Abstract
In plants, vascular stem cells continue to give rise to all xylem and phloem cells, which constitute the plant vascular system. During plant vascular development, the peptide, tracheary element differentiation inhibitory factor (TDIF), regulates vascular stem cell fate in a non-cell-autonomous fashion. TDIF promotes vascular stem cell proliferation through up-regulating the transcription factor gene WUS-related HOMEOBOX4, and it suppresses xylem differentiation from vascular stem cells through the activation of Glycogen Synthase Kinase 3 proteins. VASCULAR-RELATED NAC-DOMAIN6 and 7 (VND6 and 7) are master transcription factors, and ectopic expression of VND6 and VND7 in various plants induces differentiation of different types of cells into metaxylem and protoxylem tracheary elements, respectively. These genes up-regulate genes involved in both patterned secondary cell wall formation and programmed cell death to form tracheary elements. Secondary wall patterns are formed by localized deposition of cellulose microfibrils, which is guided by cortical microtubules. Local activation of the small G-protein, Rho-type 11 determines distribution of cortical microtubules.
Keywords: cell communication, microtubules, plant vascular development, transcription factor, xylem, stem cell
Introduction
The plant vascular system consists of xylem and phloem, which function in long-distance transport of water and nutrients, as well as signaling molecules, through the plant body.1) The establishment of vascular cells begins with the formation of procambial cells during embryogenesis. Procambial cells function as vascular stem cells and continue to give rise to all xylem and phloem cells during embryogenesis and postembryogenesis. The regulation of this plant-specific stem cell fate has been the focus of much research. However, many questions remain. For example, how is the vascular stem cell population maintained? What determines the fates of vascular stem cells? Is there a common regulatory mechanism for vascular stem cells and other types of stem cells in plants?
To elucidate the mechanism underlying vascular stem cell fate determination, we focused on xylem cell differentiation from procambial cells. The xylem consists of tracheary elements, xylem parenchyma cells, and xylem fibers. Tracheary elements are characterized by the formation of secondary cell walls, in which cellulose, hemicellulose, and lignin are deposited in annular, spiral, reticulate, or pitted patterns.2) At maturity, tracheary elements are emptied of all cell contents to form hollow tubes constituting vessels and tracheids, which serve as a pathway for fluids. The xylem is also important from an applied perspective, because biomaterials such as cellulose and lignin in the xylem represent the predominant components of terrestrial biomass.
This review summarizes advances in the field of xylem development, with a focus on our studies of the past few decades. First, xylem-inducing culture systems are described, which allow us to study xylem cell differentiation in detail. Second, key factors that regulate xylem development discovered using these cultures are presented. Finally, the current understanding of xylem development is discussed, focusing on the regulatory networks that function during each cell differentiation process.
Experimental systems
1). Zinnia mesophyll cell culture.
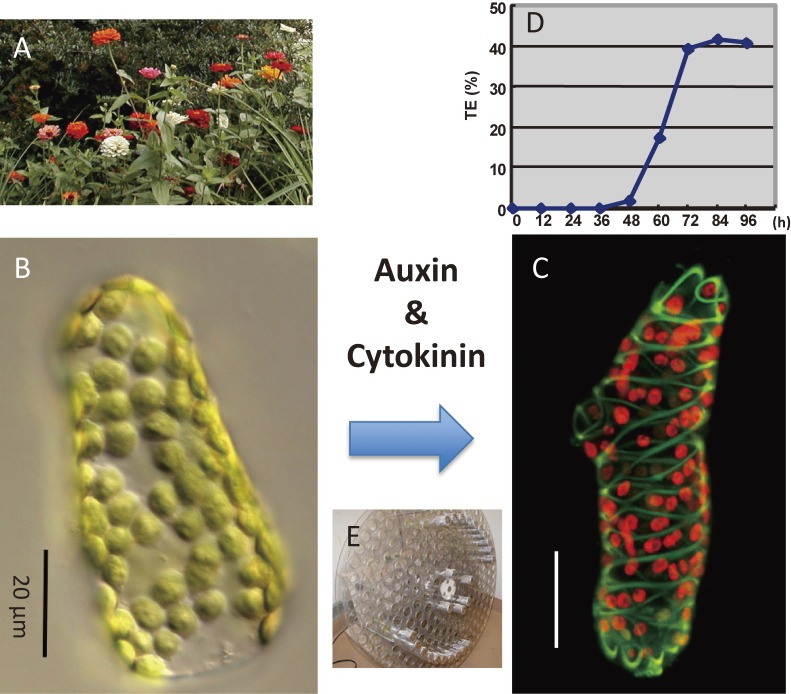
Since xylem tissue is located deep within plant organs, it is quite difficult to analyze xylem development at the cellular and molecular level. To overcome this difficulty, we established various cell culture systems in which tracheary elements form synchronously and at high frequency.3–6) In 1980, we established a xylogenic culture in which single mesophyll cells isolated from Zinnia elegans leaves differentiate into tracheary elements synchronously and at high frequency in the presence of two plant hormones, auxin and cytokinin (Fig. 1, Table 1).3) Serial observation of single cells revealed that mesophyll cells trans-differentiate into tracheary elements without intervening cell division.7,8) Comprehensive gene expression analysis of Zinnia culture revealed that mesophyll cells differentiate into tracheary elements via the vascular stem cell stage.9) Thus, Zinnia culture allowed us to identify various key factors that regulate xylem differentiation such as TDIF.10)
Figure 1.
