Highlights
-
•
Muscle dissociation protocols yield cell suspensions with different composition.
-
•
A panel of antibodies discriminating different skeletal muscle cell populations.
-
•
Mass cytometry, a new approach to characterize heterogeneous muscle cell populations.
Abstract
Biological processes that are mediated by cell–cell interactions in heterogeneous populations are best approached by methods that have single cell resolution. Most of these methods rely on the preparation, from solid tissues, of cell suspensions by enzymatic digestion, followed by analysis of single cell reactivity to an antibody panel that allows the discrimination of cell populations and characterization of their activation state. Thus for any specific biological problem, both efficient and at the same time mild, protocols for cell separation, together with tissue specific panels of antibodies, need to be developed and optimized. Here we characterize an antibody panel that permits the discrimination of mononuclear muscle cell populations by mass cytometry and use it to characterize the cell populations obtained by three different cell extraction procedures from muscle fibers. We show that our panel of antibodies, albeit limited and incomplete, is sufficient to discriminate most of the mononuclear muscle cell populations and that each cell extraction method yields heterogeneous cell populations with a different relative abundance of the distinct cell types.
Introduction
Multiparametric single cell analysis is the method of choice for studying biological phenomena in heterogeneous cell samples. Traditional single cell approaches include fluorescence microscopy and flow cytometry. These technologies, however, are limited by the number of available fluorophores and by the overlap in their emission spectra. Thus, only a limited number of readouts can be measured simultaneously.
To overcome this problem, mass cytometry, a novel single cell technology, has been recently developed. Mass cytometry is a highly multi-parametric technology that enables probing of single cell events, by labelling cell surfaces and intracellular antigens with up to 40 antibodies tagged with stable heavy metal isotopes [1]. This technology exploits the possibility to label cells with antibodies as in flow cytometry, but it adds the spectral resolution of Time-of-Flight (TOF) mass spectrometry. Isotopes of the same element differing by a mass unit can be reliably distinguished [2]. In fact, the sharp mass peaks obtained by TOF inductively coupled plasma mass spectrometry eliminate the problems of spectral overlap typical of fluorescence based flow cytometry. Based on these characteristics, mass cytometry enables the detection and characterization of rare and heterogeneous cell populations, by measuring a large number of parameters at the single cell level. This type of analysis is relatively effortless when applied to liquid tissues, but since our interest was on skeletal muscle, we had to develop a specific protocol for mass cytometry analysis of a compact, solid tissue.
Adult skeletal muscle is a relatively complex tissue, which has the ability to self-renew and to self-repair in response to mechanical or chemical damage, stress caused by genetic mutations or increased workload. The regenerative process is orchestrated by different populations of resident mononuclear cells, which directly or indirectly contribute to maintain myofiber homeostasis. The process of myofiber regeneration can be studied ex vivo by co-cultivating mononuclear cells after releasing them by enzymatic digestion from the extracellular matrix surrounding the muscle fibers.
The preparation of single cell suspensions from complex tissues, such as muscle, requires the application of proteolytic digestions, in order to free cells from the connective compartment. The enzymes commonly used have a proteolytic activity directed against the collagen and the proteoglycans, components of the connective extracellular matrix. Different protocols, relying on different enzymatic activities, for digestion of the extracellular matrix yield different distributions of mononuclear cells. An ideal extraction method should efficiently free cells from muscle fibers and extracellular matrix while limiting the modification of their physiology and of the structures of the proteins that decorate their surface. The characterization of the cell populations that are yielded by different extraction methods is of fundamental importance in standardization and optimization of experiments. The different cell populations in the muscle are defined by the combinatorial expression of CD markers on the cell surface. The identification and the abundance of the muscle populations depend on the combinations of antibodies used [3], [4].
The main players in the process of muscle regeneration are satellite cells, a progenitor cell population that represents 2–5% of the sublaminar nuclei [5]. It is commonly accepted that the paired box transcription factor Pax7 is a specific marker for satellite cells [6], [7]. However, they can also be identified by a specific reactivity pattern when challenged with other antibodies. For instance, they are stained by antibodies against cell surface receptor α-7 integrin, while they do not react with antibodies against leukocyte common antigen CD45, endothelial marker CD31 and stem cell antigen 1, Sca1 [5].
In addition to satellite cells, a variety of skeletal muscle progenitor cells have been identified residing in the muscle interstitium or recruited from other compartments following injury. These cells are either endowed with myogenic potential or have accessory function during muscle regeneration. Other mononuclear cells include interstitial cell populations, that is, myogenic and endothelial cell progenitors identified in the interstitial space of murine skeletal muscle [8], vessel-associated stem cells [9] and bone marrow-derived stem cells [10], among others.
Fibro-adipogenic progenitors (FAPs) are an important class of muscle interstitial cell, with a fibrotic and adipogenic potential, which play a positive role in muscle regeneration, while also contributing to fibro-adipogenic degeneration of skeletal muscles [11]. This heterogeneous population is characterized by the expression of Sca1, a common interstitial marker; they are also negative for CD31, α-7 integrin and CD45. Recently, a new population characterized by the same expression markers as FAPs and located in the interstitium, has been described. This population expresses the cell stress mediator PW1 and is negative for other markers of muscle stem cells such as Pax7. For these reasons the population was named PICs, for ‘PW1+/Pax7− interstitial cells’. However, differently from FAPs, PICs have a myogenic potential in vitro and contribute directly to skeletal muscle regeneration [12]. Vessel-associated stem cells are heterogeneous multipotent progenitors of mesodermal origin, which display a diverse differentiation potential, thus contributing in different ways to muscle regeneration. They include mesoangioblasts [13], pericytes [14] and myo-endothelial cells [15], which are all characterized by a specific set of surface antigen markers. Finally, immune system cells play a pivotal role in muscle regeneration, since polymorphonuclear leukocytes and monocytes are responsible for the initial phases of the process and for the activation of committed progenitors [16], [17], [18].
