Abstract
Many somatic mutations have been detected in pancreatic ductal adenocarcinoma (PDAC), leading to the identification of some key drivers of disease progression, but the involvement of large genomic rearrangements has often been overlooked. In this study, we performed mate pair sequencing (MPseq) on genomic DNA from 24 PDAC tumors, including 15 laser-captured microdissected PDAC and 9 patient-derived xenografts, to identify genome-wide rearrangements. Large genomic rearrangements with intragenic breakpoints altering key regulatory genes involved in PDAC progression were detected in all tumors. SMAD4, ZNF521, and FHIT were among the most frequently hit genes. Conversely, commonly reported genes with copy number gains, including MYC and GATA6, were frequently observed in the absence of direct intragenic breakpoints, suggesting a requirement for sustaining oncogenic function during PDAC progression. Integration of data from MPseq, exome sequencing, and transcriptome analysis of primary PDAC cases identified limited overlap in genes affected by both rearrangements and point mutations. However, significant overlap was observed in major PDAC-associated signaling pathways, with all PDAC exhibiting reduced SMAD4 expression, reduced SMAD-dependent TGFβ signaling, and increased WNT and Hedgehog signaling. The frequent loss of SMAD4 and FHIT due to genomic rearrangements strongly implicates these genes as key drivers of PDAC, thus highlighting the strengths of an integrated genomic and transcriptomic approach for identifying mechanisms underlying disease initiation and progression.
Introduction
Pancreatic cancer remains the fourth leading cause of cancer-associated mortality in the United States (1). While prognosis has improved for other major cancers due to early diagnosis, better therapeutic management strategies, and a more comprehensive knowledge of genetic factors, death rates from pancreatic cancer continue to rise. The majority (90%) of pancreatic cancers are ductal pancreatic adenocarcinomas (PDAC) that present generally in the seventh decade of life (2). Only 6% of patients survive five years postdiagnosis. Currently, only15% to 20% of pancreatic cancers are diagnosed early enough to benefit from surgical resection, with the majority of tumors having already spread to the surrounding tissues or distant organs (3).
Despite the advent of high-throughput genomic sequencing techniques, few major advances have been made in understanding the mechanisms by which pancreatic cancer progresses to invasive tumors. In the years since being identified, KRAS and TP53 remain the best-defined driver genes in the majority of tumors studied (4). Activating mutations in KRAS is one of the earliest gene alterations associated with PDAC development, followed by inactivation of CDKN2A and disruption of TP53 and SMAD4 at later stages (4–5). However, beyond these four major drivers, the broad array of other mutated genes reflects the significant heterogeneity within these tumors (6–8).
Further studies are needed to better understand the fundamental alterations that occur in PDACs, thereby leading to better diagnostic and therapeutic management. To date, the majority of genomic analyses have focused on evaluation of potential inactivating point mutations and small insertions/deletions (Indels; refs. 6–8). Large genomic rearrangements have, however, become increasingly evident as key mutagenic events in the progression of solid tumors (9–12). While recent studies have highlighted the major involvement of copy number gains and losses in key cancer driver genes (12–17), the contribution of genomic rearrangements to pancreatic tumors is not well defined. We report here an in-depth analysis of genomic rearrangements present in 24 PDAC tumors from 23 patients and contrast the results with both point mutation and transcriptome (RNA-Seq) data from subgroups of tumors.
Materials and Methods
Primary PDAC DNA/RNA isolation and sequencing
The Mayo Clinic Specialized Program of Research Excellence (SPORE) in Pancreatic Cancer identified 14 clinically and histologically confirmed PDAC patients, who provided consent for use of tissue for research, and for whom frozen PDAC and adjacent pancreatic intraepithelial neoplasia (PanIN) tissues were available. LCM was used to individually isolate tumor, PanIN, or histologically normal cells from fresh frozen tissue sections and DNA was amplified directly by a single-step procedure using the Qiagen Repli-g WGA kit, as previously described (8–11). Mate pair sequencing (MPseq) libraries were assembled from WGA DNA, as previously published (9–11) using the Illumina MP kit. Whole Exome Sequencing (WES) was performed on indexed paired-end libraries (NEB Next DNA Kit) and Agilent SureSelect Human All Exon 50 Mb kit (Agilent) as previously reported (8). RNA was isolated from separate LCM-captured cells (10 frozen sections) using Qiagen RNeasy mini-kit and established protocol. mRNA (2–10 ng, RIN>6) was amplified using NuGEN Ovation RNA-seq v2 mRNA amplification/cDNA generation kit, before library preparation using Ovation Ultralow DR Multiplex System. Indexed libraries were sequenced on the Illumina HiSeq platform 101bp paired-end reads at 2, 3, or 4 libraries per lane for MPseq, WES, and RNA-Seq, respectively.
