibrosis
Disease characterized by the formation of scar tissue in lungs. Lung function decreases over time, leading to fatality.
- Sm complex
A complex of seven small proteins (Sm B/B’, D1, D2, D3, E, F, G) that form a hetero-oligomeric ring around the Sm binding site on snRNAs.
- U12-dependent introns
A sub-class of mammalian introns (<1%) that are removed by the U11–U12 snRNPs as opposed to the canonical U1-U2 dependent splicing.
- U-bodies
Cytoplasmic non-membranous foci consisting of snRNAs and SMN complex proteins, believed to be the site of snRNP formation and snRNA quality control.
- MRP ribonuclease
RNP consisting of the MRP RNA and ten protein partners. The RNP catalyzes the cleavage of rRNA precursors.
- Cartilage Hair Hypoplasia
Rare disease characterized by sparse hair, skeletal abnormalities and immunodeficiency.
- MOPD1
Rare developmental disease characterized by extremely short stature, microcephaly and stunted growth, leading to fatality in early childhood.
- Rigid spine muscular dystrophy
Disease characterized by loss of spine flexibility, with males generally more affected.
- Fukuyama congenital muscular dystrophy
Rare form of muscular dystrophy common in Japan, characterized by muscle weakness and wasting. Also affects brain development and function, often leading to death in childhood.
- Spinocerebellar Ataxia Type 2
Muscle disorder characterized by loss of balance, co-ordination and movement, progressing to memory problems and often leading to death.
- Spinocerebellar Ataxia Type 8
Overlapping symptoms with SCA2, caused by a different mutation than SCA2.
- Limb-girdle muscular dystrophy
Disease characterized by weakness and wasting of muscles closest to the body, including those in the shoulders, upper arms and thighs.
- Wnt-dependent subclass of pediatric medulloblastoma
A rare class of brain tumor primarily affecting children, and caused by the over-activation of the Wnt signaling pathway. Overall prognosis and survival options are much better than in other classes of medulloblastomas.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singh G, et al. The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu. Rev. Biochem. 2015;84:325–354. doi: 10.1146/annurev-biochem-080111-092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Müller-McNicoll M, Neugebauer KM. How cells get the message: dynamic assembly and function of mRNA–protein complexes. Nature Reviews Genetics. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 3.Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renton AE, et al. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meola G, Cardani R. Myotonic dystrophies: An update on clinical aspects, genetic, pathology, and molecular pathomechanisms. BBA - Molecular Basis of Disease. 2015;1852:594–606. doi: 10.1016/j.bbadis.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 7.Armanios M, Blackburn EH. The telomere syndromes. Nature Reviews Genetics. 2012 doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason PJ, Bessler M. The genetics of dyskeratosis congenita. Cancer Genetics. 2011;204:635–645. doi: 10.1016/j.cancergen.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong JMY, Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes & Development. 2006;20:2848–2858. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon DH, et al. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nature Genetics. 2015;47:1482–1488. doi: 10.1038/ng.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shukla S, et al. Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nature Structural & Molecular Biology. 2016;23:286–292. doi: 10.1038/nsmb.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tseng C-K, et al. Human Telomerase RNA Processing and Quality Control. Cell Reports. 2015 doi: 10.1016/j.celrep.2015.10.075. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen D, et al. A Polyadenylation-Dependent 3′ End Maturation Pathway Is Required for the Synthesis of the Human Telomerase RNA. Cell Reports. 2015;13:2244–2257. doi: 10.1016/j.celrep.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Dhanraj S, et al. Bone marrow failure and developmental delay caused by mutations in poly(A)-specific ribonuclease (PARN) J Med Genet. 2015;52:738–748. doi: 10.1136/jmedgenet-2015-103292. [DOI] [PubMed] [Google Scholar]
- 15.Tummala H, et al. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Invest. 2015;125:2151–2160. doi: 10.1172/JCI78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuart BD, et al. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nature Genetics. 2015 doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 18.Pellizzoni L, et al. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 19.Lorson CL, et al. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proceedings of the National Academy of Sciences. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes & Development. 2010;24:438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, et al. SMN Deficiency Causes Tissue-Specific Perturbations in the Repertoire of snRNAs and Widespread Defects in Splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabanella F, et al. Ribonucleoprotein Assembly Defects Correlate with Spinal Muscular Atrophy Severity and Preferentially Affect a Subset of Spliceosomal snRNPs. PLoS ONE. 2007;2:e921–e912. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lotti F, et al. An SMN-Dependent U12 Splicing Event Essential for Motor Circuit Function. Cell. 2012;151:440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shukla S, Parker R. Quality control of assembly-defective U1 snRNAs by decapping and 5“-to-3” exonucleolytic digestion. Proceedings of the National Academy of Sciences. 2014;111:E3277–E3286. doi: 10.1073/pnas.1412614111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winkler C. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes & Development. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer U, et al. Biogenesis of spliceosomal small nuclear ribonucleoproteins. WIREs RNA. 2011;2:718–731. doi: 10.1002/wrna.87. [DOI] [PubMed] [Google Scholar]
