Abstract
Maternal genetic effects (MGEs), where genes expressed by mothers affect the phenotype of their offspring, are important sources of phenotypic diversity in a myriad of organisms. We use a single‐locus model to examine how MGEs contribute patterns of heritable and nonheritable variation and influence evolutionary dynamics in randomly mating and inbreeding populations. We elucidate the influence of MGEs by examining the offspring genotype‐phenotype relationship, which determines how MGEs affect evolutionary dynamics in response to selection on offspring phenotypes. This approach reveals important results that are not apparent from classic quantitative genetic treatments of MGEs. We show that additive and dominance MGEs make different contributions to evolutionary dynamics and patterns of variation, which are differentially affected by inbreeding. Dominance MGEs make the offspring genotype‐phenotype relationship frequency dependent, resulting in the appearance of negative frequency‐dependent selection, while additive MGEs contribute a component of parent‐of‐origin dependent variation. Inbreeding amplifies the contribution of MGEs to the additive genetic variance and, therefore enhances their evolutionary response. Considering evolutionary dynamics of allele frequency change on an adaptive landscape, we show that this landscape differs from the mean fitness surface, and therefore, under some condition, fitness peaks can exist but not be “available” to the evolving population.
Keywords: Kin selection, Social effects, adaptive landscape, dominance, frequency dependent selection
Maternal genetic effects occur when genes expressed in the mother's genome affect the phenotype of her offspring (Dickerson 1947; Willham 1963; Nordskog and Hassan 1971; Shaw and Byers 1998; Wade 1998; Cheverud and Wolf 2009; Wolf and Wade 2009). Maternal genetic effects are arguably the most common example of indirect genetic effects, wherein the genes in one individual affect the phenotype of another (Willham 1963; Wolf et al. 1998). Many of the known maternal effect genes across a diversity of organisms (including both plants and animals) act via maternal mRNA or proteins (Berleth et al. 1988; Tong et al. 2000) that are critical coordinators of development, especially prior to the maternal‐to‐zygotic transition, and often represent products from the majority of the genes in the genome (see Baroux et al. 2008; Lipshitz 2015). However, maternal effects (genetic and nongenetic) also arise from a diversity of scenarios in which mothers provide a component of the environment experienced by offspring, such as through nutritional provisioning (Dickerson 1947; Roach and Wulff 1987; Fox et al. 1999; Bowen 2009), construction of nests (Bult and Lynch 1997; Lloyd and Martin 2004), and maternal influences on seed dispersal (Donohue 1998) or oviposition site choice (Bernardo 1996). Consequently, maternal effects are an important source of variation in offspring fitness in a wide range of organisms, including insects (Mousseau and Dingle 1991), fish (Reznick et al. 1996), reptiles (Cadby et al. 2011), birds (Nordskog and Hassan 1971; Price 1998), mammals (Dickerson 1947; Chai 1956; Brumby 1960; Falconer 1960; Willham 1972; Wilson and Reale 2006; Maestripieri and Mateo 2009), and various plants (see Roach and Wulff 1987).
Like other indirect genetic effects arising from interactions with relatives (where there is a predictable relationship between the genotype of an individual and the social environment provided by relatives) (Wolf 2003; McGlothlin et al. 2010; McGlothlin et al. 2014), maternal genetic effects influence the resemblance of relatives (Willham 1963) and, as with other indirect genetic effects (Wolf et al. 1998), alter the relationship between individual genotype and phenotype compared to that expected for direct effects (Cheverud and Moore 1994; Cheverud and Wolf 2009). As a result, they can play a key role in a number of evolutionary processes, such as the response to selection (see Dickerson 1947; Falconer 1965; Willham 1972; Kirkpatrick and Lande 1989), sexual selection (Wolf et al. 1997, 1999), adaptation (e.g., Mousseau and Fox 1998; Badyaev et al. 2002), and population divergence (Perry et al. 2005). Most of the evolutionary consequences of maternal effects arise from the fact that they evolve through a kin selection process, where it is selection on offspring traits that ultimately drives the evolution of maternal effects (Falconer 1965; Nordskog and Hassan 1971; Willham 1972; Cheverud 1984, 1985; Lynch 1987; Kirkpatrick and Lande 1989; Cheverud and Moore 1994). This phenomenon is a consequence of the fact that maternal effect genes are expressed in mothers, but affect the offspring phenotype. At the same time, the offspring inherit alleles underlying the maternal effect from their mother, causing an association between offspring phenotype and offspring genotype at the maternal effect loci. As a result, selection on offspring traits leads to indirect selection on the maternal effect genes. The resulting correlated evolution of the maternal effect in response to selection on offspring can be conceptualized as the evolution of the maternally provided environment from the perspective of the offspring (Drown and Wade 2014; Wolf et al. 2014).
Our theoretical and empirical understanding of the consequences of maternal effects for trait evolution largely comes from a quantitative genetics perspective founded on variance component models (e.g., Dickerson 1947; Willham 1963; Falconer 1965; Willham 1972; Cheverud 1984; Kirkpatrick and Lande 1989; Lande and Kirkpatrick 1990; Hadfield 2012). Population genetic treatments have also been used to study a variety of problems such as the dynamics of maternal‐effect selfish genes (Wade and Beeman 1994), histocompatibility genes (Hedrick 1997; Wade 2000), mutation‐selection balance (Wade 1998; Wade et al. 2009), the evolution of genomic imprinting (Wolf and Hager 2006), maternal‐offspring interactions (Wade 1998; Wolf 2000; Wade et al. 2009), and maternal selection (Nordskog and Hassan 1971; Gavrilets 1998; Spencer 2003; Spencer and Chiew 2015). Here, we connect the quantitative and population genetic approaches to maternal effects, using a single‐locus, two‐allele model with additive and dominance effects at both the offspring (zygotic) and maternal levels. The use of a simple single‐locus model is not intended to imply that maternal effects generally have such a simple genetic architecture, but rather, it allows us to understand the dynamics of individual loci contributing to quantitative variation and evolution (where it is the variation at and dynamics of individual loci that ultimately create patterns of variation and evolution). The structure of our model arises as a direct extension of the classic single‐locus model that forms the foundation for our understanding of quantitative genetic variation (see Falconer and Mackay 1996), and therefore allows us to demonstrate similarities and differences between the contributions of direct and maternal genetic effects to evolutionary change. Many of the phenomena that we identify are not apparent from the variance component‐based quantitative genetic models and have not been previously considered in population genetic models. Because the relatedness between mothers and their offspring is a critical determinant of the evolutionary dynamics of maternal effects, we consider the influence of inbreeding throughout our analysis. When considering the evolutionary consequences of maternal effects, we conceptualize the process of evolution using the metaphor of movement of a population across the adaptive landscape (i.e., where populations evolutionarily “climb” the surface of population mean fitness in the sense that allele frequencies evolve in the direction that leads to increased mean fitness) (Wright 1932; see also Svensson and Calsbeek 2012 for a general overview of relevant concepts). Using this metaphor, we show that, when a locus has solely a direct or a maternal effect, a population will evolve to the peak on the adaptive landscape, but when a locus shows both a direct and a maternal effect, it may not evolve to the peak that maximizes mean fitness. We derive an expression that predicts the evolutionary outcome under these conditions to understand why populations do not necessarily evolve to the peak that maximizes population mean fitness when a locus has both direct and maternal effects.
