Abstract
Zfp57 is a master regulator of genomic imprinting in mouse embryos. To further test its functions, we have derived multiple Zfp57 mutant ES clones directly from mouse blastocysts. Indeed, we found DNA methylation imprint was lost at most examined imprinting control regions in these Zfp57 mutant ES clones, similar to what was observed in Zfp57 mutant embryos in the previous studies. This result indicates that these blastocyst-derived Zfp57 mutant ES clones can be employed for functional analyses of Zfp57 in genomic imprinting.
Blastocyst-derived ES clones display typical properties of ES cells
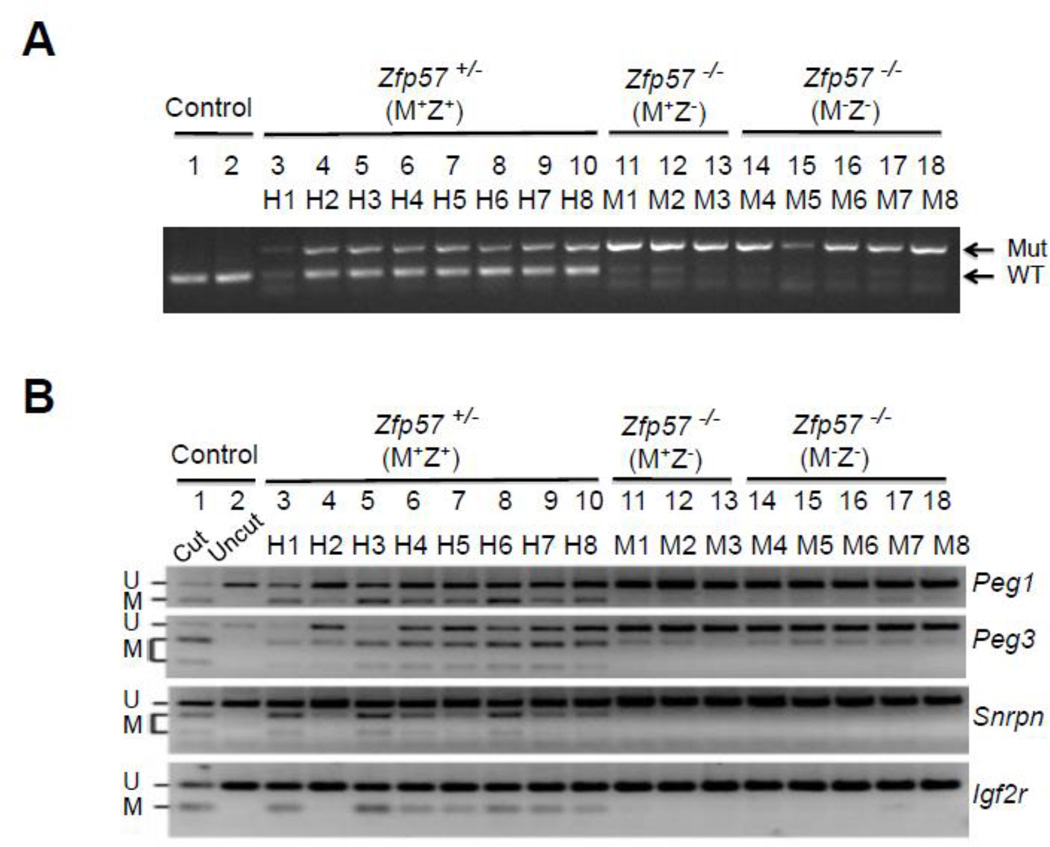
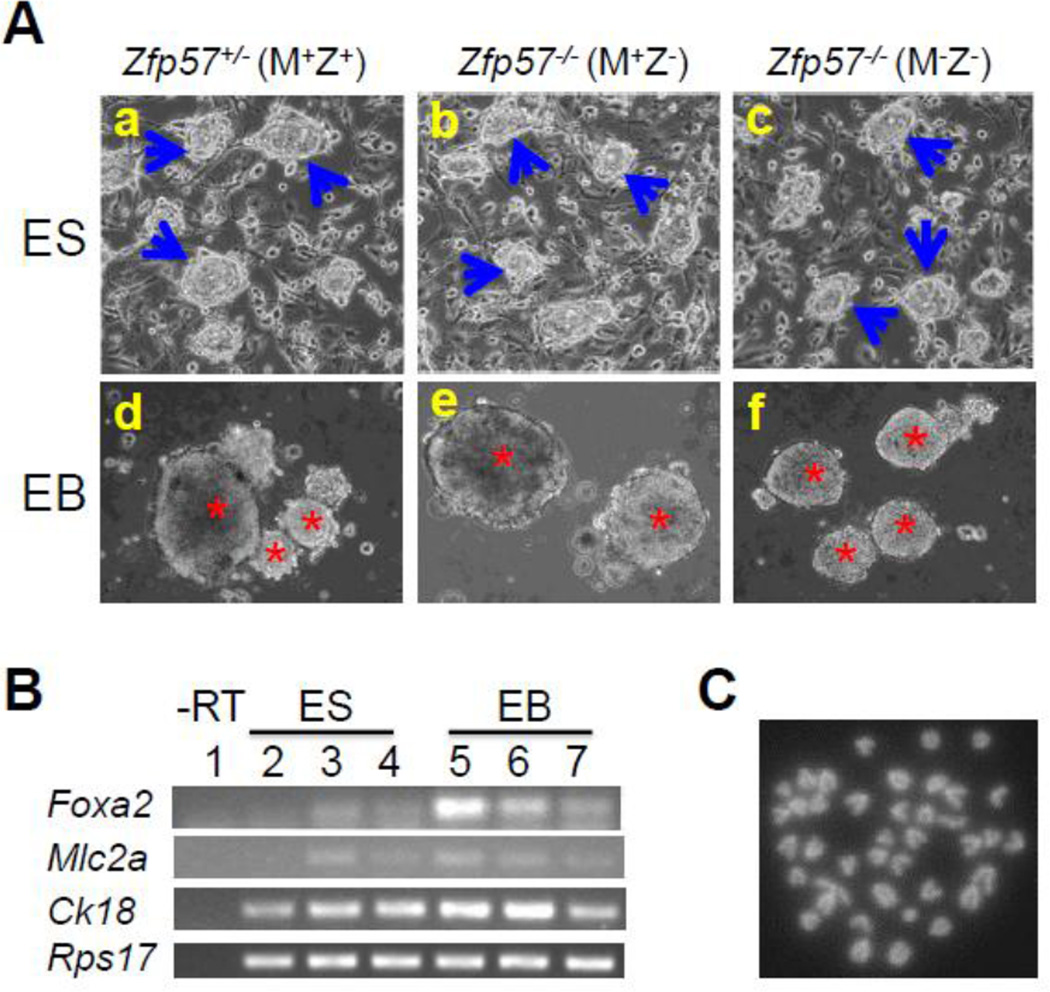
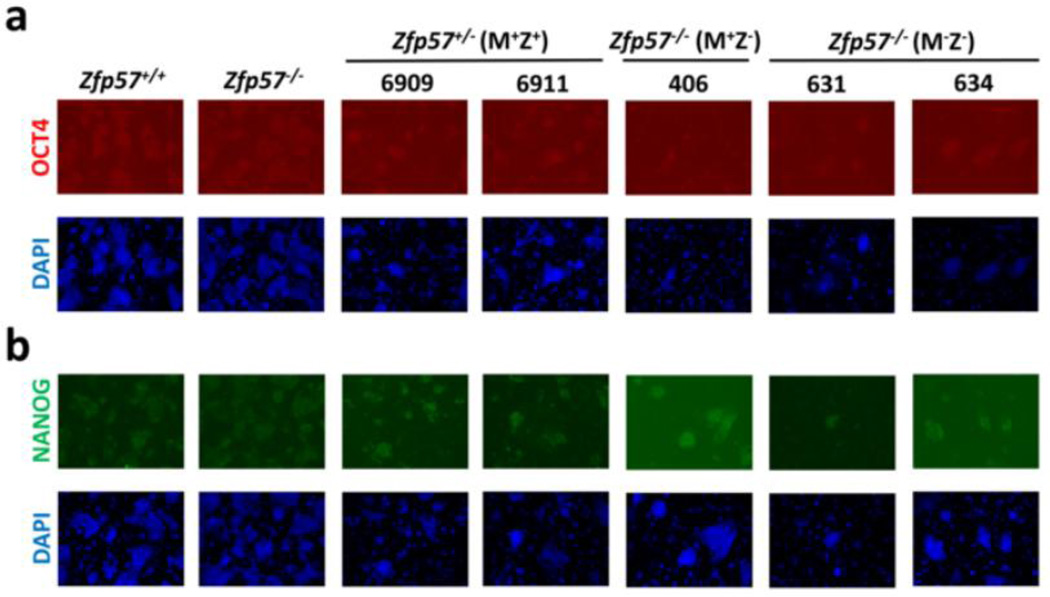
Zfp57 is a maternal-zygotic effect gene and it has both maternal (M) and zygotic (Z) functions (Li et al., 2008; Shamis et al., 2015). Eight Zfp57+/− (M+Z+) and three Zfp57−/− (M+Z−) ES clones were derived from the blastocysts generated from the cross between Zfp57+/− heterozygous female mice and Zfp57−/− homozygous male mice, whereas five Zfp57−/− (M−Z−) ES clones were derived from the cross between Zfp57−/− homozygous female mice and