- 27.Tsalikis J, et al. Intracellular Bacterial Pathogens Trigger the Formation of U Small Nuclear RNA Bodies (U Bodies) through Metabolic Stress Induction. J. Biol. Chem. 2015;290:20904–20918. doi: 10.1074/jbc.M115.659466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindahl L, et al. RNase MRP is required for entry of 35S precursor rRNA into the canonical processing pathway. RNA. 2009;15:1407–1416. doi: 10.1261/rna.1302909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu S, et al. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proceedings of the National Academy of Sciences. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lygerou Z, et al. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 31.Ridanpää M, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 32.Hermanns P, et al. Consequences of mutations in the non-coding RMRP RNA in cartilage-hair hypoplasia. Human Molecular Genetics. 2005;14:3723–3740. doi: 10.1093/hmg/ddi403. [DOI] [PubMed] [Google Scholar]
- 33.Nakashima E, et al. Cartilage hair hypoplasia mutations that lead to RMRP promoter inefficiency or RNA transcript instability. Am. J. Med. Genet. 2007;143:2675–2681. doi: 10.1002/ajmg.a.32053. [DOI] [PubMed] [Google Scholar]
- 34.Huang W, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517–522. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.He H, et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science. 2011;332:238–240. doi: 10.1126/science.1200587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edery P, et al. Association of TALS Developmental Disorder with Defect in Minor Splicing Component U4atac snRNA. Science. 2011;332:240–243. doi: 10.1126/science.1202205. [DOI] [PubMed] [Google Scholar]
- 37.Jafarifar F, et al. Biochemical defects in minor spliceosome function in the developmental disorder MOPD I. RNA. 2014;20:1078–1089. doi: 10.1261/rna.045187.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waggoner SA, Liebhaber SA. Regulation of alpha-globin mRNA stability. Exp. Biol. Med. (Maywood) 2003;228:387–395. doi: 10.1177/153537020322800409. [DOI] [PubMed] [Google Scholar]
- 39.Weiss IM, Liebhaber SA. Erythroid cell-specific mRNA stability elements in the alpha 2-globin 3' nontranslated region. Molecular and Cellular Biology. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allamand V, et al. A single homozygous point mutation in a 3′untranslated region motif of selenoprotein N mRNA causes SEPN1-related myopathy. EMBO reports. 2006 doi: 10.1038/sj.embor.7400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi K, et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. [DOI] [PubMed] [Google Scholar]
- 42.Cardo LF, et al. The screening of the 3 “UTR sequence of LRRK2 identified an association between the rs66737902 polymorphism and Parkinson”s disease. J. Hum. Genet. 2014;59:346–348. doi: 10.1038/jhg.2014.26. [DOI] [PubMed] [Google Scholar]
- 43.Buchan JR, Parker R. Eukaryotic Stress Granules: The Ins and Outs of Translation. Molecular Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 45.Sengupta MS, Boag PR. Germ granules and the control of mRNA translation. IUBMB Life. 2012;64:586–594. doi: 10.1002/iub.1039. [DOI] [PubMed] [Google Scholar]
- 46.Li YR, et al. Stress granules as crucibles of ALS pathogenesis. The Journal of Cell Biology. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alberti S, et al. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo W, et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nature Structural & Molecular Biology. 2011;18:822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Igaz LM, et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J. Clin. Invest. 2011;121:726–738. doi: 10.1172/JCI44867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barmada SJ, et al. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imbert G, et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nature Genetics. 1996;14:285–291. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 53.Simón-Sánchez J, et al. Analysis of SCA-2 and SCA-3 repeats in Parkinsonism: evidence of SCA-2 expansion in a family with autosomal dominant Parkinson's disease. Neurosci. Lett. 2005;382:191–194. doi: 10.1016/j.neulet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 54.McCann C, et al. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E655–E662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nonhoff U, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchan JR, et al. P bodies promote stress granule assembly in Saccharomyces cerevisiae. The Journal of Cell Biology. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shelkovnikova TA, et al. Multistep process of FUS aggregation in the cell cytoplasm involves RNA-dependent and RNA-independent mechanisms. Human Molecular Genetics. 2014;23:5211–5226. doi: 10.1093/hmg/ddu243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson JO, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buchan JR, et al. Eukaryotic Stress Granules Are Cleared by Autophagy and Cdc48/VCP Function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Couthouis J, et al. Exome sequencing identifies a DNAJB6 mutation in a family with dominantly-inherited limb-girdle muscular dystrophy. Neuromuscul. Disord. 2014;24:431–435. doi: 10.1016/j.nmd.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bengoechea R, et al. Myofibrillar disruption and RNA-binding protein aggregation in a mouse model of limb-girdle muscular dystrophy 1D. Human Molecular Genetics. 2015;24:6588–6602. doi: 10.1093/hmg/ddv363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li S, et al. Genetic interaction of hnRNPA2B1 and DNAJB6 in a Drosophila model of multisystem proteinopathy. Human Molecular Genetics. 2016;25:936–950. doi: 10.1093/hmg/ddv627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walters RW, et al. