Abstract
Mammary gland development during puberty and reconstruction during pregnancy and lactation is under the control of circulating endocrine hormones, such as growth hormone, which are released from the pituitary. In this study, we explored the influence of overexpression of growth hormone in the mammary gland on breast development and milk production in goats. Using transcriptome sequencing, we found that the number of highly expressed genes was greater in GH transgenic goats than non-transgenic goats. Furthermore, KEGG pathway analysis showed that the majority of the genes belonged to the MAPK signaling pathway and the ECM-receptor interaction pathway. The expression of genes related to breast development was further confirmed using qRT-PCR. Interestingly, both milk production and milk quality were increased. The results of these experiments imply that overexpression of growth hormone in the breast may stimulate breast development and enhances milk production by modulating alveolar cell proliferation or branching through the MAPK signaling pathway.
Keywords: mammary gland, growth hormone, development, IGF-1, milk production
Introduction
Physiologically, mammary glands undergo significant changes during lactation to prepare for milk production, which is controlled by hormones secreted by the endocrine system (Watson and Khaled, 2008). The serum concentration of prolactin (PRL) and GH are significantly increased, which activates PRLRs and growth hormone receptors (GHRs) to influence ductal and alveolar development and differentiation. Therefore, GH plays a critical role in mammary gland development and milk production (Shingu et al., 2004). GH can not only increase milk protein synthesis, but can also stimulate mammary gland epithelial cell differentiation and proliferation (Macias and Hinck, 2012).
Mammary glands are responsible for milk production and delivery to newborns, which is their only food source. Therefore, mammary gland growth during lactation is important for the development of newborns. Additionally, the main product of dairy animals is milk; therefore, it is necessary to determine how GH improves milk production. Mammary gland development occurs in three distinct and differentially regulated stages: embryonic, pubertal and adult. The mammary glands maintain their ability to produce milk during pregnancy, lactation and involution. In this study, we focused on changes in gene expression following GH overexpression during lactation.
Goat milk is believed to be more easily digestible and less allergenic than cow's milk. The goat milk is also high in calcium and fatty acids but low in cholesterol, which makes it an ideal food especially for the old, weak, sick and disabled (Strzalkowska et al., 2012; Gantner et al., 2015). However, the low yield of goat milk restricts it widely use. In our previous study, we produced a transgenic goat model that overexpressed GH in the mammary glands and verified increased GH expression in the breast (Zhang et al., 2014). The aim of this study was to determine the effect and mechanism of GH overexpression in the breast on breast development and milk production.
Materials and methods
Ethics statement
This study was approved by the Ethical Committee of Animal Experiments of the College of Veterinary Medicine at Nanjing Agricultural University. All animal care and use procedures were conducted in strict accordance with the Animal Research Committee guidelines of the College of Veterinary Medicine at Nanjing Agricultural University.
Experimental animals and tissue collection
Five-year-old GH transgenic (TG) and non-transgenic (NG) Saanen dairy goats were raised on the farm at the transgenic research center in Shanghai, China. All goats were healthy and fed with the same fodder. During housing, the health status of all animals was monitored twice a day. No adverse events were observed. We obtained six mammary gland tissue samples from Saanen GH transgenic goats (n = 3) and non-transgenic goats (n = 3). The tissues were collected during the goats lactation. The mammary tissue samples were harvested and immediately frozen in liquid nitrogen and preserved at −80°C.
Milk samples were collected from TG and NG Saanen dairy goats on the 1st, 7th, 14th, and 30th day of lactation, and milk production, lactose content, milk fat, protein content, density, milk solids, non-fat milk solids and conductivity were detected.
RNA isolation and transcriptome sequencing
Total RNA was extracted from the mammary gland samples of six Saanen goats using TRIzol reagent. The quantity and purity of the isolated RNA was analyzed prior to the generation of sequencing libraries using the Agilent 2200 TapeStation Systems (Agilent, USA). Sequencing was performed using the Illumina HiSeq 2500 sequencing system. The goat reference genome sequences were downloaded from NCBI. After removing the low quality reads and clipping the adapter sequences, clean reads from each sample were aligned to the goat reference genome using TopHat (Trapnell et al., 2009), which is a gapped aligner that is capable of discovering splice junctions ab initio. TopHat reports aligned results from the two mapping steps in shoot apical meristem format for further analysis. Aligned reads from TopHat mapping were subjected to Cufflinks (Trapnell et al., 2010). Gene expression was estimated using Cufflinks and normalized by calculating the reads per kilo base per million mapped reads (RPKM) for each gene and was annotated with NCBI genome assembly (Mortazavi et al., 2008).