The aim of this work was to assemble and characterize an antibody panel for the discrimination of muscle progenitor-cell populations by mass cytometry. This panel is used here to assess different mononuclear cell extraction protocols for studying the complexity of skeletal muscle tissue. In particular, we have designed a panel of eight antibodies, directed against antigens of relevance in myology. We show that this collection of antibodies, albeit limited and incomplete, is suitable for the identification of most of the main skeletal muscle populations by mass cytometry. Moreover, by comparing three different enzymatic extraction methods, we conclude that, according to the protocol utilized, the yields of the distinct cell types and their heterogeneity vary significantly.
Materials and methods
Antibody panel
From a literature search, a list of antigens was compiled that decorate the membrane of muscle mononuclear cell populations. We tested a large number of commercial antibodies. Those that showed sufficient reactivity and specificity after coupling to heavy metal isotopes are listed in Table 1. Other antibodies employed are listed in Table 2.
Table 1.
List of metal tagged antibodies used in the mass cytometry experiments.
| Antibody | Metal | Clone | Fluidigm Cat# | Population |
|---|---|---|---|---|
| Rat anti-mouse CD45 | 175 Lu | 30-F11 | 3175010B | Leukocytes (immune system cells) |
| Rat anti-mouse CD31 | 165 Ho | 390 | 3165013B | Endothelial and vessel-associated cells |
| Rat anti-mouse CD11b | 172 Yb | M1/70 | 3172012B | Granulocytes, monocytes and macrophages |
| Rat anti-mouse CD117 (c-kit) | 166 Er | 2B8 | 3166004B | Hematopoietic Stem Cells |
| Rat anti-mouse F4/80 | 146 Nd | BM8 | 3146008B | Macrophages (mature) |
| Rat anti-mouse Ly-6A/E (Sca-1) | 164 Dy | D7 | 3169015B | FAPs, PICs and endothelial cells |
| Rat anti-mouse TER-119 | 154 Sm | TER-119 | 3154005B | Erythroid cells |
| Mouse anti-PE | 156 Gd | PE001 | 3156005B | * |
The PE001 secondary antibody clone reacts with phycoerythrin (PE), which is conjugated to anti-mouse integrin α-7 primary antibody (Table 2) for the identification of satellite cells.
Table 2.
Primary antibodies.
| Antibody | Fluorophore | Clone | Source | Work dilution |
|---|---|---|---|---|
| Anti-mouse integrin α-7-PE | PE | 3C12 | Miltenyi Biotec | 1:100 (CyTOF) |
| Anti-MF20 (MyHC) | – | MF 20 | DSHB | 1:2 (IF) |
CyTOF: mass cytometry; IF: immunofluorescence, PE: phycoerythrin.
Skeletal muscle mononuclear cells isolation
Skeletal muscle mononuclear cells were isolated from hind limb muscle of 20 days old C57BL/6 mice. Experiments on animals were conducted according to the rules of good animal experimentation I.A.C.U.C. no. 432 of March 12 2006. Muscular tissue was finely minced with tweezers until no intact muscle pieces could be distinguished. The minced tissue was then completely digested employing three different enzymatic digestion protocols. In the Dispase II/Collagenase A digestion method (protocol I), the tissue was digested in DPBS with calcium and magnesium (Gibco, Catalog #14040) containing 2 μg/ml Collagenase A (Roche), 2.4 U/ml Dispase II (Roche), 10 ng/ml DNase I (Roche), for 1 h at 37°C in gentle agitation. Collagenase A is a protease with specificity for the bond between a neutral amino acid (X) and glycine in the sequence Pro-X-Gly-Pro. This peptide sequence is found at high frequency in collagen. Collagenase A alone is inefficient in dissociating the muscle tissue and it is usually used in combination with Dispase II, a neutral protease, that hydrolyzes the N-terminal peptide bounds of non-polar amino acid residues. In the Collagenase II digestion method (protocol II), tissue was digested in DPBS with calcium and magnesium with Collagenase II (0.1 mg/ml, Gibco) and 10 ng/ml DNase I (Roche) for 1 h at 37°C in gentle agitation. Collagenase II acts as Collagenase A, recognizing the same amino acid sequences, but it shows a greater activity. For this reason, it is usually used without any other enzyme. In the Trypsin digestion method (protocol III), the tissue was digested in 4 ml of Trypsin EDTA (170,000 U/l, Lonza) and 10 ng/ml DNase I (Roche). Trypsin cleaves peptides on the C-terminal side of lysine and arginine amino acid residues. Nevertheless, the employment of crude Trypsin usually is associated to an incomplete solubility, lot-to-lot variability, cell toxicity and cell surface protein/receptor damage.