Zinnia xylogenic culture. Single Zinnia mesophyll cells can differentiate directly into tracheary elements in the presence of auxin and cytokinin within 3 days. A: Zinnia elegans plants. B: A differential interference contrast (Nomarski) image of an isolated Zinnia mesophyll cell, which contains many chloroplasts. C: A confocal image of a tracheary element that is differentiating from a mesophyll cell in culture. The cell contains spiral secondary wall bands (green) and chloroplasts (red). D: Time course of tracheary element differentiation in mesophyll cell culture. E: Rotating drum for culturing mesophyll cells.
Table 1.
Xylogenic cultures
| Species | Starting material | Inducers | days required for TE differentiation after induction | frequency of TE differentiation (%) |
|---|---|---|---|---|
| Zinnia elegans | isolated mesophyll cells | auxin and cytokinin | 3 | 40 |
| Arabidopsis thaliana | cultured cells | boron and brassinosteroids | 3 | 30 |
| Arabidopsis thaliana | cultured cells harboring inducible VND6 | VND6 overexpression | 2 | 80 |
| Arabidopsis thaliana | leaves and cotyledons | bikinin, auxin and cytokinin | 3 | ND |
2). Arabidopsis culture systems.
To take advantage of the model plant Arabidopsis thaliana, for which genome information, mutants, and marker lines are available, we also established several Arabidopsis xylogenic culture systems (Table 1). First, we established a xylogenic culture system in which subcultured Arabidopsis cells are induced to differentiate into tracheary elements by the application of boron and brassinolide, a plant hormone.4) Microarray analysis with this system allows us to follow the expression of most genes during xylem differentiation, which enabled us to identify VND6 and VND7, master transcription factors that trigger tracheary element differentiation.4)
Based on this finding, we established another Arabidopsis xylogenic culture system with cells harboring an estrogen-inducible VND6 gene construct. In this system, the application of estrogen induces VND6 gene expression, which in turn initiates tracheary element differentiation from 80% of the cells within 48 h.5) Using this culture system, we performed fine expression profiling of genes involved in tracheary element differentiation, as well as live imaging of marker proteins during differentiation.5,11) Using this system, we identified various key factors that regulate microtubule organization during cell wall pattern formation.12)
Recently, we succeeded in establishing a third culture system in which bikinin, an inhibitor of plant GSK3 kinases, is used to induce xylem and phloem cell differentiation in leaf segments and cotyledons of Arabidopsis.6) This system allows us to use mutants and marker lines of Arabidopsis directly. Using this system, downstream factors of TDIF signaling have been revealed.13)
Intercellular signaling
1). Identification of intercellular signaling factors regulating vascular development.
In animals, stem cells are maintained through the support of stem cell niches. To investigate cell–cell communication in the vascular meristem, we tried to identify intercellular signaling molecules that regulate vascular cell fate. For this purpose, we used the culture medium of Zinnia cultures, because various lines of evidence suggest the presence of such factors in the medium. From this culture medium, we isolated an arabinogalactan protein that promotes tracheary element differentiation, which we designated “xylogen”.14) This protein is secreted from differentiating xylem cells to non-differentiating cells to promote their differentiation into xylem.
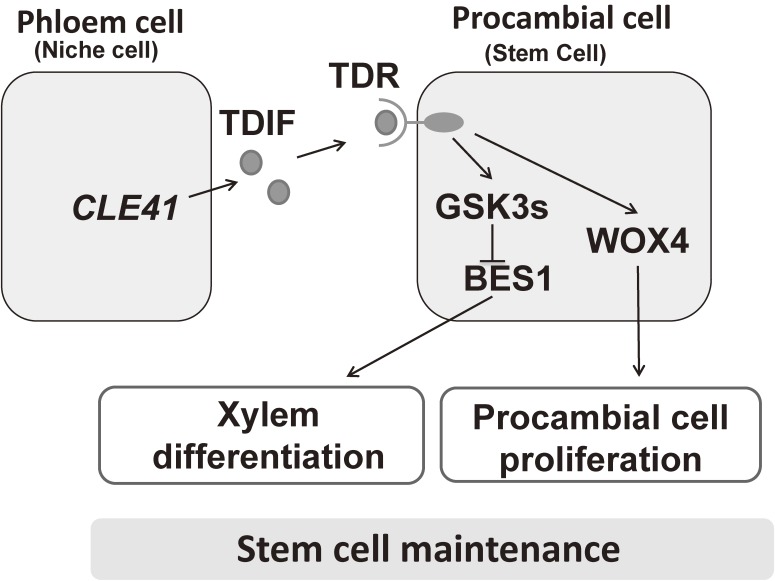
Next, we isolated a factor that inhibits tracheary element differentiation and promotes cell division from this medium, which we named tracheary element differentiation inhibitory factor (TDIF).10) TDIF is a 12-amino-acid peptide with two hydroxyproline residues that inhibits tracheary element differentiation, not only in Zinnia cultures, but also in Arabidopsis vascular bundles. Interestingly, in Arabidopsis hypocotyls, TDIF promotes procambial cell division and inhibits tracheary element differentiation (Fig. 2). Zinnia TDIF and Arabidopsis CLE41 and CLE44 encode proteins of approximately 100 amino acids with a TDIF sequence near their C-termini.10,15)
Figure 2.
