Abstract
The β-1,3-glucanase gene family is involved in a wide range of plant developmental processes as well as pathogen defense mechanisms. Comprehensive analyses of β-1,3-glucanase genes (GLUs) have not been reported in cotton. Here, we identified 67, 68, 130 and 158 GLUs in four sequenced cotton species, G. raimondii (D5), G. arboreum (A2), G. hirsutum acc. TM-1 (AD1), and G. barbadense acc. 3–79 (AD2), respectively. Cotton GLUs can be classified into the eight subfamilies (A–H), and their protein domain architecture and intron/exon structure are relatively conserved within each subfamily. Sixty-seven GLUs in G. raimondii were anchored onto 13 chromosomes, with 27 genes involved in segmental duplications, and 13 in tandem duplications. Expression patterns showed highly developmental and spatial regulation of GLUs in TM-1. In particular, the expression of individual member of GLUs in subfamily E was limited to roots, leaves, floral organs or fibers. Members of subfamily E also showed more protein evolution and subgenome expression bias compared with members of other subfamilies. We clarified that GLU42 and GLU43 in subfamily E were preferentially expressed in root and leaf tissues and significantly upregulated after Verticillium dahliae inoculation. Silencing of GLU42 and GLU43 significantly increased the susceptibility of cotton to V. dahliae.
The hydrolysis of 1,3-β-D-glucosidic linkages in β-1,3-glucans is catalyzed by the enzymes of β-1,3-glucanases (E.C. 3.2.1.39), which are widely found in bacteria, fungi, viruses1,2 and various plant species, including Arabidopsis3, rice4, tobacco5 and soybean6. β-1,3-Glucanase genes (GLUs) have formed complex and diverse gene families in plants, where they play important roles in a wide range of physiological and developmental processes3. In plants, the interest in β-1,3-glucanase has focused primarily on their antifungal activity. For example, plant β-1,3-glucanases, classified as the PR-2 family of pathogenesis-related proteins, have been proposed to defend against pathogen infection in two ways: by hydrolyzing β-1,3-glucan, a major structural component of fungal cell walls; and by promoting the release of cell wall associated immune elicitors that further stimulate defense reactions7,8. In other areas, there is strong evidence that these glycosyl hydrolases have also played the important roles in pollen development4,9, seed germination10, cold response11 and regulation of plasmodesmata signaling12,13.
According to the diversity in its C-terminal domain, plant β-1,3-glucanase has been divided into different types, and each type contains an N-terminal signal peptide and a glycoside hydrolase family 17 domain3. In tobacco, β-1,3-glucanases are grouped into five classes5,14. Class I comprises basic proteins and exhibits developmental regulation. These proteins contain a short C-terminal signal sequence that is believed to be the responsible for vacuolar localization. In contrast to class I, the class II and III proteins are acidic proteins that are induced by pathogens. They appear to be localized in the extracellular spaces because they lack the C-terminal signal sequence. The class IV and V proteins are also acidic proteins with no C-terminal extension, however, they have different expression pattern when compare to class II and III proteins. The class IV proteins, including sp41a and sp41b, are expressed in the style of tobacco flowers and are not pathogen inducible. Tag1 belongs to class V and is expressed exclusively in the tobacco anther tapetum. In Arabidopsis, β-1,3-glucanases are also classified into three major clades (denoted α, β and γ) based on their evolutionary relationships3.
Cotton is one of the most important economic crops worldwide, and provides the world’s leading natural textile fiber and considerable amounts of edible oil. Among approximately 50 species in the Gossypium genus, only four, two diploids (G. arboreum and G. herbaceum) and two allotetraploids (G. hirsutum and G. barbadense), have been independently domesticated for the production of economically valuable fibers. Previous studies have shown that allotetraploid Gossypium species were formed by a polyploidization event that occurred 1–2 million years (Myr) ago, which involved a maternal A-genome species resembling G. arboreum and a paternal D-genome species resembling G. raimondii15. With its high yield properties, G. hirsutum (commonly called Upland cotton) contributes over 90% of the annual global commercial cotton production. Nevertheless, cotton production is limited by diverse biotic and abiotic stresses. Verticillium wilt is a widespread disease that occurs in a range of cotton producing areas. More than 50% of the cotton acreage is influenced by Verticillium wilt in some years, significantly reducing the fiber quality and yield (National Cotton Council, 2012 http://www.cotton.org/). β-1,3-Glucanases have been proposed to play important roles in physiological and developmental processes, as well as in the response of plants to microbial pathogens, and show great functional diversity between the members of this large gene family7. In order to obtain an integrated image of the evolutionary characteristics and possible functions of the β-1,3-glucanase gene family in Gossypium, it is necessary to develop genome-wide analyses of the gene family at various levels. The publically available genomic information from four sequenced cotton species, G. raimondii16, G. arboreum17, G. hirsutum18 and G. barbadense19, provides us with a data source to identify candidate GLUs at a genome-wide level in Gossypium, and to mine key genes for the genetic improvement of yield and fiber quality, as well as disease resistance.
In this study, we individually identified GLUs from four sequenced cotton species, and analyzed their chromosomal location, sequence phylogeny, genomic structure and expansion pattern. We carried out a genome-wide analysis of the temporal and spatial expression profiles of GLUs in G. hirsutum, and investigated their molecular evolution and subgenome expression divergence. Further, specific focus was placed on the GLUs in subfamily E and virus-induced gene silencing (VIGS) analysis confirmed that silencing of the two GLUs in subfamily E significantly increased the susceptibility of cotton to V. dahlia. Our study provides a basis for systematically elucidating the evolutionary and functional characteristics of GLUs in cotton, for the effective clarification of the precise biological roles of GLUs and their utilization in future cotton-breeding programs.
Results
Genome-wide identification of the β-1,3-glucanase gene family in Gossypium
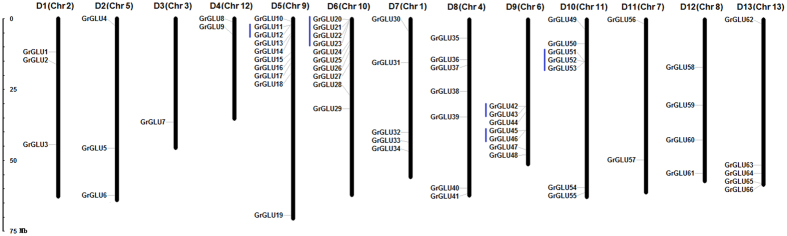
The whole genome sequence scaffolds of four sequenced cotton species (G. raimondii, G. arboreum, G. hirsutum acc. TM-1 and G. barbadense acc. 3–79) were used for the genome-wide exploration of the β-1,3-glucanase gene family in Gossypium. With data from a query on the glycoside hydrolase family 17 domain (PF00332), we searched the protein databases of four cotton species using HMMER v3.0 software. In total, 67 GLUs in G. raimondii were obtained after confirming the “Glyco_hydro_17” domain with a blastp program and these were named GrGLU1 through GrGLU67 by combining their chromosome order (D1 to D13)20 with their location on the chromosome (Fig. 1). The chromosomal distribution patterns of these GrGLUs were uneven. Chrs D5 and D6 contained the most GLUs (10 genes), while Chr D3 contained the fewest (one gene). Further, we identified 68 GLUs in G. arboreum. As these had an orthologous relationship to the GLUs in G. raimondii, the 67 GLUs in G. arboreum were named GaGLU1-67, corresponding to GrGLU1-67. Notably, GaGLU54 had undergone a tandem duplication event in G. arboreum, and the two tandem duplication genes were named GaGLU54a and GaGLU54b.
Figure 1. Chromosomal distribution of β-1,3-glucanase genes in G. raimondii.
The chromosome numbers were consistent with the interspecific genetic map (D1 to D13) in allotetraploid cultivated cotton species20 and the scaffolds (Chr.1 to Chr.13) in the genomic data of G. raimondii16. The nomenclature of GLUs was based on the order of the chromosomes in G. raimondii. Lines were drawn to indicate the tandem duplicated genes.
From a phylogenetic point of view, there were two homologous genes (homeologs from the A and D subgenomes) in the tetraploid cotton species. In total, we identified 130 GLUs in G. hirsutum acc. TM-1 (named GhGLU1-67A/D; Supplementary Fig. 1) and 158 in G. barbadense acc. 3–79, and these candidate GLUs in G. barbadense experienced more tandem duplications at the subgenome level. More than 90% of the GLUs had homoeologous genes in the two tetraploid cotton species, indicating the independent evolution of the A- and D-subgenomes after polyploid formation. Other inconsistencies might result from different sequencing methods, assembly error in partial chromosomal regions, or different degrees of colonization during the evolutionary process of Gossypium, and need to be further confirmed. In G. hirsutum acc. TM-1, the chromosomal location of GhGLUs in the D subgenome shows good collinearity with that in the D genome (G. raimondii), however, two reciprocal translocations between the A2 (Chr2) and A3 (Chr3) and between the A4 (Chr4) and A5 (Chr5) chromosomes were detected in G. hirsutum18 (Supplementary Fig. 1). Information on the GLUs in two diploid and two allotetraploid cotton species is summarized in Supplementary Table 1.