Patient-derived xenografts of PDAC tumors
An additional nine patient-derived xenografts (PDX) of surgically resected PDAC tumors propagated in immunodeficient NOD/SCID mice (18) were available for this study, from which DNA was isolated using the Qiagen Blood and Tissue Kit (#69504, Qiagen). PDX tumors were confirmed of pancreatic origin by pathologist histologic review. MP libraries were prepared from 1 µg DNA using Illumina Nextera reagents and sequencing was performed as described above.
Data analysis
MPseq
Paired reads were mapped to the human Hg19 reference genome using 32-bit binary indexing of the genome as previously described (9–11, 19). Discordant mate pairs reads mapping >15 kb apart or in different chromosomes were selected for further analysis. A mask was used to eliminate common variants and discordant MPs from experimental or algorithmic errors (19).
WES
Samples were compared to corresponding "normal" samples using SomaticSniper14 for somatic SNVs or GATK's Somatic INDEL Detector15 as previously described (9). A minimum somatic score of 20 and >8× coverage was required in reference normal sample. Somatic variants with <30× read depth ≥3 alternate reads supporting the variant call. For variants at ≥30×, alternate alleles exceeding 4% of all reads were required.
RNA-Seq
RNA-Seq reads were aligned using TopHat. Gene counts were assessed by HTSeq and normalized using conditional quantile, with differential expression analysis conducted with edgeR (20).
Validation of genomic rearrangements and KRAS Sanger sequencing
Primers spanning detected fusion junctions were used in PCR validations on tumor DNA, associated PanINs, adjacent normal (NL), and an independent human Genomic DNA control (C; G304A; Promega) using the EasyA high fidelity polymerase (Stratagene; #600404). Sanger sequencing was utilized to determine the G12 KRAS mutation status of each case from PCR amplicons derived using primers GGACCCTGACATACTCCCAAGGAAAG and GGTGAGTTTGTATTAAAAGGTACTGGTGGAG. TP53 mutations were assessed using a non-published capture approach.
Results
Genomic rearrangements in PDAC
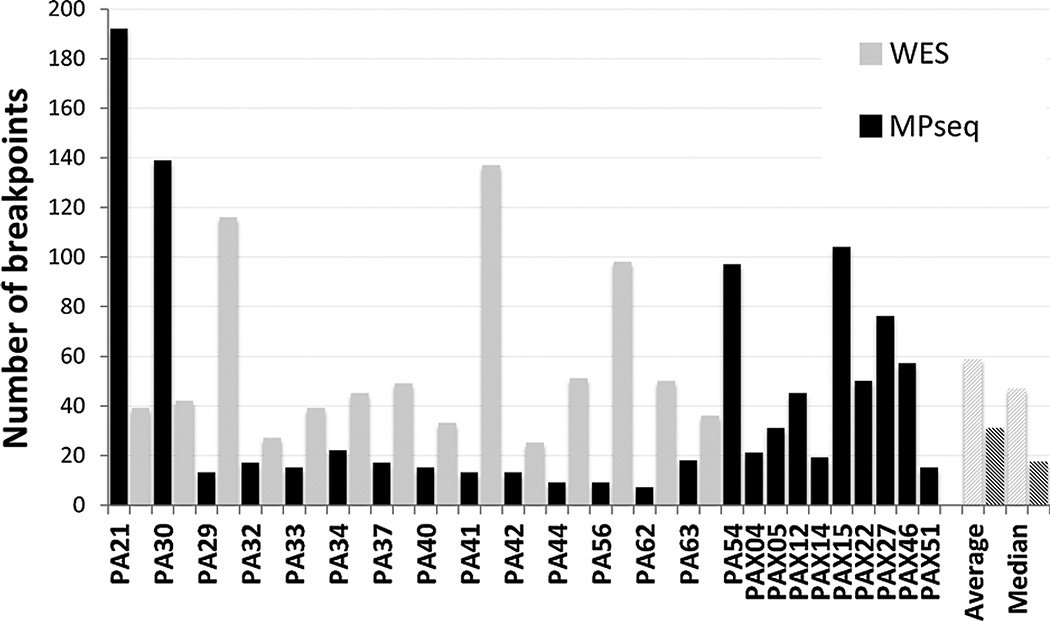