TDIF signaling pathway regulates vascular stem cell maintenance. TDIF is produced in phloem cells and secreted. TDIF is perceived by TDR in procambial cells, which both promotes procambial cell proliferation via central transcription factor WOX4 and inhibits xylem cell differentiation through activation of GSK3s and subsequent suppression of BES1. The two downstream pathways contribute to the maintenance of the procambial cell population.
2). TDIF signaling.
Arabidopsis genome encodes over 1000 ligands and over 600 putative receptors, suggesting that immobile plant cells, which are encased in cell walls, actively communicate with other cells. Circumstantial data suggest that a receptor for TDIF might be a leucine-rich repeat-receptor-like kinase (LRR-RLK) located in the plasma membrane. Over 200 LRR-RLK genes have been annotated in the Arabidopsis genome.16) We therefore performed in silico analysis and identified 56 genes whose expression is expected to occur in procambial cells, and we obtained approximately 130 T-DNA insertion lines for these genes.15) Screening of these lines revealed mutants with different alleles of a gene conferring TDIF insensitivity; we designated their causal protein TDR (TDIF receptor). Genetic and biochemical analyses demonstrated that TDR is indeed a receptor for TDIF.
Loss-of-function mutants of TDIF and its receptor exhibit a similar phenotype, in which procambial layers between xylem and phloem are absent due to inhibited procambial cell proliferation and the promotion of tracheary element differentiation from procambial cells.15,17) These results confirm that TDIF and TDR function in the same signaling pathway, and they indicate that the TDIF–TDR signaling pathway functions in the maintenance of vascular stem cells by increasing their population size and inhibiting tracheary element differentiation from these cells.
The expression profiles of CLE41 and CLE44 and analysis of the cellular localization of TDIF indicate that TDIF is synthesized in the phloem and its neighboring cells and secreted toward procambial cells, in which TDR is specifically produced. This finding implies that TDIF functions as a phloem-derived non-cell-autonomous signal that controls stem cell fate in the procambium (Fig. 2).
In the shoot meristem, WUSCHEL (WUS), a homeodomain transcription factor, is a key target of CLV3 (a CLE peptide)-CLV1 (an LRR-RLK) signaling and is essential for stem cell maintenance.18) CLV3 represses the expression of WUS, and WUS promotes the expression of CLV3. This negative feedback loop functions in the maintenance of meristem size.19) WOX4, a member of the WUS-related HOMEOBOX (WOX) gene family, acts downstream of the TDIF–TDR signaling pathway.17) The TDIF signal increases the expression of WOX4 in procambial cells in a TDR-dependent manner. WOX4 functions in procambial cell proliferation but not in xylem differentiation from procambial cells, indicating that procambial cell proliferation and its differentiation into xylem cells are independent and occur downstream of the TDIF–TDR signaling pathway.
To identify the factor that suppresses xylem cell differentiation downstream of TDIF–TDR, we performed yeast two-hybrid screening with the TDR cytoplasmic kinase domain, and found BRASSINOSTEROID-INSENSITIVE2 (BIN2), a member of the plant Glycogen Synthase Kinase 3 protein (GSK3) family.13) GSK3s have been implicated in multiple signaling pathways involved in developmental regulation in animals as well as in plants.20) Mammals contain only two isoforms of GSK3, alpha and beta, which play crucial roles in various signal transduction pathways.21,22) On the other hand, Arabidopsis GSK3s comprise four subfamilies containing ten genes.20) A model for the function of GSK3s was proposed based on genetic and cellular analyses (Fig. 2) in which six GSK3s, including BIN2, redundantly mediate TDIF–TDR signaling to suppress xylem differentiation.13) In the absence of TDIF, GSK3s bind to the kinase domain of TDR and are inactivated. TDIF activates the GSK3s via its release from TDR. The GSK3s target a transcription factor, BES1, negatively regulating its activity.
