Abstract
In insects, neuropeptides play important roles in the regulation of multiple physiological processes by binding to their corresponding receptors, which are primarily G protein-coupled receptors (GPCRs). The genes encoding neuropeptides and their associated GPCRs in the rice stem borer Chilo suppressalis were identified by a transcriptomic analysis and were used to identify potential targets for the disruption of physiological processes and the protection of crops. Forty-three candidate genes were found to encode the neuropeptide precursors for all known insect neuropeptides except for arginine-vasopressin-like peptide (AVLP), CNMamide, neuropeptide-like precursors 2-4 (NPLP2-4), and proctolin. In addition, novel alternative splicing variants of three neuropeptide genes (allatostatin CC, CCHamide 1, and short neuropeptide F) are reported for the first time, and 51 putative neuropeptide GPCRs were identified. Phylogenetic analyses demonstrated that 44 of these GPCRs belong to the A-family (or rhodopsin-like), 5 belong to the B-family (or secretin-like), and 2 are leucine-rich repeat-containing GPCRs. These GPCRs and their likely ligands were also described. qRT-PCR analyses revealed the expression profiles of the neuropeptide precursors and GPCR genes in various tissues of C. suppressalis. Our study provides fundamental information that may further our understanding of neuropeptidergic signaling systems in Lepidoptera and aid in the design of peptidomimetics, pseudopeptides or small molecules capable of disrupting the physiological processes regulated by these signaling molecules and their receptors.
Neuropeptides play an important role in the regulation of the development, reproduction, feeding, courtship, aggression, olfaction, locomotor activity, circadian rhythm, and many other physiological processes in insects1. Biologically active neuropeptides are generated by large precursors that are cleaved and further modified to produce mature peptides2. In insects, mature peptides are secreted into the extracellular environment, where they perform their physiological roles by binding to corresponding receptors, primarily G protein-coupled receptors (GPCRs), which are a large superfamily of proteins characterized by the presence of seven transmembrane domains3. Neuropeptide GPCRs belong to the A-family (or rhodopsin-like), the B-family (or secretin-like), or leucine-rich repeat-containing GPCRs (LGRs)4. Recent advances in genomic and transcriptomic analyses have led to the continual discovery of neuropeptides and their putative GPCRs in insects, and many insect neuropeptide GPCRs have been deorphanized through pharmacological screening5.
The rice stem borer Chilo suppressalis (Walker) is one of the most economically important rice pests in Asia, northern Africa, and southern Europe. This insect causes serious crop losses every year, particularly in China because of the prevalence of rice cultivation and the popularization of hybrid varieties6. C. suppressalis is a lepidopteran pest that represents a diverse and important group of agricultural insect pests that cause widespread economic damage to food and fiber crop plants, fruit trees, forests, and stored grains7,8,9. Currently, chemical pesticides represent the primary method of controlling insect pests, and the excessive use of these chemicals in the field has led to the development of insecticide resistance10. Thus, the development of alternative control agents is necessary to resolve this problem. Insect neuropeptides and their GPCRs are promising targets for a novel generation of insecticides that can offer improved specificity and environmental compatibility11. Knowledge of neuropeptidergic signaling systems provides fundamental information required for the design of peptidomimetics, pseudopeptides or small molecules capable of disrupting the physiological processes regulated by these signaling molecules and their receptors2. Indeed, the GPCR family has been successfully exploited and serves as the target of approximately 26% of all modern medicinal drugs12. Thus, the structural and functional characterization of neuropeptidomes from insect pests is the first requirement for developing strategies to replace or complement traditional neurotoxic insecticides2.
With the development of next-generation sequencing technology, RNA sequencing (RNA-seq) has become a useful tool for defining the transcriptomes of organisms, even when a reference genome is not available13. To obtain a whole transcriptome of neuropeptides and their GPCRs from C. suppressalis, we used an RNA-seq approach using Illumina HiSeq technology. In this study, we identified 43 neuropeptide genes and 51 GPCR genes for the neuropeptides of C. suppressalis by using query orthologous sequences from Bombyx mori and Drosophila melanogaster with cDNA cloning, and their expression profiles were confirmed via qRT-PCR. These results allowed us to compare the neuropeptidergic signaling systems in different insect species and provide relevant information for further functional studies in C. suppressalis. Although neuropeptides and their putative GPCRs have been reported in the silkworm B. mori7,14, their identification in C. suppressalis would further contribute to our understanding of neuropeptidergic signaling systems in Lepidoptera.
Results and Discussion
Neuropeptide and peptide hormone genes
RNA-seq data were generated using a central nervous system (CNS) cDNA library and Illumina HiSeq 2000 technology. We acquired 142,051,094 bp raw reads for C. suppressalis. After eliminating adapters, ambiguous nucleotides and low quality sequences, 138,063,130 bp clean reads remained. The total clean base pairs yielded 105,769 transcripts and 54,411 unigenes were obtained after assembling the transcripts into unigenes15. We identified 43 neuropeptide precursor genes in C. suppressalis with a local BLAST search (Table 1). The identified genes included all 38 previously identified neuropeptide genes in B. mori7. Additional neuropeptide genes were confirmed in B. mori, such as allatostatin double C (AstCC)16, CCHamide (CCH1)17, trissin (TR)18, natalisin (NTL)19, and RYamide (RY)4. Compared with the number of precursor genes in B. mori7 (Lepidoptera), D. melanogaster20 (Diptera), Nilaparvata lugens4 (Hemiptera), Apis mellifera21 (Hymenoptera), and Tribolium castaneum22 (Coleoptera), C. suppressalis (Lepidoptera) has the second-largest number of neuropeptide precursor genes, and C. suppressalis and B. mori have similar neuropeptidergic signaling systems (Table 2).
Table 1. Neuropeptides identified from C. suppressalis.
| Gene name | Accession No. | Acronym | ORF(aa) | SP(aa) | Homology search with known protein |
||
|---|---|---|---|---|---|---|---|
| Species | E-value | Protein ID | |||||
| Adipokinetic hormone 1 | KT005945 | AKH1 | 67 | 20 | Bombyx mori | 5e-29 | NP_001104825.1 |
| Adipokinetic hormone 2 | KT005946 | AKH2 | 73 | 20 | Bombyx mori | 3e-20 | NP_001124365.1 |
| AKH/corazonin-related peptide | KT005947 | ACP | 86 | 23 | Helicoverpa armigera | 8e-30 | AGH25546.1 |
| Allatostatin A | KT005948 | AstA | 220 | 18 | Helicoverpa armigera | 4e-102 | O44314.1 |
| Allatostatin B | KT005949 | AstB | 283 | 23 | Helicoverpa armigera | 9e-106 | AGH25567.1 |
| Allatostatin C | KT005950 | AstC | 125 | 27 | Mythimna unipuncta | 8e-54 | AAA93257.1 |
| Allatostatin CC splicing variant a | KT005951 | AstCCa | 140 | 18 | Bombyx mori | 7e-25 | XP_004932108.1 |
| Allatostatin CC splicing variant b | KT005952 | AstCCb | 106 | 18 | Bombyx mori | 6e-30 | XP_004932108.1 |
| Allatotropin | KT005953 | AT | 131 | 20 | Manduca sexta | 5e-72 | AAB08757.1 |
| Apis-ITG-like | KT005954 | ITG | 220 | 21 | Helicoverpa armigera | 3e-134 | AGH25548.1 |
| Bursicon alpha subunit | KT005955 | Burα | 156 | 25 | Manduca sexta | 1e-81 | Q4FCM6.1 |
| Bursicon beta subunit | KT005956 | Burβ | 137 | 24 | Manduca sexta | 1e-68 | ABB92831.1 |
| CAPA splicing variant a | KT005957 | CAPAa | 155 | 19 | Manduca sexta | 2e-60 | AAT69684.1 |
| CCHamide 1 splicing variant a | KT005958 | CCH1a | 187 | 42 | Bombyx mori | 5e-40 | XP_004930537.1 |