A total of 24 PDAC tumors were selected for structural variant analysis including 15 primary PDAC tumors, isolated through laser capture microdissection (LCM) of fresh frozen resected tissues, and nine independent bulk-extracted PDX-propagated tumors from surgically resected primary PDAC tumors (18). MP sequencing on the 24 PDAC tumors generated an average of 89 million reads per tumor. Average base pair and bridged (large fragment span) coverage were 4 and 36×, respectively (Supplementary Table S1). A total of 908 unique genomic breakpoints were detected. Numbers of detected breakpoints varied from 7 to 192 per tumor (Fig. 1), with an average of 39 and median of 18 breakpoints. Identical breakpoints were only detected between the PA21 and PA30 tumors that originated from the same patient. Of the detected breakpoints, 19% were translocations between different chromosomes, while the remaining 81% were intrachromosomal deletions, amplifications, and inversions. The majority of rearrangements (71%) had one breakpoint in a gene locus, potentially affecting expression through disruption of coding regions. Both breakpoints were located in different genes in 205 (20%) of the rearrangements, with 111 in the correct orientation for potential fusion gene products. However, only a small number of these would be expected to generate expressed fusion gene products. WES data was available for all primary tumors except PA54. A total of 760 somatic mutations altering protein-coding sequences were detected, with an average and median of 58 and 49 mutations per case, respectively. Figure 1 presents the total numbers of somatic variations detected by MPseq and WES.
Figure 1.
Mutations in PDAC tumors. Numbers of mutations detected by mate pair (MPseq; black bars) and WES (gray bars) per tumor. Average and median values across all cases represented by hatched bars.
Mutated genes in PDAC
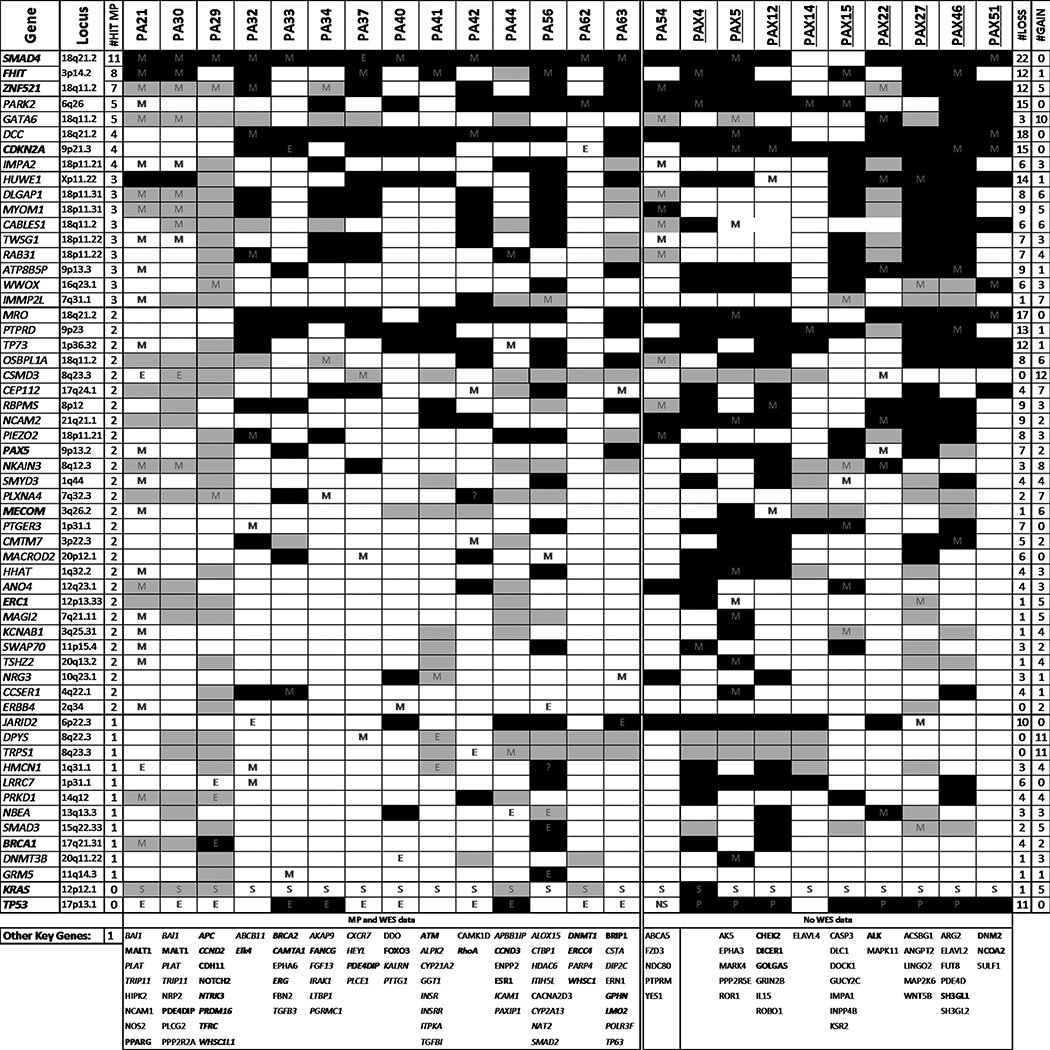