Master transcription factors initiate the development of specific xylem cells
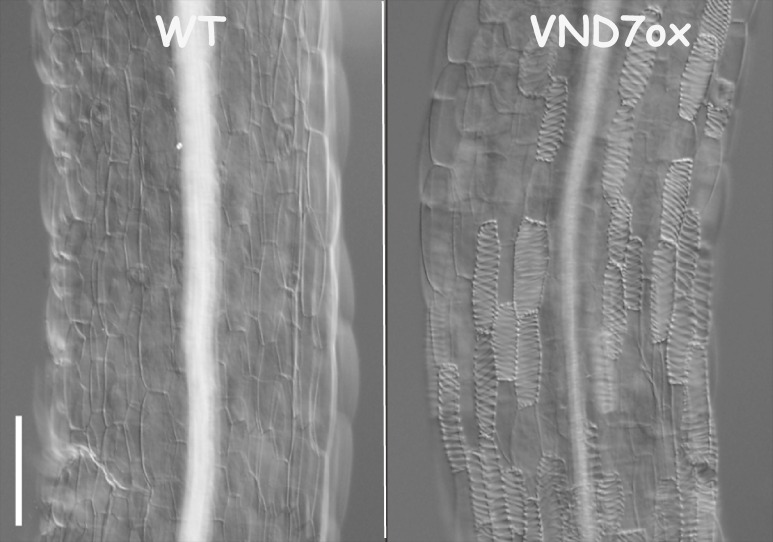
Comprehensive transcriptome analysis of xylogenic zinnia and Arabidopsis cultures revealed gene expression profiles associated with the process of tracheary element differentiation; a number of genes related to cell wall biosynthesis and programmed cell death (PCD) are expressed simultaneously before morphological changes occur.4,6,9,11) This finding suggests the presence of a master transcription factor(s) that up-regulates many genes in association with tracheary element differentiation. Specifically, VASCULAR-RELATED NAC-DOMAIN1-7 (VND1-7) genes are expressed preferentially in differentiating vascular cells.4,11) In particular, VND6 and VND7 are specifically expressed in differentiating tracheary elements of the metaxylem and protoxylem, respectively. Dominant repression of VND6 and VND7 by chimeric repressors that include the EAR motif, a repression domain inhibits the formation of metaxylem and protoxylem tracheary elements, respectively. Conversely, ectopic expression of VND6 and VND7 in not only Arabidopsis but also popular plants induces differentiation of various cells into metaxylem and protoxylem tracheary elements, respectively (Fig. 3). These results clearly indicate that VND6 and VND7 are master transcription factors that induce metaxylem and protoxylem tracheary element differentiation, respectively.4) A recent study indicated that ectopically overexpressing VND1–VND5 can also, although weakly, induce xylem tracheary element formation, probably through regulating VND7 expression.23) VNDs belong to the plant-specific NAM, ATAF1,2, and CUC2 (NAC) protein family, one of the largest groups of plant-specific transcriptional regulators, which regulate diverse plant developmental processes.24) The NAC protein VNI2 selectively binds to VND7 with high affinity and represses xylem cell specification.25)
Figure 3.
Ectopic induction of tracheary elements by overproduction of VND7. Overproduction of VND7 causes the formation of a number of tracheary elements from epidermal cells of Arabidopsis thaliana hypocotyls. Images from Kubo et al. Genes Dev. (2005).4)
NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) and NST3 (also known as SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1, SND1) belong to a subfamily of the NAC family related to the VND subfamily. These transcription factors function as master switches for inducing differentiation of xylem fiber cells.26,27) VND family genes are well conserved among angiosperms and gymnosperms, whereas NST/SND family genes are conserved only in angiosperm.28) This finding is consistent with the observation that gymnosperm wood mainly contains tracheary elements known as tracheids, whereas angiosperm wood contains xylem fibers as well as tracheary elements consisting of vessels and tracheids.
Ancestral genes of the VND family not only exist in ferns, the most primitive vascular plants, but also in the moss Physcomitrella patens, a non-vascular plant lacking xylem tissues.29) In this plant, ancestral VND genes are expressed in water-conducting and supporting cells. Loss-of-function mutants form abnormal water-conducting and supporting cells, and overexpressing these genes induces ectopic differentiation of water-conducting-like cells. Interestingly, overexpressing these moss genes in Arabidopsis induces the formation of ectopic tracheary elements with secondary wall thickenings and PCD, as does overexpressing Arabidopsis VND genes. These results reveal the conservation of transcriptional regulation and cellular function in water-conducting moss and Arabidopsis cells, which suggests that VND proteins have played a role in the adaptation of plants to land.29)
Regulation of secondary cell wall formation and PCD
1). Coordinated gene expression.
Tracheary elements are characterized by patterned secondary cell wall formation and PCD at maturity, which are tightly coupled.2) Secondary cell walls are composed of highly ordered cellulose, hemicellulose, and lignin. A number of genes for enzymes that catalyze these secondary cell wall-related biopolymers are specifically expressed just before secondary cell wall formation. In developing tracheary elements, PCD is initiated by the rupture of the central vacuole. The vacuole rupture occurs 6 h after visible secondary wall formation begins.30) Cell death-related hydrolytic enzymes accumulate before vacuole rupture. The first such enzyme isolated from the vacuole is ZEN1, an S1-type Zn2+-dependent nuclease that plays a major role in nuclear DNA degradation, hydrolyzing nuclear and organellar DNA after vacuole collapse.31,32) Genes for PCD-specific hydrolytic enzymes, including nucleases and proteases, are expressed simultaneously with secondary cell wall formation-related genes.9,11)
VND6 and VND7 directly regulate genes for both secondary cell wall formation and PCD through binding to the tracheary element-specific cis element TERE in their promoters.11,33,34) This co-regulated expression of genes for both secondary cell wall formation and PCD by master transcription factors is one reason for the tight coupling of secondary cell wall formation and cell death during tracheary element differentiation, which differs from the process of apoptosis in animals. These master regulators also regulate many genes related to secondary cell wall formation indirectly through the induction of other transcription factors such as MYB46 and MYB83.11,34–36) Thus, VND6 and VND7 induce a hierarchical gene expression network.