Classification and structural analysis of β-1,3-glucanases
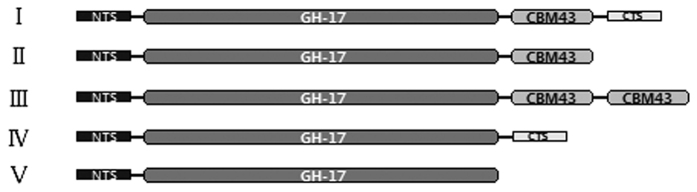
Each ortholog from the four surveyed cotton species had a similar protein structure. With the information on the GrGLUs in G. raimondii as an example of the further analysis carried out, all 67 GrGLUs contained an N-terminal signal peptide and a glycoside hydrolase family 17 domain. In detail, five types of protein domain architectures (type I to type V) were observed in the GrGLUs (Fig. 2), which was consistent with that in Arabidopsis3. The carbohydrate-binding modules family 43 (CBM43) domain, which has the ability to bind β-1,3-glucan21, was found in 39 of the 67 GrGLUs. A C-terminal hydrophobic sequence encoding a transient transmembrane domain was present in 42 GrGLUs. The transient transmembrane domain may be a vacuolar targeting peptide22 or a glycosylphosphatidylinisotol (GPI)-anchor attachment23,24. Further, we compared the number of GrGLUs with that in A. thaliana, Theobroma cacao and Vitis vinifera for each protein domain architectural type (Table 1). Type III was the smallest, with only one member in G. raimondii and A. thaliana, and none in T. cacao or V. vinifera. Type I was the largest, with 23 members in G. raimondii, 19 in A. thaliana and 16 in T. cacao. It is reasonable to suggest that the ancestral GLUs in plants may have the protein domain architecture as type I, with a CBM43 domain and a C-terminal hydrophobic sequence after the core glycosyl hydrolase family 17 domain, and these genes may play an important role in cell division or cell wall remodeling as they have abundant expression in a variety of tissues and organs. During the evolutionary process, the loss of the C-terminal region, including the GPI anchor, and the alteration of expression patterns of duplicated genes by acquiring or losing of regulatory cis-elements, led to the formation of new GLUs, which were classified as types II to V, and resulting in their functional diversity3.
Figure 2. Protein domain architectures of β-1,3-glucanases in cotton and A. thaliana.
NTS: N-terminal sequence. GH-17: glycosyl hydrolase family 17 domain. CBM43: carbohydrate-binding modules family 43. CTS: hydrophobic C-terminal sequence. These five architectural types are based on the presence/absence of CMB43 and CTS in C-terminal.
Table 1. Size of the five protein domain architectures of β-1,3-glucanases in different species.
| Species name | Total | I | II | III | IV | V |
|---|---|---|---|---|---|---|
| Gossypium raimondii | 67 | 23 | 15 | 1 | 19 | 9 |
| Arabidopsis thaliana | 51 | 19 | 9 | 1 | 10 | 12 |
| Theobroma cacao | 44 | 16 | 10 | 0 | 7 | 10 |
| Vitis vinifera | 43 | 9 | 10 | 0 | 12 | 8 |
The number of β-1,3-glucanases in each protein domain architecture is listed in G. raimondii, A. thaliana, T. cacao and V. vinifera. Thecc1EG027171t1 in T. cacao and GSVIVT01030073001, GSVIVT01012711001, GSVIVT01022253001, and GSVIVT01025431001 in V. vinifera had not classified into the above five types due to the existence of other domains in C-terminal.
Phylogenetic analysis of β-1,3-glucanase genes
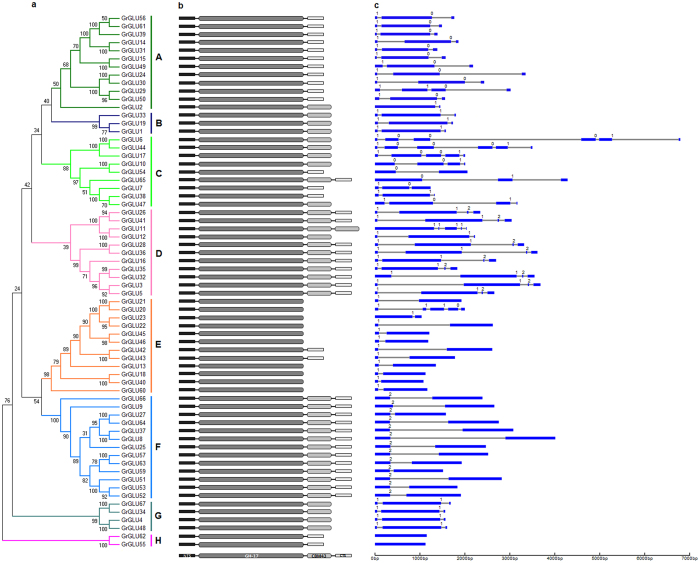
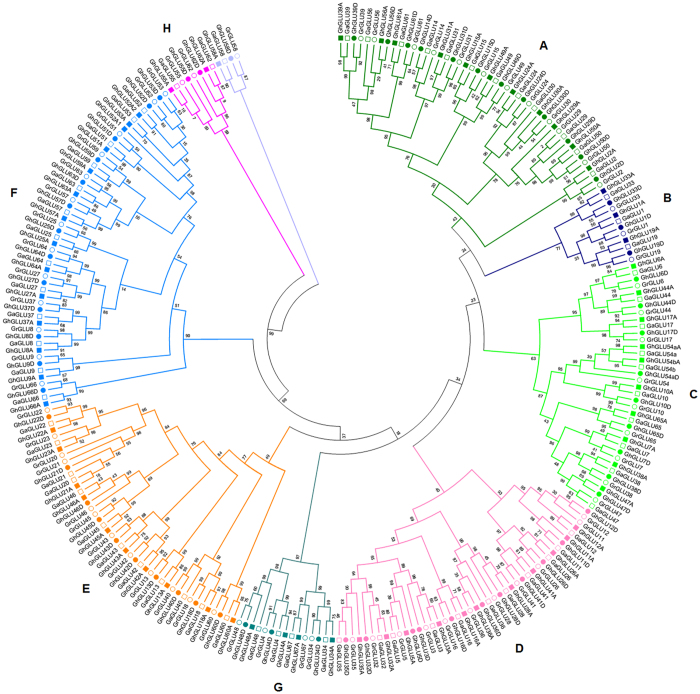
Systematic classifications of GLUs at a genome-wide level have been reported previously in Arabidopsis3 and tobacco5. To gain further insights into the evolutionary relationships, we employed MEGA v5.2 software to construct an unrooted phylogenetic tree of GLUs from G. raimondii, A. thaliana and tobacco. The phylogenetic tree clearly showed that the 123 GLUs could be clustered into eight subfamilies (A–H) (Supplementary Fig. 2), with At5g67469 and GrGLU58 (which contain a partial “Glyco_hydro_17” domain) as outgroup. All five classes of tobacco GLUs fall into subfamily E. Arabidopsis clades β and γ were consistent with subfamilies E and F, respectively, and the GLUs in clade α fall into the other subfamilies. The GrGLUs were classified into eight subfamilies, and this phylogenetic classification was consistent with the protein domain architecture and exon-intron organization (Fig. 3). Subfamilies A, C, D, E and F each contained more GrGLUs, with the number of members ranging from 9 to 13, while subfamilies B, G and H each contained only 3, 4 and 2 GrGLUs, respectively.
Figure 3. Phylogenetic relationships, protein domain architecture and gene structures of β-1,3-glucanases in cotton.
(a) The multiple alignment of the conserved glycosyl hydrolase family 17 domain were constructed with Clustal X (version 2.0)49, and gaps and poorly aligned sections were removed (Supplementary Fig. 3). Phylogenetic tree was generated using the maximum likelihood method under WAG model in MEGA v5.250, and the reliability of interior branches was assessed with 1000 bootstrap resamplings. (b) Protein domain architectures as defined in Fig. 2. (c) The gene structures were drawn using the online tool GSDS. Introns and exons were represented by black lines and blue boxes, respectively, and numbers at the exon-intron joints were intron phases.