| CCHamide 1 splicing variant b | KT005959 | CCH1b | 162 | 45 | Bombyx mori | 1e-45 | XP_004930537.1 |
| CCHamide 2 | KT005960 | CCH2 | 133 | 22 | Helicoverpa armigera | 4e-61 | AGH25550.1 |
| Corazonin | KT005961 | Crz | 105 | 19 | Bombyx mori | 4e-37 | NP_001036899.1 |
| Crustacean cardioactive peptide | KT005962 | CCAP | 128 | 23 | Helicoverpa armigera | 2e-62 | AGH25552.1 |
| Diapause hormone/phermone biosynthesis activating neruopeptide | KT005963 | PBAN | 196 | 22 | Omphisa fuscidentalis | 1e-53 | AFP87384.1 |
| Diuretic hormone 31 | KT005964 | DH31 | 111 | 24 | Helicoverpa armigera | 2e-58 | AGH25553.1 |
| Diuretic hormone 41/corticotropin-releasing factor (CRF-DH) | KT005965 | DH41 | 139 | 17 | Helicoverpa armigera | 7e-62 | AGH25554.1 |
| Diuretic hormone 34/splicing variant of CRF-DH | KT005966 | DH34 | 132 | 17 | Helicoverpa armigera | 2e-45 | AGH25555.1 |
| Diuretic hormone 45/splicing variant of CRF-DH | KT005967 | DH45 | 148 | 17 | Bombyx mori | 1e-53 | NP_001124368.1 |
| Eclosion hormone | KT005968 | EH | 89 | 26 | Helicoverpa armigera | 3e-39 | AAV69026.1 |
| Ecdysis triggering hormone | KT005969 | ETH | 107a | Danaus plexippus | 3e-29 | EHJ75233.1 | |
| FMRFamide | KT005970 | FMRF | 188 | 22 | Helicoverpa armigera | 4e-83 | AGH25556.1 |
| Glycoprotein hormone alpha 2 | KT005971 | GPA2 | 115 | 17 | Helicoverpa armigera | 2e-67 | AGH25557.1 |
| Glycoprotein hormone beta 5 | KT005972 | GPB5 | 152 | Nob | Bombyx mori | 7e-69 | NP_001124380.1 |
| IMFamide | KT005973 | IMF | 76 | 28 | Helicoverpa armigera | 7e-32 | AGH25559.1 |
| Insulin-like peptide | KT005974 | ILP | 127 | 20 | Bombyx mori | 6e-11 | NP_001233285.1 |
| Ion transport peptide-like | KT005975 | ITPL | 116 | 22 | Manduca sexta | 7e-58 | AAY29658.1 |
| Ion transport peptide | KT005976 | ITP | 112 | 22 | Manduca sexta | 7e-56 | AAY29657.1 |
| Leucokinin | KT005977 | LK | 347 | 18 | Helicoverpa armigera | 2e-154 | AGH25561.1 |
| Myosuppressin | KT005978 | MS | 97 | 25 | Helicoverpa armigera | 6e-45 | AGH25562.1 |
| Natalisin | KT005979 | NTL | 490 | 22 | Danaus plexippus | 3e-85 | EHJ74348.1 |
| Neuroparsin | KT005980 | NP | 87 | 22 | Bombyx mori | 1e-38 | NP_001124362.1 |
| Neuropeptide F 1 splicing variant a | KT005981 | NPF1a | 83 | 22 | Manduca sexta | 7e-38 | AGH20044.1 |
| Neuropeptide F 1 splicing variant b | KT005982 | NPF1b | 123 | 22 | Manduca sexta | 1e-60 | AGH20043.1 |
| Neuropeptide F 2 | KT005983 | NPF2 | 93 | 21 | Helicoverpa armigera | 1e-55 | AEE01342.1 |
| Neuropeptide-like precursor 1 | KT005984 | NPLP1 | 334 | 31 | Bombyx mori | 8e-167 | NP_001124353.1 |
| Orcokinin A | KT005985 | OKA | 195 | 21 | Bombyx mori | 4e-51 | NP_001124366.1 |
| Orcokinin B | KT005986 | OKB | 226 | 21 | Danaus plexippus | 3e-44 | EHJ77769.1 |
| Pigment dispersing factor | KT005987 | 108 | 22 | Bombyx mori | 6e-22 | NP_001036920.2 | |
| Prothoracicotropic hormone | KT005989 | PTTH | 223 | 28 | Spodoptera exigua | 4e-74 | AAT64423.2 |
| Short neuropeptide F splicing variant a | KT005990 | sNPFa | 219 | 24 | Bombyx mori | 3e-76 | NP_001127729.1 |
| Short neuropeptide F splicing variant b | KT005991 | sNPFb | 175 | 24 | Bombyx mori | 3e-94 | NP_001127729.1 |
| SIFamide | KT005992 | SIF | 69 | 22 | Helicoverpa armigera | 2e-27 | AGH25569.1 |
| Sulfakinin | KT005993 | SK | 74 | 24 | Helicoverpa armigera | 3e-18 | AGH25570.1 |
| Tachykinin | KT005994 | TK | 255 | 21 | Helicoverpa armigera | 8e-99 | AGH25571.1 |
| RYamide | KT005995 | RY | 154 | 34 | Bombyx mori | 1e-15 | XP_004925011.1 |
| Trissin | KT005996 | TR | 114 | 21 | Bombyx mori | 3e-38 | XP_004926342.1 |
ORF, open reading frame; SP, signal peptide;
anot full length; bno signal peptide.
Table 2. Neuropeptide genes in C. suppressalis and other insects.
| Peptide name(acronyms) | C. suppressalis | B. mori | D. melanogaster | N. lugens | A. mellifera | T. castaneum |
|---|---|---|---|---|---|---|
| Adipokinetic hormone 1 (AKH1) | + | + | + | + | + | + |
| Adipokinetic hormone 2 (AKH2) | + | + | nd | + | nd | + |
| AKH/corazonin-related peptide (ACP) | + | + | nd | nd | nd | + |
| Allatostatin A (AstA) | + | + | + | + | + | nd |
| Allatostatin B (AstB, MIP, PTSP) | + | + | + | + | nd | + |
| Allatostatin C (AstC) | + | + | + | + | + | + |
| Allatostatin CC splicing variant a (AstCCa) | + | + | + | + | + | + |
| Allatostatin CC splicing variant b (AstCCb) | + | |||||
| Allatotropin (AT) | + | + | nd | + | + | + |
| Apis-ITG-like (ITG) | + | + | + | + | + | + |
| Arginine–vasopressin-like peptide (AVLP) | nd | nd | nd | nd | nd | + |
| Bursicon alpha subunit (Burα) | + | + | + | + | + | + |
| Bursicon beta subunit (Burβ) | + | + | + | + | + | + |
| CAPA splicing variant a (CAPAa) | + | + | + | + | + | + |
| CCHamide 1 splicing variant a (CCH1a) | + | + | + | + | + | + |
| CCHamide 1 splicing variant b (CCH1b) | + | |||||
| CCHamide 2 (CCH2) | + | + | + | + | + | + |
| CNMamide | nd | nd | + | + | + | + |
| Corazonin (Crz) | + | + | + | + | + | nd |
| Crustacean cardioactive peptide (CCAP) | + | + | + | + | + | + |
| Diapause hormone/PBAN/Pyrokinin 2 | + | + | + | + | + | + |
| Diuretic hormone 31 (DH31)/Calcitonin-like peptide | + | + | + | + | + | + |
| Diuretic hormone 41 (DH41)/Corticotropin releasing factor (CRF-DH) | + | + | + | + | + | + |
| Diuretic hormone 34 (DH34)/splicing variant of CRF-DH | + | + | nd | nd | nd | + |
| Diuretic hormone 45 (DH45)/splicing variant of CRF-DH | + | + | nd | nd | nd | nd |
| Ecdysis triggering hormone (ETH) | + | + | + | + | + | + |
| Eclosion hormone (EH) | + | + | + | + | + | + |
| FMRFamide (FMRF) | + | + | + | + | + | + |
| Glycoprotein hormone alpha 2 (GPA2) | + | + | + | + | nd | + |
| Glycoprotein hormone beta 5 (GPB5) | + | + | + | + | nd | + |
| IMFamide (IMF) | + | + | nd | nd | nd | nd |
| Insulin-like peptide (ILP) | + | + | + | + | + | + |
| Ion transport peptide/Crustacean hyperglycemic hormone (ITP) | + | + | + | + | + | + |
| Ion transport peptide-like (ITPL) | + | + | + | + | + | + |
| Leucokinin (LK) | + | + | + | + | + | nd |
| Myosuppressin (MS) | + | + | + | + | + | + |
| Natalisin (NTL) | + | + | + | + | + | + |
| Neuroparsin (NP) | + | + | nd | + | + | + |
| Neuropeptide F 1 splicing variant a (NPF1a) | + | + | + | + | nd | nd |
| Neuropeptide F 1 splicing variant b (NPF1b) | + | + | + | + | nd | nd |
| Neuropeptide F 2 (NPF2) | + | + | nd | + | + | nd |
| Neuropeptide-like precursor 1 (NPLP1) | + | + | + | + | + | + |
| Neuropeptide-like precursor 2 (NPLP2) | nd | nd | + | nd | + | nd |
| Neuropeptide-like precursor 3 (NPLP3) | nd | nd | + | + | + | nd |
| Neuropeptide-like precursor 4 (NPLP4) | nd | nd | + | + | nd | nd |
| Orcokinin A (OKA) | + | + | + | + | + | + |
| Orcokinin A (OKB) | + | + | + | + | + | + |
| Pigment dispersing factor (PDF) | + | + | + | + | + | nd |
| Proctolin (Pro) | nd | nd | + | + | nd | + |
| Prothoracicotropic hormone (PTTH) | + | + | + | + | nd | + |
| RYamide (RY) | + | + | + | + | + | + |
| Short neuropeptide F splicing variant a (sNPFa) | + | + | + | + | + | + |
| Short neuropeptide F splicing variant b (sNPFb) | + | |||||
| SIFamide (SIF) | + | + | + | + | + | + |
| Sulfakinin (SK) | + | + | + | + | + | + |
| Tachykinin (TK) | + | + | + | + | + | + |
| Trissin (TR) | + | + | + | nd | nd | + |
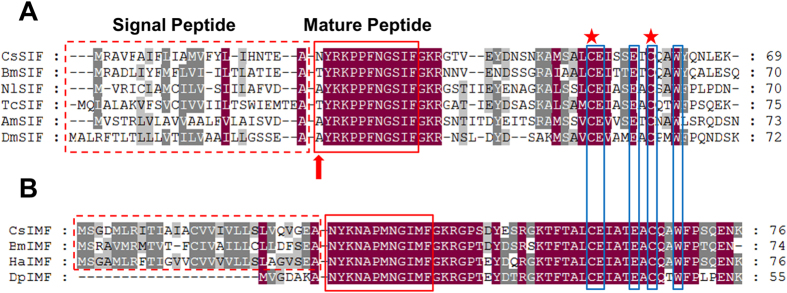
IMFamide is a unique Lepidoptera-specific neuropeptide that has not been identified in other insects (Table 2). The nomenclature for these peptides is based on the three shared C-terminal IMFamide amino acid residues (NYKNAPMNGIMFamide). IMFamide is a highly conserved neuropeptide that is identical in different Lepidoptera species (Fig. 1). In addition, another gene encoding the IMFamide paralogue that contains the C-terminal SIFamide sequence was also identified in C. suppressalis. SIFamide is one of the most highly conserved neuropeptides in insects, and it has only one substitution at the N-terminal amino acid (Fig. 1). IMFamide is similar to SIFamide, and the region between two cysteine residues separated by six amino acid residues downstream of SIFamide is particularly striking (Fig. 1). A previous study indicated that IMFamide is generated by the duplication of the SIFamide gene and suggested that the two neuropeptides have a common origin7.
