Abstract
Truncated receptor ectodomains have been described for several classes of cell surface receptors, including those that bind to growth factors, cytokines, immunoglobulins, and adhesion molecules. Soluble receptor isoforms are typically generated by proteolytic cleavage of the cell surface receptor or by alternative splicing of RNA transcripts arising from the same gene encoding the full-length receptor. Both the epidermal growth factor receptor (EGFR) and the insulin receptor (INSR) families produce soluble receptor splice variants in vertebrates and truncated forms of insulin receptor-like sequences have previously been described in Drosophila. The EGFR and INSR ectodomains share significant sequence homology with each other suggestive of a common evolutionary origin. We discovered novel truncated insulin receptor-like variants in several arthropod species. We performed a phylogenetic analysis of the conserved extracellular receptor L1 and L2 subdomains in invertebrate species. While the segregation of insulin receptor-like L1 and L2 domains indicated that an internal domain duplication had occurred only once, the generation of truncated insulin receptor-like sequences has occurred multiple times. The significance of this work is the previously unknown and widespread occurrence of truncated isoforms in arthropods, signifying that these isoforms play an important functional role, potentially related to such isoforms in mammals.
Keywords: INSR, INSRR, ectodomain, receptor L domain, Drosophila, Tribolium castaneum
The insulin receptor is a plasma membrane tyrosine kinase receptor that binds insulin and has a vital function in blood glucose homeostasis regulation (De Meyts 2004). The evolutionary history of the insulin receptor is particularly interesting because it spans a central portion of the tree of life, which coincides with a long gap in genome availability. The evolution of this vital receptor started in marine sponges (Skorokhod et al 1999), and underwent duplications in amphibians (Hernandez-Sanchez et al., 2008; Renteria et al., 2008), forming the insulin receptor-related receptor (INSRR) and the insulin-like growth factor 1 receptor (IGF1R). INSR was, unlike many other expanded receptor families and their ligands (Daza et al 2011), apparently skipped over (Collins et al 2012) during rounds 1 and 2 (R1 and R2) of the whole genome duplication events (Kuraku et al 2009). However, both insulin and EGF receptors appear to have undergone teleost-specific genome duplication (Laisney et al 2010).
The Drosophila melanogaster (Diptera: Drosophilidae) genome contains seven insulin-like peptide genes (Ilp1–7), but interestingly only one insulin receptor (“InR” – using the species-specific nomenclature) homolog. With this in mind, Pietrokovski and Shilo (2001) used bioinformatic methods to search for new signaling components within the Drosophila genome. Remarkably, they reported two genes that appear to encode truncated forms of the insulin receptor, displaying 32–45% identity to InR in the receptor L domains. These genes, CG3837 and CG10702, lack the conserved insulin receptor cytoplasmic tyrosine kinase domains, but contain the extracellular L1, cysteine-rich and L2 domains. In addition, CG10702 possesses a transmembrane domain, while CG3837 seems to encode a secreted protein. The overall amino acid similarity between CG3837 and CG10702 is only 25%. The authors hypothesized that these proteins could possibly inhibit insulin signaling by sequestering the various insulin-like peptides (Ilps), analogous to Argos function on EGFR ligands in fly (Klein et al., 2008), or potentially modulate signaling by cooperating with other receptors. What has not been clarified is the conservation of these genes within the class Insecta, or whether vertebrates produce similar truncated insulin receptor-like proteins.
While INSRR has known splice variants in rat stomach and kidney, and Mathi et al (1995) have reported 6 kb and 2 kb INSRR mRNAs in human tissues, these variants have never been fully confirmed in human. The INSRR receptor appears to have diverged approximately mid-way between the arthropod lineage, which contains truncated insulin receptor-like genes, and the higher animals that make use of truncated splice variants. Therefore, comparing the insulin receptor-like proteins, whether or not they arise from receptor-like genes or alter-native splicing, is a promising avenue towards understanding differences in conservation patterns between the functions of truncated and full-length receptors.