2). Patterned cell wall formation.
Tracheary elements have patterned thick secondary cell walls, which provide mechanical rigidity and facilitate water transport inside these vessels.2) Secondary wall patterns are formed by localized deposition of cellulose microfibrils, which results from guided movement of the cellulose synthase complex in the plasma membrane by cortical microtubules.37) Cortical microtubules also act at the delivery of trans-Golgi network vesicles to the plasma membrane that deliver cellulose synthase complexes to cell wall deposition locales.38,39)
Metaxylem tracheary elements form secondary cell walls with many ordered secondary cell wall-depleted regions known as pits. Using VND6-induced xylogenic culture, we were able to observe cortical microtubule dynamics during cell wall pit formation. We expected that cortical microtubules would form only in areas surrounding future pits. Surprisingly, cortical microtubules extended into future pit areas, but microtubules in these areas were selectively depolymerized.5) This finding suggests that some molecule(s) in future pit areas of differentiating metaxylem tracheary elements depolymerizes microtubules. Our search revealed a protein that localizes to future pit areas and is required for cortical microtubule depolymerization, which we designated microtubule depletion domain 1 (MIDD1).5) MIDD1 recruits kinesin-13A to provoke microtubule depolymerization.40)
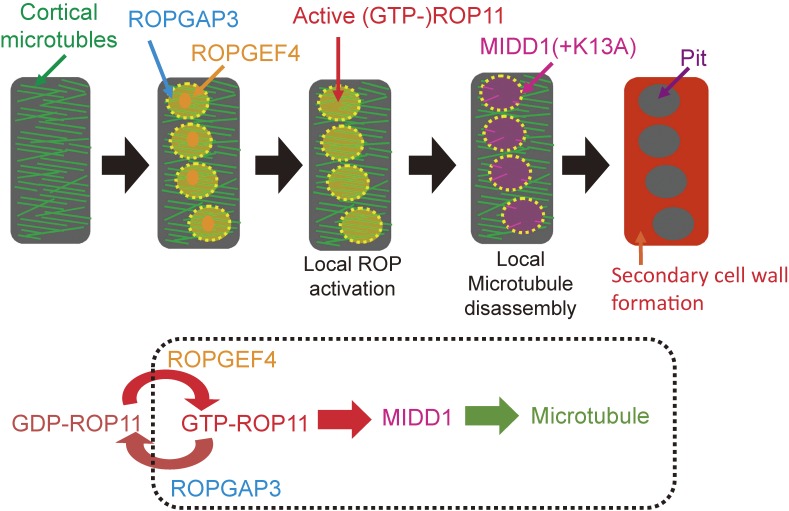
The next question to address was what determines the position and number of pits, in other words, what determines the localization of MIDD1. MIDD1 binds to plant Rho-type (ROP: Rho of plant) GTPase,41,42) suggesting that ROP is a candidate protein that localizes MIDD1 to specific membrane sites. ROP cycles between the GTP-bound (active form) and GDP-bound (inactive form) state. We found that activated ROP11 recruits MIDD1 to the plasma membrane site on which the cell wall pit will form.12) Moreover, introducing a constitutively activated form of ROP11 into differentiating xylem cells inhibits the pitted patterning of secondary cell walls, instead resulting in the formation of flat secondary cell walls. These results suggest that locally activated ROP11 initiates the pitted patterning of secondary cell wall deposition.
To investigate the local activation of ROP11 on the plasma membrane, we searched for activating and inactivating enzymes of ROP11, identifying the ROP11 activating and inactivating enzymes ROPGEF4 and ROPGAP3, respectively.12) ROPGEF4 and ROPGAP3 co-localized to future secondary cell wall pits, with wider distribution of ROPGAP3 than ROPGEF4. Indeed, loss of ROPGEF4 reduces the number of secondary cell wall pits. Surprisingly, co-introducing ROPGEF4, ROPGAP3, and ROP11 into non-xylem cells induces the formation of evenly spaced patches of active ROP11 over the plasma membrane, which resembles the localization of active ROP11 in xylem cells. This reconstruction requires intact ROP11, and GTP/GDP cycling of ROP11 is essential for its spontaneous local activation. These findings demonstrate that ROPGEF4 and ROPGAP3 mediate de novo local activation of ROP11 to establish the original patterning of secondary cell wall pits (Fig. 4).12) We suspect that the spontaneous local activation of ROP11 may be due to the breakage of symmetry through a positive feedback mechanism, as featured in Turing’s reaction-diffusion model for a slow-diffusing activator and a fast-diffusing inhibitor.43)
Figure 4.