Figure 1.
Protein alignment of the SIFamide (A) and IMFamide (B) precursor sequences from C. suppressalis (Cs), B. mori (Bm), N. lugens (Nl), T. castaneum (Tc), A. mellifera (Am), D. melanogaster (Dm), H. armigera (Ha), and D. plexippus (Dp). Identities are highlighted in dark red, and similarities are indicated by gray. The dashed boxes indicate the signal peptides, the solid red boxes indicate the mature peptides, and the solid blue boxes indicate the conserved residues between SIFamide and IMFamide. The red asterisks mark the conserved cysteine residues, and the red arrow indicates the one substitution at the N-terminal of SIFamide.
The novel neuropeptide NTL, which was named for its function in promoting reproduction (from the Latin natalis for “birth”), was recently discovered and characterized in D. melanogaster, T. castaneum, and B. mori19. NTL is an arthropod-specific neuropeptide gene encoding multiple copies of mature peptides that contain the C-terminal motif FXXXRa (Supplementary Fig. S1), which is closely related to the motif of tachykinin-related peptides (TKRPs), therefore, NTL is also a tachykinin-like signaling system. NTL modulates sexual activity and fecundity based on the RNAi phenotype in D. melanogaster and T. castaneum19. Thirteen peptides carry the FXXXRa motif, and one peptide contains the YXXXRa motif in the predicted NTL precursor of C. suppressalis (Supplementary Fig. S1).
The first periviscerokinin (PVK) was isolated from the perisympathetic organs of the American cockroach Periplaneta americana based on myotropic activity23. In D. melanogaster, a PVK/CAP2b-like peptide was isolated and a gene was later identified and named capability (CAPA). The CAPA precursor encodes three types of peptides: two PVK-like peptides (CAPA-PVK1 and CAPA-PVK2), a pyrokinin (PK)-like peptide (CAPA-PK, also known as PK1), and the CAPA precursor peptide B (CPPB)24 (Supplementary Fig. S2). The CAPA-PVK peptides have a C-terminal AFPRVamide and PKs have FXPRLamide motifs, whereas CAPA-PVK1 has a C-terminal PFPRVamide motif in B.mori and C. suppressalis (Supplementary Fig. S2). Two neuropeptides, diapause hormone (DH) and pheromone biosynthesis activating neuropeptide (PBAN), are encoded by a single mRNA that also encodes three additional neuropeptides (α-, β- and γ-SGNP (subesophageal ganglion neuropeptide)), which all possess the FXPRLamide motif (Supplementary Fig. S3), and are widespread among moths as well as in other invertebrate species25. In D. melanogaster, FXPRLamides are located on two different genes, namely the capa gene and the hugin gene, which contain two peptides CAPA-PK (PK1) and hug-PK (PK2) are the members of the DH/PBAN/PK family (Supplementary Fig. S2).
Ion transport peptide (ITP) was first isolated from the locust Schistocerca gregaria based on its antidiuretic activity in the ileum26. ITPs is released from the corpora cardiaca and stimulates the ileum to transport Cl− ions from the lumen to the hemolymph, thus forming an electrochemical gradient that drives water resorption27. ITP has an alternatively splicing variant named ITP-like (ITPL), which is distinguished by a lack of C-terminal amidation that occurs in ITP (Supplementary Fig. S4). Six spatially conserved cysteines were found in the mature peptide of ITP and ITPL, and these cysteines form three intramolecular disulfide bonds that stabilize the hormone (Supplementary Fig. S5)28,29. In C. suppressalis, ITP encodes 112 amino acids and ITPL encodes 116 amino acids, and they are similar to the ITP of M. sexta (AAY29657.1, BLAST E-value 7e-56) and the ITPL of M. sexta (AAY29658.1, BLAST E-value 7e-58), respectively (Table 1). ITPs are members of a large neuropeptide superfamily designated as the crustacean hyperglycemic hormone (CHH)27.
Previously unreported splicing variants of three neuropeptide genes
Alternative splicing occurs among different structural and functional gene products. Allatotropin (AT), neuropeptide F1 (NPF1), ion transport peptide (ITP), corticotrophin-releasing factor-like diuretic hormone (CRF-DH), CAPA and orcokinin (OK) transcripts have been reported to undergo differential expression of alternatively splicing products in Lepidoptera7. In this study, we reported the splicing variants of three neuropeptide genes.
AstCC is an allatostatin C (AstC)-like peptide and encodes a PISCF-related peptide16 (Supplementary Fig. S6). AstCC and AstC are similar peptides, and the two genes were likely generated by gene duplication, and their receptor genes likely have a common ancestor as well16. However, the AstCC gene is not a classical neuropeptide gene, and it may well be expressed in cells that do not contain the regulated secretory pathway16. In this study, AstCC was found to have two splicing variants: AstCCa, which encodes 140 amino acids; and AstCCb, which encodes 106 amino acids (Supplementary Fig. S7A).
CCH was first identified as CCH2 in B. mori7, and it is 13 amino acid residues long and contains two cysteines (forming a cystine bond) and a C-terminal histidine-amide group17. CCHamide regulates feeding motivation in blowflies30 and sensory perception and olfactory behavior in starved Drosophila31. All insects with sequenced genomes contain two CCH genes, and they each code for a specific CCH: CCH1 (hallmark sequence: SCHSYGHSCWGAHamide), and CCH2 (hallmark sequence: GCQ [or A, or S] AFGHSCY [or F] GGHamide) (Supplementary Fig. S8)32. In C. suppressalis, two CCHamide genes (CCH1 and CCH2) have been identified. Additionally, in C. suppressalis, CCH1 has two splicing variants: CCH1a, which encodes 187 amino acids; and CCH1b, which encodes 162 amino acids (Supplementary Fig. S7B).
Short neuropeptide F (sNPF) was first isolated from the horse shoe crab Limulus polyphemus33, and the first insect sNPF was identified from the midgut of the cockroach P. americana34. Because of its length, these neuropeptides are referred to as sNPF to distinguish them from the so-called long neuropeptide F (NPF). sNPF peptides are cleaved from a larger prepropeptide precursor that typically yields multiple sNPF isoforms. In D. melanogaster, the sNPF precursor encodes four mature peptides, whereas the sNPF prepropeptide of A. mellifera produces only one sNPF isoform35. We identified three mature peptides produced from the sNPF precursor of C. suppressalis: SVRSPSRRLRFamide, ESRTPVRLRFamide and APSMRLRFamide (Supplementary Fig. S9). sNPF can regulate food intake, body size, growth, insulin production, sleep, and olfactory memory in insects1. Here, sNPFa (219 amino acids) and sNPFb (175 amino acids) were identified in C. suppressalis (Supplementary Fig. S7C). Interestingly, sNPFa yields one more mature peptide, DARSPVRLRYamide (Supplementary Fig. S7C).
Lost neuropeptide genes in C. suppressalis
In this study, the amino acid sequences of the neuropeptide precursors of B. mori, D. melanogaster and other insects were used as queries for a local BLAST analysis to search for candidate sequences of neuropeptides from our C. suppressalis transcriptomic data. However, we failed to find some neuropeptide precursors (Table 1).
Arginine-vasopressin-like peptide (AVLP) was initially isolated from the CNS of L. migratoria36, and it has also been identified in T. castaneum, Nasonia vitripennis, social ants, and N. lugens4. In this study, we could not find a sequence encoding AVLP in C. suppressalis.
CNMamide is a novel insect neuropeptide that was recently discovered in D. melanogaster and named after its C-terminal consensus motif37. Although CNMamide is conserved in most arthropods, Lepidoptera lack the CNMamide gene. However, the CNMamide receptor (CNMaR) occurs in certain lepidopteran species, including B. mori and Danaus plexippus (neuropeptide receptor A18)37, and CNMaR also occurs in C. suppressalis. A recent study also revealed that CNMaR in B. mori only shows a weak sensitivity to CNMamide, suggesting that CNMaR has evolved as a receptor for another unknown ligand in B. mori37.
Neuropeptide-like precursor 2 (NPLP2), NPLP3, and NPLP4 have been identified in D. melanogaster38, NPLP2 and NPLP3 have been identified in A. mellifera21, and NPLP3 and NPLP4 have been identified in N. lugens4 (Table 2). In C. suppressalis, a BLAST search of our transcriptomic data failed to find NPLP2, NPLP3, and NPLP4.