In total, 746 different genes contained intragenic rearrangements in the 24 sequenced tumors. Together with the WES data, 1,387 genes were affected, with only 40 genes involving both rearrangements and SNVs/Indels (Supplementary Table S2). The key genes somatically mutated in this study, selected as genes altered in multiple tumors or with previously reported involvement in cancer, are presented in Fig. 2. As expected, KRAS and TP53 were the most frequently mutated driver genes, with somatic variations detected in 100% and 74% of tumors, respectively. KRAS G12V and G12D mutations were the most prominent, in ten and seven tumors, respectively (Supplementary Fig. S1). Somatic mutations in COSMIC census cancer driver genes (http://cancer.sanger.ac.uk/census) were detected in all tumors. From MPseq data alone, 23 (96%) tumors detected a rearrangement hitting directly with an intragenic breakpoint in at least one COSMIC census cancer driver gene (Supplementary Table S2), with an average of two per case.
Figure 2.
Commonly mutated genes in PDAC. Genes selected according to presence in multiple cases or previously reported association with cancer. Mutation types detected by M, MPseq; E, WES; S, Sanger; or P, TP53 panel are indicated. Copy losses and gains at specific gene loci are marked by black or gray shading, respectively. The gene locus, the number of tumors hit directly by breakpoints (#Hit MP) and the number of cases involving loss (#LOSS) and gain (#GAIN) are listed for each commonly hit gene. Genes listed in COSMIC cancer gene censor are in bold text. Other key single hit genes in the study detected by MP or WES (italics) are presented at the bottom of the table for each case. Tumors evaluated by MP and WES data, or from MP alone, in the absence of WES data, are also indicated.
While focal intragenic breakpoints within a gene region are a direct indicator of altered gene expression, loss of a gene can occur when it lies within a larger chromosomal deletion. Conversely, gene gains can occur when contained within larger chromosomal region duplications or amplifications. Frequency coverage of concordant mapping reads across chromosomes from the MPseq data (19) were used to assess chromosomal gains and losses. In addition to rearrangements present within a gene locus, Fig. 2 additionally integrates data for cases where loss or gain is predicted.