The CBM43 domain and the hydrophobic C-terminal sequence were found to be subfamily dependent. All members of subfamilies B, D, F and G contained the CBM43 domain, whereas it was absent from the members of subfamily E. The C-terminal hydrophobic sequence was present in members of subfamilies A, F and H, but was absent from members of subfamilies B and G. With the exception of subfamily C, and some members of subfamily D, such as GrGLU11 and GrGLU12, and subfamily E, such as GrGLU20 and GrGLU23, the introns in most of the GrGLUs were highly subfamily-specific (Fig. 3), indicating that GLUs in different subfamilies may have undergone independent gene duplication in G. raimondii.
β-1,3-Glucanase gene family expansion in the Gossypium genus
Previous studies have suggested that there was at least one additional whole genome duplication (WGD) after the pan-eudicot triplication in the Gossypium genome, however, the frequency and timing of the event are still being debated16,25. To investigate the β-1,3-glucanase gene family expansion pattern in cotton, we download the syntenic data of G. raimondii, A. thaliana, V. vinifera and T. cacao from the Plant Genome Duplication Database (PGDD). A. thaliana has undergone at least three WGD events, including two doublings (α and β events) since its divergence from other members of the Brassicales clade26 and one tripling (γ event) which was shared with most if not all eudicots27. The V. vinifera genome has experienced no WGD since the pan-eudicot triplication, and it is regarded as an excellent reference for determining the numbers of WGDs in eudicots28. Of the whole genome sequenced plants, T. cacao is most closely related to cotton and had no WGD after the ancient hexaploidization event16,29.
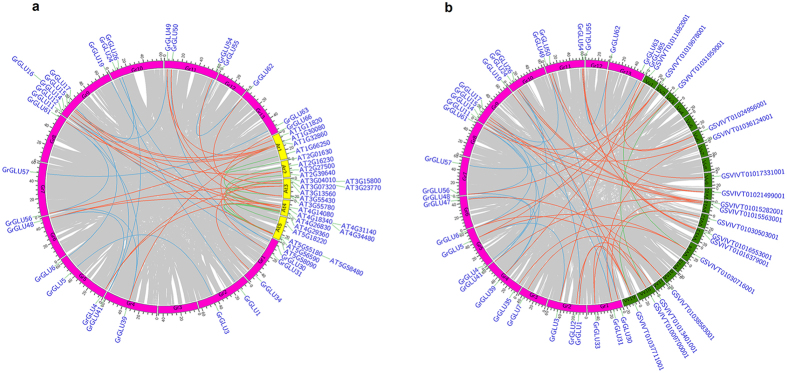
The WGD and tandem duplication of GLUs were analyzed to elucidate the gene family expansion events in G. raimondii. Among the 67 GrGLUs in G. raimondii, we identified 27 genes within 17 pairs of syntenic blocks (Fig. 4; Supplementary Table 2). The Ks values for each pair of genes within a syntenic block were used to interpret duplication events (Table 2). For two pairs of paralog genes, GrGLU6 vs GrGLU17 and GrGLU30 vs GrGLU50, the Ks values were 2.260 and 2.034, respectively, and these two paralog gene pairs may be derived from the ancient hexaploidization event. For the remaining 15 paralog gene pairs, the Ks values ranged from 0.436 to 0.806, implying that these paralog gene pairs originated from the Gossypium lineage WGD events16,25. We also analyzed the adjacent genes to investigate whether tandem duplication had taken place. 13 of the 67 GrGLUs showed tandem duplication, with a cluster of 4 members (GrGLU20, GrGLU21, GrGLU22 and GrGLU23) present in Chr D6 (Fig. 1).
Figure 4. The intra- and inter-genomic comparison showed gene synteny of β-1,3-glucanase genes in G. raimondii (Gr), A. thaliana (At) and V. vinifera (Vv).
The gray lines indicated whole genome duplication blocks. The light blue and green lines indicated the intra-genomic synteny of β-1,3-glucanase genes in G. raimondii, A. thaliana (a) and V. vinifera (b), and the orange lines indicated the inter-genomic synteny of β-1,3-glucanase genes between species.
Table 2. Ks values of gene pairs in syntenic blocks.
| Gene 1 | Gene 2 | Anchors* | Ks |
Gene 1 | Gene 2 | Anchors* | Ks |
||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||||||
| GrGLU56 | GrGLU61 | 65 | 0.806 | 0.343 | GrGLU11 | GrGLU41 | 54 | 0.771 | 0.383 |
| GrGLU56 | GrGLU39 | 15 | 0.713 | 0.294 | GrGLU3 | GrGLU5 | 34 | 0.557 | 0.206 |
| GrGLU39 | GrGLU61 | 56 | 0.634 | 0.338 | GrGLU57 | GrGLU63 | 32 | 0.436 | 0.119 |
| GrGLU14 | GrGLU31 | 34 | 0.684 | 0.460 | GrGLU67 | GrGLU34 | 15 | 0.631 | 0.367 |
| GrGLU15 | GrGLU49 | 55 | 0.605 | 0.418 | GrGLU4 | GrGLU48 | 35 | 0.701 | 0.360 |
| GrGLU24 | GrGLU30 | 9 | 0.593 | 0.213 | GrGLU62 | GrGLU55 | 22 | 0.706 | 0.335 |
| GrGLU1 | GrGLU19 | 33 | 0.701 | 0.411 | |||||
| GrGLU26 | GrGLU41 | 27 | 0.718 | 0.257 | GrGLU30 | GrGLU50 | 9 | 2.034 | 0.232 |
| GrGLU26 | GrGLU11 | 115 | 0.534 | 0.289 | GrGLU6 | GrGLU17 | 15 | 2.260 | 1.016 |
*Numbers of gene pairs in syntenic blocks.
Furthermore, we analyzed the gene duplication pattern of GLUs from different phylogenetic subfamilies. Inter-genomic synteny analysis showed that GLUs with syntenic relationships were detected in all subfamilies with the exception of subfamily E, and this indicated that the gene orders of the blocks in subfamily E were less conserved during the evolution of diploid cotton. In total there were 12 pairs of genes with syntenic relationships in subfamilies A, B, C, G and H, but these were not generated by tandem duplication events. In subfamily E, which contained 12 GLUs, there were no genes with syntenic relationships but 8 tandem duplicated genes were present. In subfamilies D and F, there were 7 genes within different syntenic blocks and 5 tandem duplicated genes (Figs 1 and 4; Table 2). These results show that GLUs from different phylogenetic subfamilies have different expansion patterns. In summary, Gossypium lineage WGD event contributed to the expansion of subfamilies A, B, C, G and H, while subfamily E expanded through tandem duplication events. WGD and tandem duplication worked together to promote subfamily D and F expansion.
It has been suggested that allotetraploid cotton species appeared in the last 1–2 Myr through hybridization and subsequent polyploidization events that involved a maternal A-genome species and a paternal D-genome species. As a relatively young polyploidy species, genes from parental genomes were mostly preserved in the G. hirsutum subgenomes18. Here, we identified 66 and 64 GLUs in the A and D subgenomes of G. hirsutum acc. TM-1, respectively (Fig. 5; Supplementary Table 1). The number of GLUs in allotetraploid cotton has nearly doubled when compared with that in diploid cotton, although a handful of homologs have been lost.
Figure 5. Phylogenetic relationships of β-1,3-glucanase genes in diploid and allotetraploid cotton.
Amino acid sequences of conserved glycosyl hydrolase family 17 domain were aligned using ClustalX, and gaps and poorly aligned sections were removed. Phylogenetic tree was generated using the maximum likelihood method under WAG model in MEGA v5.2, and the reliability of interior branches was assessed with 1000 bootstrap resamplings.