KEGG pathway analysis
To gain an overview of gene pathway networks, KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis was performed using the online KEGG automatic annotation server (https://david.ncifcrf.gov/summary.jsp).
Confirmation of RNA sequencing results
According to related research and the RNA-seq results, 15 representative genes were verified using qRT-PCR. The assays were performed in samples following RNA preparation. Total RNA was extracted and assessed in RNA sequencing experiments, and DNA was digested using DNase. First strand cDNA was synthesized using a PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, China) according to the manufacturer's instructions. Real-time PCR was conducted using the 7500 Real-Time PCR System (ABI, USA) and EvaGreen qPCR MasterMix-Low Rox (Abm, Canada). PCR amplification was conducting using the following conditions: 95°C for 10 min, followed by 35 cycles of 95°C for 30 s and 60°C for 34 s. Each sample and negative controls (no template) were run in triplicate. The gene-specific primers are listed in Table 1. The transcriptome sequencing data of the 15 identified genes is shown in the Supplementary Material.
Table 1.
QRT-PCR primers used to evaluate the RNA-seq data.
| Primer names | Type | Sequences | Product length |
|---|---|---|---|
| GHR-F | Forward | CATAGTGCGGTCTGCTTCCA | 250 bp |
| GHR-R | Reverse | GGTGTGGCTTCACTCCCAG | |
| IGF1R-F | Forward | GTGGTGACAGGCTACGTGAA | 186 bp |
| IGF1R-R | Reverse | CCCAGCTTTGATGGTCAGGT | |
| ACTB-F | Forward | CACCACACCTTCTACAA | 106 bp |
| ACTB-R | Reverse | TCTGGGTCATCTTCTCAC | |
| IGF1-F | Forward | GGATGCTCTCCAGTTCGTGT | 161 bp |
| IGF1-R | Reverse | TGAGAGGCGCACAGTACATC | |
| IGF2-F | Forward | CAACAGCTGACCTCATTTCCC | 243 bp |
| IGF2-R | Reverse | TGCGTATTGCAAACCGAACAG | |
| IGFBP3-F | Forward | TTCCTGAACACACTCAGCCC | 212 bp |
| IGFBP3-R | Reverse | GTGGCCACAGTCTACTTGCT | |
| MST1R-F | Forward | CCGATGTGTGGTCGTATGGT | 213 bp |
| MST1R-R | Reverse | CTGCTAGCTCTCCGAAGGTG | |
| TGFA-F | Forward | CGCGCTGGGTATTTTCTTGG | 129 bp |
| TGFA-R | Reverse | AACTGACTGTGGGAAGCTGG | |
| EGFR-F | Forward | TCCTTCACACGTACTGCACC | 172 bp |
| EGFR-R | Reverse | AAAACTGGCCGTGTTGCTTC | |
| FGF7-F | Forward | AAGTTGCACAGGGCAGACAA | 83 bp |
| FGF7-R | Reverse | GCTGTTTGCTACTTGACTTTTGTTC | |
| FGFR2-F | Forward | AAGCTGACCAAGCGTATCCC | 207 bp |
| FGFR2-R | Reverse | TTTGCCCAGCGTCAGCTTAT | |
| TGFB1-F | Forward | AACCTGTGTTGCTCTCTCGG | 110 bp |
| TGFB1-R | Reverse | GAGGTAGCGCCAGGAATTGT | |
| WNT5A-F | Forward | CACTGGTTCACCCTGTAGCC | 189 bp |
| WNT5A-R | Reverse | TCTCTAGGCACAGCCTCTCA | |
| COL4A5-F | Forward | CGGGACTTCCAGGACTTTCT | 230 bp |
| COL4A5-R | Reverse | GTGGACCTTGAAAACCTGTAGGA | |
| COL6A1-F | Forward | ATCGGGCCAAAAGGATACCG | 119 bp |
| COL6A1-R | Reverse | TCTTCACCCCTCTCACCCAT | |
| COL6A5-F | Forward | GCATTTGCATCCTGGCTCTG | 176 bp |
| COL6A5-R | Reverse | AAAGCAGTTGGTTTTCCCGC |
Morphology observation of mammary gland
The mammary gland was fixed in Bouin's fluid for 48 h at room temperature. After fixation, samples were cut consecutively into five blocks by using a scalpel, in parallel with the vertical section and cross-section. The blocks were dehydrated and then embedded in paraffin. Six micrometer sections were cut from the paraffin block using a rotary microtome. All sections were mounted on slides and stained with hematoxylin-eosin (HE).