Therefore, in the present study we analyzed the evolutionary relationship of the recently discovered truncated variants of the insulin receptor family. We present phylogenetic evidence for similarity between the mammalian INSRR and truncated variants in insects, new evidence for truncated variants in other arthropods, and preliminary findings of a putative truncated splice variant of the INSRR in humans.
1 Materials and Methods
1.1 Sequence search method
The bioinformatics search method was implemented as a Perl script (available on request). We used a query consisting of Drosophila InR ectodomain (C4G-LC –Furin– C4G-LC) and the same region from Drosophila EGFR (not including the second Furin domain). The TBLASTN cutoff was 1e–5. If a hit was found, it was categorized as kinase-positive if there was a Drosophila InR kinase hit within 10,000 bps, at a hit quality 1e–5, in the same orientation. If the original hit was closer than 10 kbp to the contig end (depending on hit direction), the hit was disqualified as uninformative (“overshoot” error).
The following genomes were analyzed: AAAB01 (Anopheles gambiae str. PEST), AAGE02 (Aedes aegypti, strain Liverpool), AAWU01 (Culex quinquefasciatus strain JHB), AAJJ01 (Tribolium castaneum strain Georgia), ACJG01 (Daphnia pulex). In addition, Glossina morsitans (Diptera: Calyptratae) was searched manually.
1.2 Phylogenetic analysis
MAFFT v7.023b (2013/02/03) E-INS-i alignment of 46 sequences was tested in ProtTest 3.2 (Darriba et al 2011), calculating a comparison of seven different model selection frameworks, including AIC (Akaike Information Criteria). The best scoring substitution model, across the seven frameworks, was LG+I+Γ (Le & Gascuel, 2008). We used RAxML 7.0.4 to run a rapid Bootstrap analysis and search for best-scoring ML tree in one program run using the Le & Gascuel model +I +Γ, with 1 000 bootstraps. To increase bootstrap values, we added INSR sequences from Gallus gallus, Takifugu rubripes (INSR-1 and INSR-2), Branchiostoma floridae, Saccoglossus kowalevskii, Strongylocentrotus purpuratus, Biomphalaria glabrata, Bombyx mori, and truncated homologues from Drosophila grimshawi. We repeated the phylogenetic analysis for the ectodomain (L1-CR-L2).
1.3 PCR and sequencing of cDNA
Human whole-brain RNA (Yorkshire Bioscience Ltd, York, UK) was used as a template for complementary DNA (cDNA) synthesis. The total RNA was derived from a pool of brain tissue from five adult male donors (Batch B209031). cDNA was synthesized using MLV reverse transcriptase (Invitrogen AB, Lidingö) and random hexamers (Roche Diagnostics Scandinavia AB, Bromma) as primers for reverse transcription. Reactions were run according to the manufacturer’s specifications.
Polymerase chain reaction (PCR) was used to amplify cDNA sequences, using Unocycler (VWR International AB, Stockholm). Reactions were run using a primer pair specific for the human INSRR gene but placed to be able to detect truncated variants: forward primer 5′-CGTGCCTGTGTAGCTTGC-3′ (coding exon 3); reverse primer 5′-GGAGTGTAATAGAAGGAGCTGGTC-3′ (located 18 bp from the start of intron 5); and Biotools DNA polymerase (Techtum Lab AB, Umeå) at the following conditions: initial denaturation at 95° C for 3 minutes followed by 50 cycles at 95° C denaturation for 30 seconds, 58.6° C annealing temperature for 30 seconds and 72° C elongation temperature for 1 minute followed by a final elongation at 72° C for 3 minutes. Products were visualized on agarose gel. Gel fragments containing the products were cut out and products purified using a QIAEX II gel extraction kit (QIAGEN AB, Solna). Products were then sequenced at the Uppsala Genome Center, using Sanger sequencing.
