Respected Editor,
Hb HPLC is an important tool in the diagnosis of various haemoglobinopathies. However, in some cases, it has its diagnostic limitations, when used alone and it is judicious to reconfirm by a second independent technique, as per current guidelines [1]. To illustrate this, we are presenting two cases of haemoglobinopathies reported in Haematology Department of a tertiary care hospital.
In one case (Case 1), a 24 years old North Indian Punjabi female presented in our Haematology Lab for routine antenatal Hb HPLC referred by Obstetrics and Gynaecology Department her Hb analysis by HPLC demonstrated HbF (0.7 %), HbA0 (49.8 %) and a variant Hb (43.1 %) in the HbA2 window with a retention time of 3.62 min (Fig. 1). Her Hb was 11.8 Gm%, RBC indices were normal and blood picture was normocytic normochromic.
Fig. 1.
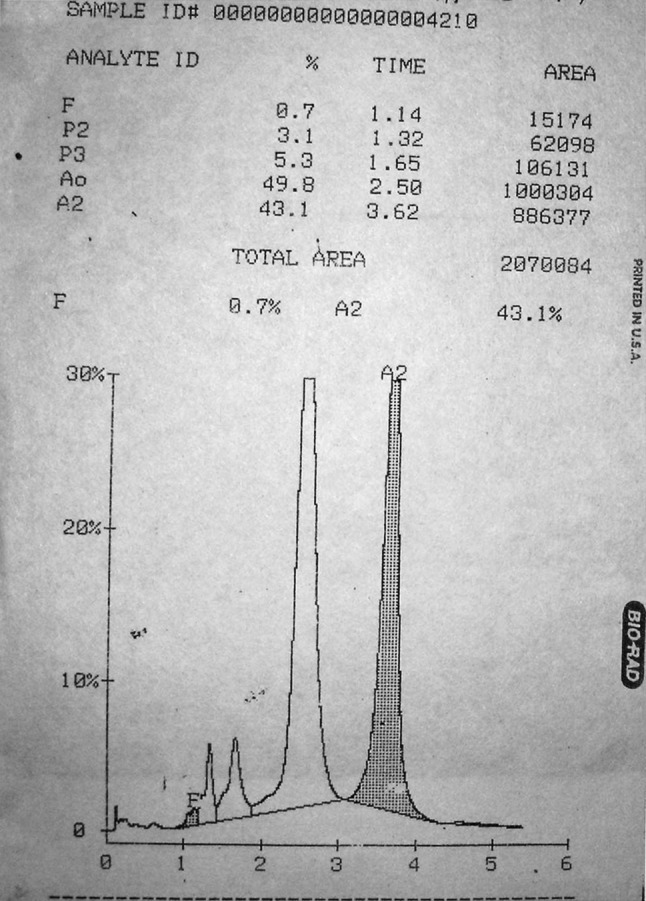
Hb HPLC of Case 1
In another case (Case 2), a 26 years old North Indian Punjabi male, presented for routine Hb HPLC. His Hb analysis by HPLC demonstrated HbF (0.7 %), HbA0 (49.8 %) and a variant Hb (42.1 %) in the HbA2 window with a retention time of 3.62 min. His Hb was 15.4 Gm% with normal red cell indices and normocytic normochromic blood picture.
The haemoglobins which can elute by HPLC in the HbA2 window with the retention time range of 3.3–3.9 min are HbA2 (R.T.3.63), HbE (R.T.3.69), Hb Lepore (R.T.3.37), HbD-Iran (R.T.3.49), HbG Copenhagen (R.T.3.69), Hb OsuChristianborg (R.T.3.77) and HbG Honolulu (R.T.3.86). The possibility of variant Hb to be HbA2 with retention time of 3.63 was considered, but HbA2 is usually not over 7 % in heterozygous β thalassemia [2]. The possibility of the variant Hb to be HbE (R.T.3.69) was considered but HbE is usually not over 30 % in HbE trait [1]. Hb electrophoresis was performed in both the samples on agarose gel at alkaline pH of 8.6 and possibility of variant Hb being HbA2 or HbE was further ruled out, as a well formed band was seen for this variant Hb in the S/D/G region in both the cases and there was no band in the A2/E region (Fig. 2).