After sequential filtration through a 100 μm, 70 μm and 40 μm cell strainer and intermediate washing with Hank's Balanced Salt Solution with calcium and magnesium (HBSS Gibco, Catalog #14025-092) supplemented with 0.2% BSA and 1% Penicillin–Streptomycin (P/S, 10,000 U/ml), lysis of red blood cells with RBC Lysis Buffer (Santa Cruz Biotechnology) and centrifugation at 600 × g for 5 min, cells were magnetically sorted and seeded for the immunofluorescence or directly stained for mass cytometry analysis.
Cell surface labelling for mass cytometry analysis
Enzymatically dissociated muscle mononuclear cells were suspended in Maxpar Cell Staining Buffer (Fluidigm, Cat# 201068) and were divided into aliquots of 6 × 106 cells, in a volume of 100 μl, into 15 ml polystyrene tubes for each sample to be stained. 100 μl of the antibody cocktail were added to each tube. The final staining volume was 200 μl (100 μl of cell suspension plus 100 μl of antibody cocktail, with final dilution of 1:100 for each antibody). The cocktail contained the following antibodies: rat anti-mouse CD45-175Lu, rat anti-mouse CD31-165Ho, rat anti-mouse CD11b-172Yb, rat anti-mouse CD117 (c-kit)-166Er, rat anti-mouse F4/80-146Nd, rat anti-mouse Ly-6A/E (Sca-1)-164Dy, rat anti-mouse TER-119-154Sm, mouse anti-PE-156Gd and anti-mouse integrin α-7-PE.
Samples were gently vortexed and incubated for 30 min at room temperature (RT). After incubation, samples were washed twice by adding 2 ml Maxpar Cell Staining Buffer to each tube, centrifuged for 5 m at 600 × g and the supernatant discarded. Cells were resuspended in residual volume by gently vortexing and incubated for 1 h at RT in the intercalation solution, composed of Cell-ID Intercalator-Ir (Fluidigm, Cat# 201192A, 125 μM) into Maxpar Fix and Perm Buffer (Fluidigm, Cat# 201067) to a final concentration of 125 nM (a 1000× dilution of the 125 μM stock solution). Cells were washed twice by adding 2 ml of Maxpar Cell Staining Buffer and centrifuged for 5 m at 800 × g. Pellets obtained were resuspended once with 2 ml of Milli-Q water (Millipore) and centrifuged for 5 m at 800 × g. The pellets were left until ready for the analysis. Immediately prior to mass cytometry data acquisition, the cell concentration was adjusted to 2.5–5 × 105/ml with Milli-Q water and the cell suspension was filtered into 5 ml round bottom polystyrene tubes with 30 μm-cell strainer cap. Data were analyzed using mass cytometry platform, of DVS Sciences (CyTOF2), after stabilization and calibration of the instrument. viSNE, a computational approach suitable for the visualization of high-dimensional data, such as the mass cytometry output [19], [20], was used for data analysis.
CD45+ cells depletion
For the ex vivo differentiation studies, skeletal muscle mononuclear cells, isolated with the protocols described above, were magnetically sorted as CD45− cells, in order to deplete the hematopoietic compartment and enrich the myogenic fraction. The CD45+ cells were magnetically labelled with anti-CD45 MicroBeads (Miltenyi) and the cell suspension was loaded onto a MACS column (Miltenyi), placed in the magnetic field of a MACS separator. The magnetically labelled CD45+ cells were retained within the column and discarded. The flow through of unlabeled cells were then used for the differentiation studies.
Culture conditions for differentiation experiments
Selected CD45− muscle mononuclear populations were plated in Dulbecco's modified Eagle medium (DMEM) GlutaMAX (Gibco) supplemented with 20% heat-inactivated foetal bovine serum (FBS, EuroClone), 1% sodium pyruvate (Sigma, 100 mM), 1% penicillin (10,000 U/ml) and streptomycin (10 mg/ml) on Matrigel-pre-coated 12-wells plates, until cells reached the desired confluence.
Adipogenic differentiation was induced in two different stages. At 80% of confluence, cells were induced to differentiate using an induction medium, composed of DMEM GlutaMAX high-glucose (Gibco), supplemented with 10% FBS, 1% penicillin and streptomycin, 1% sodium pyruvate, 1 μM dexamethasone (Sigma), 0.5 mM isobutylmethylxanthine (IBMX, Sigma) and insulin 1 μg/ml (Sigma). After 48 h, the induction medium was replaced by the differentiation medium for 4 days, that is, DMEM GlutaMAX high-glucose, 1% penicillin and streptomycin, 1% sodium pyruvate supplemented with 20% FBS, and insulin (1 μg/ml). Osteogenic differentiation was induced by lowering FBS concentration to 5% in DMEM GlutaMAX, 1% penicillin and streptomycin, 1% sodium pyruvate and treating cells with bone morphogenetic protein 2, BMP2 (0.3 μg/ml, PeproTech) for 5 days.
Immunofluorescence and alkaline phosphatase staining
Isolated muscle mononuclear populations were cultured in Matrigel-coated 12-well plates in appropriate culture media. After fixation with 2% paraformaldehyde (PFA) for 15 m and permeabilization with 0.5% Triton-X 100 in DPBS (Gibco) for 5 m, cells were incubated for 1 h at RT with the anti-myosin heavy chain (MF20; DHSB) primary antibody, diluted in accordance with the manufacturer's instructions. Cells were then incubated with the appropriate secondary fluorophore-conjugated antibodies. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). For the detection of lipid droplets typical of adipocytes, 0.5% filtered solution of Oil Red-O (Sigma–Aldrich) in isopropanol was used, followed by DPBS washing and DAPI staining. Fluorescence photomicrographs were taken with the Leica DMI6000 B inverted microscope equipped with a fluorescence detection system at 20× magnification.