Statistical analysis
All statistical analyses were performed by using SPSS 17.0. The statistical analysis for gene analysis and milk composition analysis was performed using independent t-test. The comparison of the means was performed, and the differences were considered significant if P < 0.05
Results
Global analysis of gene expression in the mammary glands of NG and TG
To assess the mammary gland transcriptome in GH transgenic goats, we constructed cDNA libraries for the different groups. After filtering, the sequences of the six libraries were mapped to the goat reference genome and the number of mapped genes was complied with the requirements. The RAW data were submitted to NCBI as SRA, and the accession numbers were SRR3659132 and SRR3659145 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR3659132). We observed that 85.64 and 81.10% of the genes mapped uniquely to only one location and could be assigned to a single annotated goat gene in the NG and TG libraries, respectively. We evaluated the expression of genes using RPKM based on methods described by Mortazavi (Mortazavi et al., 2008). To further assess the transcriptome data, the RPKM values were divided into five groups (Table 2): RPKM < 0.1, 0.1 < RPKM < 1, 1 < RPKM < 10, 10 < RPKM < 100, and RPKM > 100. The transcriptome results revealed that the most frequent group in the NG was 1 < RPKM < 10, whereas in the TG, the most frequent group was 10 < RPKM < 100. Moreover, the RPKM > 100 group in the TG was higher compared to that in the NG. We analyzed the genes in the RPKM > 1 group in the following experiments.
Table 2.
Distribution of the expression values of the differentially expressed genes.
| RPKM range | NG (frequency) | TG (frequency) | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | |
| (0, 0.1) | 5733 | 5372 | 5407 | 4014 | 3767 | 3685 |
| (0.1, 1) | 3455 | 3816 | 3825 | 2361 | 2137 | 2189 |
| (1, 10) | 7491 | 7368 | 7464 | 5928 | 5327 | 5403 |
| (10, 100) | 2494 | 2608 | 2474 | 6493 | 7421 | 7446 |
| (100, INF) | 205 | 214 | 209 | 584 | 668 | 657 |
Differentially expressed genes in TG and NG
Genes were determined to be differentially expressed based on the false discovery rate of the adjusted RPKM > 1 based on P < 0.01 and fold-change in expression >2. This resulted in the identification of 4451 differentially expressed genes; 2274 were expressed higher and 2217 were expressed lower in the TG.
KEGG pathway enrichment analysis
KEGG pathway analysis, which is an alternative approach to categorize gene functions with a focus on biochemical pathways, was performed on the annotated genes. A total of 991 genes were assigned one or more KEGG annotations and were mapped to KEGG pathways. A total of 37 pathways were significantly enriched, including the MAPK signaling pathway, calcium signaling pathway, cell adhesion molecules, ECM-receptor interaction pathway, and p53 signaling pathway (Figure 1A).
Figure 1.
Gene expression analysis. (A) KEGG pathway analysis of the RNA sequencing data. The x-axis represents the number of genes for each pathway. (B) Heat map of the genes expressed in the TG and NG groups. NG, non-transgenic goat. TG, transgenic goat (n = 3 per group). Red represents high expression and green represents low expression. (C–F) Differences in gene expression between the NG and TG groups as detected using qRT-PCR. The x-axis represents the name of the detected genes and the y-axis represents the relative mRNA expression for each detected gene. The data are expressed as the mean ± SE (n = 3). *P < 0.05; **P < 0.01.