Proctolin is a pentapeptide with the mature peptide of RYLPT, and it was the first insect neuropeptide to be sequenced and chemically characterized39. The first identification of a proctolin precursor gene was CG7105 in D. melanogaster40. Although a previous study showed that proctolin is absent in B. mori7, this pentapeptide was recently identified in a proteomic analysis of B. mori wings41. However, the Bombyx proctolin gene does not produce a mature peptide because cleavage sites are not present at the N-terminal and C-terminal of the RYLPT sequence, and a similar gene is observed in C. suppressalis (Supplementary Fig. S10). Therefore, a true proctolin has been considered to be not observed in B. mori and C. suppressalis.
G protein-coupled receptors (GPCRs) for neuropeptides
A total of 51 putative neuropeptide GPCRs were identified in C. suppressalis by using all of the homologs of previously categorized B. mori and D. melanogaster neuropeptide GPCRs as queries (Table 3 and Supplementary Table S1). Based on the predicted amino acid sequences, phylogenetic trees were constructed for the neuropeptide GPCRs of Chilo, Bombyx, Drosophila, and other insects. Of these GPCRs, 44 belonged to the A-family (Fig. 2), 5 belonged to the B-family (Fig. 3), and 2 belonged to the LGRs (Fig. 4). However, the receptors for eclosion hormone (EH), insulin-like peptides (ILPs), neuroparsin (NP), and prothoracicotropic hormone (PTTH) are not GPCRs42,43,44,45. In addition, the receptors for Apis-ITG-like (ITG), NPLPs, and OKs have not been currently identified in insects.
Table 3. G protein-coupled receptors for neuropeptides identified from C. suppressalis.
| Gene name | Accession No. | Length | ORF | Putative identification | Species | Accession No. | E-value |
|---|---|---|---|---|---|---|---|
| Neuropeptide receptor A1 | KT030998 | 2170 | 1248 | Allatostatin receptor | Manduca sexta | ADX66345.1 | 0 |
| Neuropeptide receptor A2 | KT030999 | 1135 | 1014 | Neuropeptide receptor A2 | Bombyx mori | NP_001127737.1 | 5e-128 |
| Neuropeptide receptor A3 | KT031000 | 1386 | 1314 | Neuropeptide receptor A3 | Bombyx mori | NP_001127738.1 | 0 |
| Neuropeptide receptor A4 | KT031001 | 1888 | 1146 | Neuropeptide receptor A4 | Danaus plexippus | EHJ70829.1 | 0 |
| Neuropeptide receptor A5 | KT031002 | 2679 | 1692 | Neuropeptide receptor A5 | Bombyx mori | NP_001127740.1 | 0 |
| Neuropeptide receptor A6-A | KT031003 | 2170 | 1737 | Ecdysis triggering hormone receptor subtype A | Manduca sexta | AAX19163.1 | 0 |
| Neuropeptide receptor A6-B | KT031004 | 2235 | 1695 | Ecdysis triggering hormone receptor isoform B | Bombyx mori | NP_001165737.1 | 0 |
| Neuropeptide receptor A7 | KT031005 | 1422 | 1323 | Neuropeptide receptor A7 | Bombyx mori | NP_001127742.1 | 0 |
| Neuropeptide receptor A8 | KT031006 | 2869 | 1272 | Neuropeptide receptor A8 | Bombyx mori | NP_001127743.1 | 0 |
| Neuropeptide receptor A9 | KT031007 | 3592 | 1374 | Neuropeptide receptor A9 | Bombyx mori | NP_001127744.1 | 0 |
| Neuropeptide receptor A10 | KT031008 | 2423 | 1302 | Neuropeptide receptor A10 | Bombyx mori | NP_001127707.1 | 0 |
| Neuropeptide receptor A11 | KT031009 | 2842 | 1392 | Neuropeptide receptor A11 | Bombyx mori | NP_001127708.1 | 0 |
| Neuropeptide receptor A12 | KT031010 | 1431 | 1332 | Neuropeptide receptor A12 | Bombyx mori | NP_001127709.1 | 0 |
| Neuropeptide receptor A13 | KT031011 | 2042 | 1128 | Neuropeptide receptor A13 | Bombyx mori | NP_001127710.1 | 0 |
| Neuropeptide receptor A14 | KT031012 | 1879 | 1230 | Neuropeptide receptor A14 | Bombyx mori | NP_001127711.1 | 0 |
| Neuropeptide receptor A15 | KT031013 | 1463 | 1152 | Neuropeptide receptor A15 | Bombyx mori | NP_001127712.1 | 0 |
| Neuropeptide receptor A16 | KT031014 | 1932 | 1542 | Allatotropin receptor | Manduca sexta | ADX66344.1 | 0 |
| Neuropeptide receptor A17 | KT031015 | 1808 | 1122 | Neuropeptide receptor A17 | Bombyx mori | NP_001127715.1 | 9e-169 |
| Neuropeptide receptor A18 | KT031016 | 1952 | 1302 | Neuropeptide receptor A18 | Bombyx mori | NP_001127716.1 | 7e-147 |
| Neuropeptide receptor A19 | KT031017 | 1418 | 1347 | Neuropeptide receptor A19 | Bombyx mori | NP_001127717.1 | 0 |
| Neuropeptide receptor A20 | KT031018 | 1992 | 1269 | Neuropeptide receptor A20 | Danaus plexippus | EHJ69284.1 | 0 |
| Neuropeptide receptor A21 | KT031019 | 1812 | 1302 | Neuropeptide receptor A21 | Bombyx mori | NP_001127719.1 | 0 |
| Neuropeptide receptor A22 | KT031020 | 2788 | 1341 | Neuropeptide receptor A22 | Bombyx mori | NP_001127720.1 | 0 |
| Neuropeptide receptor A23 | KT031021 | 2134 | 1449 | Neuropeptide receptor A23 | Bombyx mori | NP_001127721.1 | 0 |
| Neuropeptide receptor A24 | KT031022 | 1780 | 1254 | Neuropeptide receptor A24 | Bombyx mori | NP_001127722.1 | 0 |
| Neuropeptide receptor A25 | KT031023 | 1017 | Neuropeptide receptor A25 | Bombyx mori | NP_001127723.1 | 2e-105 | |
| Neuropeptide receptor A26 | KT031024 | 1712 | 1314 | Neuropeptide receptor A26 | Bombyx mori | NP_001127724.1 | 0 |
| Neuropeptide receptor A27 | KT031025 | 1644 | 1473 | Neuropeptide receptor A27 | Bombyx mori | NP_001127725.1 | 0 |
| Neuropeptide receptor A28 | KT031026 | 1822 | 1329 | Neuropeptide receptor A28 | Bombyx mori | NP_001127726.1 | 9e-171 |
| Neuropeptide receptor A29 | KT031027 | 1190 | Neuropeptide receptor A29 | Bombyx mori | NP_001127745.1 | 0 | |
| Neuropeptide receptor A30 | KT031028 | 1652 | 1275 | Neuropeptide receptor A30 | Bombyx mori | NP_001127746.1 | 0 |
| Neuropeptide receptor A31 | KT031029 | 1293 | Neuropeptide receptor A31 | Bombyx mori | NP_001127747.1 | 0 | |
| Neuropeptide receptor A32 | KT031030 | 1169 | Neuropeptide receptor A32 | Bombyx mori | NP_001127748.1 | 0 | |
| Neuropeptide receptor A33 | KT031031 | 2681 | 1194 | Neuropeptide receptor A33 | Bombyx mori | NP_001127749.1 | 0 |
| Neuropeptide receptor A34 | KT031032 | 828 | Neuropeptide receptor A34 | Bombyx mori | NP_001127750.1 | 5e-154 | |
| Neuropeptide receptor A35 | KT031033 | 2151 | 1275 | Neuropeptide receptor A35 | Bombyx mori | NP_001127751.1 | 0 |
| Adipokinetic hormone receptor | KT031034 | 2275 | 1182 | Adipokinetic hormone receptor | Manduca sexta | ACE00761.1 | 0 |
| Allatostatin A receptor | KT031035 | 2260 | 1083 | Allatostatin A receptor | Spodoptera littoralis | ACJ06649.1 | 0 |
| Diapause hormone receptor | KT031036 | 1914 | 1353 | Diapause hormone receptor | Ostrinia nubilalis | AGL12069.1 | 4e-165 |
| FMRFamide receptor | KT031037 | 2217 | 1284 | FMRFamide receptor | Bombyx mori | NP_001037007.1 | 0 |
| Myosuppressin receptor | KT031038 | 1272 | 1140 | Myosuppressin receptor | Bombyx mori | NP_001036929.1 | 0 |
| Pheromone biosynthesis activating neuropeptide receptor-A | KT031039 | 1672 | 1041 | Pheromone biosynthesis activating neuropeptide receptor-A | Ostrinia nubilalis | AGL12066.1 | 2e-176 |
| Pheromone biosynthesis activating neuropeptide receptor-B | KT031040 | 1563 | Pheromone biosynthesis activating neuropeptide receptor-B | Ostrinia nubilalis | AGL12067.1 | 0 | |
| SIFamide receptor | KT031041 | 1888 | 1425 | SIFamide receptor | Bombyx mori | NP_001266380.1 | 0 |
| Sex peptide receptor | KT031042 | 2477 | 1242 | Sex peptide receptor | Spodoptera litura | AGE92037.1 | 0 |
| Neuropeptide receptor B1 | KT031043 | 5860 | 1197 | Neuropeptide receptor B1 | Danaus plexippus | EHJ71642.1 | 0 |
| Neuropeptide receptor B2 | KT031044 | 951 | Neuropeptide receptor B2 | Bombyx mori | NP_001127733.1 | 1e-128 | |
| Neuropeptide receptor B3 | KT031045 | 2716 | 2361 | Neuropeptide receptor B3 | Bombyx mori | NP_001127734.1 | 0 |
| Neuropeptide receptor B4 | KT031046 | 1395 | 1248 | Neuropeptide receptor B4 | Danaus plexippus | EHJ67831.1 | 0 |
| Diuretic hormone receptor | KT031047 | 1345 | Diuretic hormone receptor | Manduca sexta | P35464.1 | 0 | |
| Leucine-rich repeat G protein-coupled receptor 1 | KT031048 | 2334 | 2211 | Leucine-rich repeat G protein-coupled receptor | Bombyx mori | NP_001037033.1 | 0 |
| Leucine-rich repeat G protein-coupled receptor 2 | KT031049 | 2692 | Putative Leucine-rich transmembrane protein | Danaus plexippus | EHJ76329.1 | 0 |
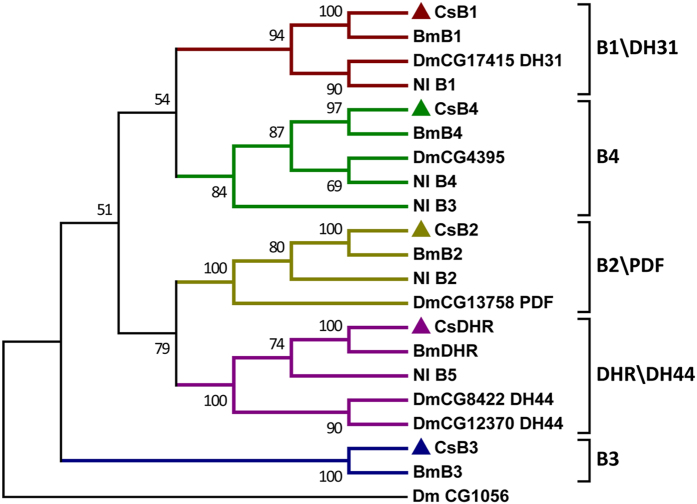
Figure 2. Phylogenetic tree of the A-family neuropeptide GPCRs from C. suppressalis (Cs), B. mori (Bm), and D. melanogaster (Dm).