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA. 2015;21:1660–1671. doi: 10.1261/rna.053116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones DTW, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Epling LB, et al. Cancer-associated mutants of RNA helicase DDX3X are defective in RNA-stimulated ATP hydrolysis. Journal of Molecular Biology. 2015;427:1779–1796. doi: 10.1016/j.jmb.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hilliker A, et al. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Molecular Cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arnold ES, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E736–E745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arimoto K, et al. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- 71.Liquori CL, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 72.Mahadevan M, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 73.Brook JD, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- 74.Fu YH, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- 75.Timchenko LT, et al. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Research. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Philips AV, et al. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 77.Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kanadia RN, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 79.Jiang H, et al. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Human Molecular Genetics. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 80.Kuyumcu-Martinez NM, et al. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Molecular Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mizielinska S, et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013;126:845–857. doi: 10.1007/s00401-013-1200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zu T, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cooper-Knock J, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137:2040–2051. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haeusler AR, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mizielinska S, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ash PEA, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Farg MA, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Human Molecular Genetics. 2014;23:3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kwon I, et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O'Rourke JG, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–1329. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y-J, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016 doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chew J, et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348:1151–1154. doi: 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Freibaum BD, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang K, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jovičić A, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koob MD, et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nature Genetics. 1999;21:379–384. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]
- 98.Day JW, et al. Spinocerebellar ataxia type 8: Clinical features in a large family. Neurology. 2000;55:649–657. doi: 10.1212/wnl.55.5.649. [DOI] [PubMed] [Google Scholar]
- 99.Moseley ML, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nature Genetics. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 100.Daughters RS, et al. RNA Gain-of-Function in Spinocerebellar Ataxia Type 8. PLoS Genet. 2009;5:e1000600–e1000613. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zu T, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U.S.A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Houseley J, Tollervey D. The Many Pathways of RNA Degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 103.Song M-G, et al. Multiple mRNA Decapping Enzymes in Mammalian Cells. Molecular Cell. 2010;40:423–432. doi: 10.1016/j.molcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Song MG, et al. Multiple Nudix family proteins possess mRNA decapping activity. RNA. 2013;19:390–399. doi: 10.1261/rna.037309.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiao X, et al. A Mammalian Pre-mRNA 5′ End Capping Quality Control Mechanism and an Unexpected Link of Capping to Pre-mRNA Processing. Molecular Cell. 2013;50:104–115. doi: 10.1016/j.molcel.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Y, et al. Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA. 2011;17:419–428. doi: 10.1261/rna.2439811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Miller TM, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hu J, et al. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wheeler TM, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lagier-Tourenne C, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Staats KA, et al. Rapamycin increases survival in ALS mice lacking mature lymphocytes. Mol Neurodegener. 2013;8:31. doi: 10.1186/1750-1326-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang X, et al. Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–425. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- 114.Childs-Disney JL, et al. Induction and reversal of myotonic dystrophy type 1 pre-mRNA splicing defects by small molecules. Nature Communications. 2013;4:2044. doi: 10.1038/ncomms3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rzuczek SG, et al. A toxic RNA catalyzes the in cellulo synthesis of its own inhibitor. Angew. Chem. Int. Ed. Engl. 2014;53:10956–10959. doi: 10.1002/anie.201406465. [DOI] [PubMed] [Google Scholar]
- 116.Childs-Disney JL, et al. Structure of the Myotonic Dystrophy Type 2 RNA and Designed Small Molecules That Reduce Toxicity. ACS Chem. Biol. 2014;9:538–550. doi: 10.1021/cb4007387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mitchell JR, et al. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 118.Vulliamy T, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 119.Moghadaszadeh B, et al. Mutations in SEPN1 cause congenital muscular dystrophy with spinal rigidity and restrictive respiratory syndrome. Nature Genetics. 2001;29:17–18. doi: 10.1038/ng713. [DOI] [PubMed] [Google Scholar]