Confirmation of the RNA-seq results using qRT-PCR
To validate our RNA-seq results, we subjected 15 differentially expressed genes to qRT-PCR. The qRT-PCR results largely confirmed our RNA-seq data (Figure 1C). Although the fold-change values in the qRT-PCR and RNA-seq data were not directly related, they displayed similar tendencies (Figure 1B). The expression of eight genes was increased (IGF1R, IGF1, IGF2, IGFBP3, FGF7, FGFR2, COL4A5 and COL6A5) and that of four genes were decreased (GHR, MST1R, EGFR and WNT5A). There was no significant change in the expression of TGFA, TGFB1 and COL6A1.
The effect of GH on mammary gland structure
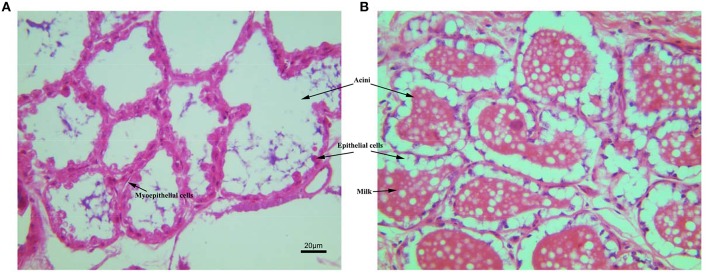
The tissue sections of mammary gland stained with HE were shown in Figure 2. The Mammary glands of NG and TG were collected in the same lactation period. Compared with the NG, more goat milk with clear lipid droplet was visible in the acini of TG. The epithelial cells in the NG were rounded, while the shape of epithelial cells in TG was more flat with abundant milk. The myoepithelial cells underneath the epithelial cells were also detected around the epithelial cells.
Figure 2.
The histological structure of mammary gland stained with HE. (A) Mammary gland of non-transgenic goat (NG); (B) mammary gland of GH transgenic goat (TG). The Mammary glands of NG and TG were collected in the same lactation period, then fixed and cut into paraffin section. The tissues were stained with HE. The milk, aicni and myoepithelial cells underneath the epithelial cells were labeled.
Milk detection
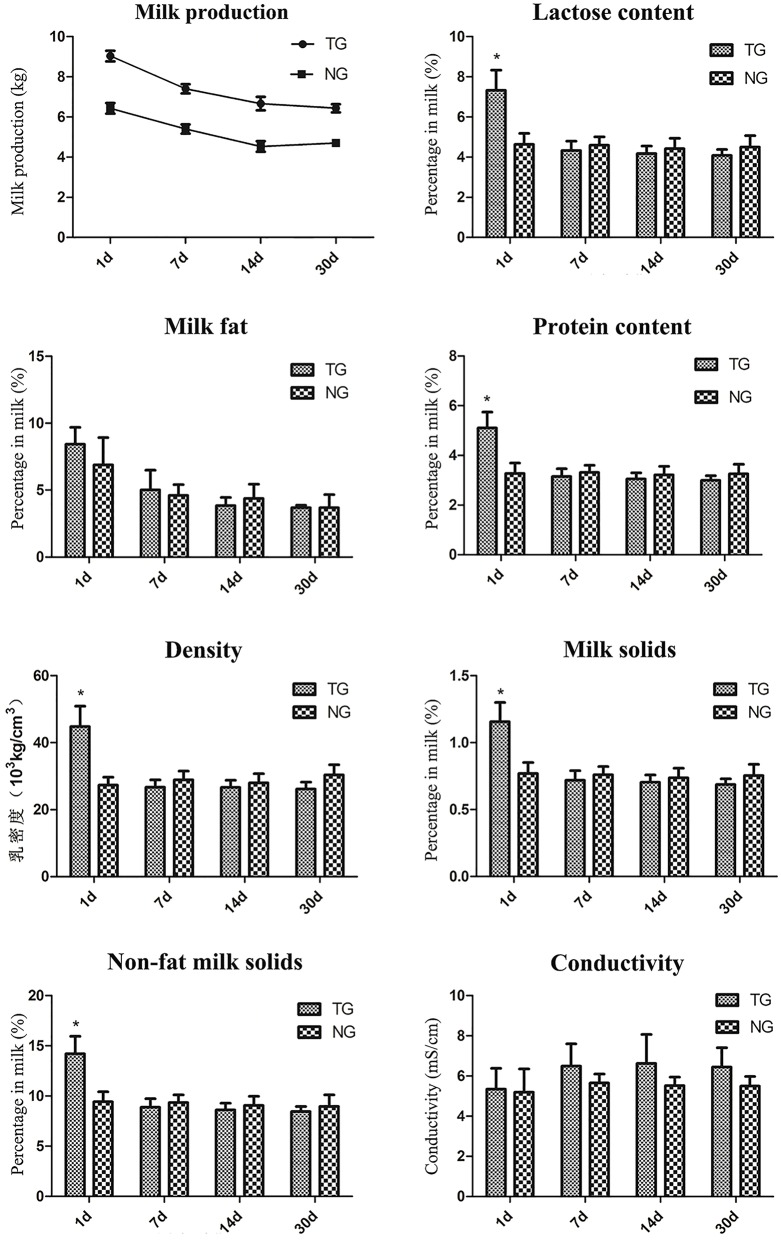
Milk production was significantly increased in the TG group compared with the NG group during the lactation detection period (Figure 3). There were no significant changes in milk fat and conductivity in the TG group during the lactation detection period. The lactose content, protein content, density, milk solids and non-fat milk solids were significantly increased in the TG group compared with the NG group on the first day of lactation. However, there were no significant differences in these measures during the remaining three lactation detection time periods.
Figure 3.
Milk sample detection. Eight milk samples were assessed for different measures, including milk production, lactose content, milk fat, protein content, density, milk solids, non-fat milk solids and conductivity. The x-axis represents the date that each sample was collected and the y-axis represents the value of each measurement. *P < 0.05.