Zfp57−/− homozygous male mice. The genotypes for these ES clones were confirmed by PCR-based genotyping (see Fig. 3A below). These ES clones displayed typical ES cell morphology as exemplified by one ES clone for each genotype shown in Fig. 1A, suggesting that undifferentiated ES clones can be properly established without maternal (M−) or zygotic (Z−) Zfp57. They formed embryoid bodies (EBs) when grown in suspension (Fig. 1A). Based on semi-quantitative RT-PCR analysis, expression of the markers for endoderm (Foxa2), mesoderm (Mlc2a) and ectoderm (Ck18) seems to be increased in the EB samples compared with the ES cell samples in three tested blastocyst-derived ES clones (Fig. 1B). We also counted the metaphase chromosome numbers in five ES clones (Table 1), as exemplified by one metaphase chromosome spread (Fig. 1C). All five ES clones appeared to have relatively normal chromosome numbers, with over 60% or more euploid cells with 40 chromosomes in these ES clones (Table 1). Based on immunostaining, these five ES clones express OCT4 and NANOG, two pluripotency markers (Fig. 2).
Figure 3. COBRA analysis of maternally inherited DNA methylation imprint.
A, PCR genotyping of these ES clones, similar to our previously published study (Li et al., 2008). Mut and WT, the marked gel positions for the PCR product of the mutant (Mut) and wild-type (WT) alleles of Zfp57. B, COBRA analysis was carried out for analyzing DNA methylation imprint at the maternally inherited Peg1, Peg3, Snrpn and Igf2r imprinted regions. Restriction enzyme (RE) digestion was performed for the bisulphite PCR product of all ES samples, and “Cut” (Lane 1) but not “Uncut” (Lane 2) control wild-type mouse tail DNA sample. U and M, unmethylated (U) and methylated (M) product after RE digestion, respectively. Lanes 3–10, eight Zfp57+/− (M+Z+) ES clones derived from the blastocysts generated from the cross between Zfp57+/− heterozygous female mice and Zfp57−/− homozygous male mice. Lanes 11–13, three Zfp57−/− (M+Z−) ES clones derived from the blastocysts generated from the cross between Zfp57+/− heterozygous female mice and Zfp57−/− homozygous male mice. Lanes 14–18, five Zfp57−/− (M−Z−) ES clones derived from the cross between Zfp57−/− homozygous female mice and Zfp57−/− homozygous male mice.
Figure 1. Blastocyst-derived ES clones display normal characters of ES cells.