Expression patterns of β-1,3-glucanase genes in G. hirsutum acc. TM-1
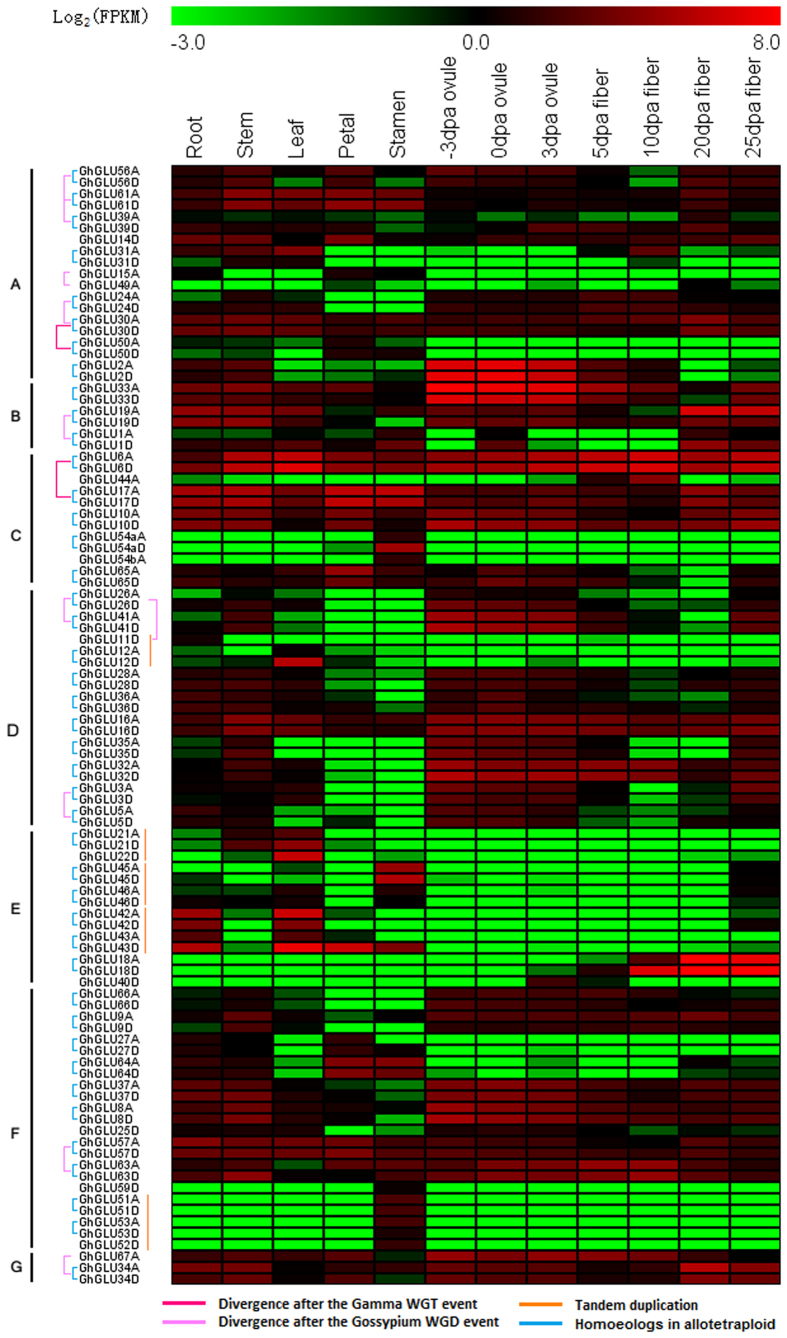
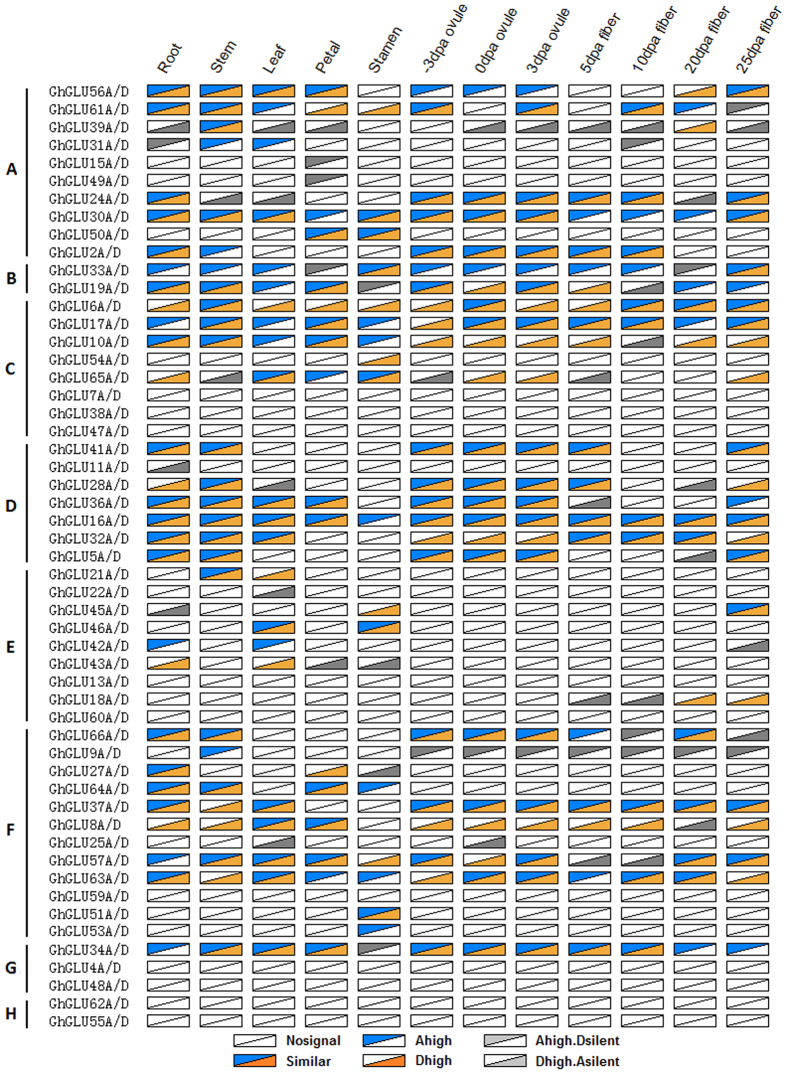
Previous studies have suggested that GLUs are expressed in all plant tissues and are regulated temporally and spatially depending on environmental conditions and developmental stage3,7. Recently-published research18, including G. hirsutum acc. TM-1 gene expression profiles, allowed us to investigate the expression of the β-1,3-glucanase gene family in different organs and developmental stages. The FPKM method was employed to normalize the total short read sequences. Among the 130 GhGLUs, there were 98 genes with an FPKM > 1 in at least one of the 12 investigated organs and developmental stages, and these 98 genes were used to gauge the relative expression of each β-1,3-glucanase gene (Fig. 6). The remaining 32 GhGLUs may be pseudogenes, or are only expressed at specific developmental stages, or under special environmental conditions that were not analyzed in this study. We observed that the accumulation of the 98 GhGLUs transcripts was associated with different organs and developmental stages, and that the expression patterns differed among each subfamily (Fig. 6). Notably, the expression of the individual member of GLUs in subfamily E was limited to roots, leaves, floral organs or cotton fibers, not all tissues we analyzed.
Figure 6. Expression profiles and their evolutional pattern of β-1,3-glucanase genes in cotton.
The RNA-seq relative expression data of 12 tissues was used to re-construct expression patterns of β-1,3-glucanase genes in G. hirsutum acc. TM-1. The lines showed the blocks containing the corresponding β-1,3-glucanase genes experienced the WGD events, or tandem duplication events.
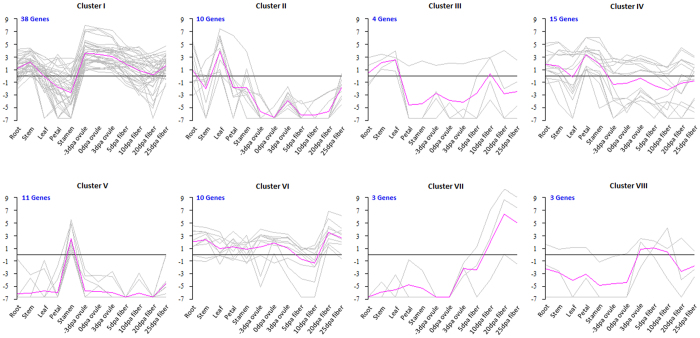
A hierarchical clustering analysis of expression profiles in the 12 tissues produced 8 clusters (Cluster I–VIII) comprising a total of 94 GhGLUs, and the remaining 4 genes could not be divided into any of the above clusters (Fig. 7, Supplementary Table 3). Thirty-eight GhGLUs were highly expressed in −3, 0 and 3 days post anthesis (DPA) ovules and fell into Cluster I. Members in this cluster also had high expression in stems but low expression in stamens. There were 14 GhGLUs in Clusters II and III and these were highly expressed in leaves. The GhGLUs in Cluster III were also highly expressed in stems. The expression of 26 genes was highest in petals or stamens and these fell into Clusters IV and V, respectively. The remaining 16 genes were highly expressed in the fibers and these fell into Clusters VI, VII and VIII. Expression of the GhGLUs in Clusters VI and VII was highest in the 20 DPA fibers, although transcripts of the genes in Cluster VI were also detected in vegetative and reproductive organs.