Neighbor-joining trees were constructed using MEGA 5 software with 1000-fold bootstrap re-sampling. The numbers at the nodes of the branches represent the level of bootstrap support for each branch.
Figure 3. Phylogenetic tree of the B-family neuropeptide GPCRs from C. suppressalis (Cs), B. mori (Bm), N. lugens (Nl), and D. melanogaster (Dm).
Neighbor-joining trees were constructed using MEGA 5 software with 1000-fold bootstrap re-sampling. The numbers at the nodes of the branches represent the level of bootstrap support for each branch.
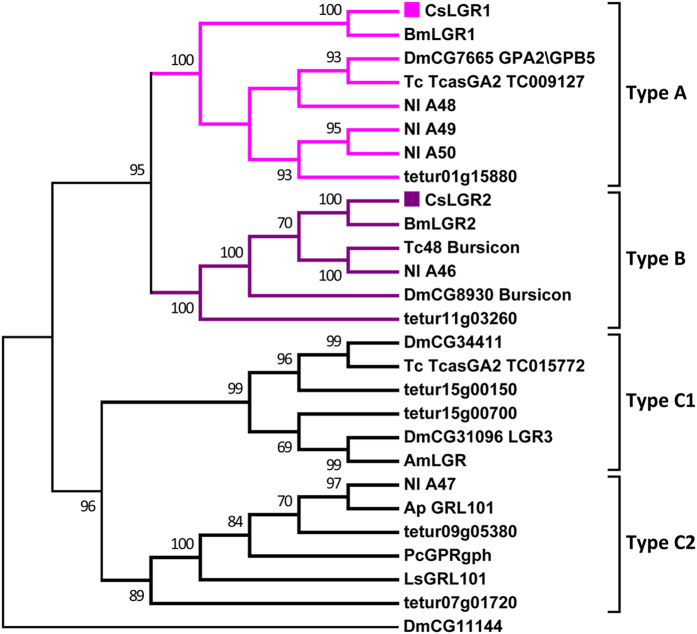
Figure 4. Phylogenetic tree of the leucine-rich repeat-containing GPCRs from C. suppressalis (Cs), B. mori (Bm), N. lugens (Nl), D. melanogaster (Dm), T. castaneum (Tc), A. mellifera (Am), A. pisum (Ap), Pediculus humanus corporis (Pc), Lymnaea stagnalis (Ls), and Tetranychus urticae (tetur).
Neighbor-joining trees were constructed using MEGA 5 software with 1000-fold bootstrap re-sampling. The numbers at the nodes of the branches represent the level of bootstrap support for each branch.
A-family GPCRs
In C. suppressalis, 44 A-family neuropeptide GPCRs were identified (Fig. 2). Because C. suppressalis and B. mori are both lepidopteran insects, they have the same number of A-family neuropeptide GPCRs, including neuropeptide receptor A1-A35, adipokinetic hormone receptor (AKHR), allatostatin A receptor (AstAR), diapause hormone receptor (DpHR), FMRFamide receptor (RFaR), myosuppressin receptor (MSR), pheromone biosynthesis activating neuropeptide receptor (PBANR), sex peptide receptor (SPR), and SIFamide receptor (SIFR) (Fig. 2).
In B. mori, neuropeptide receptor A2 and A34 have been recently deorphanized as ITP receptors, whereas A24 acts as an ITPL receptor5. Additionally, A24 has also been identified as a receptor for tachykinin (TK)46. A previous study demonstrated that the TKs and ITPL in B. mori have a stimulatory effect on feeding, it is possible that ITPL and TK signaling during the regulation of feeding behavior may directly affect one another by interacting with the receptor, neuropeptide receptor A245.
In the phylogenetic tree, the high-confidence clade shows the relationships between the various GPCRs for RY, LKs, TKs, and NTL, including the neuropeptide receptor A19, A22, A23, A24, A32, and A33 (Fig. 2). NTL is the most recently identified neuropeptide, and its receptors have been deorphanized. In B. mori, A32 and A33 have been deorphanized as NTLRs and distinguished because A32 is specific to the FXXXRa motif of NTL and A33 is specific to the YXXXRa motif19. The two different motifs are conserved in the NTL precursor in lepidopteran species, and the presence of two receptors that differentiate the two ligand motifs suggests that the two signaling systems diverged at an early evolutionary stage in Lepidoptera19.
The clade of the NPF and sNPF receptors is shown in the phylogenetic tree (Fig. 2). Neuropeptide receptor A4 has been identified as a NPF receptor47, whereas A10 and A11 are sNPF receptors14. Although A7 clusters nicely with A10 and A11, the ligand binding specificity of A7 has not been determined.
Our phylogenetic analysis identified the GPCR clade for the receptors of the GnRH-related peptides and CCAP, including the neuropeptide receptor A21, A26, A28, A29, A30, and AKHR (Fig. 2). A21 acts as a corazonin receptor48, whereas A26 and A30 are located near the previously characterized Drosophila CCAP receptor CG6111. In B. mori, AKHR is activated by AKH1 and AKH2 with a high affinity and by ACP with a low affinity, whereas A28 and A29 are activated by ACP at a high affinity and by AKH1 and AKH2 at a low affinity49.
A subtree containing the PK and PVK receptors is shown in the phylogenetic tree (Fig. 2). Neuropeptide receptor A25 and A27 are orthologous to the Drosophila CAPA-PVK receptor CG14575. DpHR is fairly exclusive to the Drosophila CAPA-PK1 receptor CG9918, whereas PBANR clusters with the Drosophila PK2 receptor CG8784 and CG8795.
In the phylogenetic tree, the high-confidence clade contains the GPCRs for CNMamide, FMRFamide, myosuppressin (MS), and allatostatin B (AstB) as well as a number of orphan GPCRs (Fig. 2). Neuropeptide receptor A18 has been identified as a CNMamide receptor (CNMaR)37, which is placed near Drosophila CNMaR CG33696 and Drosophila proctolin receptor CG6986. MSR and A13 cluster nicely with Drosophila MSR CG8985 and CG13802. Unfortunately, the neuropeptide receptor A3, A8, and A20 have not been deorphanized.
Additionally, the neuropeptide receptor A12, A17, and A35 cluster with Drosophila CG13995 (Fig. 2), and they are all orphan neuropeptide GPCRs.
B-family GPCRs
Five putative B-family GPCRs were identified in C. suppressalis (Fig. 3). Neuropeptide receptor B1 and B2 are orthologous to Drosophila CG17415 and CG13758, which have been annotated as diuretic hormone 31 (DH31) receptor and pigment-dispersing factor (PDF) receptor, respectively50. These receptors are both calcitonin-like receptors that are involved in the regulation of Ca2+ homeostasis. Diuretic hormone-like receptor was also identified in this family, and it is close to the Drosophila diuretic hormone 44 (DH44) receptor CG8422 and CG12370, which are corticotropin releasing factor-related receptors. Additionally, the neuropeptide receptor B4 that is homologous to Drosophila CG4395 and B3 have not been deorphanized.