A, ES clones on feeder cells (a-c) or EBs on non-adherent petri dish plates (d-f). One Zfp57+/− (M+Z+) ES clone (a, d) and one Zfp57−/− (M+Z−) ES clone (b, e) were shown here as the examples for the ES clones derived from the blastocysts generated from the cross between Zfp57+/− heterozygous female mice and Zfp57−/− homozygous male mice, whereas one Zfp57−/− (M−Z−) ES clone (c, f) shown here was an example for the ES clones derived from the cross between Zfp57−/− homozygous female mice and Zfp57−/− homozygous male mice. Blue arrows in a-c, undifferentiated ES cell colonies grown on feeder cells. Red asterisks in d-f, embryoid bodies (EBs) in suspension formed by ES cells plated on non-adherent petri dish plates. B, RT-PCR expression analysis of the marker genes in three ES clones and the EBs derived from these three ES clones after growing in suspension culture for 7–8 days. Lane 1, negative control without reverse transcription (-RT) of the same total RNA sample in Lane 4. Lane 2, ES cells of one Zfp57−/− (M−Z−) ES clone. Lanes 3–4, the ES cells of two Zfp57+/− (M+Z+) ES clones. Lane 5, EBs of the Zfp57−/− (M−Z−) ES clone. Lanes 6–7, EBs of two Zfp57+/− (M+Z+) ES clones. Foxa2, Mlc2a and Ck18 are markers for endoderm, mesoderm and ectoderm, respectively. Rps17 is a house-keeping gene that was used as the loading control. C, DAPI-stained metaphase chromosome spread derived from a cell of one Zfp57−/− (M−Z−) ES clone. An example is provided here for the metaphase chromosome spread of five ES clones.
Table 1.
Counting of metaphase chromosome spreads of five ES clones
| ES clone | 6909 | 6911 | 406 | 631 | 634 |
| Genotype |
Zfp57+/− (M+Z+) |
Zfp57+/− (M+Z+) |
Zfp57−/− (M+Z−) |
Zfp57−/− (M−Z−) |
Zfp57−/− (M−Z−) |
| # of counted metaphase spreads | 16 | 20 | 20 | 20 | 20 |
| # of spreads with 40 chromosomes | 10 | 12 | 14 | 14 | 13 |
| % of euploid cells | 62.5% | 60% | 70% | 70% | 65% |
Figure 2. Blastocyst-derived ES clones express pluripotent markers OCT4 and NANOG.
Immunostaining was performed to analyze the expression of OCT4 (a) and NANOG (b) in five blastocyst-derived ES clones. Two leftmost columns, control wild-type TC1 (Zfp57+/+) and a Zfp57−/− mutant ES clone generated by homologous recombination in vitro (Li et al., 2008; Zuo et al., 2012). 6909, 6911, 406, 631 and 634 are five blastocyst-derived ES clones generated in this study. Blue signal, DAPI staining. Red signal, OCT4 immunostaining. Green signal, Nanog immunostaining.
Maintenance of maternally inherited DNA methylation imprint
We also analyzed DNA methylation imprint inherited on the maternal chromosomes at four imprinted regions (Peg1, Peg3, Snrpn and Igf2r) in these ES clones. Similar to what had been observed in Zfp57+/− (M+Z+) and Zfp57−/− (M−Z−) mouse embryos in our previous study (Li et al., 2008), DNA methylation imprint was lost at these four imprinted regions in five Zfp57−/− (M−Z−) ES clones (M4-M8 of Fig. 3B) in comparison with eight Zfp57+/− (M+Z+) ES clones (H1-H8 of Fig. 3B). DNA methylation imprint was also lost at these four imprinted regions in three Zfp57−/− (M+Z−) ES clones (M1-M3 of Fig. 3B), whereas partial loss of DNA methylation was observed at these four imprinted regions in Zfp57−/− (M+Z−) zygotic mutant embryos lacking just zygotic Zfp57. Again these results suggest that maternal Zfp57 appears to have more lasting effect on the maintenance of DNA methylation imprint inherited on maternal chromosomes in mouse embryos than in blastocyst-derived ES cells.