Fig. 2.
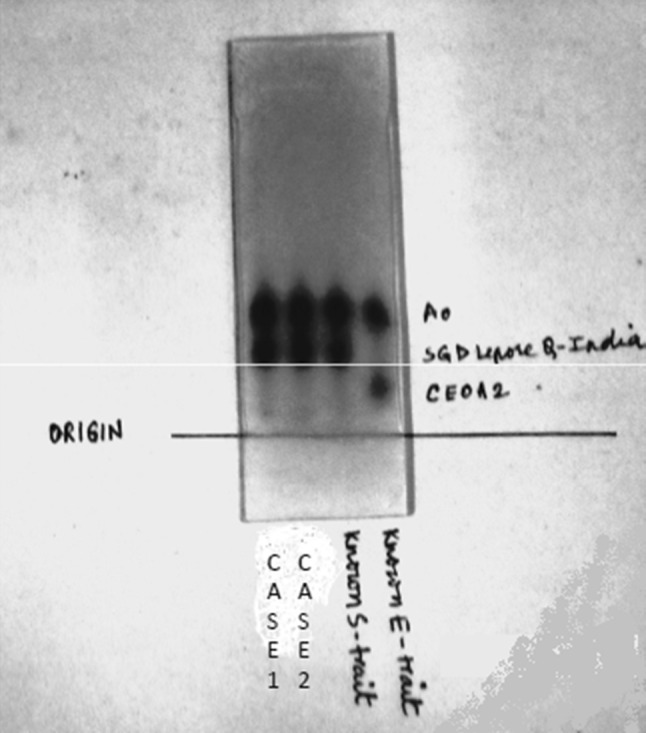
Hb electrophoresis of Case 1 and Case 2
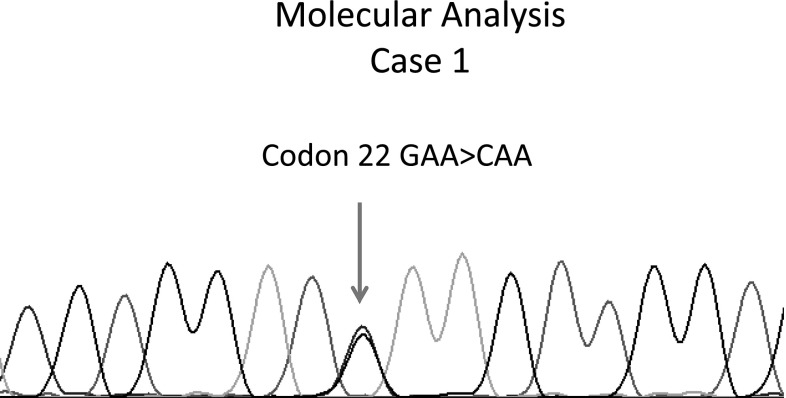
Keeping in view, the HPLC findings, the concentration of variant Hb, retention time, Hb electrophoresis findings and after going through the literature [1–4], a possibility of HbG/HbD-Iran was considered in both these cases. A subtle way to distinguish HbD-Iran from HbG (like G-Philadelphia) is that the latter, by virtue of being an alpha chain variant will show an additional tiny G2 peak (variant A2 formed by β-G-Philadelphia chains and normal delta globin chains) [5], but conclusive identification of the exact variant was only possible after DNA analysis. Therefore, both the samples were subjected to DNA sequencing, being the existing gold standard of point mutations testing to detect the exact nucleotide change in HBB gene. Bidirectional sequencing using The BigDye® Terminator v3.1 Cycle Sequencing Kit on ABI 3100 automated sequencer was performed using PCR fragment containing first two exons and first intron amplified from DNA extracted from peripheral blood. Single base substitution GAA > CAA (indicative of HbD-Iran) in the heterozygous form was seen (Fig. 3) by sequencing in both the cases and a final diagnosis of HbD-Iran was made in both these cases. The haematological and molecular details of both the cases are shown in the tabulated form (Table 1). The replacement of glutamic acid by glutamine at β22 (β22Glu → Gln) is responsible for the formation of HbD-Iran. Gene Sequencing was done in our cases since it was available in our institution and out of academic interest. In clinical terms, HbD-Iran, even in the homozygous state is an innocuous incidental finding. Once HPLC and Hb electrophoresis are done, most of these cases only require spousal Hb HPLC for reassurance about any genetic consequences.