The Model
We consider a single‐locus (the A locus) that has two alleles (A 1 and A 2). We assume the locus can have direct and/or maternal effects on offspring phenotypes (including fitness). Thus, our model allows for the presence of pleiotropy between direct and maternal effects, while providing a model that considers only either direct or maternal effects by simply setting one set of parameters to zero). There is a large body of evidence suggesting that genes with both direct and maternal genetic effects are likely to be common in eukaryotes. In early embryos it is common to find that a large fraction of the protein‐coding genome is represented as maternal mRNAs (ca. 40% in the mouse, 50% in the flour beetle, Tribolium castaneum [Preuss et al. 2012], and 65% in the fruit fly, Drosophila melanogaster [Tadros and Lipshitz 2009]), providing opportunities for a large fraction of the genome to have both direct and maternal effects. Likewise, empirical estimates of genetic correlations between direct and maternal effects, which presumably primarily reflect loci with pleiotropic direct and maternal effects, are typically large (Robinson 1996; Roff 1997; Wolf and Brodie III 1998; Wilson and Reale 2006), and mapping studies of maternal effects have directly demonstrated that such pleiotropy can be common (Casellas et al. 2009; Wolf et al. 2011; Wolf and Cheverud 2012).
Although we use a single‐locus system to explore the properties of direct and maternal effects, in the Supplementary Material we demonstrate that, as with the classic single locus model of direct effects, the results extend to a multilocus system when selection is weak enough that the population remains in “quasi‐linkage‐equilibrium” (QLE) (Kimura 1965; Nagylaki 1976, 1993).
Throughout, we view the genotype‐phenotype relationship and fitness from the perspective of the “offspring” because it determines how selection at the level of the individual (i.e., offspring) drives evolutionary dynamics (Kirkpatrick and Lande 1989; Cheverud and Wolf 2009). The effects of the locus are summarized using two classic parameters, the additive (a) and dominance (d) genotypic values or effects (see Falconer and Mackay 1996). Following Wolf and Cheverud (2012), we define the direct and maternal effects of the locus in an idealized scenario where each is measured in the absence of the other effect. We take this approach (of defining each in the absence of the other) because, in the traditional quantitative genetic approach, direct, and maternal effects are partially confounded due to relatedness (i.e., the fact that some genotypes of mothers can only produce a subset of possible offspring genotypes; e.g., A 1 A 1 mothers cannot have A 2 A 2 offspring). Our definitions of direct and maternal effects are therefore equivalent to the values that one might recover through cross‐fostering, where offspring genotypes are randomized across maternal genotypes (Wolf and Cheverud 2012). We use the subscripts “o” and “m” to indicate direct (“offspring”) and maternal effects, respectively. The additive direct effect genotypic value, ao, is defined as half the difference between the phenotypic values of the A 1 A 1 and A 2 A 2 genotypic classes (Falconer and Mackay 1996) and the dominance direct effect genotypic value, do, as the deviation of the heterozygote phenotypic value from the unweighted average of the phenotypic values of the homozygotes in the absence of maternal effects. Analogously, the additive maternal effect genotypic value, am ,, is half the difference between the mean phenotypes of the offspring of A 1 A 1 and A 2 A 2 mothers, while the dominance maternal effect genotypic value, dm, is defined as the deviation of the average offspring phenotype of heterozygous mothers from the average of the offspring means of homozygous mothers, in the absence of direct effects (cf. Wolf and Cheverud 2012). The offspring phenotypes as a function of these genetic effects are summarized in Table 1.
Table 1.
The phenotype (or fitness) of offspring as a function of their genotype and the genotype of their mother
| Offspring genotype | |||||
|---|---|---|---|---|---|
| A 1 A 1 | A 1 A 2 | A 2 A 1 | A 2 A 2 | Maternal litter average | |
| Maternal genotype | |||||
| A 1 A 1 | 1 + am + ao | 1 + am + do | — | — | μ + aop 1 + dop 2 + am |
| A 1 A 2 | 1 + dm + ao | 1 + dm+ do | 1 + dm+ do | 1 + dm – ao | μ + ½do + ½ao(p 1 – p 2) + dm |
| A 2 A 2 | — | — | 1 –am+ do | 1 –am – ao | μ – aop 2 + dop 1 – am |
| Offspring average | μ + amp 1 + dmp 2 + ao | μ + amp 1 + dmp 2 + do | μ –amp 2 + dmp 1 + do | μ –amp 2 + dmp 1 – ao | |
Offspring genotypes have alleles ordered by their parent‐of‐origin, with the maternally inherited allele appearing first, whereas maternal genotypes are unordered (since the values are identical for the maternal genotype reciprocal heterozygotes). Marginal means are given for each maternal and offspring genotype for the case without inbreeding (see Table S1 for the values under inbreeding). Cells containing a “—” indicate combinations that do not exist (after Table 4 in Wolf and Cheverud 2012).
For simplicity, we let the parameters p 1 and p 2 describe allele frequencies of the A 1 and A 2 alleles, respectively, in both parents and offspring, and therefore they can be used to calculate the frequencies of the various maternal‐offspring genotype combinations (Table 2). Hence, in the presence of selection, they describe the frequency of alleles after selection in the parental generation and before selection in the offspring generation. Initially, we assume random mating but relax this assumption later in our modeling when we consider the influence of inbreeding (the frequency values in Table 2 apply for both the case with and without inbreeding). We assume selection is weak enough that genotypes remain in Hardy–Weinberg proportions (i.e., “quasi Hardy–Weinberg proportions,” QHW, see Nagylaki 1976, Table 1, and Supplementary Material), except where we model deviations from Hardy–Weinberg proportions associated with inbreeding. This allows us to describe the distributions of parental and offspring genotypes as a simple function of allele frequencies (p 1 and p 2) with minimal loss of generality. By doing so we can formalize the problem and provide simple analytical results that capture the basic properties of genes with direct and maternal effects. Those basic properties do not change substantively as we add loci or deviate from H–W equilibrium (HWE)—those processes simply add a small degree of error around predictions. In the Supplementary Material we analyse the nature of the error associated with the assumptions of “quasi‐Hardy–Weinberg equilibrium” (QHW) and “quasi‐linkage‐equilibrium” (QLE).
Table 2.
The frequencies of the maternal‐offspring genotype combinations before selection
| Offspring genotype | ||||||||
|---|---|---|---|---|---|---|---|---|
| A 1 A 1 | A 1 A 2 | A 2 A 1 | A 2 A 2 | |||||
| Maternal genotype | ||||||||
| A 1 A 1 | f 11 u 11 | f 11 u 12 | — | — | ||||
| A 1 A 2 | ½ f 12 u 11 | ½f 12 u 12 | ½f 12 u 21 | ½f 12 u 22 | ||||
| A 2 A 1 | ½f 12 u 11 | ½f 12 u 12 | ½f 12 u 21 | ½f 12 u 22 | ||||
| A 2 A 2 | — | — | f 22 u 21 | f 22 u 22 | ||||
| Genotype Frequencies |
|
|
|
|
||||
Genotypes have alleles ordered by their parent‐of‐origin, with the maternally inherited allele appearing first. The combinations are a function of the frequency of the four ordered genotypes (fij, where i is the maternally inherited allele) and the frequencies of the offspring genotypes within the maternal genotypes (uij, where again i is the maternally inherited allele). The frequencies of the ordered genotypes (fij) appear at the bottom of the columns for the offspring genotypes (the genotype frequencies are the same as those of the four possible maternal genotypes and so are only listed under the offspring genotypes for simplicity) and the frequencies of the offspring genotypes within the maternal genotypes (uij) are: , ,,, and cells containing a “—” indicate combinations that do not exist.
COMPONENTS OF VARIATION
The components of genetic variance resulting from direct effects and maternal effects, in the absence of the other, are given in Table 3. These expressions are equivalent to the classic expressions of the quantitative genetics literature (Falconer and Mackay 1996) and are presented here for comparison. Using the phenotypes in Table 1 and the frequencies in Table 2 we can derive the additive genetic covariance (owing to pleiotropy) between the direct effect of an individual's own genes and the maternal effect it experiences arising from its mother's genes (i.e., the covariance between direct and indirect genetic effects on the offspring's phenotype) (Willham 1963; Riska et al. 1985; Kirkpatrick and Lande 1989; Wolf and Brodie III 1998),
| (1) |
Table 3.