2 Results
2.1 Sequence search method
We identified new insulin receptor-like ectodomain sequences on the following genomic contigs (direction +/−): Anopheles gambiae (African malaria mosquito; Diptera: Culicidae) AAAB01008984.1 (+) 11.28 Mb; Aedes aegypti (Yellow fever mosquito; Diptera: Culicidae) AAGE02003510.1 (+) 23.82 kb; Aedes aegypti (Diptera: Culicidae) AAGE02003511.1 (+) 112.64 kb; Aedes aegypti (Diptera: Culicidae) AAGE02009010.1 (−) 27.13 kb; Culex quinquefasciatus (Southern house mosquito; Diptera: Culicidae) AAWU01008821.1 (−) 56.80 kb; Tribolium castaneum (Red flour beetle; Coleoptera: Tenebrionidae) AAJJ01000005.1 (+) 167.72 kb; Daphnia pulex (Water flea; Cladocera: Daphniidae) ACJG01002238.1 (+) 49.51 kb. The findings are summarized in Figs. 1 – 2, Table 1, and Supplementary Text S1 and S2.
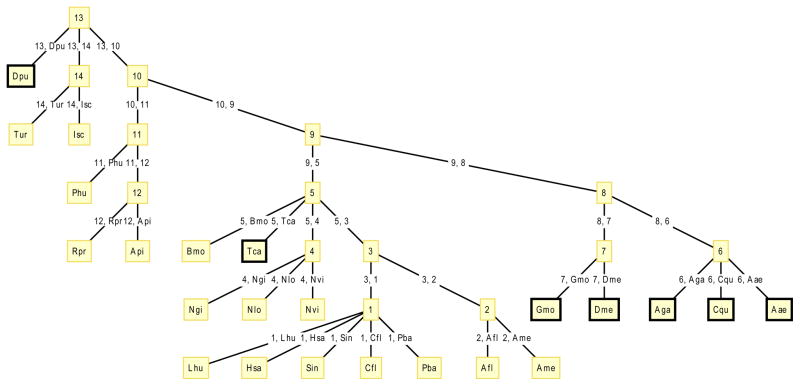
Fig. 1. Arthropod lineage tree, showing lineages mined and searched for insulin receptor-like genes without tyrosine kinase domains.
Black boxes indicate lineages containing ectodomain-only findings: Tenebrionidae (Tca), Glossinidae (Gmo), Drosophilidae (Dme), Culicidae (Aae, Aga, Cqu) and Daphniidae (Dpu). The findings in Diptera could be linked to the finding in Coleoptra, which both stem from Endopterygota. However, no findings have been made in Hymenoptera or Lepidoptera. The fact that the positive findings are spread out over the tree suggest multiple formation events.
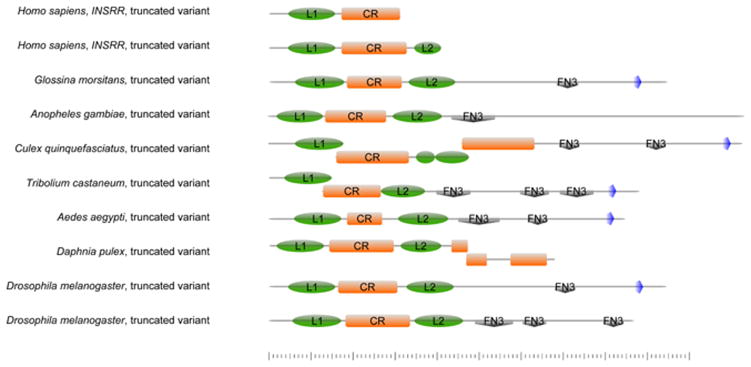
Fig. 2. Schematic drawing of the domain structure of ectodomain-only sequences.
The truncated insulin receptor-like sequences in arthropods span the receptor L1 domain (green oval), the furin-like domain (orange) and the receptor L2 domain (green oval). In most cases, they also contain between 1 and 3 fibronectin domains (grey diamond) and a transmembrane domain (blue pentagon). The putative truncated splice variants of INSRR identified in human brain cDNA are much shorter, encompassing only the L1 domain (green oval), the furin-like domain (orange), and in the longer variant, half of the L2 domain (green oval). Created using http://prosite.expasy.org/cgi-bin/prosite/mydomains/.
Table 1.