Discussion
Mammary gland development is dynamic and divided into three stages: embryonic, pubertal, and adult (Gjorevski and Nelson, 2011). Adult mammary gland growth is an important stage of preparation during pregnancy and lactation in mammals. GH plays a critical role in this stage (Molik et al., 2010; Macias and Hinck, 2012). Higher exogenous overexpression of GH was detected in mammary gland in TG goats (Zhang et al., 2014). This allows us to study the effects of GH on mammary gland growth.
Mammary gland development during lactation requires growth factors that increase the proliferation and survival of mammary epithelial cells. Overexpression of GH stimulates the expression of growth factors. KEGG pathway analysis showed that these genes were involved in mammary gland physiology, including the MAPK signaling pathway, calcium signaling pathway, cell adhesion molecules, ECM-receptor interaction pathway and p53 signaling pathway. Based on these results, we selected 15 genes and examined differences in the expression levels between the TG and NG.
GH binds to GHRs, which activate its effects. Our results showed that overexpression of GH altered the expression of GHR in the mammary glands. GH induces paracrine signaling by activating GHRs on stromal cells. This process could induce the expression of insulin-like growth factor 1 (IGF1) in stromal cells (Perez et al., 2016). IGF1 binds to cell surface receptors (IGF1R) on epithelial cells (Perez et al., 2016) to stimulate mammary gland epithelial cell growth and development, as well as cellular DNA synthesis (Kleinberg et al., 2000; Perez et al., 2016). Similar to IGF1, insulin-like growth factor 2 (IGF2) also increases cell proliferation and survival by binding to IGF1Rs (Nawathe et al., 2016) (Figure 4). Although in our study expression of GHR was decreased, the expression of IGF1, IGF2 and IGF1R was significantly increased in the TG group. Insulin-like growth factor-binding protein 3 (IGFBP3) exerts antiproliferative effects in many cell types by blocking the ability of IGF1 and IGF2 to activate IGF1Rs (Nawathe et al., 2016). Our results showed that IGFBP3 was significantly increased in the TG group. The upregulation of IGF1, IGF2, and IGF1R might lead to excessive proliferation of epithelial cells. Increasing IGFBP3 and decreasing GHR might prevent the excessive growth of the mammary glands, which could damage the organism (Figure 1C).
Figure 4.