Alkaline phosphatase (AP) assay was performed by diluting the substrates nitro-blue tetrazolium chloride (NBT, 0.33 mg/ml) and 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (BCIP, 0.165 mg/ml) in AP Buffer (TRIS 100 mM; NaCl 10 mM; MgCl2 1 mM). Cells were incubated with the solution for 15–30 m at room temperature and photomicrographs were taken with the Leica DMI6000 B inverted microscope.
Results
Characterization of muscle mononuclear cells preparations by differentiation assays
Three digestion protocols were tested, differing in the proteolytic activities used to dissociate the muscle fibers from the extracellular matrix in order to release resident mononuclear cells. One method uses the proteolytic enzymes Dispase II and collagenase A (protocol I) for the isolation of muscle progenitors [21]. A second method (protocol II), which is utilized in our laboratory for the isolation of porcine pericytes [22], makes use of collagenase II. Finally, these dissociation methods were compared with trypsin treatment (protocol III), which is commonly used in cell culture practice to remove adherent cells from plastic surfaces and in dissociation protocols for the isolation of alive cells from embryo tissues [23].
The pool of mononuclear cells freshly extracted from hind limb muscles, was depleted of CD45+ cell fraction, an antigen on the membranes of cells of the immune system, and tested for their potential to differentiate into different cell types of mesodermal origin. Skeletal muscle mononuclear cells were cultured in growth medium until the desired confluence (about 80%) and tested for myogenic, adipogenic and osteogenic differentiation potential by growing in specific differentiation media. Myotubes were identified by staining with a fluorescent antibody against the myosin heavy chain (MyHC) while adipocyte and osteogenic precursors were recognized by Oil Red O staining or alkaline phosphatase (AP), respectively.
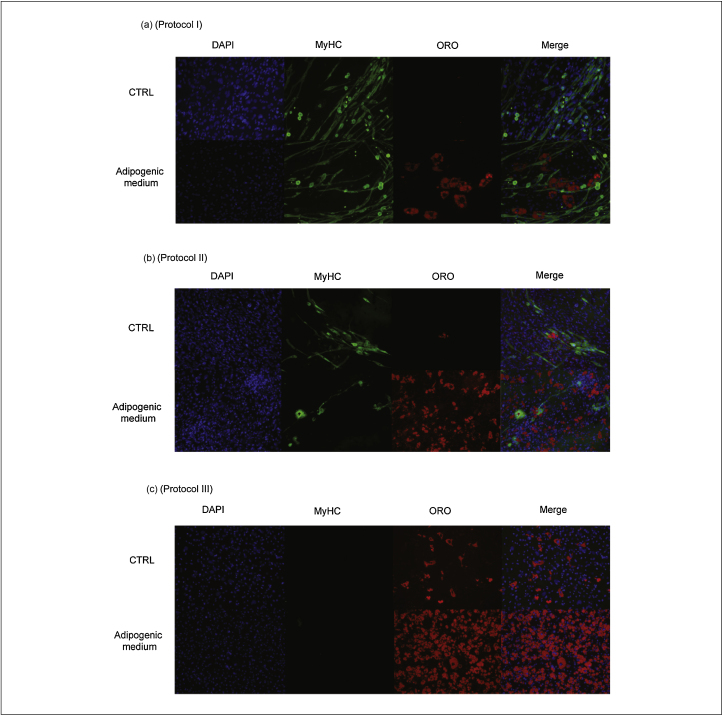
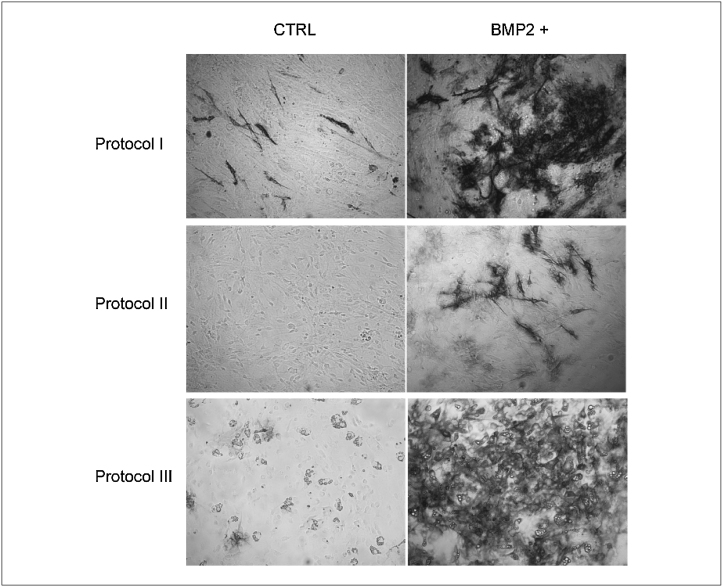
Cells extracted with protocols I and II, under specific stimulations, expressed markers of myogenic, osteogenic and adipogenic lineages with equal efficiency (Figure 1, Figure 2). Furthermore, it was interesting to note that after adipogenic stimuli, some cell populations maintained their myogenic differentiation potential, highlighting the heterogeneity of the cell populations isolated from the muscle and the differential population response to external stimuli. Cells prepared with protocol I showed a more robust staining with the MyHC marker while cells prepared with protocol II differentiated more readily into adipocytes. In addition, cells prepared with protocol I stained more efficiently with AP when exposed to the pro-osteogenic factor, BMP2.