Patterns of direct and maternal effect variation in randomly mating and inbreeding populations (with the degree of inbreeding given by F)
| Random mating | ||||
|---|---|---|---|---|
| Variance component | Direct genetic effects (ao, do only) | Maternal genetic effects (am, dm only) | ||
| V Additive |
|
|
||
| V Dominance |
|
|
||
| V Total | V direct = VA ( o ) + VD ( o ) | V maternal = VA ( m ) + VD ( m ) | ||
| Inbreeding to degree F | ||||
|---|---|---|---|---|
| V Additive |
|
|
||
| V Dominance |
|
|
||
| V Total |
|
|
||
In each case, the variation associated with direct or maternal effects is defined in the absence of the other type of effect, so that the confounding of the effects does not contribute to the pattern of variation (see the main text for the case where both occur simultaneously). Variation under both random mating and inbreeding is decomposed in additive (V Additive) and dominance (V Dominance) genetic variation, where the total is simply the sum of these components.
Equation (1) also represents half of the additive genetic covariance between direct and maternal effects of the locus at the individual level (i.e., the additive genetic relationship between the influence that individuals’ genes have on their own phenotype and the influence of their genes on the phenotype of their offspring is 2covmo) (Riska et al. 1985; Cheverud and Moore 1994). It is important to keep in mind that the covmo is the covariance between effects (direct and maternal) on the phenotypes of an individual, not a measure of the maternal‐offspring covariance (i.e., it is not a measure of the resemblance of mothers and offspring). Variance component models of maternal effects have shown that this covariance plays an important role in determining how maternal effects contribute to the offspring genotype‐phenotype relationship and, thereby, to patterns of offspring phenotypic variation (cf. the parameter Gmo in Kirkpatrick and Lande [1989] and cov(Ao, Am) in Cheverud [1984]).
Maternal effects contribute to the offspring genotype–phenotype relationship because offspring experience a maternal effect arising from their mother's genome while also inheriting half their genes from their mother. With both direct and maternal effects, the variance among unordered (VG) offspring genotypes (i.e., combining the A 1 A 2 and A 2 A 1 heterozygotes into a single heterozygote class) equals:
| (2) |
From equation (2) we see that the variation among offspring genotypes, VG, includes the variance arising from additive (VA ( o )) and dominance direct effects (VD ( o )), as well as one fourth of the additive genetic maternal‐effect variance (VA ( m )) , and the additive genetic covariance between direct and maternal effects, which is twice the direct‐maternal genetic covariance (covmo).
The contribution of a single locus to the total genetic variance (eq. 2) can be partitioned into additive and dominance components of genetic variation (Fisher 1918; Falconer and Mackay 1996). With maternal effects, the additive genetic variance among offspring genotypes contains a component contributed by maternal effects and the direct‐maternal genetic covariance (covmo) (Dickerson 1947). In equation (2), the total additive genetic variance appears as the first term in brackets, where , while the dominance genetic variance is the remainder or simply the direct dominance variance (VD = VD ( o )). This result demonstrates that, in addition to the contribution of direct effects (VA ( o )) to the additive genetic variance, both additive maternal effects and the additive maternal‐offspring covariance (twice the direct‐maternal genetic covariance, eq. 1) contribute to the additive genetic variance among offspring (cf. Kirkpatrick and Lande 1989). As a result a trait can have heritable variation (i.e., VA > 0) even when the locus does not directly affect the expression of the trait (i.e., VA ( o ) = 0) because maternal effects can contribute a component of heritable variation. Likewise, the trait can be affected by both direct and maternal genetic effects (i.e., VA ( o ) > 0 and VA ( m ) > 0), but have no heritable (additive genetic) variation overall because these two effects oppose each other (i.e., show a negative genetic covariance) and therefore cancel each other out (which occurs when ) (see also Dickerson 1947 for a discussion of trait heritability). Interestingly, equation (2) also demonstrates that maternal effects contribute only to additive variation but not to dominance variation among offspring genotypes.
Because the two types of homozygous mothers (A 1 A 1 and A 2 A 2) can each produce only one of the two types of reciprocal heterozygotes (A 1 A 2 and A 2 A 1, respectively, with the maternally inherited allele appearing first; see Table 1), the phenotypes of the reciprocal heterozygotes will differ when there are maternal effects (Hager et al. 2008). As a result, maternal genetic effects contribute a component of variation between the reciprocal heterozygotes that is equal to . Because this variation is between the heterozygote classes that are genetically identical, but differ in the parent of origin of their alleles, it appears as a type of parent‐of‐origin dependent variation, analogous to the imprinting variance (Spencer 2002).
Although maternal genetic effects contribute to the variance among offspring genotypes, they also contribute variation that is not associated with the offspring genotype (cf. Byers et al. 1997). For example, consider an idealized scenario where a maternal effect is caused by a single locus in a population of families of full‐siblings and accounts for all variation in some trait—all offspring in the same family experience the same maternal effect because they share the same mother, and so they all have the same phenotype. However, offspring with the same phenotype owing to maternal effects may have different genotypes at the maternal effect locus because there is Mendelian segregation variance within families. As a result, the offspring genotype at the maternal effect locus cannot fully account for variation in offspring phenotype, despite the fact that the phenotype was determined by that locus albeit in the mother (i.e., through a maternal effect). Therefore, while a maternal effect can be wholly genetically determined and fully account for offspring trait variation, when we measure variation among offspring, their genotype at the maternal effect locus only accounts for a component of that trait variation. The rest of the phenotypic variation contributed by maternal genetic effects is, therefore, uncorrelated to offspring genotype, and consequently, from the perspective of the offspring a component of phenotypic variation arising from maternal genetic effects appears to be a random environmental source of variation. This is not meant to imply that maternal genetic effects are “environmental” variation in the sense of being caused by the ecological environment, but rather, it follows from the standard partitioning perspective in quantitative genetics in which variation that is not explained by an individual's genotype (i.e., variation that is random with respect to genotype) is lumped together in the “environmental” variance. As a result, even when all phenotypic variation arises from genetic influences (i.e., from direct or maternal genetic effects), there can still be a component of random “environmental” variation among offspring genotypes (i.e., phenotypic variation among offspring that is not associated with their genotype) corresponding to:
| (3) |
Equation (3) illustrates that half of the additive genetic maternal‐effect variance and, all of the dominance maternal‐effect variation appear as environmental components of variation from the perspective of the offspring genotype–phenotype relationship (i.e., these maternal genetic effects contribute variation that appears within, not among, classes of offspring genotypes, and hence is conceptually the same as other sources of environmental variation). This result is implied in most models of maternal effects (e.g., Willham 1963; Kirkpatrick and Lande 1989), where the contribution of maternal effects to patterns of variation and response to selection in offspring are typically weighted by a factor of ½, but these earlier models considered only the contribution of additive maternal effects and not maternal dominance effects that predominate in VE.
Because maternal effects are attributed to the genotype of the mother, they contribute a component of variation among maternal families (i.e., among the groups of offspring produced by the different types of mothers) (Dickerson 1947). Therefore, the entire maternal effect variance (V maternal), shown in Table 3, appears as an among‐maternal‐family component of variation, while the contribution of direct effects to among family variation depends on the mating structure (e.g., whether offspring are full‐siblings or half‐siblings etc.).