Accession numbers (DDBJ, VectorBase, FlyBase, GenBank) of truncated insulin receptor-like sequences in arthropods and truncated variants in human brain cDNA
| Accession number | Length (AAs) | Species | Domain structure | Method of finding |
|---|---|---|---|---|
| BR000948(†) | 679 | D. pulex | L1-Furin-L2-Furin | Mining (‡) |
| BR000971 (†) | 881 | T. castaneum | L1-Furin-L2-3 × Fn3-TM | Mining (‡) |
| AGAP009262 | 613 | A. gambiae | L1-Furin-L2-1 × Fn3 | Mining (‡) |
| BR000961 (†) | 849 | A. aegypti | L1-Furin-L2-2 × Fn3-TM | Mining (‡) |
| CPIJ005469 | 1,130 | C. quinquefasciatus | L1-Furin-L2-Furin-2 × Fn3-TM | Mining (‡) |
| TMP002939 | 947 | G. moristans | L1-Furin-L2-1 × Fn3-TM | Database search |
| CG3837 | 868 | D. melanogaster | L1-Furin-L2-3 × Fn3 | Previously known |
| CG10702 | 946 | D. melanogaster | L1-Furin-L2-1 × Fn3-TM | Previously known |
| Not submitted | 313 | H. sapiens | L1-Furin | cDNA sequencing |
| JQ991924 (§) | 410 | H. sapiens | L1-Furin-L2 | cDNA sequencing |
DDBJ Third party annotation
Automated mining using a Perl script
Submitted to GenBank
2.2 Phylogenetic analysis
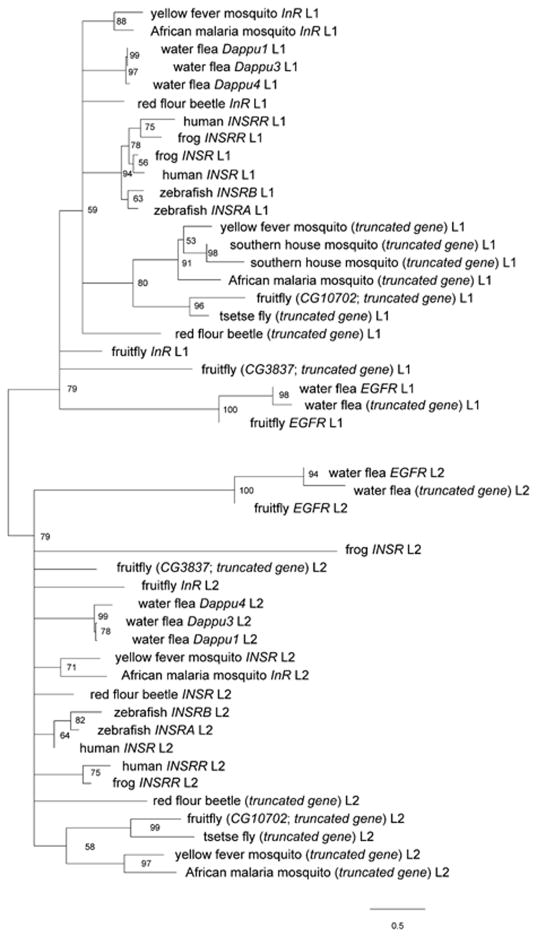
The L1 and L2 domains of the sequences identified in our mining were analyzed through maximum likelihood. The tree was rooted on the edge that bipartitioned the receptor L1 and L2 sub-trees, which in turn supports the nodes that segregates EGFR and InR, displaying significant bootstrap (BP) support of 79% in both sub-trees (Fig. 3). The segregation between receptor L1 and L2 domains was consistent with a single, ancient domain duplication prior to the formation of both the lineages of INSR and EGFR. The truncated gene in D. pulex was clearly clustering with EGFR, displaying 94–98% BP (Fig. 3), indicating that it had formed in a separate event from the other truncated sequences. These results were confirmed using the ectodomain (L1-CR-L2) (Fig. S1), which also showed a 100% segregation between the INSR/INSRR and EGFR lineages.
Fig. 3. Maximum likelihood phylogenetic tree of L1 and L2 based on amino acid sequences.