The different pathways altered by overexpression of GH. Cellular crosstalk between the epithelial and stromal compartments is mediated by growth factors, including IGF1, EGF, and FGFs, which bind to their receptors to induce cell proliferation and branching. The WNT5A pathway can also influence epithelial cells proliferation. High tension from cell-generated mechanical stress can promote the binding of growth factors to their receptors.
Several receptor tyrosine kinases have profound effects on mammary gland development during puberty, including RON, which negatively regulates mammary gland branching morphogenesis (Meyer et al., 2009). RON is encoded by MST1R, which was significantly decreased in our study. The kinetic profile of MAPK activity may determine the final morphogenetic response. Sustained MAPK activation downstream of transforming growth factor-α (TGFα) and epidermal growth factor receptor (EGFR) induces branching. Moreover, transient MAPK activation downstream of fibroblast growth factor 7 (FGF7) and fibroblast growth factor receptor 2 (FGFR2) induces proliferation (Sternlicht et al., 2005; Fata et al., 2007) (Figure 4). Our results show that there was no difference in the expression of TGFA between the two groups and EGFR was slightly downregulated in the TG group. However, FGF7 and FGFR2 were significantly increased in the TG group (Figure 1D). These results suggest that overexpression of GH might promote mammary gland proliferation through FGF7 and FGFR2, but does not promote mammary gland branching through TGFα and EGFR. In addition, inhibition of RON might promote mammary gland branching.
Transforming growth factor-β (TGFβ) plays an important role in mammary gland development, and its concentration profile is determined by tissue geometry. In microfabricated gland mammary tissue, branching is inhibited at sites of high concentrations of TGFβ (Nelson et al., 2006). Overexpression of TGFβ1 leads to inhibition of mammary gland development in vivo (Pierce et al., 1993), whereas TGFβ-deficient mice exhibit accelerated ductal proliferation and lateral branching (Ewan et al., 2002; Crowley et al., 2005). WNT5A acts downstream of TGFβ in vivo (Roarty and Serra, 2007) and is required for TGFβ-mediated inhibition of mammary branching morphogenesis (Roarty and Serra, 2007; Macias et al., 2011; Kumawat and Gosens, 2016) (Figure 4). We detected the expression of TGFB1 and WNT5A in the two groups and found that the expression of WNT5A was significantly downregulated in the TG group, whereas there was no change in the expression of TGFB1 between the TG and NG groups (Figure 1E). These results suggest that overexpression of GH might ameliorate the inhibition of mammary development by reducing WNT5A expression and not TGFB1 expression.
Cell-generated mechanical stress can remodel the surrounding matrix and release extracellular matrix (ECM)-bound regulatory molecules, such as growth factors (Vogel and Sheetz, 2006). The binding of these growth factors to their receptors under high tension conditions results in increased growth factor signaling (Sternlicht et al., 2006; Muschler and Streuli, 2010). Cryptic binding sites in the ECM may be unable to access their receptors under low tension conditions; however, under high tension, the ECM may expose these binding sites, which allows them to engage more integrin receptors and trigger enhanced downstream signaling (Vogel and Sheetz, 2006; Insua-Rodriguez and Oskarsson, 2016) (Figure 4). Collagen IV is an important component of the ECM in the mammary glands (Siu and Cheng, 2008). Therefore, we detected three genes related to the expression of collagen IV, including COL4A5, COL6A1, and COL6A5 (Khoshnoodi et al., 2008; Meuwissen et al., 2015). Two of these genes were significantly upregulated, which suggests that overexpression of GH in the mammary glands enhances mechanical stress and promotes growth factor activation (Figure 1F).
Goats' milk is highly nutritious, contains essential vitamins and minerals and is an ideal food for the whole family to enjoy. However, limited goat milk yield makes it expensive and not easy to get. To determine whether overexpression of GH could influence milk composition during mammary gland growth, we assessed several indicators of milk composition. Our results showed that milk production was significantly increased during the lactation period. This result was also supported by the mammary gland development observed by the HE staining. We found that more goat milk was visible in the acini of TG and the shape of epithelial cells in TG was more flat with abundant milk, which is inconsistent with the high milk yield in TG. There were no changes in milk fat or conductivity during our detection period. Moreover, the lactose content, protein content, density, milk solids and non-fat milk solids were increased on the first day of lactation. However, similar levels of these measures were observed between the two groups in the subsequent detection time periods. These results indicate that overexpression of GH increase milk production, but do not change the composition of milk. This phenomenon will be promising for the goat milk consumption.