Leucine-rich repeat-containing GPCRs (LGRs)
Three distinct types of LGRs (type A, B, and C) have been defined based on their structural characteristics, and they are distinguished by the number of leucine-rich repeat (LRR) motifs, the absence or presence of an LDLa motif (low density lipoprotein receptor domain class A) and their type-specific hinge region51. Two putative LGRs (LGR1 and LGR2) were identified in C. suppressalis (Fig. 4). LGR1 clusters with Drosophila CG7665 (DLGR1), which is an insect receptor for glycoprotein hormones. LGR1 contains eight LRRs, which are a characteristic feature of type A LGRs. Glycoprotein hormones are heterodimers consisting of α-subunit (GPA2) and β-subunit (GPB5) assembled by noncovalent bonds52. In D. melanogaster, GPA2 and GPB5 function via DLGR152, which is located in the epithelial cells of the hindgut where it increases the production of cAMP when stimulated by GPA2/GPB5. Hindgut cAMP stimulates the reabsorption of water53, and DLGR1 has been reported to play a critical role in development because its role in the regulation of pupariation54. LGR2 clusters with Tc48 and Drosophila CG8930 (DLGR2), which are receptors for bursicon. LGR2 belongs to type B LGRs characterized by the presence of 16–18 LRRs, which is roughly twice the number of LRRs found in the other types of LGRs. In D. melanogaster, bursicon mediates the tanning process in newly emerged adults via DLGR2, which is encoded by the rickets gene. Once activated, DLGR2, elicits the cAMP/PKA signaling pathway, which activates tyrosine hydroxylase, a key enzyme responsible for tanning agent synthesis55.
Expression profiles of neuropeptide precursors and neuropeptide GPCRs
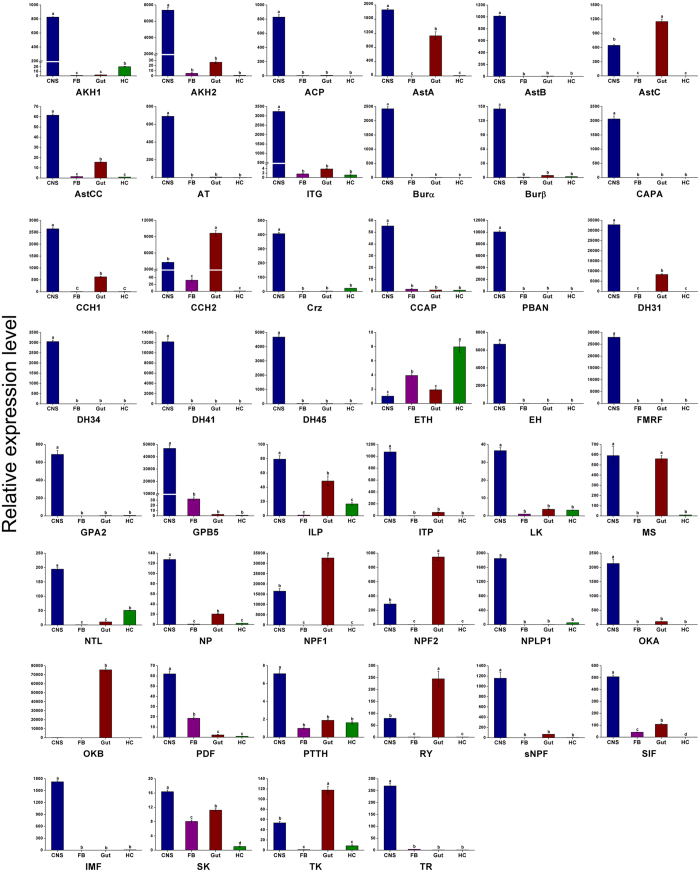
The tissue-specific expression profiles of the neuropeptide precursors and neuropeptide GPCRs in C. suppressalis were determined by qRT-PCR from various tissues, including the CNS (brain, suboesophageal ganglion, thoracic ganglion, and abdominal ganglion), the gut (foregut, midgut, hindgut, and Malpighian tubes), hemocytes (HC), and fat body (FB). Of the expression profiles for the neuropeptide precursors, only ecdysis triggering hormone (ETH) was most strongly expressed in the HC, whereas AstC, CCH2, NPF1, NPF2, RY, and TK were predominately expressed in the gut. Interestingly, OKB was expressed only in the gut. Most of the other neuropeptide precursors were preferentially expressed in the CNS (Fig. 5). AstC, NPF, OK, and TK are conserved brain-gut peptides, and their functions include myotropic effects and regulation of feeding behavior56. Our findings indicate that they may play a significant role in the digestive system.
Figure 5. qRT-PCR results showing the relative expression levels of the neuropeptides in various tissues of C. suppressalis.
Standard errors are represented by the error bars, and significant differences are represented by the different letters above each bar (p < 0.05).
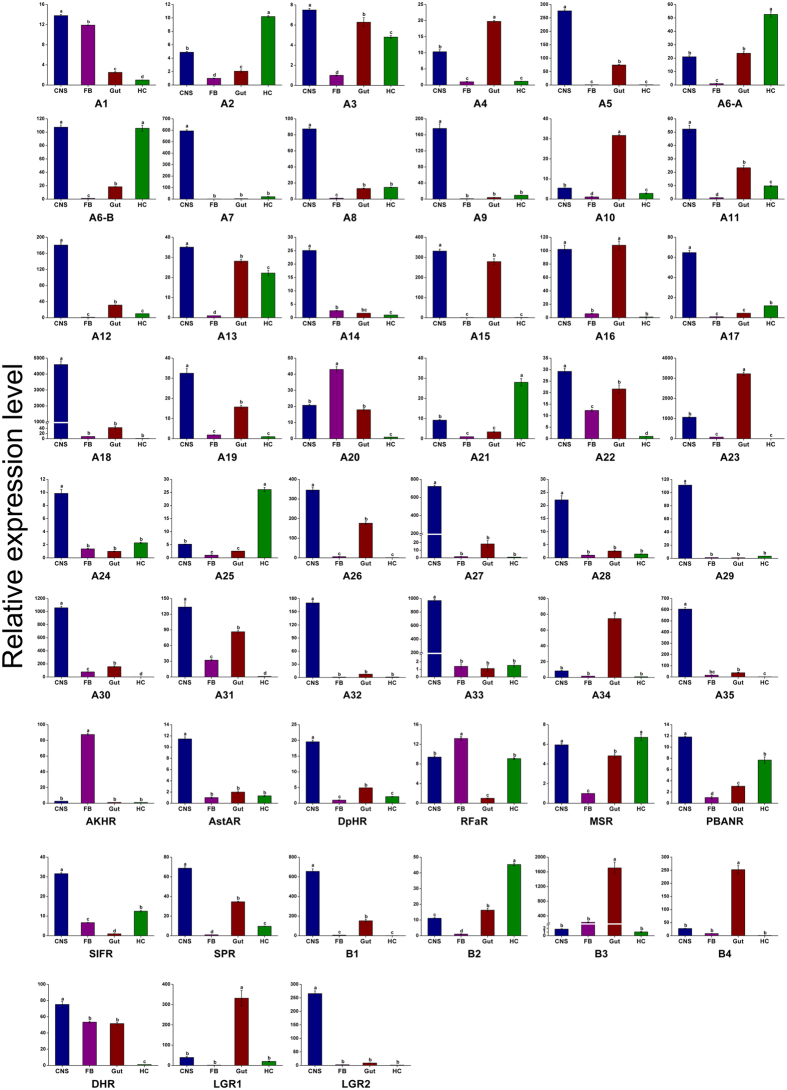
The neuropeptide GPCRs, A20, AKHR and RFaR had the highest expression in the FB, whereas A2, A6-A, A6-B, A21, A25, MSR, and B2 were predominately expressed in the HC and A4, A10, A16, A23, A34, B3, B4, and LGR1 had the highest expression in the gut. Additionally, most of the other neuropeptide GPCRs were predominately expressed in the CNS (Fig. 6). Our results showed that most of neuropeptide precursors and their GPCR genes were limited to the CNS, and numerous previous studies have indicated that neuropeptides and their GPCRs play crucial roles in neuromodulation and many other physiological processes in insects.
Figure 6. qRT-PCR results showing the relative expression levels of G protein-coupled receptors for the neuropeptides in various tissues of C. suppressalis.
Standard errors are represented by the error bars, and significant differences are represented by the different letters above each bar (p < 0.05). Neuropeptide receptor A1-A35 are abbreviated as A1-A35, respectively; Neuropeptide receptor B1-B4 are abbreviated as B1-B4, respectively.
Conclusions
RNA-seq is a useful tool for defining the transcriptome of an organism, even when a reference genome is not available. Our transcriptomic analysis of neuropeptide precursors and their putative GPCRs revealed the neuropeptidergic signaling systems in C. suppressalis. Our results elucidated the specific characteristics and expression profiles of the neuropeptide precursors and their putative GPCRs in C. suppressalis, and provided fundamental information to enhance our understanding of neuro-hormone mechanisms. The data from this study provide the first comprehensive description of neuropeptides and neuropeptide GPCRs in lepidopteran pests. We believe that this data will contribute to pharmacological research into the design of peptidomimetics, pseudopeptides or small molecules capable of disrupting physiological processes regulated by signaling molecules and their GPCRs and will provide beneficial insights into insect pest control.