This figure shows a collapsed version (at 50% BP) of our maximum likelihood analysis. We used RAxML 7.0.4 to run a rapid Bootstrap analysis and search for best-scoring ML tree using the best scoring substitution model, Le & Gascuel model, +I +Γ, with 1000 bootstraps. Scale bars indicate substitutions per site.
2.3 PCR and sequencing of cDNA
In a wet lab effort to identify potential splice variants of INSRR, two PCR-products, approximately 600 and 900 bp in length were amplified from human brain cDNA. DNA sequencing showed the shorter of these products to be principally identical to sIRR-1 in rat. It was comprised of exons 3–5. At the end of exon 5, the donor splice site was not used. There was an in-frame stop codon in the beginning of intron 5. Four unknown positions in the cDNA sequence were replaced by comparison to reference sequence in Ensembl. This sequence was submitted: JQ991924. The protein translation of this variant would contain receptor L domain 1, the Furin-like domain, and the first half of receptor L domain 2.
The longer product started at the same position in exon 3. It then retained all of intron 3, exon 4, all of intron 4, and almost all of exon 5. It appeared that this PCR product may have been derived from either genomic DNA or an unspliced RNA molecule since it retained the intervening intron sequences. However, if this cDNA accurately represents an INSRR splice variant, then the resulting protein would be truncated just before the end of the Furin-like domain, because of a stop codon located in the beginning of intron 3. Thus, in summary, we identified two new potential splice variants of INSRR, one ending just after coding exon 3, and one ending just after coding exon 5 (Fig. S2).
3 Discussion
For the truncated variants of insulin receptor-like receptors lacking the intracellular tyrosine kinase domain, there are four known examples: 1) truncated insulin receptor-like genes in the fly genome (Pietrokovski and Shilo, 2001), 2) IGF2R in all organisms, which lacks a tyrosine kinase domain in its native form (Oshima et al., 1988), 3) truncated splice variants of insulin related receptor (INSRR) in rat (Itoh et al 1993), and 4) truncated splice variants of epidermal growth factor receptor in humans (EGFR) (Reiter & Maihle, 1996; Reiter et al., 2001).
When Pietrokovski and Shilo (2001) discovered the truncated insulin receptor-like genes in fly CG3837 and CG10702, genome sequences were not yet available and it was not appreciated that such sequences were actually widespread in arthropods. These protein products may have been encountered already in 1985, in reports of a 100 kDa protein considered at the time an evolutionary precursor of EGFR and InR in fly (Thompson et al 1985). It was recently discovered that the crustacean Daphnia pulex contains four duplications of the insulin receptor (Boucher et al 2010), which differ in their domain structure, but all contain the tyrosine kinase domain. Here, we report previously unknown truncated variants, which all lack the tyrosine kinase domain, in several arthropod species, including tsetse fly, malaria mosquito, yellow fever mosquito, southern house mosquito, red flour beetle, and water flea.
Long considered an orphan receptor (Civelli et al 2013), in 2011 it was shown that the L1-furin domain of INSRR works as an alkali sensor in kidney, triggering signaling at high pH (Deyev et al., 2011). We may have found two putative truncated isoforms of this receptor in human brain cDNA. The first truncated form ends just after coding exon 5, making it highly similar to sIRR-1 in rat (Itoh et al 1993). The second truncated form retains both of introns 3 and 4, but terminates at a stop codon just after coding exon 3. In fact, a truncated isoform was previously known in human kidney tumor tissue. An in-silico PCR, using our primers, identified a previously reported cDNA from human kidney (FLJ17802), corresponding to our longer product. Some highly localized brain expression of INSRR had also been observed in the past, such as in adult rat trigeminal and dorsal root ganglion neurons (Reinhardt et al 1993). While truncated splice variants of INSRR in human brain cDNA have never before been documented, a caveat enjoining from our methodology is the risk of amplifying incompletely spliced, full-length transcripts or even genomic DNA. However, we emphasize that we have used the RefSeq INSRR sequence in the maximum likelihood analysis, to determine a putative truncation point and the validity of these putative human INSRR splice variants does not alter our results.