Figure 7. Expression clusters of β-1,3-glucanase genes in G. hirsutum.
The gray line shows expression profile of each gene, and the purple line is the average expression of one cluster.
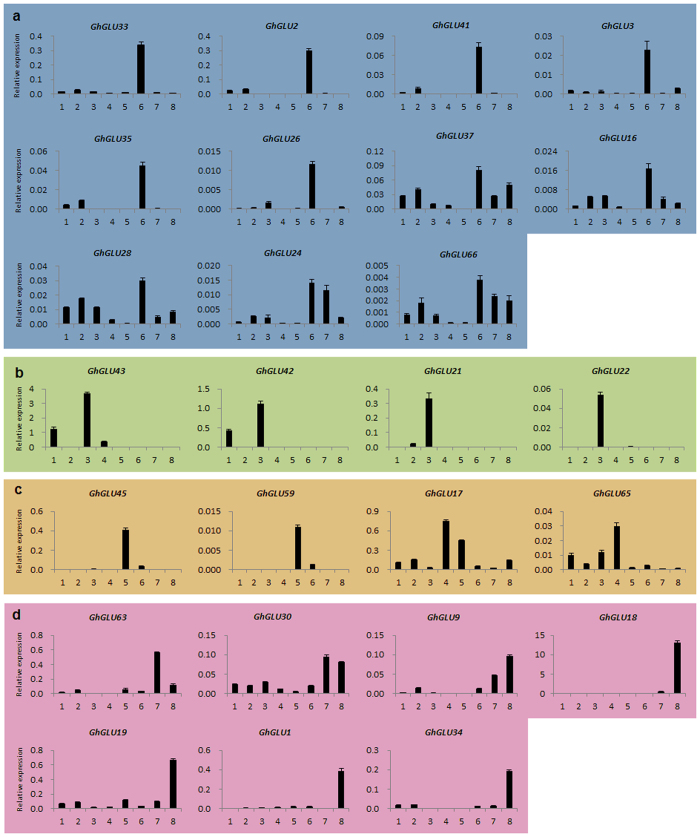
To confirm the expression profiles above, qRT-PCR was conducted for 26 genes in 8 organs and developmental stages of TM-1 (Fig. 8). The qRT-PCR primers used (Supplementary Table 3) were gene-specific but not homoeolog-specific. Firstly, 11 GhGLUs showed expression peaks in 0 DPA ovules, and these were divided into two groups. GhGLU33, GhGLU3, GhGLU41, GhGLU3, GhGLU35 and GhGLU26 were predominantly found to accumulate in ovules at 0 DPA ovules, and GhGLU37, GhGLU16, GhGLU24, GhGLU28 and GhGLU66 were also detected in vegetative organs and fibers, with low expression levels in petals and anthers. Secondly, four GhGLUs, GhGLU42, GhGLU43, GhGLU21 and GhGLU22, were predominantly found to accumulate in roots and leaves. Thirdly, the expression peaks of four GhGLUs, GhGLU45, GhGLU59, GhGLU17 and GhGLU65, occurred mainly in petals or anthers. Lastly, seven GhGLUs were found to accumulate at high levels in fibers at 10 and 20 DPA. GhGLU63 showed preferential expression in fibers at 10 DPA, and GhGLU30 had similar expression levels in 10 and 20 DPA fibers, while GhGLU9, GhGLU18, GhGLU19, GhGLU1 and GhGLU34 were highly expressed in 20 DPA fibers (Fig. 8). The results of our qRT-PCR analysis of GhGLUs are consistent with the RNA-Seq data.
Figure 8. Expression patterns of GhGLUs in different tissues and developmental stages by qRT-PCR.
1: root; 2: stem; 3: leaf; 4: petal; 5: anther; 6: ovule at 0 DPA; 7: fiber at 10 DPA; 8: fiber at 20 DPA. (a) High expression in the ovule at 0 DPA (b) Expression peaks in the leaf (c) High expression in the flowers (d) High expression in the fibers.
Molecular evolution and subgenome expression divergence of β-1,3-glucanase genes
The whole genome sequencing of G. raimondii, G. arboreum, G. hirsutum, and G. barbadense has provided us with an opportunity to explore the molecular evolutionary properties of GLUs in diploids and their allotetraploid derivatives. We calculated the nonsynonymous substitution rate (Ka), synonymous substitution rate (Ks) and Ka/Ks ratios within and between species30 to explore the divergence and selection pressures that have accumulated in GLUs (Table 3). Average Ka and Ks values were higher in inter-genomic contrasts (A vs. D or At vs. Dt) than in intra-genomic contrasts (A vs. At or D vs. Dt), and this is consistent with the hypothesis that the A–D genome divergence occurred well before the formation of the allotetraploid species. To our surprise, the average Ka of GLUs in subfamily E was higher than that in other subfamilies, indicating that members of subfamily E experienced quicker protein evolution. The overall low Ka/Ks ratios in the inter- and intra-genomic contrasts suggested negative or purifying selection of GLUs in both diploid and polyploid cotton species, however, subfamily E had a higher Ka/Ks ratio in At vs. Dt than other subfamilies, with A vs. D as the control. Therefore, GLUs in subfamily E experienced more protein evolution in both diploid and polyploid cotton, and they also underwent more selection pressures when compared to other subfamilies.
Table 3. Molecular evolutionary rates of β-1,3-glucanase genes in various comparisons among Gossypium taxa.
| Subfamily | Ka/Ks/Ka:Ks ratio |
|||
|---|---|---|---|---|
| A vs Ata | D vs Dt | A vs D | At vs Dt | |
| A | 0.0026/0.0077/0.3469 | 0.0037/0.0120/0.5216 | 0.0088/0.0412/0.2368 | 0.0079/0.0398/0.2371 |
| B | 0.0033/0.0109/0.3281 | 0.0042/0.0226/0.2055 | 0.0098/0.0590/0.1582 | 0.0115/0.0455/0.2577 |
| C | 0.0031/0.0098/0.5355 | 0.0027/0.0124/0.3123 | 0.0106/0.0496/0.2201 | 0.0097/0.0506/0.1900 |
| D | 0.0035/0.0037/0.7288 | 0.0041/0.0105/0.3837 | 0.0110/0.0487/0.2152 | 0.0112/0.0467/0.2259 |
| E | 0.0044*/0.0107/0.4989 | 0.0063*/0.0156/0.4986 | 0.0162*/0.0489/0.3312* | 0.016*2/0.0503/0.3305* |
| F | 0.0019/0.0081/0.2733 | 0.0033/0.0148/0.3064 | 0.0095/0.0455/0.2301 | 0.0103/0.0436/0.2406 |
| G | 0.0032/0.0112/0.4037 | 0.0032/0.0134/0.2446 | 0.0090/0.0506/0.1788 | 0.0090/0.0505/0.1746 |
| H | 0.0028/0.0136/0.2855 | 0.0036/0.0133/0.6975 | 0.0049/0.0417/0.1607 | 0.0071/0.0355/0.2927 |
aGenome codes are as follows: G. arboreum (A), G. raimondii (D), G. hirsutum “A homoelog” (At), G. hirsutum “D homoeolog” (Dt).
“*”: t test at P < 0.05.
Through the genome-wide analysis of GLU expression levels in 12 TM-1 (G. hirsutum) tissues (53 homoeologs × 12 tissues = 636 combinations), we found that 147 (23.1%) homoeologous gene pairs showed similar expression levels, 340 (53.5%) pairs were undetectable, and 149 (23.4%) pairs were subgenome-biased (Fig. 9). Of the 149 subgenome-biased pairs, 52 pairs showed one gene expressed while the other was undetectable, 45 pairs showed higher levels in A than D subgenome (A subgenome bias), and 52 pairs showed higher levels in D than A subgenome (D subgenome bias). In particular, only 19 homoeologous gene pairs in subfamily E were found to have transcripts in the tested tissues, of them, there were 15 (78.9%) pairs showed subgenome bias expression, implying specific roles for members in subfamily E during the evolutionary process.
Figure 9. Ratios of β-1,3-glucanase homoeologs transripts in G. hirsutum.
Nosignal: FPKM of both homoeologs was less than one. Similar: No expression divergence detected between homoeologs. Ahigh: A homoeolog expression was higher than D. Dhigh: D homoeolog expression was higher than A. Ahigh.Dsilent: A homoeolog expression was higher than D, and FPKM of D homoeolog was less than one. Dhigh.Asilent: D homoeolog expression were higher than A, and FPKM of A homoeologs was less than one. The higher expression level of homoelogs were detected through pairwise t-test (P value < 0.01, FDR < 0.05 and at least 1.5-fold difference in expression levels).