Fig. 3.
Molecular analysis of Case 1
Table 1.
Haematological and molecular details of Case 1 and Case 2
| Parameters | Case 1 | Case 2 |
|---|---|---|
| Age (years)/sex | 24/F | 26/M |
| Hb (g/dL) | 11.8 | 15.4 |
| Hct (%) | 38.0 | 49.0 |
| MCV (fL) | 82.8 | 86.4 |
| MCH (pg) | 29.3 | 30.6 |
| MCHC (g/dL) | 31.5 | 32.4 |
| RBC count (×106/µL) | 4.9 | 5.4 |
| RDW-CV (%) | 14.0 | 14.5 |
| TLC (×103/µL) | 6.8 | 7.2 |
| Platelets count (×103/µL) | 234 | 280 |
| HPLC | ||
| HbF% | 0.7 | 0.7 |
| HbA0% | 49.8 | 49.8 |
| Variant Hb% (in HbA2 window) | 43.1 | 42.1 |
| Results of gene sequencing in both cases | ||
| Gene sequencing | Single base substitution GAA > CAA (indicative of HbD-Iran in the heterozygous form) | Single base substitution GAA > CAA (indicative of HbD-Iran in the heterozygous form) |
Conclusion
The retention times obtained by HPLC, the percentages of the variant Hbs obtained and the appearance of the HPLC chromatograms, are useful sources of information, which can help in the identification of many Hb variants, but not in all the cases. In some cases, we have to analyze the results in combination with Hb electrophoresis and in selected cases we may require molecular analysis to reach at the final diagnosis. All current guidelines [1] state that any variant hemoglobin detected by any technique should be reconfirmed by a second independent technique and our cases demonstrate the judiciousness of this recommendation.
References
- 1.Ryan K, Bain BJ, Worthington D, et al. Significant haemoglobinopathies: guidelines for screening and diagnosis. Br J Haematol. 2010;149(1):35–49. doi: 10.1111/j.1365-2141.2009.08054.x. [DOI] [PubMed] [Google Scholar]
- 2.Bain BJ. Other significant haemoglobinopathies. In: Bain BJ, editor. Haemoglobinopathy diagnosis. 2. Oxford: Blackwell Publishing; 2006. pp. 190–233. [Google Scholar]
- 3.Joutovsky A, Hadzi-Nesic J, Nardi MA. Retention time as a diagnostic tool for hemoglobin variants and hemoglobinopathies: a study of 60,000 samples in a clinical diagnostic laboratory. Clin Chem. 2004;50:1736–1747. doi: 10.1373/clinchem.2004.034991. [DOI] [PubMed] [Google Scholar]
- 4.Colah RB, Surve R, Sawant P, et al. HPLC studies in hemoglobinopathies. Indian J Pediatr. 2007;74(7):657–662. doi: 10.1007/s12098-007-0117-8. [DOI] [PubMed] [Google Scholar]
- 5.Kumar M, Sharma P, Kumar V, et al. Continuing diagnostic relevance of the sickling test in the era of CE-HPLC. Indian J Hematol Blood Transfus. 2013;29(1):58–60. doi: 10.1007/s12288-012-0144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]