Figure 1.
Adipogenic and myogenic differentiation potential of mononuclear cell preparations. The three panels (a, b and c) are immunofluorescence microphotographs of mononuclear cells extracted from mouse muscles by the three different protocols described in ‘Materials and Methods’. From left to right the four panels on each row are microphotographs of the same sample preparation stained with DAPI to reveal nuclei (blue, nuclei), an antibody against the Myosin Heavy Chain (MF20, myotubes, green), and Oil Red O (ORO) to label adipocytes (red); the fourth panel combines and merges the signals. Cells seeded in control wells (CTRL) were maintained in growth medium. The adipogenic and myogenic differentiation was observed 7 days after incubation in the adipogenic medium.
Figure 2.
Osteogenic differentiation of isolated mononuclear cell populations. Phase contrast images revealing alkaline phosphatase positive staining (black) upon BMP2 exposure of cell culture samples derived from the three extraction methods (described in ‘Materials and Methods’). Osteogenic differentiation was induced by lowering FBS concentration to 5% and by treating cells with BMP2 (0.3 μg/ml) for 5 days. In the control (CTRL), cells were grown in the same medium conditions, without addition of BMP2.
In contrast, cells isolated using trypsin digestion (protocol III) were less heterogeneous, the vast majority showing a fibroblast-like phenotype, characterized by a spindle-shape morphology. Strikingly, this cell preparation did not differentiate into myotubes under any conditions tested, suggesting that viable precursors with myogenic potential could not be extracted from the muscle by this protocol. However, in adipogenic and osteogenic induction media, they differentiated effectively into cell types that stained efficiently with Oil Red O or AP. Remarkably, this preparation contains cells that spontaneously differentiate into adipocytes, albeit with a low efficiency. When cultivated in the specific ‘adipogenic medium’, the differentiation potential increased dramatically to almost 100%. This functional evidence suggests that interstitial cell populations are the predominant populations in this preparation. This initial analysis based on differentiation assays provides a picture of the diversity of the cell preparations obtained by the three different extraction methods.
An antibody panel that allows the identification of the main muscle mononuclear populations by mass cytometry
To gain further insight into the diversity of the cell preparations obtained by the three different methods, we set out to characterize the different cell populations by mass cytometry. After literature mining, searching for antibodies recognizing antigens found on muscle resident cell membranes, and after testing several, we assembled a collection of eight antibodies showing sufficient reactivity and specificity in the mass cytometry analysis (Table 1). These include antibodies that recognize antigens expressed both by muscle progenitors and by cells of the hematopoietic compartment. This antibody panel was validated by labelling skeletal muscle mononuclear cells, extracted from mouse hind limb muscles, by applying the Dispase II/collagenase A protocol, commonly employed for the isolation of muscle populations [24], [25]. Cells were analyzed without further purification in a CyTOF2 instrument.
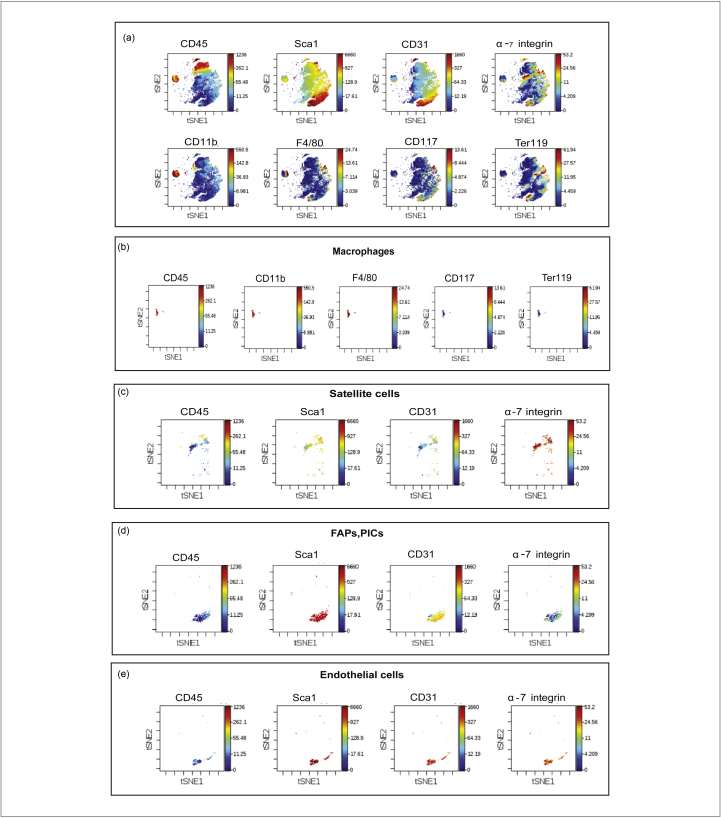
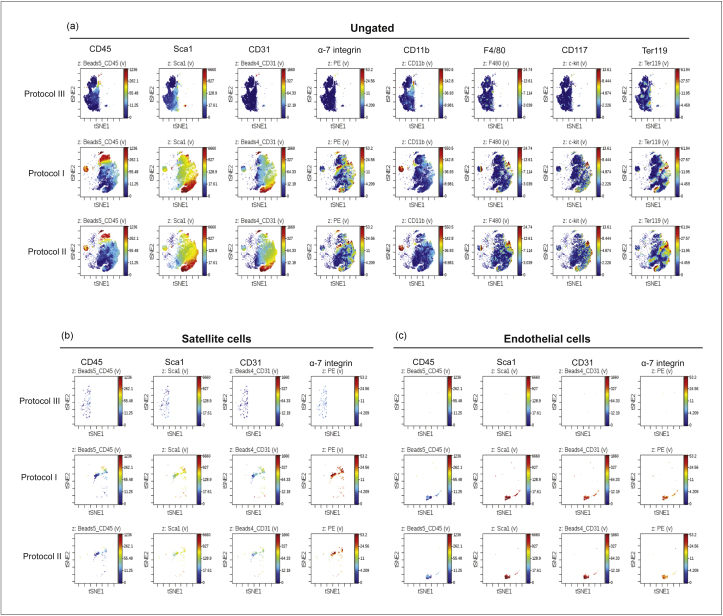
Figure 3 shows an analysis of the mass cytometry results obtained by applying the viSNE algorithm. Each map represents a bi-dimensional projection of the eight-dimensional space of the multiparametric analysis. Each event in the figure represents a cell expressing at specific levels the antigens monitored by the antibody panel. Cells expressing similar levels of the eight antigens are close in the bi-dimensional map. A third dimension can be added to the viSNE map by representing the level of expression of any given antigen according to a colour scale (from blue to red). By this analysis we first identified two clusters of cell populations that differ in expression