Potential function of two tandem duplicated β-1,3-glucanase genes in Verticillium dahliae resistance
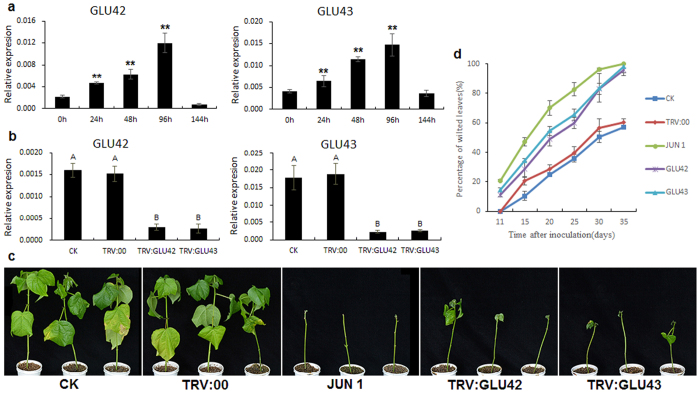
Four tandem duplicated GLUs from subfamily E, GLU42 and GLU43, and GLU21 and GLU22, were found to be predominantly accumulated in leaves, and GLU42 and GLU43 were also detected in roots (Fig. 8). qRT-PCR showed that GLU42 and GLU43 were significantly induced after Verticillium dahliae inoculation, with the highest expression observed at 96 h post-inoculation (Fig. 10a).
Figure 10. Silencing of GLU42 and GLU43 enhances cotton susceptibility to Verticillium dahlia infection.
(a) Expression analysis of GLU42 and GLU43 after inoculation by V. dahliae. Asterisks indicate significant expression differences compared inoculated time with 0 h (“*”: t test at P < 0.05, “**”: t test at P < 0.01). (b) Expression of GLU42 and GLU43 in silenced and control plant leaves by qRT-PCR analysis. Different letters indicate the significant expression difference each other (Tukey’s multiple comparison test, P < 0.01). (c) Plant phenotypes at 30 days after V. dahlia inoculation. (d) Percentage of diseased leaves after V. dahlia inoculation. The experiments were repeated using 20 plants per treatment. The error bars were calculated based on three independent experiments using standard deviation. Percentage of diseased leaves for each treatment was showed in Supplementary Table 4. Different letters indicate the significant difference each other (Tukey’s multiple comparison test, P < 0.05).
To investigate the function of GLU42 and GLU43 in Verticillium resistance, we constructed TRV2:GLU42 and TRV2:GLU43 vectors to silence the endogenous genes in Hai7124, with TRV:00 as the mock treatment and TRV2:GhCLA1 (Cloroplastos alterados 1) as an indicator. An Agrobacterium culture containing all recombinant pTRV vectors was infiltrated into the cotyledons of cotton cultivar Hai7124. Three weeks later, the plants infiltrated with CLA1 exhibited the photobleaching phenotype (Supplementary Fig. 4a), and GLU42 and GLU43 were silenced in TRV2:GLU42- and TRV2:GLU43-infiltrated plants when compared to no-infiltration control (CK) and mock treated plants (Fig. 10b). Further, we found that GLU42 was silenced in TRV2:GLU43-silenced plants, and GLU43 was also silenced in TRV2:GLU42 silenced plants. This may be due to the 90.98% nucleotide identity between the two tandem duplicated genes (Supplementary Fig. 5). We also performed qRT-PCR to examine the expression of other GLUs in subfamily E. Only two GLUs, GLU21 and GLU22, were detected to express in leaf tissue, however their expression had not been interfered in GLU42/43 silenced plants (Supplementary Fig. 6). After the GLU42/43 were silenced, we inoculated the cotton seedlings by dip-infection with V. dahliae isolate V991, with the final concentration adjusted to 1 × 107 spores per milliliter. Eleven days after inoculation, yellow leaf veins and wilting were found in cotyledons of GLU42/43-silenced plants. Similar phenotypes were observed in susceptible plants (G. hirsutum cv. Junmian 1), but not in no-infiltration control and mock treated plants. Twenty days later, the true leaves of GLU42/43-silenced plants became withered and etiolated, and thirty days later, most of the true leaves were defoliated (Fig. 10c). In general, no-infiltration control and mock treated plants exhibited a weak partial leaf wilting phenotype. However, silencing of GLU42/43 enhanced the plants’ susceptibility to V. dahlia: over 83% of GLU42/43 silenced plant leaves exhibited severe wilting symptoms inoculated thirty days later, it’s higher than that of mock treated plants. Further, more than 96% GLU42/43 silenced plant inoculated thirty-five days later were severely infected, similar to the results observed in susceptible control plants (G. hirsutum cv. Junmian 1) (Fig. 10c,d, Supplementary Table 4). No-inoculation control with silenced GLU42/43 showed the normal growth and development (Supplementary Fig. 4b). The VIGS results indicated that GLU42 and GLU43 play important roles in resistance to V. dahliae infection in cotton, and have a potential utilization in cotton disease-resistant breeding.
Discussion
Whole-genome duplication, followed by massive silencing and elimination of duplicated genes, has long been recognized as a pervasive force in plant evolution31. Complete genome analyses support the idea that two polyploidy events (ρ and σ) occurred in monocots, and one triplication event (γ) was probably shared by all core eudicots26,27,32. The Gossypium lineage underwent a WGD after the γ event, although the frequency and timing of this event are still being debated16,25. Meanwhile, the fate of the duplicated genes derived from WGD events is still poorly understood, both evolutionarily and functionally. Several studies have shown that some duplicate genes were more prone to retention than others. For example, transcription factors, signal proteins, and membrane proteins were preferentially retained after duplication in Arabidopsis33. The duplicate genes retained after an ancestral WGD event enhanced root nodule symbiosis in the Papilionoideae34.
In this study, we identified 67 GrGLUs in G. raimondii. Of them, 27 GrGLUs were detected within 17 pairs of syntenic blocks (Fig. 4; Table 2), with two paralogs derived from the ancient hexaploidization, and 15 paralogs from the Gossypium lineage WGD events. These paralogs were retained after the WGD events. We also detected 21 GrGLUs that showed there was only one β-1,3-glucanase gene remaining in the syntenic blocks (Supplementary Table 2), and further, their corresponding orthologs were also found in A. thaliana (Fig. 4a), V. vinifera (Fig. 4b) or T. cacao (Supplementary Table 2). This indicated the duplicated genes in the syntenic blocks were lost in Gossypium after WGD event. The retained paralogs might have gained new functions (neofunctionalization), lost functions (pseudogenization), or partitioned aggregate ancestral function (subfunctionalization)35,36. In G. raimondii, 99.4% of the duplicated genes were reported to exhibit differential expression in at least one of three tissues (petal, leaf, and seed)37. In this study, we also found that GhGLU41 and GhGLU26 showed different expression to GhGLU11, indicating the functional diversity of these paralogs. However, evolutionary mechanisms underlying the preservation and diversification of duplicated genes followed by WGD remain largely unknown.
According to parsimony reconstruction, the ancestral GLUs most probably displayed a somewhat nonspecific expression pattern and were involved in cell division or cell wall remodeling-like functions3. After several rounds of ancient WGD events in seed plants and angiosperms, the GLUs with ancestral functions may have diverged in expression; leading to the production of new functions in pathogen resistance, pollen development and tube growth, seed germination, and other processes3,31.
The sequence-based phylogeny of GLUs is not entirely consistent with the expression divergence in Arabidopsis3 and cotton in our study, possibly because duplicated genes acquired or lost tissue or developmental cis-regulatory elements during the evolutionary process. However, the GLUs in subfamily E were specifically expressed in particular tissues or organs. For example, the expression of GhGLU18 (Accession number: D88416.1) specific to fibers at 20 DPA played an important role in reopening plasmodesmata by degrading callose during cotton fiber development38. An Arabidopsis β-1,3-glucanase gene, AtBG_pap, is considered as a plasmodesmal gate keeper for intercellular communication12,13. Due to the temporary deposition of callose, a transient closure of plasmodesmata occurs during the rapid phase of cotton fiber elongation. The expression of GhGLU18 highly correlates with the timing of plasmodesmata opening, and could play a role in cotton fiber development by degrading callose in plasmodesmata38,39,40. The expression of GhGLU21, GhGLU22, GhGLU42 and GhGLU43 was highly specific to roots and leaves. The expression peaks of GhGLU45 occurred mainly in anthers, which may indicate a role in cotton flowering. At the onset of meiosis, a secondary callose wall composed of β-1,3-glucan is deposited between the primary wall and the plasma membrane of the pollen mother cells41. It is reported that the timing of the callose wall degradation by β-1,3-glucanase is important for proper microsporegenesis, and the premature dissolution and delayed degradation of the callose walls caused male sterility in tobacco9 and rice4, respectively.