Methods
Insect rearing
The C. suppressalis colony has been continuously reared in our laboratory. Larvae were originally collected from a rice field in Fuyang, Zhejiang Province, China, in 2012. The larvae were reared on an artificial diet57 and maintained at a temperature of 25 ± 1 °C, relative humidity of 80%, and a light:dark cycle of 14:10.
RNA-seq
To identify the genes encoding neuropeptides and their putative GPCRs in C. suppressalis, RNA samples of the fifth instar larval CNS were prepared for RNA-seq, because larvae are the targets for control and most neuropeptide-related genes are predominately expressed in the CNS. CNS samples (the brain, the suboesophageal ganglion, the thoracic ganglion, and the abdominal ganglion) of 100 fifth instar larvae were individually dissected in a saline solution containing an RNase inhibitor (TaKaRa, Japan). Transcriptome sequencing was performed on an Illumina HiSeq 2000 platform (Novogene Bioinformatics Technology Co.Ltd, Beijing, China), and resulted in 142,051,094 bp raw reads, and these raw reads were then subjected to de novo assembly using Trinity software13. The transcriptomic data were submitted to the Sequence Read Archive (SRA) database under the accession number SRX102269115.
Identification of the neuropeptides and their putative G protein-coupled receptors
We used the amino acid sequences of the neuropeptides and GPCRs of the silkworm B. mori, the fruit fly D. melanogaster and other insects as BLAST queries to search for the candidate sequences of neuropeptides and GPCRs from our C. suppressalis transcriptomic data. The BLAST + 2.2.23 software (downloadable from the National Center for Biotechnology Information, Bethesda, MD, USA; ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/) was used for local BLAST search of the assembled unigenes. After gene identification, we used the BLASTX and BLASTN programs against the non-redundant protein (Nr) and non-redundant nucleotide (Nt) NCBI database to identify homologous sequences in other insects.
Structure and domain analyses and sequence alignments
To identify neuropeptide signal peptide, we used SignalP 4.0 58 (http://www.cbs.dtu.dk/services/SignalP/). The predicted transmembrane domains of the putative neuropeptide GPCRs were verified using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). For the domain analysis, we used the NCBI Conserved Domain (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Multiple alignments of the amino acid sequences were performed with ClustalX2 59 and edited with GeneDoc software.
Phylogenetic analysis
We compared the neuropeptide GPCRs of C. suppressalis with the neuropeptide GPCRs from D. melanogaster, B. mori and other arthropods. For the Drosophila sequences, the CG numbers were used, whereas for the other species, the originally published names of the GPCRs were used, and the names of identified ligands were added4. The phylogenetic trees were constructed with MEGA5.0 60 using the neighbor-joining method. The reliability of each tree node was evaluated by bootstrap proportions using 1000 times.
Tissue-specific expression analysis
To study the tissue-specific expression profiles of the neuropeptides and their putative GPCRs, total RNA was extracted from various tissues, including the CNS, gut (including the foregut, midgut, hindgut, and Malpighian tubes), hemocytes (HC) and fat body (FB) of fifth instar larvae using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. For the hemocyte collection, the fifth instar larvae were surface-sterilized with 75% ethanol and the total hemolymph was collected with a 20 μl sterilized pipette by cutting its proleg, and then centrifuged at 200 × g for 10 min at 4 °C to collect the hemocyte precipitate. Other tissues were dissected from the fifth instar larvae on ice. The TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kits (Transgen, Beijing, China) were used to synthesize cDNA from 1 μg RNA. Specific primers for the qRT-PCR analysis were designed with Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/) (Supplementary Table S2 and Table S3). The CFX Connect™ Real-Time Detection System (Bio-rad, USA) was used to conduct the qRT-PCR analysis. The reference gene elongation factor 1 alpha (EF-1) was used to normalize the expression of the target genes. The qRT-PCR procedure was performed in a 25 μl reaction containing 12.5 μl SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa, Japan), 1 μl of each primer (10 μM), 5 μl of cDNA template, and 5.5 μl of sterile H2O. The conditions for the qRT-PCR procedure were as follows: 95 °C for 30 s, and then 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The PCR products were then heated to 95 °C for 15 s, cooled to 60 °C for 1 min and heated to 95 °C for 30 s and cooled to 60 °C for 15 s to measure the dissociation curves. Three biological replicates of each tissue were used to ensure the reliability and reproducibility of the results.
The relative quantification of each tissue was calculated using the comparative 2−ΔΔCT method61. All of the data were normalized to the endogenous EF-1 levels from the same individual samples. In the analysis of the relative expression level in different tissues, the lowest expression level was used as the calibrator. Thus, the relative expression level in different tissues was assessed by comparing the expression level of each target gene in other tissues with that in the tissue with the lowest expression. The results were presented as the mean of the expression level of three biological replicates. The relative expression levels in the various tissues were analyzed using a one-way analysis of variance (ANOVA) followed by Tukey’s honestly significant difference (HSD) test to determine whether significant differences occurred. All of the statistical analyses were performed by the Data Processing System (DPS) software package (Version 9.5)62.
Additional Information
How to cite this article: Xu, G. et al. Identification and expression profiles of neuropeptides and their G protein-coupled receptors in the rice stem borer Chilo suppressalis. Sci. Rep. 6, 28976; doi: 10.1038/srep28976 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Special Agricultural Research Projects for Public Welfare, China (201303017), National Science Fund for Innovative Research Groups of Biological Control (Grant 31321063) and National High-Tech R&D Program of China (863 Program, 2011AA10A204). The authors sincerely thank Shuang-Yang Wu, Pi-Hua Zhou, Fang Liu, and Lu-Lu Gu for assistance in collecting and feeding the rice stem borer.
Footnotes
The authors declare no competing financial interests.
Author Contributions G.X., S.F.W., J.H., G.Y.Y. and Q.F. conceived and designed the experimental plan. G.X., G.X.G. and Z.W.T. performed the experiments. G.X. analyzed and interpreted the sequence data and experimental data. G.X., J.H., G.Y.Y. and Q.F. drafted the manuscript. Q.S.S. provided some suggestions for revising and polishing the manuscript. All of the authors read and approved the final manuscript.
References
- Nassel D. R. & Winther A. M. Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92, 42–104 (2010). [DOI] [PubMed] [Google Scholar]
- Sterkel M. et al. OKB, a novel family of brain-gut neuropeptides from insects. Insect Biochem. Mol. Biol. 42, 466–473 (2012). [DOI] [PubMed] [Google Scholar]
- Zandawala M. & Orchard I. Identification and functional characterization of FGLamide-related allatostatin receptor in Rhodnius prolixus. Insect Biochem. Mol. Biol. 57, 1–10 (2015). [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Suetsugu Y., Yamamoto K., Noda H. & Shinoda T. Transcriptome analysis of neuropeptides and G-protein coupled receptors (GPCRs) for neuropeptides in the brown planthopper Nilaparvata lugens. Peptides 53, 125–133 (2014). [DOI] [PubMed] [Google Scholar]
- Nagai C., Mabashi-Asazuma H., Nagasawa H. & Nagata S. Identification and characterization of receptors for ion transport peptide (ITP) and ITP-like (ITPL) in the silkworm Bombyx mori. J. Biol. Chem. 289, 32166–32177 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. et al. Molecular characterization and expression profiles of nicotinic acetylcholine receptors in the rice striped stem borer, Chilo suppressalis (Lepidoptera: Crambidae). Insect Sci., doi: 10.1111/1744-7917.12324 (2016). [DOI] [PubMed] [Google Scholar]
- Roller L. et al. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38, 1147–1157 (2008). [DOI] [PubMed] [Google Scholar]
- Wu S. F., Xu G., Stanley D., Huang J. & Ye G. Y. Dopamine modulates hemocyte phagocytosis via a D1-like receptor in the rice stem borer, Chilo suppressalis. Sci. Rep. 5, 12247 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Z. W. et al. Effects of the endoparasitoid Cotesia chilonis (Hymenoptera: Braconidae) parasitism, venom, and calyx fluid on cellular and humoral immunity of its host Chilo suppressalis (Lepidoptera: Crambidae) larvae. J. Insect Physiol. 85, 46–56 (2016). [DOI] [PubMed] [Google Scholar]
- Su J. Y., Zhang Z. Z., Wu M. & Gao C. F. Changes in insecticide resistance of the rice striped stem borer (Lepidoptera: Crambidae). J. Econ. Entomol. 107, 333–341 (2014). [DOI] [PubMed] [Google Scholar]
- Scherkenbeck J. & Zdobinsky T. Insect neuropeptides: structures, chemical modifications and potential for insect control. Bioorg. Med. Chem. 17, 4071–4084 (2009). [DOI] [PubMed] [Google Scholar]