Maintenance of paternally inherited DNA methylation imprint
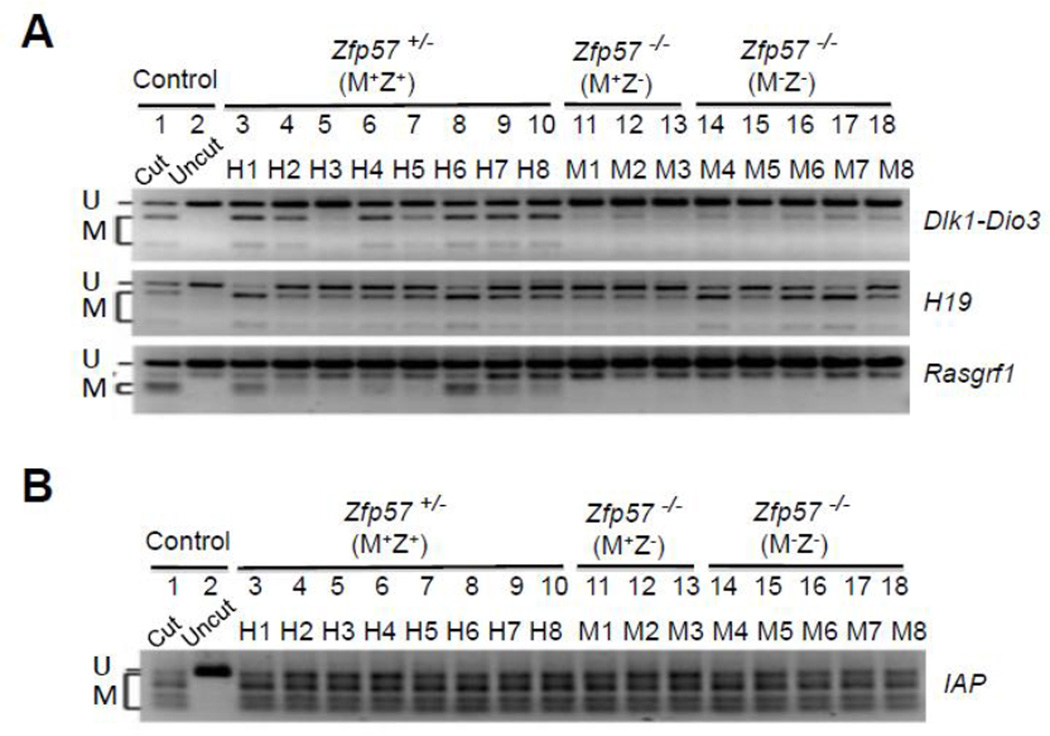
We examined paternally inherited DNA methylation imprint at three imprinted regions (Dlk1-Dio3, H19 and Rasgrf1) in these ES clones by COBRA. Similar to what was found in Zfp57+/− (M+Z+) and Zfp57−/− (M−Z−) mouse embryos in our previous study (Li et al., 2008), DNA methylation imprint was lost at the Dlk1-Dio3 imprinted region but not at the H19 imprinted region in five Zfp57−/− (M−Z−) ES clones (M4-M8 of Fig. 4A) in comparison to eight Zfp57+/− (M+Z+) ES clones (H1-H8 of Fig. 4A). Unlike partial loss of DNA methylation imprint observed in Zfp57−/− (M+Z−) zygotic mutant embryos lacking just zygotic Zfp57 in our previous study (Li et al., 2008), DNA methylation imprint was almost completely lost at the Dlk1-Dio3 imprinted region but not at the H19 imprinted region in three Zfp57−/− (M+Z−) ES clones (M1-M3 of Fig. 4A), indicating that maternal Zfp57 has more lasting effect on the maintenance of DNA methylation imprint inherited on paternal chromosomes in mouse embryos than in blastocyst-derived ES cells. We also found that DNA methylation imprint was similarly lost at the Rasgrf1 imprinted region in three Zfp57−/− (M+Z−) and five Zfp57−/− (M−Z−) ES clones in comparison with eight Zfp57+/− (M+Z+) ES clones (Fig. 4A). This is the first experimental evidence demonstrating that DNA methylation imprint at the Rasgrf1 imprinted region is lost without Zfp57.