The protein domain architecture and intron/exon structure of the genes in subfamily E were highly conserved (Fig. 3). Most of these genes lost the C-terminal region, including the CBM43 domain and the hydrophobic C-terminal sequence, and this change may enable them to be secreted extracellularly. Furthermore, GLUs in subfamily E experienced quicker protein evolution in both diploid and polyploid cotton, and showed subgenome-biased expression. Our findings suggest that the specific expression, loss of the C-terminal region, fast evolution and subgenome expression bias of these members together led to the functional divergence of GLUs in subfamily E in cotton.
As a member of the pathogenesis-related (PR) protein group, the interest in β-1,3-glucanases has focused primarily on their antifungal activity. This glucanohydrolase can degrade β-1,3-glucan in the cell walls of pathogens, and the release of cell-wall derived elicitors can further stimulate defense reactions7,8. In soybean, β-1,3-glucanases have been demonstrated to release active elicitors from fungal cell walls, and then induce the accumulation of the phytoalexin glyceollin42. Constitutive expression of a β-1,3-glucanase gene in tobacco plants led to reduced symptoms when infected with the glucan-containing fungi Peronospora tabacina and Phytophthora parasiticavar nicotiana43. Further, expression of the β-1,3-glucanase gene in combination with chitinase transgenes led to a remarkable synergic effect. Transgenic tomato plants that were overexpressing β-1,3-glucanase in class I and chitinase from tobacco, showed strong antifungal activity against Fusarium oxysporum44. Tobacco plants expressing both β-1,3-glucanase and chitinase showed reduced susceptibility to infection by Fusarium solani45.
Verticillium wilt is a serious disease that significantly reduces the yield and quality of cotton. The availability of the whole genome sequences of Gossypium species provide us with an opportunity to identify certain GLUs that are involved in V. dahliae resistance. In this study, based on the genome-wide analyses, we found two tandem duplicated GLUs in subfamily E, GLU42 and GLU43, which were significantly induced in cotton roots after V. dahliae inoculation. VIGS assays showed that silencing of these two genes significantly increased the susceptibility of cotton plants to V. dahlia, while the control and mock treated plants exhibited a weak and partial leaf wilting phenotype (Fig. 10). Our finding implies that GLU42 and GLU43 play crucial roles in resistance to V. dahliae in cotton, and that overexpression of GLU42/43 will help defend cotton against fungal infection.
Materials and Methods
Identification and chromosomal mapping
The gene files of G. raimondii, A. thaliana, V. vinifera, and T. cacao were downloaded from Phytozome v9.0 (http://www.phytozome.net/). The gene information of G. arboreum, G. hirsutum acc. TM-1, and G. barbadense acc. 3–79, were downloaded from http://cgp.genomics.org.cn, http://mascotton.njau.edu.cn and http://cotton.cropdb.org, respectively. The Hidden Markov Model (HMM) profile of the glycoside hydrolase family 17 domain (PF00332) was obtained from the Pfam website (http://pfam.xfam.org/), and was employed as a query to identify all possible GLUs using HMMER (V3.0) software46. To validate the HMM search, all candidate sequences were used as queries to search the NCBI non-redundant (nr) protein database with the blastp program, and the results with the best hits of “Glyco_hydro_17” were retained for further analysis. For prediction of GPI-anchor attachment sites, the BGI-PI47 and GPI-SOM48 algorithms were used.
Positional information on GLUs was parsed from the General Feature Format (GFF) files downloaded from Phytozome v9.0, and the locations of GLUs in G. raimondii and G. hirsutum were drafted by MapInspect software (http://www.plantbreeding.wur.nl/uk/software-mapinspect.html).
Phylogenetic and exon-intron structural analysis
A multiple alignment of the sequences encoding the conserved glycosyl hydrolase family 17 domain were constructed with ClustalX (version 2.0)49, and gaps and poorly aligned sections were removed. A phylogenetic tree was generated using the maximum likelihood method and WAG model in MEGA v5.250 software, and the reliability of interior branches was assessed with 1000 bootstrap resamplings.
The gene structure of the GLUs was parsed from the General Feature Format (GFF) files, and diagrams of the exon-intron structures were drawn using the online program Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn/).
Genome synteny and gene duplication
The syntenic information of G. raimondii, A. thaliana, V. vinifera and T. cacao was downloaded from the Plant Genome Duplication Database (PGDD; http://chibba.agtec.uga.edu/duplication/). GLUs were mapped to the syntenic blocks for intra- and inter-genomic comparison. The syntenic diagram was drawn using Circos software51.
The timing of segmental duplication events can be estimated by computing mean Ks values for all anchor points located in the corresponding syntenic block16,25, and all the Ks values were parsed from PGDD syntenic data. Genes separated by five or fewer genes within a 100-kb region on a chromosome may have resulted from tandem duplication52.
Plant materials and treatments
The genetic standard line, G. hirsutum acc. TM-1, was used for tissue/organ expression analysis. Roots, stems and leaves were collected from two-week-old seedlings grown in a greenhouse. Petals, anthers and ovules were collected from plants grown under standard field conditions on the day of flowering, and fibers were excised from developing bolls on selected days post anthesis (DPA). The tissue/organ materials were quick-frozen in liquid nitrogen and stored at −70 °C before RNA extraction.
G. barbadense cv. Hai7124 with Verticillium resistance, was used for fungal pathogen (V. dahliae) inoculation. We hand-inoculated three-week-old Hai7124 seedlings with conidial suspensions carrying 1 × 107 spores of V. dahliae strain V991 through dip-infection. The roots were harvested, with three repeats at different time points (0, 24, 48, 96 and 144 hours) after V991 treatment, and then quick-frozen in liquid nitrogen and stored at −70 °C before RNA extraction. G. hirsutum cv. Junmian 1 with Verticillium susceptibility was used as a susceptible control.
RNA isolation and quantitative reverse transcription PCR
Total RNA was isolated using the CTAB-acidic Phenolic method53. Each RNA sample was treated with DNase I to remove the genomic DNA. Total RNA samples (2 μg per reaction) from different tissues/organ were reversely transcribed into cDNA by M-MLV reverse transcriptase.
The expression of GLUs was analyzed using an ABI 7500 real-time PCR system with the HiScript Q RT SuperMix (Vazyme, Nanjing, China). Gene-specific primers were designed based on the β-1,3-glucanase gene sequences using Oligo 6.0. Cotton histone3 (AF024716) was used as the reference gene54. The amplification parameters were as follows: 95 °C hold for 10 min, followed by 40 cycles at 95 °C for 15 s, 58 °C for 15 s and 72 °C for 15 s. For the melting curve stage, the default settings were chosen. Nonspecific products were identified by inspecting melting curves. The primer pairs used for real-time PCR were listed in Supplementary Table 5.
Estimation of the evolution rates of β-1,3-glucanase genes
Estimation of the rates of nonsynonymous substitutions per nonsynonymous site (Ka) and synonymous substitutions per synonymous site (Ks) was performed within and between Gossypium species using DnaSP version 555. Based on the definition of Ka/Ks, a value of 1 represented neutral evolution, and a value less than 1 indicated negative or purifying selection, whereas a value greater than 1 indicated positive selection acting on amino acids.
Construction of VIGS vectors and agro-infiltration
The binary TRV vectors pTRV1 and pTRV2 were kindly provided by Dr. Libo Shan at Texas A&M University (College Station, TX, USA). pTRV2:GhCLA1, where GhCLA1 (Cloroplastos alterados 1) encodes 1-deoxy-D-xylulose-5-phosphate synthase, was used as a control56. The sequences of GLU42 and GLU43 were amplified using the primers listed in Supplementary Table 5, and the PCR fragments cloned from Hai7124, with 334 bp for GLU42 and 326 bp for GLU43, were inserted into pTRV2. These pYL156 derivatives were individually transformed into Agrobacterium GV3101.
Agrobacterium cultures containing recombinant TRV vectors were prepared and then infiltrated into two fully expanded cotyledons of eight-day-old seedlings, as previously described by Zhang et al.57. The seedlings were cultivated in a greenhouse at 23/22 °C (day/night) with a 16 h light/8 h dark photoperiod. Agrobacterium-mediated VIGS assays were repeated three times with more than 20 plants for each construct per repeat.
Additional Information
How to cite this article: Xu, X. et al. Genome-wide characterization of the β-1,3-glucanase gene family in Gossypium by comparative analysis. Sci. Rep. 6, 29044; doi: 10.1038/srep29044 (2016).