EVOLUTIONARY DYNAMICS
To understand the evolutionary consequences of maternal genetic effects for selection on offspring, we assume that the genetic parameters, ao, do, am, and dm, correspond to effects on offspring fitness. As above, we assume that selection stemming from these fitness differences is sufficiently weak that it does not produce significant deviations from the Hardy–Weinberg expectations. (See Supplementary Material, where we show that the deviations from H–W proportions are generally ∼ O(s 2), where s represents the strength of selection)
The mean fitness of the population equals:
| (4) |
where μ is the baseline (expected) fitness value that is not associated with variation at the A locus. This equation for population mean fitness is often described as the “adaptive landscape” (Wright 1932; see also; Svensson and Calsbeek 2012); it represents the relationship between allele frequencies and expected fitness. In standard theory, one of the assumptions underlying the utility of the adaptive landscape is that “….the fitness of individuals is affected by their own trait values, but not by the trait values of other individuals in the population” (Arnold 2003, p. 368). That assumption is explicitly violated here where genes in the maternal genome affect the offspring trait value and fitness. Regardless of whether this assumption is met, peaks on the adaptive landscape occur where the partial derivative of mean fitness () with respect to the frequency of either of the alleles is zero (e.g., when ). Taking the partial derivative of mean fitness in equation (3) with respect to p 1 and setting it equal to zero yields:
| (5) |
Thus, we see that direct and maternal effects have symmetrical effects on population mean fitness (i.e., their contributions are weighted equally in eq. 4) (Nordskog and Hassan 1971), and hence symmetrically affect the shape of the mean fitness surface. However, although they have symmetrical effects on mean fitness, they do not contribute symmetrically to evolutionary changes in allele frequencies (illustrated by , where ):
| (6) |
The difference in the weighting of direct and maternal effects in equations (5) and (6) reflects the differential influence on the offspring genotype‐phenotype relationship (see eq. 2). Consequently, populations may not evolve to the peak on the mean fitness surface when there are both direct and maternal effects because of the asymmetrical weighting of maternal and direct genetic effects in the equation for allele frequency change (eq. 6). Thus, while the direct and maternal parameters, am and dm, are weighted equally in the mean fitness surface (eq. 4), maternal effects are weighted by a factor of ½ in the equation for evolutionary change (eq. 6) because they are an indirect genetic effect (so the weighting of ½ here reflects the coefficient of relatedness between mothers and their offspring). As a result, the realized peak on the adaptive landscape (corresponding to the evolutionary equilibrium) occurs where the change in allele frequencies (eq. 6) is zero (not counting the trivial equilibria where p 1 = 1 or 0):
| (7) |
(where ).
In order to reconcile the fact that the dynamics of evolutionary change (eq. 6) and location of equilibria are not reflected in the shape of the mean fitness surface (eq. 4) we define the realized adaptive landscape (i.e., the equation that governs how a population will evolve in terms of allele frequencies), , as a fitness surface with direct and maternal effects weighted by their contribution to the genotype‐phenotype relationship:
| (8) |
The partial derivative of equation (8) with respect to p 1 is zero at the same equilibrium given by eq. (7). Equation (8) demonstrates that, because of their differential contributions to the genotype–phenotype relationship, direct effects (ao and do) are twice as important in a randomly mating population as maternal effects (am and dm) in determining the evolutionary dynamics and ultimate evolutionary equilibrium for a locus.
THE FREQUENCY‐DEPENDENT OFFSPRING G–P RELATIONSHIP
In the classic quantitative genetic model of direct effects, the phenotype or fitness is a “fixed” property of the genotype independent of allele frequencies. With maternal effects, the expected phenotype of an offspring of a given genotype depends on the probability that it was produced by each of the different possible maternal genotypes (Table 2; see Wade 1978). This frequency dependence is apparent from the expected (average) offspring phenotypes given in Table 1, where the maternal effect contributions are all weighted by allele frequencies. To understand how this frequency‐dependent genotype‐phenotype relationship affects evolutionary dynamics, we can examine the realized additive genotypic value, aR ( o ) of the A locus, which ultimately determines how selection on offspring phenotypes drives evolution of maternal effects. Using the classic definition of the additive genotypic value as half the difference between the phenotypes of the offspring homozygotes (calculated as the mean of the A 1 A 1 minus the mean of the A 2 A 2 offspring), the realized additive genotypic value is:
| (9) |
(i.e., this is the value we could calculate if we simply examined the difference between the average phenotypes of the two offspring homozygotes, ignoring whether the difference arose as a consequence of a direct or a maternal effect). Equation (9) demonstrates that half of the additive maternal effect (am) appears like an offspring direct effect because it makes the two homozygous offspring genotypes phenotypically distinct irrespective of allele frequencies (Wolf and Cheverud 2012). We also see that the dominance maternal effect (dm) weighted by (p 2 – p 1) contributes to the realized additive effect in offspring genotypes. Most importantly, we see that, when the A 1 allele is more common that the A 2 allele, then the weighting term, (p 2 – p 1), is negative, whereas when the A 1 allele is less common that the A 2 allele, the weighting is positive, and consequently the contribution of the dominance maternal effect to the additive genotypic value changes sign depending on which allele is more common. This phenomenon means that a maternal effect showing positive dominance (dm > 0), which might arise from overdominance for the maternal effect, would lead to negative frequency dependence wherein the rarer allele will appear to have a positive effect. In contrast, a maternal effect showing negative dominance (dm < 0), which might arise from underdominance for the maternal effect, would lead to positive frequency dependence.
INBREEDING ALTERS RELATIVE CONTRIBUTION OF MATERNAL EFFECTS
Under random mating, the correlation between maternal and offspring genotypes, rmo, is one‐half, that is it equals the coefficient of relatedness (i.e., the correlation between alleles present in mothers and their offspring, not between their phenotypes). This correlation governs the contribution of maternal effects to the evolution of allele frequencies (see eqs. 6, 7, 8, 9) because they evolve through kin selection (Cheverud 1984). Therefore, inbreeding modifies the evolution of maternal effects because it alters the correlation between maternal and offspring genotypes. With inbreeding of degree F (which corresponds to the reduction in the frequency of heterozygotes from that expected under random mating), the correlation between maternal and offspring genotypes is increased from ½ to (1+F)/2 (cf. Harris 1964).
Assuming that a population reaches an equilibrium level of inbreeding defined by F (i.e., where F is the same in parents and their offspring) (following Wright 1969; Wade 2000), the expected phenotypes of each of the different maternal‐offspring genotype combinations remain the same as in the random mating case (i.e., follows Table 1), but the frequencies of the maternal‐offspring genotype combinations differ from the random mating case (Table 2), and consequently, the expected phenotypes of each offspring genotype differ (see Table S1 in the Supplementary Material).
Inbreeding has differential influences on the contribution of additive versus dominance effects to components of variation because it resorts variation from heterozygote into homozygote classes, increasing the contribution of additive effects and diminishing those of dominance (Crow and Kimura 1970). We see this in our model where the contribution of additive effects to the genetic variance (either direct or maternal effect) is increased by a factor of (1 + F) while the contribution of dominance effects is decreased by a factor of (1 – F)2/(1 + F) (cf. Harris 1964; Weir and Cockerham 1977). In addition to shifting the relative heterozygote‐homozygote frequencies, inbreeding also changes the relationship between direct and maternal effects, reflected in the direct‐maternal genetic covariance (where, as with eq. 1, this covariance reflects the relationship between the direct effect of the individual's own genotype and the maternal effect they experience from their mother's genotype):
| (10) |
If there are only additive effects (i.e., do = dm = 0), then the direct‐maternal covariance is increased by a factor of (1 + F)2, whereas, if there are only dominance effects, it is diminished by a factor of (1 – F)2.
Although inbreeding has equivalent influences on direct and maternal effect variation (Table 3), they make differential contributions to the additive genetic variance among offspring genotypes, which ultimately determines their contribution to heritable variation in and response to selection on offspring phenotypes:
| (11) |
It is clear from equation (11) that, when F = 1 (a fully inbred population), direct and maternal effects both make the same relative contribution to the offspring additive genetic variance. Therefore, although additive direct effects contribute twice as much as additive maternal effects in outbreeding populations, with increasing inbreeding, the difference diminishes to the point where the two make equivalent relative contributions.