A model of local disassembly of cortical microtubules. ROPGEF4 and ROPGAP3 mediate local activation of ROP11 at the plasma membrane. Activated ROP11 (GTP-ROP11) recruits MIDD1 to the plasma membrane domains. Kinesin 13A (K13A), which is recruited by MIDD1, promotes disassembly of cortical microtubules in the domain. Secondary wall formation does not occur on the microtubule-depleted domains, and cell wall pits are formed.
Stabilizing microtubules with Taxol causes the formation of elongated, oblique secondary cell wall pits in xylem vessels.44) We found that cortical microtubules restrict the borders of the plasma membrane domains of active ROP11 via MIDD1.12) Together, pit patterning is established by a mutual inhibition between microtubule assembly and the ROP11/MIDD1/kinesin-13A pathway.
Perspectives
Xylem cell differentiation represents a typical plant developmental pathway. Intensive studies in the past few decades have revealed the outline and key steps of xylem cell differentiation from vascular stem cells. Further sophisticated analysis is required to elucidate the detailed molecular mechanism of xylem differentiation. On the other hand, vascular stem cells differentiate not only into xylem cells, but also into phloem cells. Therefore, to understand how vascular cell fate is regulated, we must also understand the mechanism of phloem differentiation from vascular stem cells. However, no suitable culture system is currently available to study phloem differentiation at the molecular level. Recently, we found that GSK3s regulate phloem differentiation as well as xylem differentiation, and that inhibition of GSK3 activity by bikinin can induce phloem differentiation in the VISUAL system (Y. Kondo, unpublished). This phloem-inducing system will allow us to follow the phloem differentiation process at the cellular and molecular level and identify key factors regulating phloem differentiation. Indeed, we succeeded in isolating a novel regulator of phloem differentiation using this system (Y. Kondo, unpublished). Another important aspect of vascular development is the establishment of vascular stem cells. Interestingly, bikinin also plays a role in vascular stem cell initiation from mesophyll cells in the VISUAL system.6) Therefore, further studying the functions of GSK3s with VISUAL is a promising strategy for elucidating vascular stem cell initiation. Ohashi-Ito’s and Weijers’s groups have revealed that heterodimers of two bHLH transcription factors, LONESOME HIGHWAY and TARGET OF MONOPTEROS5 (TOM5)/TOM5-LIKE1, function in coordinated cell division and differentiation of vascular stem cells via negative feedback loops with their target genes in roots.45–49) Importantly, this process is tightly coupled with the local biosynthesis and functioning of two plant hormones, auxin and cytokinin. Therefore, exploring the crosstalk among plant hormones and peptide signaling pathways in specific vascular cells and future vascular cells is crucial for further elucidating the process of vascular stem cell initiation.
Acknowledgments
This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology in Japan (15H05958 and NC-CARP project), the Japan Society for the Promotion of Science (23227001), and the Naito Foundation.
Abbreviations
- BIN2
BRASSINOSTEROID-INSENSITIVE 2
- CLE
CLV3/ESR-related
- GSK3
Glycogen Synthase Kinase 3
- LRR-RLK
leucine-rich repeat-receptor-like kinase
- MIDD1
microtubule depletion domain 1
- NAC
NAM, ATAF1,2, and CUC2
- PCD
programmed cell death
- ROP
Rho of plants
- ROPGEF4
ROP GDP/GTP exchange factor 4
- ROPGAP3
ROP GTPase-activating protein 3
- TDIF
tracheary element differentiation inhibitory factor
- TDR
TDIF receptor
- NST
NAC SECONDARY WALL THICKENING PROMOTING FACTOR
- SND1
SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1
- VND
VASCULAR-RELATED NAC-DOMAIN
- VISUAL
Vascular cell Induction culture System Using Arabidopsis Leaves
- WOX
WUS-related HOMEOBOX
- ZEN1
zinnia endonuclease 1
- TOM5
TARGET OF MONOPTEROS 5
Profile
Hiroo Fukuda was born in Hamamatsu city in 1953. He graduated from the University of Tokyo in 1977 and received a Ph.D. from the University of Tokyo in 1982. He worked as an Assistant Professor at Osaka University beginning in 1983, an Associate Professor and Full Professor at Tohoku University beginning in 1989, and a Full Professor in the Department of Biological Sciences, School of Science, the University of Tokyo since 1995. For 10 months in 1987/1988, Dr. Fukuda was a visiting scientist at the Max-Plank-Institute for Plant Breeding, Cologne, Germany. He is now Dean of the School of Science, the University of Tokyo and a Council Member, Science Council of Japan. Dr. Fukuda is a plant physiologist. Using several original cell culture systems, he has uncovered various key factors regulating plant cell differentiation, particularly vascular cell differentiation, including the following: VND6 and VAN7, master transcription factors involved in xylem vessel differentiation; TDIF–TDR, a peptide ligand and its receptor involved in vascular stem cell maintenance; and MIDD1, which functions in cell wall patterning. Recently, he demonstrated that plant GSK3 proteins play a major role in intracellular signaling downstream of TDIF–TDR. Dr. Fukuda has also contributed to the plant field as the Editor-in-Chief of Plant and Cell Physiology (2004–2007) and President of the Botanical Society of Japan (2009–2012). He was recently appointed to serve as President of the Japan Society of Plant Physiologists beginning in 2016.