Supplementary Material
Acknowledgments
This program was financially supported in part by National Science Foundation in China (31171590), the National Transgenic Program (2011ZX08005-004), Key R&D program in Jiangsu Province (BE2015360), Six talent peaks project in Jiangsu province (2015-NY-002), the Priority Academic Program Development of Jiangsu Higher Education Institutions (010-809001), and JCIC-MCP project (No. 10).
Footnotes
Author Contributions Experiments were designed by W.Z.G. with suggestion from X.Y.W. Experiments were performed by X.Y.X., Y.F., S.F., J.X. and X.Y.W., X.Y.X. and W.Z.G. drafted the manuscript, W.Z.G. and X.Y.W. revised the manuscript. All authors read and approved the final manuscript.
References
- Sun L., Gurnon J. R., Adams B. J., Graves M. V. & Van Etten J. L. Characterization of a beta-1,3-glucanase encoded by chlorella virus PBCV-1. Virology 276, 27–36 (2000). [DOI] [PubMed] [Google Scholar]
- Bachman E. S. & McClay D. R. Molecular cloning of the first metazoan beta-1,3 glucanase from eggs of the sea urchin Strongylocentrotus purpuratus. Proc Natl Acad Sci USA 93, 6808–6813 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxey A. C., Yaish M. W., Moffatt B. A., Griffith M. & McConkey B. J. Functional divergence in the Arabidopsis beta-1,3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol Biol Evol 24, 1045–1055 (2007). [DOI] [PubMed] [Google Scholar]
- Wan L. et al. A rice beta-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta 233, 309–323 (2011). [DOI] [PubMed] [Google Scholar]
- Linthorst H. J. et al. Analysis of gene families encoding acidic and basic beta-1,3-glucanases of tobacco. Proc Natl Acad Sci USA 87, 8756–8760 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Horner H. T., Palmer R. G. & Shoemaker R. C. Analysis and mapping of gene families encoding beta-1,3-glucanases of soybean. Genetics 153, 445–452 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G. & Meins F. Jr. Functions and regulation of plant β-1,3-glucanases (PR-2) (eds Datta S. K. et al.) 49–76 (CRC Press, 1999).
- Boller T. Chemoperception of Microbial Signals in Plant-Cells. Annu Rev Plant Physiol Plant Mol Biol 46, 189–214 (1995). [Google Scholar]
- Worrall D. et al. Premature dissolution of the microsporocyte callose wall causes male sterility in transgenic tobacco. Plant Cell 4, 759–771 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G. & Meins F. Jr. Sense transformation reveals a novel role for class I beta-1,3-glucanase in tobacco seed germination. Plant J 23, 215–221 (2000). [DOI] [PubMed] [Google Scholar]
- Hincha D. K., Meins F. & Schmitt J. M. beta-1,3-glucanase is cryoprotective in vitro and is accumulated in leaves during cold acclimation. Plant Physiol 114, 1077–1083 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A., Erlanger M., Rosenthal M. & Epel B. L. A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J 49, 669–682 (2007). [DOI] [PubMed] [Google Scholar]
- Levy A., Guenoune-Gelbart D. & Epel B. L. beta-1,3-Glucanases: Plasmodesmal gate keepers for intercellular communication. Plant Signal Behav 2, 404–407 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciaglia P. A. & Smith A. G. Cloning and characterization of Tag 1, a tobacco anther beta-1,3-glucanase expressed during tetrad dissolution. Plant Mol Biol 24, 903–914 (1994). [DOI] [PubMed] [Google Scholar]
- Wendel J. F. & Cronn R. C. Polyploidy and the evolutionary history of cotton. Adv Agron 78, 139–186 (2003). [Google Scholar]
- Paterson A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427 (2012). [DOI] [PubMed] [Google Scholar]
- Li F. et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46, 567–572 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33, 531–537 (2015). [DOI] [PubMed] [Google Scholar]
- Yuan D. et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep 5, 17662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol 16, 108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral P. et al. An olive pollen protein with allergenic activity, Ole e 10, defines a novel family of carbohydrate-binding modules and is potentially implicated in pollen germination. Biochem J 390, 77–84 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticher L., Hinz U., Meyer A. D. & Meins F. Jr. Intracellular transport and processing of a tobacco vacuolar beta-1,3-glucanase. Planta 188, 559–565 (1992). [DOI] [PubMed] [Google Scholar]
- Henrissat B. & Davies G. J. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol 124, 1515–1519 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G. H., Sherrier D. J., Stevens T. J., Arkin I. T. & Dupree P. Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol 129, 486–499 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44, 1098–1103 (2012). [DOI] [PubMed] [Google Scholar]
- Bowers J. E., Chapman B. A., Rong J. & Paterson A. H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438 (2003). [DOI] [PubMed] [Google Scholar]
- Jaillon O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007). [DOI] [PubMed] [Google Scholar]
- Lin L. et al. Comparative analysis of Gossypium and Vitis genomes indicates genome duplication specific to the Gossypium lineage. Genomics 97, 313–320 (2011). [DOI] [PubMed] [Google Scholar]
- Argout X. et al. The genome of Theobroma cacao. Nat Genet 43, 101–108 (2011). [DOI] [PubMed] [Google Scholar]
- Senchina D. S. et al. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol Biol Evol 20, 633–643 (2003). [DOI] [PubMed] [Google Scholar]
- Jiao Y. et al. Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (2011). [DOI] [PubMed] [Google Scholar]
- Tang H., Bowers J. E., Wang X. & Paterson A. H. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci USA 107, 472–477 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G. & Wolfe K. H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16, 1679–1691 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. G., Zhang L., Li C., Dunwell J. M. & Zhang Y. M. Comparative genomics suggests that an ancestral polyploidy event leads to enhanced root nodule symbiosis in the Papilionoideae. Mol Biol Evol 30, 2602–2611 (2013). [DOI] [PubMed] [Google Scholar]
- Moore R. C. & Purugganan M. D. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8, 122–128 (2005). [DOI] [PubMed] [Google Scholar]
- Adams K. L. & Wendel J. F. Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8, 135–141 (2005). [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S. et al. Ancient gene duplicates in Gossypium (cotton) exhibit near-complete expression divergence. Genome Biol Evol 6, 559–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y. L., Xu S. M., White R. & Furbank R. T. Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol 136, 4104–4113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y. L. Goldacre paper: Rapid cell expansion and cellulose synthesis regulated by plasmodesmata and sugar: insights from the single-celled cotton fibre. Funct Plant Biol 34, 1–10 (2007). [DOI] [PubMed] [Google Scholar]
- Ruan Y. L., Llewellyn D. J. & Furbank R. T. The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 13, 47–60 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterkeyn L. Les parois microsporocytaires de nature callosique chez Helleborus et Fadescantia. Cellule 62, 225–255 (1962). [Google Scholar]
- Umemoto N. et al. The structure and function of a soybean beta-glucan-elicitor-binding protein. Proc Natl Acad Sci USA 94, 1029–1034 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusso M. & Kuc J. The effect of sense and antisense expression of the PR-N gene for β-1,3-glucanase on disease resistance of tobacco to fungi and viruses. Physiol Mol Plant P 49, 267–283 (1996). [Google Scholar]
- Jongedijk E. et al. Synergistic activity of chitinases and β-1,3-glucanases enhances fungal resistance in transgenic tomato plants. Euphytica 85, 173–180 (1995). [Google Scholar]
- Sela-Buurlage M. B. et al. Only Specific Tobacco (Nicotiana tabacum) Chitinases and β-1,3-glucanases Exhibit Antifungal Activity. Plant Physiol 101, 857–863 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J. & Eddy S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39, W29–W37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B. et al. Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol 133, 1691–1701 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser N. & Maser P. Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics 21, 1846–1852 (2005). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol 10, 282 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. & Zhang T. Extraction of total RNA in cotton tissues with CTAB-acidic phenolic method. Cotton Sci 15, 166–167 (2003). [Google Scholar]
- Xu Y. H., Wang J. W., Wang S., Wang J. Y. & Chen X. Y. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol 135, 507–515 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Wang X., Lv K., Cai C., Xu J. & Guo W. Establishment and application of TRV-mediated virus-induced gene silencing in cotton. Acta Agron Sin 40, 1356–1363 (2014). [Google Scholar]
- Zhang X., Wang L., Xu X., Cai C. & Guo W. Genome-wide identification of mitogen-activated protein kinase gene family in Gossypium raimondii and the function of their corresponding orthologs in tetraploid cultivated cotton. BMC Plant Biol 14, 345 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.