of CD45, a leukocyte marker (Fig. 3). Among the cell populations that were positive for CD45, we identified mature macrophages, characterized by both expression of CD11b, a marker of granulocytes, monocytes and macrophages, and F4/80, a specific marker of mature macrophages. Among the CD45− cells, we identified satellite cells as α-7 integrin+, Sca1− and CD31−. Moreover, we also discriminated the fibro-adipogenic progenitors (FAPs) and PW1+ Interstitial cells (PICs) as Sca1+, CD45−, CD31−, α-7 integrin−, thus confirming that the selected antibody panel is suitable for the identifications of the main skeletal muscle populations. Interestingly, we also identified another mononuclear cell population, whose presence has also been reported in the literature, namely endothelial or myo-endothelial cells [26]. They express endothelial markers such as CD31 and Sca1, and are also positive for the myogenic marker α-7 integrin but negative for immune cell markers (CD45, CD11b, F4/80). Furthermore, additional events in the viSNE maps have an antigenic profile that cannot be matched to populations already described, for example, CD31+ and Sca1+ cells, which are probably vessel-associated stem cells. More antibodies, not yet included in our panel, are needed for their complete characterization.
Figure 3.
viSNE analysis of cell heterogeneity in suspensions prepared by protocol I (Dispase II/collagenase A). Panel a: The diagrams represent viSNE analysis of a mass cytometry experiment on cells extracted using the Dispase II/Collagenase A extraction method (protocol I). The viSNE algorithm allows the representation in a bi-dimensional projection of the events in the eight-dimensional space, having as coordinates the eight values of the signals revealed by the antibody panel. Each event in the figure represents a cell expressing at specific levels the antigens monitored in the experiment. Cells expressing similar levels of the eight antigens are close in the bi-dimensional map. The colour scale (from blue to red) represents the level of expression of the selected antigen and can be considered a supplementary dimension added to the bi-dimensional plots. Panels b to e: show in the two dimensional viSNE map subpopulations obtained by a gating approach. Macrophages are identified as cells expressing the antigens CD45, CD11b, F4/80 with low signal for CD117 and Ter119. Moreover, satellite cells are selected as CD45−, Sca1−, CD31−, α-7 integrin+ events; FAP and PICs as CD45−, Sca1+, CD31−, α-7 integrin− cells; and endothelial cells as CD45−, Sca1+, CD31+, α-7 integrin+.
Comparison of mononuclear cells extraction protocols by mass cytometry
In order to evaluate whether and how the extraction protocol influences the composition of the isolated mononuclear populations, we stained, with the panel of eight antibodies in Table 1, the cell suspensions obtained after the digestion of the muscle tissue.
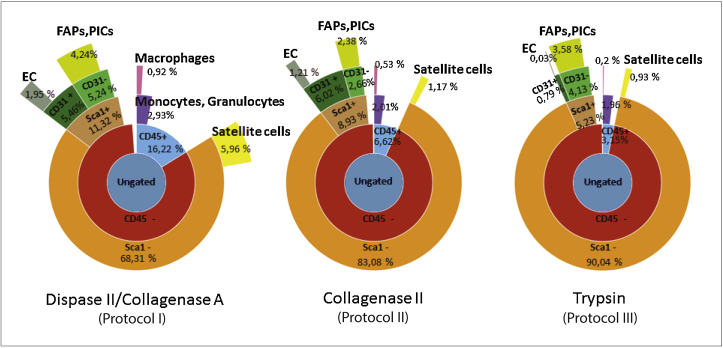
The diagrams in Fig. 4 are ‘sunburst representations’ of population hierarchies as identified by the single cell analysis of the heterogeneous cell populations obtained by mass cytometry. The concentric rings represent the level of population hierarchies and the size of the wedges is proportionate to the number of events in the population. This representation clearly highlights differences in the relative abundance of each identified cell type obtained by the three different extraction protocols. The major differences were observed comparing the cell populations obtained by protocol III (trypsin), with those obtained by the ‘muscle specific’ protocols (Fig. 4). The protocol relying on trypsin digestion was less effective in releasing cells from the muscle fibers. More specifically this protocol yielded comparably fewer myogenic and Sca1+ interstitial cell populations. In particular, the reduction in abundance of endothelial cells and satellite cell populations was evident, the latter representing the myogenic fraction responsible for myotube formation. In addition, we observed a general reduction of the hematopoietic cell populations, identified as the CD45+ fraction.