Figure 4. COBRA analysis of paternally inherited DNA methylation imprint.
Restriction enzyme (RE) digestion was performed for the bisulphite PCR product of all ES samples, and “Cut” (Lane 1) but not “Uncut” (Lane 2) control wild-type mouse tail DNA sample. U and M, unmethylated (U) and methylated (M) product after RE digestion, respectively. Lanes 3–10, eight Zfp57+/− (M+Z+) ES clones derived from the blastocysts generated from the cross between Zfp57+/− heterozygous female mice and Zfp57−/− homozygous male mice. Lanes 11–13, three Zfp57−/− (M+Z−) ES clones derived from the blastocysts generated from the cross between Zfp57+/− heterozygous female mice and Zfp57−/− homozygous male mice. Lanes 14–18, five Zfp57−/− (M−Z−) ES clones derived from the cross between Zfp57−/− homozygous female mice and Zfp57−/− homozygous male mice.
A, COBRA analysis was carried out at the IG-DMR of the Dlk1-Dio3 imprinted region, H19 DMR of the Igf2-H19 imprinted region and Rasgrf1 DMR.
B, COBRA analysis of the IAP repeats.
DNA methylation at the IAP repeats
Previously, we did not observe any loss of DNA methylation at the IAP repeat regions in mouse embryos lacking Zfp57 (Li et al., 2008). Similarly, IAP repeat regions were highly methylated in eight Zfp57+/− (M+Z+) (H1-H8), three Zfp57−/− (M+Z−) (M1-M3) and five Zfp57−/− (M−Z−) (M4-M8) ES clones (Fig. 4B). Thus, Zfp57 is not required for the maintenance of DNA methylation at the IAP repeats in blastocyst-derived ES clones.
Materials and Methods
Zfp57 mutant mouse strain
Zfp57 mutant mouse was generated in our previous study (Li et al., 2008). The mouse strain carrying the Zfp57 mutant allele was maintained in the animal facility of Icahn School of Medicine at Mount Sinai. The animal work was performed in full compliance with the animal care protocol approved by IACUC of the institution.
Derivation of the ES clones from blastocysts
Mouse blastocytsts were isolated from the timed pregnancy mating between Zfp57+/− heterozygous female mice and Zfp57−/− homozygous male mice or between Zfp57−/− homozygous female mice and Zfp57−/− homozygous male mice. These blastocysts were used for derivation of the ES clones following the protocol described in this paper (Meissner et al., 2009). Basically, isolated blastocysts were plated on irradiated MEF cells (feeder cells) until undifferentiated ES cell colonies formed on the plate. They were picked individually to a well of 24-well plate to establish the stable ES cell lines. Then the established ES cell lines were expanded in 6-well plates seeded with feeder cells.
ES cell culture
ES cells were cultured in the presence of feeder cells in the ES cell growth medium made of the high-glucose DMEM medium supplemented with 15% fetal bovine serum (FBS). They were passaged once every 3–4 days before the ES cells became too confluent. The ES cell medium was changed daily.
Generation of embryoid bodies (EBs)
Roughly 1–2 million of ES cells grown on a well of 6-well plate with irradiated SNL feeder cells were separated by trypsin digestion before being added to a non-adherent 10-cm petri dish plates pre-treated with poly-hema (Sigma). ES cell growth medium without LIF was used for the EB culture. The medium was changed once every 2–3 days without removing the aggregated EBs. The floating EBs were harvested for total RNA sample preparation after growing in suspension for 7–8 days. Usually 2 ml of Trizol reagent (Invitrogen) was added to the EBs collected from a 10- cm dish plate after removing the medium. Total RNA samples were prepared according to the manufacturer’s manual.
Chromosome number counting of ES clones
Metaphase chromosome spread was prepared for five ES clones. Karyomax (Invitrogen Cat# 15210-040) was added to the ES cells grown in a well of 6-well plates with a final concentration of Colcemid at 1 µg/ml. After incubation for one hour with Karyomax, the ES cells were harvested by trypsin digestion followed by centrifugation. The cell pellets were gently mixed with 5 ml of ice-cold 0.56% KCl solution in sterile distilled water. After incubation for six minutes at room temperature, the ES samples were precipitated by centrifugation and the pellets were mixed with the fixative solution of acetic acid and methanol (1:3). The pellets were resuspended in this fixative solution before being spotted onto the slides for examination under microscope. After photography, the chromosome numbers of about 20 good spreads were counted for each of these five ES clones.