References
- 1).Lucas W.J., Groover A., Lichtenberger R., Furuta K., Yadav S.R., Helariutta Y., He X.Q., Fukuda H., Kang J., Brady S.M., Patrick J.W., Sperry J., Yoshida A., Lopez-Millan A.F., Grusak M.A., Kachroo P. (2013) The plant vascular system: evolution, development and functions. J. Integr. Plant Biol. 55, 294–388. [DOI] [PubMed] [Google Scholar]
- 2).Fukuda H. (1996) Xylogenesis: Initiation, progression and cell death. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 299–325. [DOI] [PubMed] [Google Scholar]
- 3).Fukuda H., Komamine A. (1980) Establishment of an experimental system for the tracheary element differentiation from single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol. 65, 57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005) Transcriptional switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Oda Y., Iida Y., Kondo Y., Fukuda H. (2010) Wood cell-wall structure requires local 2D-microtubule disassembly by a novel plasma membrane-anchored protein. Curr. Biol. 20, 1197–1202. [DOI] [PubMed] [Google Scholar]
- 6).Kondo Y., Fujita T., Sugiyama M., Fukuda H. (2015) A novel system for xylem cell differentiation in Arabidopsis thaliana. Mol. Plant 8, 612–621. [DOI] [PubMed] [Google Scholar]
- 7).Fukuda H., Komamine A. (1980) Direct evidence for cytodifferentiation to tracheary elements without intervening mitosis in a culture of single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol. 65, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Fukuda H., Komamine A. (1981) Relationship between tracheary element differentiation and the cell cycle in single cells isolated from the mesophyll of Zinnia elegans. Physiol. Plant. 52, 423–430. [DOI] [PubMed] [Google Scholar]
- 9).Demura T., Tashiro G., Horiguchi G., Kishimoto N., Kubo M., Matsuoka N., Minami A., Nagata-Hiwatashi M., Nakamura K., Okamura Y., Sassa N., Suzuki S., Yazaki J., Kikuchi S., Fukuda H. (2002) Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc. Natl. Acad. Sci. U.S.A. 99, 15794–15799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. (2006) Dodeca-CLE peptides as suppressors of plant stem cell. Science 313, 842–845. [DOI] [PubMed] [Google Scholar]
- 11).Ohashi-Ito K., Oda Y., Fukuda H. (2010) Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates genes that govern programmed cell death and secondary wall formation in a coordinated way during xylem differentiation. Plant Cell 22, 3461–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Oda Y., Fukuda H. (2012) Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337, 1333–1336. [DOI] [PubMed] [Google Scholar]
- 13).Kondo Y., Ito T., Nakagami H., Hirakawa Y., Saito M., Tamaki T., Shirasu K., Fukuda H. (2014) Plant GSK3s regulate stem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 5, 4505. [DOI] [PubMed] [Google Scholar]
- 14).Motose H., Sugiyama M., Fukuda H. (2004) A proteoglycan mediates inductive interaction during plant vascular development. Nature 429, 873–878. [DOI] [PubMed] [Google Scholar]
- 15).Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y., Fukuda H. (2008) Non-cell-autonomous control of vascular stem cell fates by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. U.S.A. 105, 15208–15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.H., Mayer K.F., Li W.H. (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Hirakawa Y., Kondo Y., Fukuda H. (2010) TDIF peptide signaling regulates vascular stem cell proliferation via the wox4 homeobox gene in Arabidopsis. Plant Cell 22, 2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- 19).Mayer K.F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- 20).Jonak C., Hirt H. (2002) Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends Plant Sci. 7, 457–461. [DOI] [PubMed] [Google Scholar]
- 21).Forde J.E., Dale T.C. (2007) Glycogen synthase kinase 3: A key regulator of cellular fate. Cell. Mol. Life Sci. 64, 1930–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Hur E.M., Zhou F.Q. (2010) GSK3 signalling in neural development. Nat. Rev. Neurosci. 11, 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Endo H., Yamaguchi M., Tamura T., Nakano Y., Nishikubo N., Yoneda A., Kato K., Kubo M., Kajita S., Katayama Y., Ohtani M., Demura T. (2015) Multiple Classes of Transcription Factors Regulate the Expression of VASCULAR-RELATED NAC-DOMAIN7, a Master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254. [DOI] [PubMed] [Google Scholar]
- 24).Ooka H., Satoh K., Doi K., Nagata T., Otomo Y., Murakami K., Matsubara K., Osato N., Kawai J., Carninci P., Hayashizaki Y., Suzuki K., Kojima K., Takahara Y., Yamamoto K., Kikuchi S. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. [DOI] [PubMed] [Google Scholar]