Figure 4.
Quantitative analysis of cell populations yielded by the different extraction protocols. Diagrams are sunburst charts representing the heterogeneous muscle cell populations as identified by mass cytometry. They show the relative abundance (percentage of total events) of skeletal muscle mononuclear populations extracted using different protocols (as described in ‘Materials and Methods’). The concentric rings represent the gating hierarchies adopted and the size of the wedges is proportional to the abundance of the population examined.
Comparing the collagenase II and the Dispase II/collagenase A digestion protocols, minor differences were observed. In particular, in the collagenase II extraction method, we noticed a reduction in the abundance of fibro-adipogenic progenitors and PW1+ interstitial populations, and an increase in the Sca1− interstitial compartment.
The observed differences in cell population abundances can be confirmed by comparing the viSNE plots of the total events for each protocol (Fig. 5 and Supplementary Figure S1). In fact, the single cell events distributed differently in the two-dimensional space of the viSNE maps, and events also differed for the specific expression of different markers. This analysis supports the conclusion that protocol I (Dispase II/collagenase A) is the most effective method for obtaining high yields of heterogeneous cell populations from muscle. Although protocols II and III have an inferior yield, they may be considered for enriching specific muscle populations, since they favour release of cells of the interstitial compartment (i.e. FAPs, PICs, endothelial cells, Sca1− cells).
Figure 5.
viSNE comparison of cell purification protocols. Panel a. The diagrams show the viSNE analysis of mass cytometry experiments on cell suspensions extracted using the three different enzymatic extraction methods (Protocols I, II and III in ‘Materials and Methods’). The blue to red colour scale on the right of each panel represents the level of expression of the antigen indicated at the top of each map. The maps generated can be superimposed and, by adopting the gating strategy described in the legend to Fig. 3, the main populations can be identified by combining the expression of the markers that characterize the cell type. The viSNE maps in a (ungated) represents the expression of the markers in each recorded event, for each protocol. Two examples of gating approaches are reported in b and c: satellite cells, selected as CD45−, Sca1−, CD31−, α-7 integrin+ events; and endothelial cells as CD45−, Sca1+, CD31+, α-7 integrin+.
Discussion
Many biological phenomena are regulated by the interaction of different cell types via an intricate cross-talk causing a complex response to a variety of stimuli. The intrinsic cell heterogeneity of tissues and organs and the complexity of their physiology and pathology cannot be approached with methods based on measurement of average values of bulk populations. In recent years this growing awareness has stimulated the development and application of methods based on single cell technologies. Many of these approaches rely on the preparation of cell suspensions and on the separation of single cells by microfluidic methods.
Among single-cell approaches that take advantages of microfluidic methods, mass cytometry (CyTOF, CyTOF2, Helios platforms) is the most recent and most powerful addition. Mass cytometry is a multi-parametric technology, which enables the study of heterogeneous populations with higher resolution than flow cytometry. The platform is used for studying diverse cell types and tissues, including peripheral blood mononuclear cells, bone marrow cells, splenocytes, lymph node cells and solid tissue cancer cells [27], [28]. However, when the biological system to be analyzed is a solid tissue, specific protocols need to be developed to release the cell populations of interest as single cell suspensions. The protocols need to be sufficiently vigorous to generate high yields of the cell populations forming the tissue while, at the same time, sufficiently mild to preserve the physiological properties of the cells. In addition, each biological system requires the development of specific antibodies, suitable for mass cytometry analysis, to permit the identification of the different cell populations in the tissue.
Here we report an initial effort to assemble an antibody panel for the characterization of muscle cell populations by mass cytometry. Since many of the antibodies that we have tested (not shown) did not display sufficient specificity or were ‘resistant’ to metal labelling, the panel that we are currently using does not exploit the full potential of the mass cytometry technology. However, we have shown here that this incomplete collection of antibodies is sufficient to identify most mononuclear muscle cell populations thus offering a toolbox for the characterization of an additional solid tissue by the powerful mass cytometry approach.
Furthermore, we additionally tested three diverse protocols of tissue dissociation for the isolation of mononuclear muscle cells in suspension and we show that each different cell extraction method yields heterogeneous cell populations with a different relative abundance of the distinct cell types. Additional effort is required to expand the antibody panel described in the present work, in order to develop an approach that can provide a complete overview of the complexity of the muscle tissue and to identify yet uncharacterized populations.
Acknowledgements
This work was supported by a grant of the European Research Council (grant N 322749) and a grant of the Italian Association for Cancer Research, AIRC (grant N 14135) to GC.
Footnotes
Supplementary material related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.nbt.2015.12.007.
Contributor Information
Cesare Gargioli, Email: cesare.gargioli@uniroma2.it.
Gianni Cesareni, Email: cesareni@uniroma2.it.