From elementary considerations, inbreeding must increase the among‐family genetic variance and diminish the within‐family genetic variance (Dickerson 1947). With inbreeding, the variance among families of full‐siblings depends on the exact mating scheme, so we consider families of half siblings sharing the same mother. With inbreeding the among‐maternal‐half‐sib family variance is
| (12) |
which demonstrates that inbreeding converts additive offspring variation to an among‐family component of variation. As a result, in a fully inbred population (F = 1), both direct and maternal effects are exclusively among‐family components of variation. The within‐family variance is simply: .
Inbreeding also alters the evolutionary dynamics, affecting mean fitness through the decline in the frequency of heterozygotes (and hence only affects the contribution of dominance to mean fitness):
| (13) |
Inbreeding also influences the allele frequency dynamics in response to selection (see Wright 1969):
| (14) |
Inbreeding changes the evolutionary dynamic by increasing the contribution of additive direct effects by a factor (1 + F) (cf. Harris 1964; Weir and Cockerham 1977) and increasing that of the additive maternal effects by a factor of (1 + F)2. The contribution of direct dominance effects is decreased by a factor of (1 – F), while dominance maternal effects are reduced by (1 – F2), which is smaller.
With inbreeding, the allele frequency equilibria differ from those expected under random mating (eq. 7) for two reasons. First, inbreeding increases the correlation between the maternal and offspring genotypes (rmo). Second, inbreeding alters the relative contribution of additive effects to gene frequency change, but not their relative contribution to mean fitness. Consequently, the adaptive landscape that determines the evolutionary equilibrium is one on which additive effects are weighted by a factor of (1 + F), giving the realized adaptive landscape:
| (15) |
The derivative of the realized adaptive landscape in equation (15) is zero under the same conditions that the change in allele frequency (eq. 14) is zero (not counting the trivial equilibria where p 1 = 1 or 0). Hence, the realized adaptive landscape predicts the evolutionary equilibrium while the mean fitness surface does not. This equilibrium occurs when:
| (16) |
which differs from that expected under random mating (cf. eq. 7).
Discussion
The quantitative genetics framework built on variance component models has demonstrated that maternal effects contribute to evolutionary change through kin selection, because the response to selection on maternal effects is proportional to the relatedness between mother and offspring (Falconer 1965; Willham 1972; Cheverud 1984, 1985; Lynch 1987; Kirkpatrick and Lande 1989). Here, we use a single‐locus model to link these results from quantitative genetics to population genetic expressions for allele frequency change. Our approach allows us to understand how maternal genetic effects influence the offspring genotype‐phenotype relationship and thereby influence evolutionary dynamics of maternal effects resulting from selection on offspring phenotypes (i.e., resulting from kin selection). By doing so we identify important results that do not appear in the classic quantitative genetic treatments of maternal effects.
Maternal genetic effects are a component of phenotypic variation that covaries with the genotype of the offspring due to the mother‐offspring relatedness, a covariance that is enhanced by inbreeding. This covariance allows maternal genetic effects to evolve when selection acts on offspring phenotypes and is the underlying reason why models of maternal effects evolution are fundamentally kin selection models (Cheverud 1984). Variance component models of maternal effects (see Kirkpatrick and Lande 1989) have found that half of the additive genetic variation in maternal effects must appear as phenotypic variation among the different offspring genotypes. We too find that half of the additive genetic variance in maternal effects appears as variation among offspring. However, we show further that half of this variation (i.e., ¼ of the total additive genetic variation in maternal effects) arises because of the differences between the reciprocal heterozygotes, which have mothers of different genotype and therefore experience different maternal effects. For this reason, half of the total variance contributed by maternal genetic effects to differences among offspring genotypes appears as an “epigenetic” difference between reciprocal heterozygotes, a finding typically associated with genomic imprinting (see also Santure and Spencer 2006; Hager et al. 2008). Consequently, maternal genetic effects and genomic imprinting effects cause similar patterns of phenotypic variation in populations that could well be confounded for one another unless their independent contributions are recognized (Hager et al. 2008; Wolf and Cheverud 2012).
In general, the contribution of additive and dominance maternal effects to the evolutionary response to selection on offspring phenotypes is similar to that of direct effects, but is weighted by a factor of ½ in an outbred population (eq. 5). However, they contribute to gene frequency change by a fundamentally different process with respect to the offspring genotype‐phenotype relationship. Consider the stable equilibrium (eq. 7) that occurs when alleles at the A‐locus are overdominant in their direct effects but have no additive effects. This is the classic case of a fitness peak at a frequency of 0.5 (Fig. 1A). In this case, populations evolve to the peak (at an allele frequency of 0.5) and remain there because offspring heterozygotes have the highest mean fitness (Fig. 1B). In contrast, consider a maternal effect showing the same pattern of overdominance but with no additive maternal effect. Here, the relative shape of the mean fitness surface is the same as that for the direct effects only case, but the realized adaptive landscape is flatter (Fig. 1A) because of the reduced strength of the genotype‐phenotype relationship for maternal effects compared to that of direct effects (i.e., maternal effects experience indirect selection, where selection on offspring results in selection on maternal effects in proportion to the relatedness of mothers and their offspring, and hence their contribution to evolutionary change is lower than that of direct effects) (Wolf 2000). Although the evolutionary outcome is the same, evolution proceeds more slowly (Fig. 1B), which leads to greater sequence variation at the equilibrium between mutation and balancing selection, a theoretical prediction supported by patterns of sequence variation in genes with maternal effects (Wade 1998; Barker et al. 2005; Cruickshank and Wade 2008). However, the evolutionary process is fundamentally different because maternal effects introduce frequency dependence into the offspring genotype‐phenotype relationship (Cheverud and Wolf 2009). With maternal effects, offspring genotypes no longer have a fixed phenotypic (fitness) value as they do in the direct effects model. Rather, the expected phenotype (fitness) of an offspring genotype depends on allele frequencies because, as allele frequencies change, so too does the probability that a particular offspring genotype was produced by a given maternal genotype. With overdominant maternal effects (Fig. 2) we see that at no allele frequency does the heterozygote class have the highest fitness; one of the two homozygote classes always has the highest fitness (except at the equilibrium, where all genotypes have equal fitness). This pattern of genotypic fitness is manifested as a negative frequency‐dependent additive effect that is (cf. eq. 9). Therefore, although the structure of the dynamical equation for change in allele frequency (eq. 6) when there is an overdominant maternal effect is the same as for a direct effect, the evolutionary process differs from the perspective of selection on the offspring.
Figure 1.
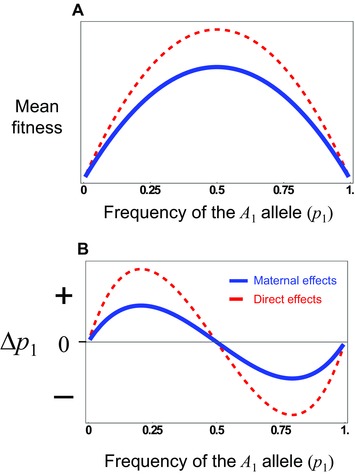
Evolution of overdominance with direct and maternal effects. These figures illustrate a case where there is a direct and a maternal effect of the same size showing only dominance (values are arbitrary, so the y axis is shown as unit free). (A) The red (dashed) line shows the “surface” of mean fitness, while the blue line (solid) shows the “realized adaptive landscape” that determines how the population evolves. (B) The change in allele frequency for a locus showing a direct or maternal dominance effect across allele frequency space. With either type of effect the population evolves to an internal equilibrium at p 1 = 0.5 when there is allelic variation, but the direct effect (red, dashed) evolves to the equilibrium faster than the maternal effect (blue, solid), when started at the same gene frequency.
Figure 2.