Overall, our results showed that overexpression of GH stimulate mammary gland development. This process involves the upregulation of several growth factors, which activate mammary gland proliferation and branching. In addition, inhibition of mammary gland development was decreased. Moreover, mechanical stress was increased, which could improve the function of growth factors. Although overexpression of GH might alter mammary gland development, the milk composition remains stable.
Author contributions
ZB and JL: substantially contributed to the conception and implementation of the experiment; JC and QZ: raised the goats; ZB and LY: performed sample collection and analysis; QY and QY: contributed to the concept, design of the work and financial support.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Animal Transgenic Breeding Grant Project (No. 2014ZX08008-004 and 2016ZX08008002-005) and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2016.00278
References
- Crowley M. R., Bowtell D., Serra R. (2005). TGF-beta, c-Cbl, and PDGFR-alpha the in mammary stroma. Dev. Biol. 279, 58–72. 10.1016/j.ydbio.2004.11.034 [DOI] [PubMed] [Google Scholar]
- Ewan K. B., Shyamala G., Ravani S. A., Tang Y., Akhurst R., Wakefield L., et al. (2002). Latent transforming growth factor-beta activation in mammary gland: regulation by ovarian hormones affects ductal and alveolar proliferation. Am. J. Pathol. 160, 2081–2093. 10.1016/S0002-9440(10)61158-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata J. E., Mori H., Ewald A. J., Zhang H., Yao E., Werb Z., et al. (2007). The MAPK(ERK-1,2) pathway integrates distinct and antagonistic signals from TGFalpha and FGF7 in morphogenesis of mouse mammary epithelium. Dev. Biol. 306, 193–207. 10.1016/j.ydbio.2007.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner V., Mijic P., Baban M., Skrtic Z., Turalija A. (2015). The overall and fat composition of milk of various species. Mljekarstvo 65, 223–231. 10.15567/mljekarstvo.2015.0401 [DOI] [Google Scholar]
- Gjorevski N., Nelson C. M. (2011). Integrated morphodynamic signalling of the mammary gland. Nat. Rev. Mol. Cell Biol. 12, 581–593. 10.1038/nrm3168 [DOI] [PubMed] [Google Scholar]
- Insua-Rodriguez J., Oskarsson T. (2016). The extracellular matrix in breast cancer. Adv. Drug Deliv. Rev. 97, 41–55. 10.1016/j.addr.2015.12.017 [DOI] [PubMed] [Google Scholar]
- Khoshnoodi J., Pedchenko V., Hudson B. G. (2008). Mammalian collagen IV. Microsc. Res. Tech. 71, 357–370. 10.1002/jemt.20564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberg D. L., Feldman M., Ruan W. (2000). IGF-I: an essential factor in terminal end bud formation and ductal morphogenesis. J. Mammary Gland Biol. Neoplasia 5, 7–17. 10.1023/A:1009507030633 [DOI] [PubMed] [Google Scholar]
- Kumawat K., Gosens R. (2016). WNT-5A: signaling and functions in health and disease. Cell. Mol. Life Sci. 73, 567–587. 10.1007/s00018-015-2076-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias H., Hinck L. (2012). Mammary gland development. Wiley Interdiscipl. Rev. Dev. Biol. 1, 533–557. 10.1146/annurev-animal-031412-103632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias H., Moran A., Samara Y., Moreno M., Compton J. E., Harburg G., et al. (2011). SLIT/ROBO1 signaling suppresses mammary branching morphogenesis by limiting basal cell number. Dev. Cell 20, 827–840. 10.1016/j.devcel.2011.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen M. E. C., Halley D. J. J., Smit L. S., Lequin M. H., Cobben J. M., de Coo R., et al. (2015). The expanding phenotype of COL4A1 and COL4A2 mutations: clinical data on 13 newly identified families and a review of the literature. Genet. Med. 17, 843–853. 10.1038/gim.2014.210 [DOI] [PubMed] [Google Scholar]