- 25).Yamaguchi M., Ohtania M., Mitsuda N., Kubo M., Ohme-Takagi M., Fukuda H., Demura T. (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Zhong R., Demura T., Ye Z.-H. (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Mitsuda N., Iwase A., Yamamoto H., Yoshida M., Seki M., Shinozaki K., Ohme-Takagi M. (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Nakano Y., Yamaguchi M., Endo H., Rejab N.A., Ohtani M. (2015) NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Xu B., Ohtani M., Yamaguchi M., Toyooka K., Wakazaki M., Sato M., Kubo M., Nakano Y., Sano R., Hiwatashi Y., Murata T., Kurata T., Yoneda A., Kato K., Hasebe M., Demura T. (2014) Contribution of NAC transcription factors to plant adaptation to land. Science 343, 1505–1508. [DOI] [PubMed] [Google Scholar]
- 30).Obara K., Kuriyama H., Fukuda H. (2001) Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in Zinnia. Plant Physiol. 125, 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Aoyagi S., Sugiyama M., Fukuda H. (1998) BEN1 and ZEN1 cDNAs encoding S1-type DNases that are associated with programmed cell death in plants. FEBS Lett. 429, 134–138. [DOI] [PubMed] [Google Scholar]
- 32).Ito J., Fukuda H. (2002) ZEN1 is a key enzyme in degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 14, 3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Pyo H., Demura T., Fukuda H. (2007) TERE; a novel cis-element responsible for a coordinated expression of genes related to programmed cell death and secondary wall formation during tracheary element differentiation. Plant J. 51, 955–965. [DOI] [PubMed] [Google Scholar]
- 34).Yamaguchi M., Mitsuda N., Ohtani M., Ohme-Takagi M., Kato K., Demura T. (2011) VASCULAR-RELATED NAC-DOMAIN7 directly regulates expression of a broad range of genes for xylem vessel formation. Plant J. 66, 579–590. [DOI] [PubMed] [Google Scholar]
- 35).Zhong R., Lee C., Zhou J., McCarthy R.L., Ye Z.H. (2008) A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).McCarthy R.L., Zhong R., Ye Z.H. (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. [DOI] [PubMed] [Google Scholar]
- 37).Watanabe Y., Meents M.J., McDonnell L.M., Barkwill S., Sampathkumar A., Cartwright H.N., Demura T., Ehrhardt D.W., Samuels A.L., Mansfield S.D. (2015) Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350, 198–203. [DOI] [PubMed] [Google Scholar]
- 38).Crowell E.F., Bischoff V., Desprez T., Rolland A., Stierhof Y.D., Schumacher K., Gonneau M., Hofte H., Vernhettes S. (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21, 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806. [DOI] [PubMed] [Google Scholar]
- 40).Oda Y., Fukuda H. (2013) Rho of plant GTPase signaling regulates the behavior of Arabidopsis Kinesin-13A to establish secondary cell wall patterns. Plant Cell 25, 4439–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Lavy M., Bloch D., Hazak O., Gutman I., Poraty L., Sorek N., Sternberg H., Yalovsky S. (2007) A Novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr. Biol. 17, 947–952. [DOI] [PubMed] [Google Scholar]
- 42).Li S., Gu Y., Yan A., Lord E., Yang Z. (2008) RIP1 (ROP Interactive Partner 1)/ICR1 marks pollen germination sites and may act in the ROP1 pathway in the control of polarized pollen growth. Mol. Plant 1, 1021–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Oda Y., Fukuda H. (2013) Spatial organization of xylem cell walls by ROP GTPases and microtubule-associated proteins. Curr. Opin. Plant Biol. 16, 743–748. [DOI] [PubMed] [Google Scholar]
- 44).Oda Y., Mimura T., Hasezawa S. (2005) Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol. 137, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Ohashi-Ito K., Obuchi M., Kojima M., Sakakibara H., Fukuda H. (2013) Auxin-associated initiation of vascular cell differentiation by LONESOME HIGHWAY. Development 140, 765–769. [DOI] [PubMed] [Google Scholar]
- 46).Ohashi-Ito K., Saegusa M., Iwamoto K., Oda Y., Katayama H., Kojima M., Sakakibara H., Fukuda H. (2014) A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr. Biol. 24, 2053–2058. [DOI] [PubMed] [Google Scholar]
- 47).Katayama H., Iwamoto K., Kariya Y., Asakawa T., Kan T., Fukuda H., Ohashi-Ito K. (2015) A negative feedback loop controlling bHLH complexes is involved in vascular cell division and differentiation in the root apical meristem. Curr. Biol. 25, 3144–3150. [DOI] [PubMed] [Google Scholar]
- 48).De Rybel B., Möller B., Yoshida S., Grabowicz I., Barbier de Reuille P., Boeren S., Smith R.S., Borst J.W., Weijers D. (2013) A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell 24, 426–437. [DOI] [PubMed] [Google Scholar]
- 49).De Rybel B., Adibi M., Breda A.S., Wendrich J.R., Smit M.E., Novák O., Yamaguchi N., Yoshida S., Van Isterdael G., Palovaara J., Nijsse B., Boekschoten M.V., Hooiveld G., Beeckman T., Wagner D., Ljung K., Fleck C., Weijers D. (2014) Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345, 1255215. [DOI] [PubMed] [Google Scholar]