The offspring genotype–phenotype relationship for a maternal effect showing only dominance. (A) The expected offspring phenotype (fitness) for the three unordered offspring genotypes as a function of the frequency of the A 1 allele. The heterozygote line represents the mean of the two reciprocal heterozygotes. It is always the case with maternal effect dominance that the mean fitness of the offspring heterozygote is midway between the two homozygotes. The fitness values are on an arbitrary scale and are not labeled (but are all positive). (B) The realized additive effect of the locus is shown as a function of the frequency of the A 1 allele.
Perhaps the most important consequence of the frequency‐dependent offspring genotype‐phenotype relationship created by maternal effects is that, at the allele frequency equilibrium, the genotype‐phenotype relationship can disappear (eq. 9) (Dickerson 1947). If a locus only has a maternal effect, and no direct effect, then the change in allele frequencies and the realized additive effect are both zero when . Hence, with maternal genetic effects, evolution proceeds to an allele frequency where the offspring genotype‐phenotype relationship disappears, and consequently a population stops evolving because the locus is effectively neutral (Wade 2000). If the locus also shows an additive direct effect, then both the change in allele frequency and the realized additive effect will be zero when . This means that the direct and maternal effects cancel one another out, so that the locus appears to have no effect on the phenotype at the equilibrium allele frequency when in reality it has contrasting maternal and direct effects (e.g., Wade 2000). When displaced from this equilibrium, the locus will appear to have an additive effect that favors one of the two alleles, depending on the side of the peak from which it is displaced (i.e., this is the manifestation of the negative frequency dependence discussed above). Thus, from an empirical perspective, this implies that the additive effect will change temporally within populations as allele frequencies evolve, and spatially across populations that differ in allele frequencies either by drift or selection. Furthermore, if a population resides at an equilibrium allele frequency, the locus may appear to have no effect on the phenotype, which could be in conflict with an observation that the locus might show a molecular signature consistent with selection favoring a polymorphism.
Because of the differential weighting of direct and maternal effects to the realized adaptive landscape, when a locus has both a direct and a maternal effect there can be cases where a population is unable to “find” the peak on the mean fitness surface because selection on direct effects overwhelms the contribution of maternal effects. For example, if there is both an additive direct effect and a dominance maternal effect then there is significant parameter space where a peak on the mean fitness surface exists, but that peak is not “available” to the evolving population. Instead, the population evolves to fix the allele with the positive direct effect on fitness (Fig. 3). In short, the evolution at a pleiotropic locus, one with both a direct and a maternal effect, does not necessary evolve to maximize mean fitness. The genetic but nonheritable components of maternal effects (i.e., those not reflected in the offspring genotype‐phenotype relationship) are unavailable to selection.
Figure 3.

Evolution of a locus showing an additive direct effect and a dominance maternal effect that produce a peaked fitness surface. (A) The red (dashed) line shows the “surface” of mean fitness, where there is a peak at an allele frequency of 0.75. The blue (solid) line shows the “realized adaptive landscape” for maternal effects, which determines how the population evolves. Note that the realized adaptive landscape does not have a peak. (B) The change in allele frequencies at the A locus (Δp 1) as a function of the frequency of the A 1 allele, demonstrating that the population evolves to fixation of the A 1 allele.
We note, however, that changing the level of selection from the individual offspring phenotype to the full‐sib family mean makes the mean fitness surface equivalent to the adaptive landscape. Thus, the fitness peak seen in Figure 3, unattainable by individual selection, becomes attainable by among‐family selection and maximizes mean fitness. These results show why there are situations in which selection on group means (here acting at the family level, where selection is based on the mean of a family) could lead to higher mean fitness than individual selection (e.g., Dickerson 1947; Griffing 1967).
Inbreeding can play an important role in the evolution of maternal effects (Wade 2000) because it increases the correlation (rmo) between maternal and offspring genotypes (i.e., the correlation between alleles in the mother and her offspring). It increases the contribution of maternal effects to variation among offspring genotypes and, consequently, the evolutionary response to selection (Dickerson 1947). Inbreeding increases the contribution of additive maternal effects to the additive offspring genetic variance by a factor (eq. 11) (which is equal to ¼(1+F)2). As F increases, the relative contributions to the offspring additive variance of direct offspring and additive maternal effects equalize (eq. 11). Differently put, maternal effects become more heritable with inbreeding.
Inbreeding also changes the evolutionary response to selection. The contribution of additive maternal effects is weighted by a factor of (1 + F)2 (eq. 14) while the contribution of additive direct effects is weighted by a factor of (1 + F). Thus, inbreeding makes maternal genetic effects more available for an evolutionary response to selection, with the potential to double their contribution to adaptation when a population is fully inbred (see also Wade 2000).
Supporting information
Figure S1. Comparison of genotype frequencies after selection to the Hardy‐Weinberg expectation.
Figure S2. Evolutionary trajectories at the A locus (illustrated as the frequency of the A1 allele, p1, through generations) comparing the QHW approximation with the exact dynamics.
Figure S3. Evolutionary trajectories at the A locus (illustrated as the frequency of the A1 allele, p1, through generations) for the single locus case and the two locus case for a series of arbitrary scenarios.
Table S1. The expected phenotypes of offspring as a function of the offspring genotype or maternal genotype.
ACKNOWLEDGMENTS
We thank Nick Priest and Hamish Spencer for discussions during the development of these models and ideas and anonymous reviewers for input that greatly improved the presentation of this work. This work was supported by funding from the Biotechnology and Biological Sciences Research Council (BBSRC), UK, to J.B.W., the NIH R01 GM65414‐04A1 and R01GM084238‐07 to M.J.W., and the Esther Parkin Trust.
To derive the additive genetic variance we need to derive the average effect and average excess of the two alleles (Fisher 1930). For a general background on this derivation see Crow and Kimura (Crow and Kimura 1970) and Templeton (Templeton 1987, 2006). The average excess of the alleles are measures of the average phenotype or fitness associated with an allele, measured as a deviation from the population mean:
| (A1) |
| (A2) |
The average effects an allele substitution (α, measured as a substitution of an A 1 allele for an A 2 allele) is defined as the slope of the least‐squares regression of the phenotypic or fitness deviations (i.e., deviations from the mean) of a genotype on the number of copies of the A 1 allele possessed by a genotype (Lynch and Walsh 1998). The average effects of the alleles are then defined from this regression as α1 = p 2α and α2 = –p 1α. The least‐squares approach has a simple solution when we consider the influence of inbreeding (i.e., F), where the average effect is a function of the average excess and the degree of inbreeding (Lynch and Walsh 1998; Templeton 2006):
| (A3) |
| (A4) |
The average effect and average excess can be used to derive the additive genetic variance (Va) as:
| (A5) |