In summary, while several well supported clusters (i.e. those with BP >50%) have been identified in our phylogenetic analysis, their relative order cannot be resolved, due to the long evolutionary distances as well as the inherent similarity of the sequences we are comparing (Fig. 3). Including new INSR sequences from mollusks or echinoderms helped to stabilize the basal nodes (Fig. S1). The key nodes from which we draw our conclusions all received reasonable bootstrap values.
Our phylogenetic analysis shows a clear bi-partitioning between the sub-phylogenies of receptor L1 and L2 domains, as well as a 79% bootstrap (BP) support for the nodes segregating INSR/INSRR and EGFR, consistent with a single domain duplication (Leach et al., 1992; Kugelberg et al., 2010) occurring prior to the formation of the INSR and EGFR lineages. However, our evolutionary model (Fig. S3) also shows that the truncated sequences we have discovered were very likely generated by multiple duplication events. In particular, the D. pulex truncated sequence is EGFR-like (94–98 BP), unlike the other truncated sequences, which are INSR-like (Fig. 1). The phylogenetic analysis of the whole ectodomain (Fig. S1) confirmed these findings, and showed a 100% BP for segregating INSR/INSRR and EGFR.
Supplementary Material
Un-collapsed maximum likelihood (ML) tree for ectodomain (L1-CR-L2). The red stars indicate nodes that are supported by less than 50% BP. Scale bars indicate substitutions per site.
Screenshot from Fancy Gene 1.4 of the human INSRR locus; the coding exons of isoforms 1 and 2 are indicated below.
Schematic drawing of the evolution of the insulin receptor family from its formation in sponges to the splitting of INSR and INSRR in amphibians.
Details of third party-annotations of truncated insulin receptor-like sequences in arthropods.
Sequence set of L1 and L2.
Acknowledgments
We thank Takehide KOSUGE (DDBJ) for help with TPA annotations.
References
- Boucher P, Ditlecadet D, Dube C, Dufresne F. Unusual duplication of the insulin-like receptor in the crustacean Daphnia pulex. BMC Evol Biol. 2010;10:305. doi: 10.1186/1471-2148-10-305. 1471-2148-10-305 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelli O, et al. G Protein-Coupled Receptor Deorphanizations. Annu Rev Pharmacol Toxicol. 2012 doi: 10.1146/annurev-pharmtox-010611-134548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JE, White S, Searle SM, Stemple DL. Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome Res. 2012;22:2067–2078. doi: 10.1101/gr.137901.112. gr.137901.112 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza DO, Sundstrom G, Bergqvist CA, Duan C, Larhammar D. Evolution of the insulin-like growth factor binding protein (IGFBP) family. Endocrinology. 2011;152:2278–2289. doi: 10.1210/en.2011-0047. en.2011–0047 [pii] [DOI] [PubMed] [Google Scholar]
- De Meyts P. Insulin and its receptor: structure, function and evolution. Bioessays. 2004;26:1351–1362. doi: 10.1002/bies.20151. [DOI] [PubMed] [Google Scholar]
- Deyev IE, et al. Insulin receptor-related receptor as an extracellular alkali sensor. Cell Metab. 2011;13:679–689. doi: 10.1016/j.cmet.2011.03.022. S1550-4131(11)00180-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Sanchez C, Mansilla A, de Pablo F, Zardoya R. Evolution of the insulin receptor family and receptor isoform expression in vertebrates. Mol Biol Evol. 2008;25:1043–1053. doi: 10.1093/molbev/msn036. msn036 [pii] [DOI] [PubMed] [Google Scholar]
- Itoh N, Jobo K, Tsujimoto K, Ohta M, Kawasaki T. Two truncated forms of rat insulin receptor-related receptor. J Biol Chem. 1993;268:17983–17986. [PubMed] [Google Scholar]