SMAD4 mutations
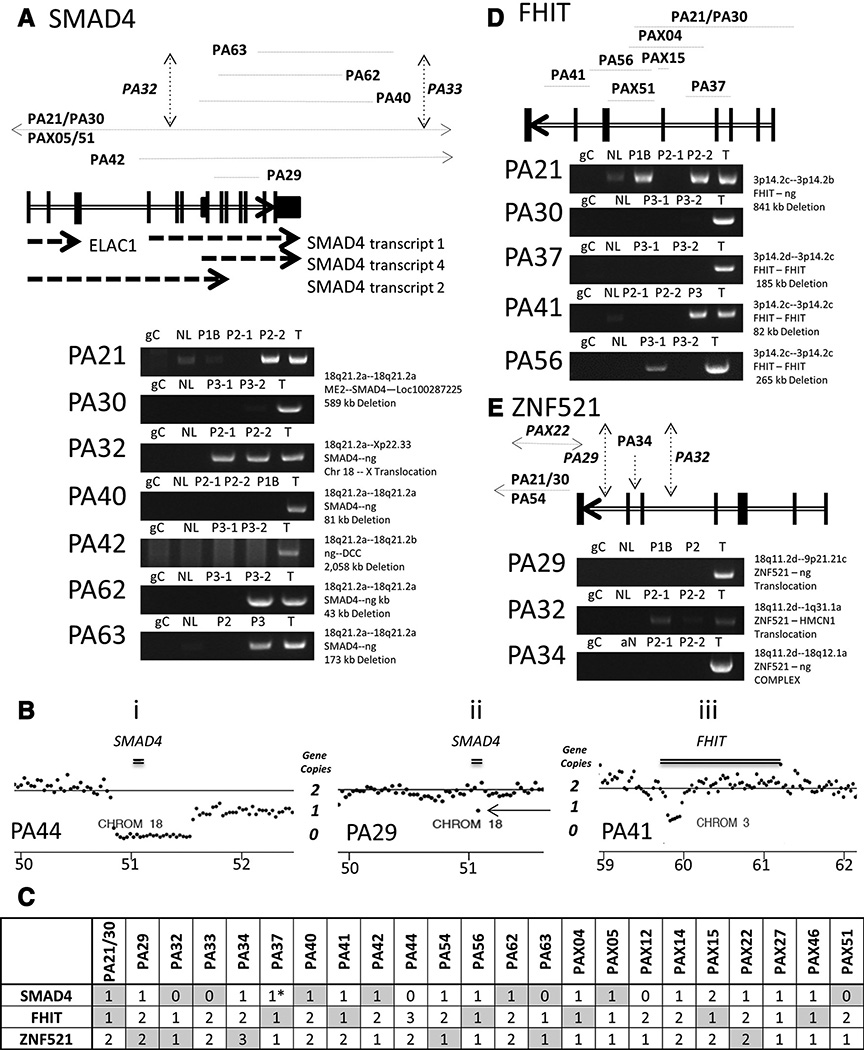
SMAD4 was most frequently altered by genomic rearrangements with eleven tumors (46%) containing breakpoints within the SMAD4 gene locus. Intrachromosomal deletions ranged from ~25 to 2,000 kb in eight cases, with two additional cases (PA32 and PA33) presenting with interchromosomal translocations at the SMAD4 locus (Fig. 3A). Events were validated by PCR in seven selected tumors for which DNA was additionally available from laser capture microdissected PanIN located adjacent to the tumors (Fig. 3A; ref. 8). Three of the SMAD4 rearrangements were restricted to tumors, whereas three were also detected in adjacent PanIN 2 lesions and two were observed in adjacent PanIN 3 lesions.
Figure 3.
Deletions at the SMAD4/FHIT/ZNF521 loci. Schematics of predicted breakpoints and deletions, together with PCR validations at the SMAD4 18q21.2a (A), FHIT 3p14.2c (D), and ZNF521 18q11.2d gene loci (E). Exons (vertical black lines) are marked on each gene (double lined arrows), with breakpoints (vertical black dashed-arrows) or deletion spans (horizontal black dotted-lines) marked above. PCR validation gels presented with labeling of tumors (T), PanINs 1B, 2, or 3 (P1B, P2 or P3), adjacent normal (NL), and a mixed population human genomic DNA control (gC). B, examples of copy number loss for SMAD4 in PA44 (i) and PA29 (ii) and for FHIT in PA41 (iii). Central gray lines indicate 2-gene copy levels from normalized values across the whole genome. Lower black base line indicates the hg38 chromosomal coordinates (Mb). Black dots mark the frequency of coverage across 30 kb windows. The SMAD4 and FHIT genes positions are presented as horizontal double black lines. C, heterozygosity in each case for SMAD4, FHIT, and ZNF521 is summarized as 3, 2, 1, or 0 copies. The gray shaded boxes indicate cases where large genomic rearrangements were detected. *, case where SMAD4 gene mutated by SNV.
Frequency coverage data confirmed the loss of genomic sequence at the SMAD4 locus in all directly hit tumors (Fig. 3B and Supplementary Figs. S2A and S2E) but demonstrated additional losses in the majority of other cases. In total at least one allelic copy of SMAD4 was lost in 22 (92%) of the 24 tumors. The remaining two cases, however, lacked integrated SNV data for SMAD4. The absence of discordant mapping MPseq reads predicting many of these events were due to larger q-arm deletions of chromosome 18 with no detectable fusion junctions (Supplementary Fig. S2B). In total, large chr18q deletions were observed in 18 (67%) tumors, and were often in combination with predicted smaller focal SMAD4 deletions in the second allele. Homozygous loss was predicted in six cases (Fig. 3C), with an additional case PA37, containing a validated damaging Q1348S mutation (8) in the remaining allele. In addition to the larger deletions, PA29 predicted a smaller approximately 25 kb microdeletion across the SMAD4 locus (arrow; Fig. 3B, ii), below the standard 30 kb bioinformatics filter applied in this analysis. No further microdeletions were detected at the SMAD4 locus in additional cases.