LITERATURE CITED
Associate Editor: J. Etterson
Handling Editor: R. Shaw
- Arnold, S. J. 2003. Performance surfaces and adaptive landscapes. Integr. Comp. Biol. 43:367–375. [DOI] [PubMed] [Google Scholar]
- Badyaev, A. V. , Hill G. E., Beck M. L., Dervan A. A., Duckworth R. A., McGraw K. J., Nolan P. M., and Whittingham L. A.. 2002. Sex‐biased hatching order and adaptive population divergence in a passerine bird. Science 295:316–318. [DOI] [PubMed] [Google Scholar]
- Barker, M. S. , Demuth J. P., and Wade M. J.. 2005. Maternal expression relaxes constraint on innovation at the anterior determinant, bicoid . Plos Genet. 1:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux, C. , Autran D., Gillmor C. S., Grimanelli D., and Grossniklaus U.. 2008. The maternal to zygotic transition in animals and plants. Cold Spring Harb. Symp. Quant. Biol. 73:89–100. [DOI] [PubMed] [Google Scholar]
- Berleth, T. , Burri M., Thoma G., Bopp D., Richstein S., Frigerio G., Noll M., and Nüsslein‐Volhard C.. 1988. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. Embo J. 7:1749–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo, J. 1996. Maternal effects in animal ecology. Am. Zool. 36:83–105. [Google Scholar]
- Bowen, W. D. 2009. Maternal effects on offspring size and development in pinnipeds Pp. 104–132 in Maestripieri D., and Mateo J. J., eds. Maternal effects in mammals. Chicago Univ. Press, Chicago. [Google Scholar]
- Brumby, P. J. 1960. The influence of the maternal environment on growth in mice. Heredity 14:1–18. [Google Scholar]
- Bult, A. and Lynch C. B.. 1997. Nesting and fitness: lifetime reproductive success in house mice bidrectionally selected for thermoregulatory nest‐building behavior. Behav. Genet. 27:231–240. [DOI] [PubMed] [Google Scholar]
- Byers, D. L. , Platenkamp G. A. J., and Shaw R. G.. 1997. Variation in seed characters in Nemophila menziesii: evidence of a genetic basis for maternal effect. Evolution 51:1445–1456. [DOI] [PubMed] [Google Scholar]
- Cadby, C. D. , Jones S. M., and Wapstra E.. 2011. Potentially adaptive effects of maternal nutrition during gestation on offspring phenotype of a viviparous reptile. J. Exp. Biol. 214:4234–4239. [DOI] [PubMed] [Google Scholar]
- Casellas, J. , Farber C. R., Gularte R. J., Haus K. A., Warden C. H., and Medrano J. F.. 2009. Evidence for maternal QTL affecting growth and obesity in adult mice. Mamm. Genome 20:269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, C. K. 1956. Analysis of quantitative inheritance of body size in mice. I. Hybridization and maternal influence. Genetics 41:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud, J. M. 1984. Evolution by kin selection: a quantitative genetic model illustrated by maternal performance in mice. Evolution 38:766–777. [DOI] [PubMed] [Google Scholar]
- Cheverud, J. M. . 1985. A quantitative genetic model of altruistic selection. Behav. Ecol. Sociobiol. 16:239–243. [Google Scholar]
- Cheverud, J. M. and Moore A. J.. 1994. Quantitative genetics and the role of the environment provided by relatives in behavioural evolution . Pp. 67–100 in Boake C. R. B., ed. Quantitative genetic studies of behavioural evolution. Chicago Univ. Press, Chicago. [Google Scholar]
- Cheverud, J. M. and Wolf J. B.. 2009. Genetics and evolutionary consequences of maternal effects Pp. 11–37 in Maestripieri D., and Mateo J. J., eds. Maternal effects in mammals. Chicago Univ. Press, London. [Google Scholar]
- Crow, J. F. and Kimura M.. 1970. An introduction to population genetics theory. Harper and Row, New York. [Google Scholar]
- Cruickshank, T. E. and Wade M. J.. 2008. Microevolutionary support for a developmental hourglass: gene expression patterns shape sequence variation and divergence in Drosophil. Evol. Dev. 10:583–590. [DOI] [PubMed] [Google Scholar]
- Dickerson, G. E. 1947. Composition of hog carcasses as influenced by heritable differences in rate and economy of gain. Res. Bull. Iowa Agric. Exp. Station. 354:489–524. [Google Scholar]
- Donohue, K. 1998. Maternal environmental effects in plants Pp. 137–177 in Mousseau T. A., and Fox C. W., eds. Maternal effects as adaptations. Oxford Univ. Press, New York. [Google Scholar]
- Drown, D. M. and Wade M. J.. 2014. Runaway coevolution: adaptation to heritable and non‐heritable environments. Evolution 68:3039–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer, D. S. 1960. The genetics of litter size in mice. J. Cell. Physiol. 56:153–167. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S. . 1965. Maternal effects and selection response Pp. 763–774 in Geerts S. J., ed. Genetics today, Proc. XI Int. Cong. Genet. Pergamon Press, Oxford, UK. [Google Scholar]
- Falconer, D. S. and Mackay T. F. C.. 1996. Introduction to quantitative genetics, 4th ed Longman, Essex. [Google Scholar]
- Fisher, R. A. 1918. The correlation among relatives on the supposition of Mendelian inheritance. Trans. R Soc. Edinburgh 52:399–433. [Google Scholar]
- Fisher, R. A. . 1930. The genetical theory of natural selection. Oxford Univ. Press, London. [Google Scholar]
- Fox, C. W. , Czezak E., Mousseau T. A., and Roff D. A.. 1999. The evolutionary genetics of an adaptive maternal effect: egg size plasticity in a seed beetle. Evolution 53:1999. [DOI] [PubMed] [Google Scholar]
- Gavrilets, S. 1998. One‐locus two‐allele models with maternal (parental) selection. Genetics 149:1147–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing, B. 1967. Selection in reference to biological groups. I. Individual and group selection applied to populations of unordered groups. Aust. J. Biol. Sci. 20:127–139. [PubMed] [Google Scholar]
- Hadfield, J. D. 2012. The quantitative genetic theory of parental effects Pp. 267–284 in Royle N., Smiseth P., and Kolliker M., eds. The evolution of parental care. Oxford Univ. Press, UK. [Google Scholar]
- Hager, R. , C. J. M., Roseman C., and Wolf J. B.. 2008. Maternal effects as the cause of parent‐of‐origin effects that mimic genomic imprinting. Genetics 178:1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, D. L. 1964. Genotypic covariances between inbred individuals. Genetics 50:1319–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, P. W. 1997. Neutrality or selection? Nature 387:138. [DOI] [PubMed] [Google Scholar]
- Kimura, M. 1965. Attainment of quasilinkage equilibrium when gene frequencies are changiung by natural selection. Genetics 52:875–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick, M. and Lande R.. 1989. The evolution of maternal characters. Evolution 43:485–503. [DOI] [PubMed] [Google Scholar]
- Lande, R. and Kirkpatrick M.. 1990. Selection response in traits with maternal inheritance. Genet. Res. 55:189–197. [DOI] [PubMed] [Google Scholar]
- Lipshitz, H. D. , ed. 2015. The maternal‐to‐zygotic transition. Academic Press, London. [Google Scholar]
- Lloyd, J. D. and Martin T. E.. 2004. Nest‐site preference and maternal effects on offspring growth. Behav. Ecol. 15:816–823. [Google Scholar]
- Lynch, M. 1987. Evolution of intrafamilial interactions. Proc. Nat. Acad. Sci USA 84:8507–8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M. and Walsh B.. 1998. Genetics and analysis of quantitative traits. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Maestripieri, D. and Mateo J. J., eds. 2009. Maternal effects in mammals. Chicago Univ. Press, London. [Google Scholar]
- McGlothlin, J. W. , Moore A. J., Wolf J. B., and Brodie E. D. III. 2010. Interacting phenotypes and the evolutionary process. III. Social evolution. Evolution 64:2558–2574. [DOI] [PubMed] [Google Scholar]
- McGlothlin, J. W. , Wolf J. B., Brodie E. D. III, and Moore A. J.. 2014. Quantitative genetic versions of Hamilton's rule with empirical applications. Phil. Trans. Royal Soc. B 369:20130358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousseau, T. A. and Dingle H.. 1991. Maternal effects in insect life histories. Ann. Rev. Entomol. 36:511–534. [Google Scholar]
- Mousseau, T. A. and Fox C. W.. 1998. Maternal effects as adaptations. Oxford Univ. Press, New York. [Google Scholar]
- Nagylaki, T. 1976. The evolution of one‐ and two‐locus systems. Genetics 83:583–600. [PMC free article] [PubMed] [Google Scholar]