- Garland S. L. Are GPCRs still a source of new targets? J. Biomol. Screen. 18, 947–966 (2013). [DOI] [PubMed] [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N. et al. Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS One 3, e3048 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. et al. De novo assembly and characterization of central nervous system transcriptome reveals neurotransmitter signaling systems in the rice striped stem borer, Chilo suppressalis. BMC Genomics 16, 525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra J. A. & Allatostatin C. and its paralog allatostatin double C: the arthropod somatostatins. Insect Biochem. Mol. Biol. 39, 161–170 (2009). [DOI] [PubMed] [Google Scholar]
- Hansen K. K., Hauser F., Williamson M., Weber S. B. & Grimmelikhuijzen C. J. The Drosophila genes CG14593 and CG30106 code for G-protein-coupled receptors specifically activated by the neuropeptides CCHamide-1 and CCHamide-2. Biochem. Biophys. Res. Commun. 404, 184–189 (2011). [DOI] [PubMed] [Google Scholar]
- Ida T. et al. Identification of the endogenous cysteine-rich peptide trissin, a ligand for an orphan G protein-coupled receptor in Drosophila. Biochem. Biophys. Res. Commun. 414, 44–48 (2011). [DOI] [PubMed] [Google Scholar]
- Jiang H. B. et al. Natalisin, a tachykinin-like signaling system, regulates sexual activity and fecundity in insects. Proc. Natl. Acad. Sci. USA 110, E3526–3534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Broeck J. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides 22, 241–254 (2001). [DOI] [PubMed] [Google Scholar]
- Hummon A. B. et al. From the genome to the proteome: uncovering peptides in the Apis brain. Science 314, 647–649 (2006). [DOI] [PubMed] [Google Scholar]
- Li B. et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res. 18, 113–122 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Predel R., Linde D., Rapus J., Vettermann S. & Penzlin H. Periviscerokinin (Pea-PVK): a novel myotropic neuropeptide from the perisympathetic organs of the American cockroach. Peptides 16, 61–66 (1995). [DOI] [PubMed] [Google Scholar]
- Predel R. & Wegener C. Biology of the CAPA peptides in insects. Cell. Mol. Life Sci. 63, 2477–2490 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. H. & Denlinger D. L. Identification of a cDNA encoding DH, PBAN and other FXPRL neuropeptides from the tobacco hornworm, Manduca sexta, and expression associated with pupal diapause. Peptides 25, 1099–1106 (2004). [DOI] [PubMed] [Google Scholar]
- Audsley N., McIntosh C. & Phillips J. E. Isolation of a neuropeptide from locust corpus cardiacum which influences ileal transport. J. Exp. Biol. 173, 261–274 (1992). [DOI] [PubMed] [Google Scholar]
- Begum K., Li B., Beeman R. W. & Park Y. Functions of ion transport peptide and ion transport peptide-like in the red flour beetle Tribolium castaneum. Insect Biochem. Mol. Biol. 39, 717–725 (2009). [DOI] [PubMed] [Google Scholar]
- Stewart M. J. et al. Cloning of the crustacean hyperglycemic hormone and evidence for molt-inhibiting hormone within the central nervous system of the blue crab Portunus pelagicus. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 164, 276–290 (2013). [DOI] [PubMed] [Google Scholar]
- Liu M. Q., Pan L. Q., Li L. & Zheng D. B. Molecular cloning, characterization and recombinant expression of crustacean hyperglycemic hormone in white shrimp Litopenaeus vannamei. Peptides 53, 115–124 (2014). [DOI] [PubMed] [Google Scholar]
- Ida T. et al. Isolation of the bioactive peptides CCHamide-1 and CCHamide-2 from Drosophila and their putative role in appetite regulation as ligands for G protein-coupled receptors. Front. Endocrinol. 3, 177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan A. et al. The CCHamide 1 receptor modulates sensory perception and olfactory behavior in starved Drosophila. Sci. Rep. 3, 2765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. et al. Expression patterns of the Drosophila neuropeptide CCHamide-2 and its receptor may suggest hormonal signaling from the gut to the brain. PLoS One 8, e76131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaus G. et al. The sequences of 5 neuropeptides isolated from Limulus using antisera to FMRFamide. Biol. Bull. 184, 322–329 (1993). [DOI] [PubMed] [Google Scholar]
- Veenstra J. A. & Lambrou G. Isolation of a novel RFamide peptide from the midgut of the American cockroach, Periplaneta americana. Biochem. Biophys. Res. Commun. 213, 519–524 (1995). [DOI] [PubMed] [Google Scholar]
- Dillen S., Verdonck R., Zels S., Van Wielendaele P. & Vanden Broeck J. Identification of the short neuropeptide F precursor in the desert locust: evidence for an inhibitory role of sNPF in the control of feeding. Peptides 53, 134–139 (2014). [DOI] [PubMed] [Google Scholar]
- Proux J. P. et al. Identification of an arginine vasopressin-like diuretic hormone from Locusta migratoria. Biochem. Biophys. Res. Commun. 149, 180–186 (1987). [DOI] [PubMed] [Google Scholar]
- Jung S. H. et al. Identification of a novel insect neuropeptide, CNMa and its receptor. FEBS Lett. 588, 2037–2041 (2014). [DOI] [PubMed] [Google Scholar]
- Baggerman G., Cerstiaens A., De Loof A. & Schoofs L. Peptidomics of the larval Drosophila melanogaster central nervous system. J. Biol. Chem. 277, 40368–40374 (2002). [DOI] [PubMed] [Google Scholar]
- Starratt A. N. & Brown B. E. Structure of the pentapeptide proctolin, a proposed neurotransmitter in insects. Life Sci. 17, 1253–1256 (1975). [DOI] [PubMed] [Google Scholar]
- Taylor C. A. et al. Identification of a proctolin preprohormone gene (Proct) of Drosophila melanogaster: expression and predicted prohormone processing. J. Neurobiol. 58, 379–391 (2004). [DOI] [PubMed] [Google Scholar]
- Shi X. F. et al. Proteomic analysis of the phenotype of the scaleless wings mutant in the silkworm, Bombyx mori. J. Proteomics 78, 15–25 (2013). [DOI] [PubMed] [Google Scholar]
- Chang J. C., Yang R. B., Adams M. E. & Lu K. H. Receptor guanylyl cyclases in Inka cells targeted by eclosion hormone. Proc. Natl. Acad. Sci. USA 106, 13371–13376 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogiolo W. et al. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11, 213–221 (2001). [DOI] [PubMed] [Google Scholar]
- Vogel K. J., Brown M. R. & Strand M. R. Ovary ecdysteroidogenic hormone requires a receptor tyrosine kinase to activate egg formation in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 112, 5057–5062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz K. F., Yamanaka N., Gilbert L. I. & O’Connor M. B. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science 326, 1403–1405 (2009). [DOI] [PubMed] [Google Scholar]
- He X. B. et al. Activation of BNGR-A24 by direct interaction with tachykinin-related peptides from the silkworm Bombyx mori leads to the Gq- and Gs-coupled signaling cascades. Biochemistry 53, 6667–6678 (2014). [DOI] [PubMed] [Google Scholar]
- Deng X. Y. et al. Activation of Bombyx neuropeptide G protein-coupled receptor A4 via a Gαi-dependent signaling pathway by direct interaction with neuropeptide F from silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 45, 77–88 (2014). [DOI] [PubMed] [Google Scholar]
- Yang J. W. et al. Specific activation of the G protein-coupled receptor BNGR-A21 by the neuropeptide corazonin from the silkworm, Bombyx mori, dually couples to the Gq and Gs signaling cascades. J. Biol. Chem. 288, 11662–11675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. et al. Identification and functional characterization of two orphan G-protein-coupled receptors for adipokinetic hormones from silkworm Bombyx mori. J. Biol. Chem. 286, 42390–42402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y. et al. The G protein-coupled receptors in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 40, 581–591 (2010). [DOI] [PubMed] [Google Scholar]
- Van Hiel M. B., Vandersmissen H. P., Van Loy T. & Broeck J. V. An evolutionary comparison of leucine-rich repeat containing G protein-coupled receptors reveals a novel LGR subtype. Peptides 34, 193–200 (2012). [DOI] [PubMed] [Google Scholar]
- Sudo S., Kuwabara Y., Park J. I., Hsu S. Y. & Hsueh A. J. Heterodimeric fly glycoprotein hormone-α2 (GPA2) and glycoprotein hormone-β5 (GPB5) activate fly leucine-rich repeat-containing G protein-coupled receptor-1 (DLGR1) and stimulation of human thyrotropin receptors by chimeric fly GPA2 and human GPB5. Endocrinology 146, 3596–3604 (2005). [DOI] [PubMed] [Google Scholar]
- Sellami A., Agricola H. J. & Veenstra J. A. Neuroendocrine cells in Drosophila melanogaster producing GPA2/GPB5, a hormone with homology to LH, FSH and TSH. Gen. Comp. Endocrinol. 170, 582–588 (2011). [DOI] [PubMed] [Google Scholar]
- Vandersmissen H. P., Van Hiel M. B., Van Loy T., Vleugels R. & Vanden Broeck J. Silencing D. melanogaster lgr1 impairs transition from larval to pupal stage. Gen. Comp. Endocrinol. 209, 135–147 (2014). [DOI] [PubMed] [Google Scholar]
- An S. et al. Insect neuropeptide bursicon homodimers induce innate immune and stress genes during molting by activating the NF-κB transcription factor Relish. PLoS One 7, e34510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L., Jiang F., Yang P. C., Wang X. H. & Kang L. Molecular characterization and expression profiles of neuropeptide precursors in the migratory locust. Insect Biochem. Mol. Biol. 63, 63–71 (2015). [DOI] [PubMed] [Google Scholar]
- Han L. Z., Li S. B., Liu P. L., Peng Y. F. & Hou M. L. New artificial diet for continuous rearing of Chilo suppressalis (Lepidoptera: Crambidae). Ann. Entomol. Soc. Am. 105, 253–258 (2012). [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G. & Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Tang Q. Y. & Zhang C. X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 20, 254–260 (2013). [DOI] [PubMed] [Google Scholar]
- Hewes R. S. & Taghert P. H. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 11, 1126–1142 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.