- Klein DE, Stayrook SE, Shi F, Narayan K, Lemmon MA. Structural basis for EGFR ligand sequestration by Argos. Nature. 2008;453:1271–1275. doi: 10.1038/nature06978. nature06978 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg E, et al. The tandem inversion duplication in Salmonella enterica: selection drives unstable precursors to final mutation types. Genetics. 2010;185:65–80. doi: 10.1534/genetics.110.114074. genetics.110.114074 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. msn222 [pii] [DOI] [PubMed] [Google Scholar]
- Laisney JA, Braasch I, Walter RB, Meierjohann S, Schartl M. Lineage-specific co-evolution of the Egf receptor/ligand signaling system. BMC Evol Biol. 2010;10:27. doi: 10.1186/1471-2148-10-27. 1471-2148-10-27 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Molecular Biology and Evolution. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Leach DR, Lloyd RG, Coulson AF. The SbcCD protein of Escherichia coli is related to two puta-tive nucleases in the UvrA superfamily of nucleotide-binding proteins. Genetica. 1992;87:95–100. doi: 10.1007/BF00120998. [DOI] [PubMed] [Google Scholar]
- Mathi SK, Chan J, Watt VM. Insulin receptor-related receptor messenger ribonucleic acid: quantitative distribution and localization to subpopulations of epithelial cells in stomach and kidney. Endocrinology. 1995;136:4125–4132. doi: 10.1210/endo.136.9.7649121. [DOI] [PubMed] [Google Scholar]
- Oshima A, Nolan CM, Kyle JW, Grubb JH, Sly WS. The human cation-independent mannose 6-phosphate receptor. Cloning and sequence of the full-length cDNA and expression of functional receptor in COS cells. Journal of Biological Chemistry. 1988;263:2553–2562. [PubMed] [Google Scholar]
- Pietrokovski S, Shilo BZ. Identification of new signaling components in the Drosophila genome sequence. Funct Integr Genomics. 2001;1:250–255. doi: 10.1007/s101420000023. [DOI] [PubMed] [Google Scholar]
- Reinhardt RR, Chin E, Zhang B, Roth RA, Bondy CA. Insulin receptor-related receptor messenger ribonucleic acid is focally expressed in sympathetic and sensory neurons and renal distal tubule cells. Endocrinology. 1993;133:3–10. doi: 10.1210/endo.133.1.8319578. [DOI] [PubMed] [Google Scholar]
- Reiter JL, Maihle NJ. A 1.8 kb alternative transcript from the human epidermal growth factor receptor gene encodes a truncated form of the receptor. Nucleic Acids Res. 1996;24:4050–4056. doi: 10.1093/nar/24.20.4050. 6g0280 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JL, et al. Comparative genomic sequence analysis and isolation of human and mouse alternative EGFR transcripts encoding truncated receptor isoforms. Genomics. 2001;71:1–20. doi: 10.1006/geno.2000.6341. S0888754300963419 [pii] [DOI] [PubMed] [Google Scholar]
- Renteria ME, Gandhi NS, Vinuesa P, Helmerhorst E, Mancera RL. A comparative structural bioinformatics analysis of the insulin receptor family ectodomain based on phylogenetic information. PLoS One. 2008;3:e3667. doi: 10.1371/journal.pone.0003667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorokhod A, et al. Origin of insulin receptor-like tyrosine kinases in marine sponges. Biol Bull. 1999;197:198–206. doi: 10.2307/1542615. [DOI] [PubMed] [Google Scholar]
- Thompson KL, Decker SJ, Rosner MR. Identification of a novel receptor in Drosophila for both epidermal growth factor and insulin. Proc Natl Acad Sci U S A. 1985;82:8443–8447. doi: 10.1073/pnas.82.24.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Un-collapsed maximum likelihood (ML) tree for ectodomain (L1-CR-L2). The red stars indicate nodes that are supported by less than 50% BP. Scale bars indicate substitutions per site.
Screenshot from Fancy Gene 1.4 of the human INSRR locus; the coding exons of isoforms 1 and 2 are indicated below.
Schematic drawing of the evolution of the insulin receptor family from its formation in sponges to the splitting of INSR and INSRR in amphibians.
Details of third party-annotations of truncated insulin receptor-like sequences in arthropods.
Sequence set of L1 and L2.