- Nagylaki, T. . 1993. The evolution of multilocus systems under weak selection. Genetics 134:627–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordskog, A. W. and Hassan G. M.. 1971. Direct and maternal effects of egg‐size genes on hatchability. Genetics 67:267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, G. M. L. , Audet C., and Bernatchez L.. 2005. Maternal genetic effects on adaptive divergence between anadromous and resident brook charr during early life history. J. Evol. Biol. 18:1348–1361.</bib type="Periodical"> [DOI] [PubMed] [Google Scholar]
- Preuss, K. M. , Lopez J. A., Colbourne J. K., and Wade M. J.. 2012. Identification of maternally‐loaded RNA transcripts in unfertilized eggs of Tribolium castaneum . Genome Biol. Evol. 4:763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, T. 1998. Maternal and paternal effects in birds Pp. 202–224 in Mousseau T. A., and Fox C. W., eds. Maternal effects as adaptations. Oxford Univ. Press, New York. [Google Scholar]
- Reznick, D. , Callahan H., and Llauredo R.. 1996. Maternal effects on offspring quality in Poeceliid fishes. Am. Zool. 36:147–156. [Google Scholar]
- Riska, B. , Rutledge J. J., and Atchley W. R.. 1985. Covariance between direct and maternal genetic effects in mice, with a model of persistent environmental influences. Genet. Res. Cambridge 45:287–297. [DOI] [PubMed] [Google Scholar]
- Roach, D. A. and Wulff R. D.. 1987. Maternal effects in plants. Ann. Rev. Ecol. Syst. 18:209–236. [Google Scholar]
- Robinson, D. L. 1996. Models which might explain negative correlations between direct and maternal genetic effects. Livestock Prod. Sci. 45:111–122. [Google Scholar]
- Roff, D. A. 1997. Evolutionary quantitative genetics. Chapman & Hall, New York, NY. [Google Scholar]
- Santure, A. W. and Spencer H. G.. 2006. Influence of mom and dad: quantitative genetic models for maternal effects and genomic imprinting. Genetics 173:2297–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, R. and Byers D.. 1998. Genetics of maternal and paternal effects In Mousseau T., and Fox C., eds. Maternal effects as adaptations. Oxford Univ. Press, New York. [Google Scholar]
- Spencer, H. G. 2002. The correlation between relatives on the supposition of genomic imprinting. Genetics 161:411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, H. G. . 2003. Further properties of Gavrilets' one‐locus two‐allele model of maternal selection. Genetics 164:1689–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, H. G. and Chiew K. X.. 2015. The maintenance of single‐locus polymorphism by maternal selection. G3: Genes, Genomes, Genetics 5:9630969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson, E. I. and Calsbeek R., eds. 2012. The adaptive landscape in evolutionary biology. Oxford Univ. Press, Oxford. [Google Scholar]
- Tadros, W. and Lipshitz H. D.. 2009. The maternal‐to‐zygotic transition: a play in two acts. Development 136:3033–3042. [DOI] [PubMed] [Google Scholar]
- Templeton, A. R. 1987. The general relationship between average effect and average excess. Genet. Res. 49:69–70. [DOI] [PubMed] [Google Scholar]
- Templeton, A. R. . 2006. Population genetics and microevolutionary theory. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- Tong, Z.‐B. , Gold L., Pfeifer K. E., Dorward H., Lee E., Bondy C. A., Dean J., and Nelson L. M.. 2000. Mater, a maternal effect gene required for early embryonic development in mice. Nat. Genet. 26:267–268. [DOI] [PubMed] [Google Scholar]
- Wade, M. J. 1978. Kin selection: a classical approach and a general solution. Proc. Nat. Acad. Sci. USA 75:6154–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, M. J. . 1998. The evolutionary genetics of maternal effects Pp. 5–21 in Mousseau T. A., and Fox C. W., eds. Maternal effects as adaptations. Oxford Univ. Press, New York. [Google Scholar]
- Wade, M. J. . . 2000. Opposing levels of selection can cause neutrality: mating patterns and maternal‐fetal interactions. Evolution 54:290–292. [DOI] [PubMed] [Google Scholar]
- Wade, M. J. and Beeman R. W.. 1994. The population dynamics of maternal‐effect selfish genes. Genetics 138:1309–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, M. J. , Priest N. K., and Cruickshank T. E.. 2009. A theoretical overview of genetic maternal effects: evolutionary predictions and empirical tests with mammalian data In Maestripieri D., and Mateo J. J., eds. Maternal effects in mammals. Chicago Univ. Press, Chicago. [Google Scholar]
- Weir, B. S. and Cockerham C. C.. 1977. Two‐locus theory in quantitative genetics Pp. 247–269 in Pollak E., Kempthorne O., and Bailey T. B., eds. Proceedings of the International Conference on Quantitative Genetics. Iowa Univ. Press, Ames, Iowa. [Google Scholar]
- Willham, R. L. 1963. The covariance between relatives for characters composed of components contributed by related individuals. Biometrics. 19:18–27. [Google Scholar]
- Willham, R. L. . 1972. The role of maternal effects in animal breeding: III. Biometrical aspects of maternal effects in animals. J. Animal Sci. 35:1288–1293. [DOI] [PubMed] [Google Scholar]
- Wilson, A. J. and Reale D.. 2006. Ontogeny of additive and maternal genetic effects: lessons from domestic mammals. Am. Nat. 167:E23–E38. [DOI] [PubMed] [Google Scholar]
- Wolf, J. B. 2000. Gene interactions from maternal effects. Evolution 54:1882–1898. [DOI] [PubMed] [Google Scholar]
- Wolf, J. B. . 2003. Genetic architecture and evolutionary constraint when the environment contains genes. Proc. Nat. Acad. Sci. USA 100:4655–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, J. B. and Brodie E. D. III. 1998. Coadaptation of parental and offspring characters. Evolution 52:535–544. [DOI] [PubMed] [Google Scholar]
- Wolf, J. B. , Brodie E. D. III, Cheverud J. M., Moore A. J., and Wade M. J.. 1998. Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. 13:64–69. [DOI] [PubMed] [Google Scholar]
- Wolf, J. B. and Cheverud J. M.. 2012. Detecting maternal effect loci by statistical cross fostering. Genetics 191:261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, J. B. and Hager R.. 2006. A maternal‐offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 4:e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, J. B. , Leamy L., Roseman J. C. C., and Cheverud J. M.. 2011. Disentangling prenatal and postnatal maternal genetic effects reveals persistent prenatal effects on offspring growth in mice. Genetics 189:1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, J. B. , Moore A. J., and Brodie E. D. III. 1997. The evolution of indicator traits for parental quality: the role of maternal and paternal effects. Am. Nat. 150:639–649. [DOI] [PubMed] [Google Scholar]
- Wolf, J. B. , Moore A. J., and Brodie E. D. III. . 1999. Maternal and paternal effects in sexual selection and the evolution of parental investment. J. Evol Biol. 12:1157–1167. [Google Scholar]
- Wolf, J. B. , Royle N. J., and Hunt J.. 2014. Genotype‐by‐environment interactions when the social environment contains genes Pp. 63–98 in Hunt J., and Hosken D. J., eds. Genotype‐by‐environment interactions and sexual selection. Wiley‐Blackwell, Chichester, West Sussex. [Google Scholar]
- Wolf, J. B. and Wade M. J.. 2009. What are maternal effects (and what are they not)? Philosophical Transactions of the Royal Society of London. Series B Biol. Sci. 364:1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. 1932. The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proc. 6th Int. Cong Genet. 1:356–366. [Google Scholar]
- Wright, S. . 1969. Evolution and the genetics of populations: vol. 2 The theory of gene frequencies. Chicago Univ. Press, Chicago. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of genotype frequencies after selection to the Hardy‐Weinberg expectation.
Figure S2. Evolutionary trajectories at the A locus (illustrated as the frequency of the A1 allele, p1, through generations) comparing the QHW approximation with the exact dynamics.
Figure S3. Evolutionary trajectories at the A locus (illustrated as the frequency of the A1 allele, p1, through generations) for the single locus case and the two locus case for a series of arbitrary scenarios.
Table S1. The expected phenotypes of offspring as a function of the offspring genotype or maternal genotype.