Appendix A. Supplementary data
The following is supplementary data to this article:
References
- 1.Ornatsky O., Bandura D., Baranov V., Nitz M., Winnik M.A., Tanner S. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;361(1–2):1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Tanner S.D., Baranov V.I., Ornatsky O.I., Bandura D.R., George T.C. An introduction to mass cytometry: fundamentals and applications. Cancer Immunol Immunotherapy. 2013;62(5):955–965. doi: 10.1007/s00262-013-1416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barberi L., Scicchitano B., De Rossi M., Bigot A., Duguez S., Wielgosik A. Age-dependent alteration in muscle regeneration: the critical role of tissue niche. Biogerontology. 2013;14(3):273–292. doi: 10.1007/s10522-013-9429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawke T.J., Garry D.J. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol. 2001;91(2):534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- 5.Yin H., Price F., Rudnicki M.A. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013;93(1):23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Maltzahn J., Jones A.E., Parks R.J., Rudnicki M.A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci U S A. 2013;110(41):16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102(6):777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 8.Tamaki T., Akatsuka A., Ando K., Nakamura Y., Matsuzawa H., Hotta T. Identification of myogenic-endothelial progenitor cells in the interstitial spaces of skeletal muscle. J Cell Biol. 2002;157(4):571–577. doi: 10.1083/jcb.200112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sancricca C., Mirabella M., Gliubizzi C., Broccolini A., Gidaro T., Morosetti R. Vessel-associated stem cells from skeletal muscle: from biology to future uses in cell therapy. World J Stem Cells. 2010;2(3):39–49. doi: 10.4252/wjsc.v2.i3.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto A., Collins-Hooper H., Patel K. The origin, molecular regulation and therapeutic potential of myogenic stem cell populations. J Anat. 2009;215(5):477–497. doi: 10.1111/j.1469-7580.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joe A.W., Yi L., Natarajan A., Le Grand F., So L., Wang J. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12(2):153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell K.J., Pannerec A., Cadot B., Parlakian A., Besson V., Gomes E.R. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat Cell Biol. 2010;12(3):257–266. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
- 13.Minasi M.G., Riminucci M., De Angelis L., Borello U., Berarducci B., Innocenzi A. The meso-angioblast: a multipotent, self-renewing cell that originates from the dorsal aorta and differentiates into most mesodermal tissues. Development (Cambridge, England) 2002;129(11):2773–2783. doi: 10.1242/dev.129.11.2773. [DOI] [PubMed] [Google Scholar]
- 14.Birbrair A., Zhang T., Wang Z.M., Messi M.L., Enikolopov G.N., Mintz A. Role of pericytes in skeletal muscle regeneration and fat accumulation. Stem Cells Dev. 2013;22(16):2298–2314. doi: 10.1089/scd.2012.0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abou-Khalil R., Mounier R., Chazaud B. Regulation of myogenic stem cell behavior by vessel cells: the menage a trois of satellite cells, periendothelial cells and endothelial cells. Cell Cycle (Georgetown, Tex) 2010;9(5):892–896. doi: 10.4161/cc.9.5.10851. [DOI] [PubMed] [Google Scholar]
- 16.Bentzinger C.F., Wang Y.X., Dumont N.A., Rudnicki M.A. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 2013;14(12):1062–1072. doi: 10.1038/embor.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shireman P.K., Contreras-Shannon V., Ochoa O., Karia B.P., Michalek J.E., McManus L.M. MCP-1 deficiency causes altered inflammation with impaired skeletal muscle regeneration. J Leuk Biol. 2007;81(3):775–785. doi: 10.1189/jlb.0506356. [DOI] [PubMed] [Google Scholar]
- 18.Brunelli S., Rovere-Querini P. The immune system and the repair of skeletal muscle. Pharmacol Res. 2008;58(2):117–121. doi: 10.1016/j.phrs.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Hinton LvdMaG. Visualizing data using t-SNE. J Mach Learn Res. 2008;1:1–48. [Google Scholar]
- 20.Amir el A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mozzetta C., Consalvi S., Saccone V., Tierney M., Diamantini A., Mitchell K.J. Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old Mdx mice. EMBO Mol Med. 2013;5(4):626–639. doi: 10.1002/emmm.201202096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuoco C., Sangalli E., Vono R., Testa S., Sacchetti B., Latronico M.V. 3D hydrogel environment rejuvenates aged pericytes for skeletal muscle tissue engineering. Front Physiol. 2014;5:203. doi: 10.3389/fphys.2014.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conner DA. Mouse embryo fibroblast (MEF) feeder cell preparation. Current protocols in molecular biology/edited by Frederick M Ausubel et al. 2001; Chapter 23:Unit 23.2. [DOI] [PubMed]
- 24.Rando T.A., Blau H.M. Methods for myoblast transplantation. Methods Cell Biol. 1997;52:261–272. doi: 10.1016/s0091-679x(08)60382-9. [DOI] [PubMed] [Google Scholar]
- 25.Sacco A., Doyonnas R., Kraft P., Vitorovic S., Blau H.M. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456(7221):502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng B., Cao B., Crisan M., Sun B., Li G., Logar A. Prospective identification of myogenic endothelial cells in human skeletal muscle. Nat Biotechnol. 2007;25(9):1025–1034. doi: 10.1038/nbt1334. [DOI] [PubMed] [Google Scholar]
- 27.Bendall S.C., Simonds E.F., Qiu P., Amir el A.D., Krutzik P.O., Finck R. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science (New York, NY) 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giesen C., Wang H.A., Schapiro D., Zivanovic N., Jacobs A., Hattendorf B. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.