RT-PCR expression analysis of the marker genes for three germ layers
Total RNA samples were prepared from three ES clones and their EBs after growing in suspension culture for 7–8 days. Approximately equal amount of total RNA samples were used for reverse transcription (RT) with Transcriptor First Strand cDNA Synthesis Kit (Roche). RT reaction was initiated with the anchored-oligo(dT)18 primer included in the kit, with a negative control without reverse transcriptase (-RT) for one total RNA sample. 1 ul of RT product was used for PCR amplification. The amplification of 25 PCR cycles was applied to Ck18 and Rps17 encoding the housekeeping ribosomal protein S17, whereas Foxa2 and Mlc2a were subjected to PCR amplification of 35 cycles. PCR product was separated on agarose gels before photography.
Immunostaining of ES clones
The ES clones grown on a 24-well plate seeded with irradiated SNL feeder cells were directly subjected to immunostaining with antibodies from Santa Cruz Biotechnology against OCT4 (sc-5279) or Nanog (sc-376915). The nuclei were stained with DAPI.
Bisulphite mutagenesis
The genomic DNA samples isolated from these ES clones and the control wild-type mouse tail were subjected to bisulphite treatment with the EZ DNA Methylation-Gold™ Kit (Zymo Research). The bisulphite-treated DNA product was used for COBRA analysis of the imprinting control regions (ICR) and the non-imprinted IAP repeats.
Combined Bisulphite Restriction Analysis (COBRA)
COBRA was used for analyzing DNA methylation imprint at the ICRs and IAP repeats in this study. After bisulphite mutagenesis, the purified mutagenized genomic DNA was subjected to PCR amplification with the primers covering a portion of the imprinting control region (ICR) for the imprinted regions or a portion of the non-imprinted IAP repeat regions (Zuo et al., 2012). The resultant PCR product was subjected to restriction enzyme digestion targeting the CpG sites located in the amplified ICR or IAP repeat regions. The restriction enzyme digested PCR product was separated by gel electrophoresis.
Acknowledgments
The work in the author’s laboratory is currently supported by the grant from NIH (R01GM093335). XL conceived the study. HTL and LL performed the experiments. XL wrote the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
| Name of Stem Cell construct | Not Applicable |
| Institution | Icahn School of Medicine at Mount Sinai |
| Person who created resource | Ho-Tak Lau, Xiajun Li |
| Contact person and email | Xiajun Li, xiajun.li@mssm.edu |
| Date archived/stock date | November, 2010 |
| Origin | Mouse blastocysts |
| Type of resource | Biological reagent: Mouse Embryonic Stem (ES) Cell Lines |
| Sub-type | Zfp57 mutant ES cell lines |
| Key transcription factors | Zfp57 |
| Authentication | Undifferentiated ES cell morphology confirmed (Figure 1) |
| Link to related literature (direct URL links and full references) | http://www.ncbi.nlm.nih.gov/pubmed/18854139 |
| Information in public databases | None |
References
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Eminli S, Jaenisch R. Derivation and manipulation of murine embryonic stem cells. Methods Mol Biol. 2009;482:3–19. doi: 10.1007/978-1-59745-060-7_1. [DOI] [PubMed] [Google Scholar]
- Shamis Y, Cullen DE, Liu L, Yang G, Ng SF, Xiao L, Bell FT, Ray C, Takikawa S, Moskowitz IP, et al. Maternal and zygotic Zfp57 modulate NOTCH signaling in cardiac development. Proc Natl Acad Sci U S A. 2015;112:E2020–E2029. doi: 10.1073/pnas.1415541112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, et al. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem. 2012;287:2107–2118. doi: 10.1074/jbc.M111.322644. [DOI] [PMC free article] [PubMed] [Google Scholar]