- Meyer S. E., Zinser G. M., Stuart W. D., Pathrose P., Waltz S. E. (2009). The Ron receptor tyrosine kinase negatively regulates mammary gland branching morphogenesis. Dev. Biol. 333, 173–185. 10.1016/j.ydbio.2009.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molik E., Misztal T., Romanowicz K., Zieba D. A. (2010). The effects of melatonin on prolactin and growth hormone secretion in ewes under different photoperiods, during the early post partum period. Small Rumin. Res. 94, 137–141. 10.1016/j.smallrumres.2010.07.014 [DOI] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Muschler J., Streuli C. H. (2010). Cell-matrix interactions in mammary gland development and breast cancer. Cold Spring Harb. Perspect. Biol. 2:a003202. 10.1101/cshperspect.a003202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawathe A. R., Christian M., Kim S. H., Johnson M., Savvidou M. D., Terzidou V. (2016). Insulin-like growth factor axis in pregnancies affected by fetal growth disorders. Clin. Epigenetics 8:11. 10.1186/s13148-016-0178-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. M., Vanduijn M. M., Inman J. L., Fletcher D. A., Bissell M. J. (2006). Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 314, 298–300. 10.1126/science.1131000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez L., Ortiz-Delgado J. B., Manchado M. (2016). Molecular characterization and transcriptional regulation by GH and GnRH of insulin-like growth factors I and II in white seabream (Diplodus sargus). Gene 578, 251–262. 10.1016/j.gene.2015.12.030 [DOI] [PubMed] [Google Scholar]
- Pierce D. F., Jr., Johnson M. D., Matsui Y., Robinson S. D., Gold L. I., Purchio A. F., et al. (1993). Inhibition of mammary duct development but not alveolar outgrowth during pregnancy in transgenic mice expressing active TGF-beta 1. Genes Dev. 7, 2308–2317. 10.1101/gad.7.12a.2308 [DOI] [PubMed] [Google Scholar]
- Roarty K., Serra R. (2007). Wnt5a is required for proper mammary gland development and TGF-beta-mediated inhibition of ductal growth. Development 134, 3929–3939. 10.1242/dev.008250 [DOI] [PubMed] [Google Scholar]
- Shingu H., Hodate K., Kushibiki S., Ueda Y., Touno E., Shinoda M., et al. (2004). Hormonal and lactational responses to growth hormone-releasing hormone treatment in lactating Japanese Black cows. J. Dairy Sci. 87, 1684–1693. 10.3168/jds.S0022-0302(04)73321-4 [DOI] [PubMed] [Google Scholar]
- Siu M. K. Y., Cheng C. Y. (2008). Extracellular matrix and its role in Spermatogenesis. Mol. Mech. Spermatogenesis 636, 74–91. 10.1007/978-0-387-09597-4_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht M. D., Kouros-Mehr H., Lu P., Werb Z. (2006). Hormonal and local control of mammary branching morphogenesis. Differentiation 74, 365–381. 10.1111/j.1432-0436.2006.00105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternlicht M. D., Sunnarborg S. W., Kouros-Mehr H., Yu Y., Lee D. C., Werb Z. (2005). Mammary ductal morphogenesis requires paracrine activation of stromal EGFR via ADAM17-dependent shedding of epithelial amphiregulin. Development 132, 3923–3933. 10.1242/dev.01966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzalkowska N., Jozwik A., Bagnicka E., Polawska E., Krzyzewski J., Pyzel B., et al. (2012). Fatty acid profiles, cholesterol concentration, and susceptibility to lipolitic fat fraction in goat's milk. Med. Weter. 68, 40–44. [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V., Sheetz M. (2006). Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7:265–275. 10.1038/nrm1890 [DOI] [PubMed] [Google Scholar]
- Watson C. J., Khaled W. T. (2008). Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development 135, 995–1003. 10.1242/dev.005439 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Chen J. Q., Lin J., Yu Q. H., Yu H. Q., Xu X. J., et al. (2014). Production GH transgenic goat improving mammogenesis by somatic cell nuclear transfer. Mol. Biol. Rep. 41, 4759–4768. 10.1007/s11033-014-3347-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.