FHIT mutations
The fragile histidine triad gene, FHIT, was the next most frequently rearranged gene, with focal deletions observed in eight tumors (Fig. 2). A large 841 kb deletion and loss of the first five coding exons was detected in PA21/30. Deletions of exons 4 and 7 were detected in PA37 and PA41, respectively. A 265 kb deletion including exon 6 was present in PA56. Breakpoints were validated by PCR in tumors with associated PanINs (Fig. 3D) and frequency coverage data supported these large deletions (Fig. 3B, iii and Supplementary Fig. S2C and S2E). The rearrangements in PA21/30 and PA37 validated solely in tumors (Fig. 3D), whereas those in PA56 and PA41, also validated in adjacent PanINs. Four additional cases (PA32, PA63, PAX15, and PAX27) presented with additional FHIT losses through larger chromosome 3 deletions. Thus, in total, the FHIT gene was deleted in 12 (50%) tumors. No point mutations were detected in the FHIT gene from the WES analysis.
ZNF521 mutations
Rearrangements hitting ZNF521, a zinc finger protein, were present in seven tumors (Fig. 2E). Rather than focal deletions, these events were more complex. Two cases, PA29 and PA32, presented with interchromosomal translocations to chromosomes 1 and 9, respectively. The remaining five cases involved more complex multiple intrachromosomal rearrangements on 18q11 where ZNF521 is located. Frequency coverage at the ZNF521 locus was also not consistent for these events, with frequency coverage data predicting both gains and losses in these seven tumors. Eight additional cases presented with ZNF521 loss without detected rearrangements (Supplementary Fig. S2D and S2E). In total 15 (63%) tumors were hit by rearrangements or chromosomal loss at the ZNF521 locus.
Additional mutated genes
A variety of cancer-related genes were hit directly with intragenic breakpoints in multiple tumors. Integrating additional frequency coverage data increased the numbers of affected cases for the majority of genes (Fig. 2). CDKN2A, heavily linked with PDAC progression, had predicted loss-of-function in a total of 15 (63%) tumors, but just four tumors had intragenic breakpoints within the gene (Fig. 2). Nine tumors had larger chromosomal deletions, spanning CDKN2A, with two additional cases hit by damaging point mutations. In addition to FHIT, three other fragile site genes, PARK2, WWOX, and IMMP2L had intragenic breakpoints in three or more tumors, with additional larger losses observed in additional cases.
A significant number of commonly rearranged genes were located on chromosome 18, focused in three major hotspots (Fig. 2). DCC and MRO are located on 18q21.2 adjacent to SMAD4, whereas OSBPL1A, GATA6, and CABBLES1 are located on 18q11.2 adjacent to ZNF521. The third hotspot was located on the p-arm of chromosome 18 (18p11.31-21) and included, IMPA2, RAB31, TWIGS1, PIEZO2, DLGAP1, and MYOM1. Additional potentially significant cancer genes with intragenic breakpoints detected in multiple tumors included HUWE1, PTPRD, TP73, PAX5, MARCOD2, CSMD3, ERBB4, ERC1, and MECOM. Just five genes were mutated in both the MPseq and WES data in the same tumor, with potential homozygous loss of function. These were EPO, ME2, MEGF6, NPAS3, and STXBP5L (Supplementary Table S2). An additional 38 genes were mutated in both the MPseq and WES data in different tumors including SMAD4, SMAD3, BRCA1, ERBB4, CSMD1, DNMT3B, PRKD1, and FOXN1. Many significant cancer genes were also observed mutated solely in individual cases, including ATM, APC, BRCA2, DNMT1, ALK, FOXO3, NCAM1, SMAD2, PPARG, and NOTCH2 have been associated with cancer initiation and progression.
Common gene losses and gains in pancreatic cancer
Genes hit directly by intragenic breakpoints from large rearrangements or point mutations infer stronger evidence for altered gene expressions. However, many genes are lost or gained through larger chromosomal rearrangements, without intragenic breakpoints. While SMAD4, FHIT, and ZNF521 are mainly affected by direct intragenic rearrangements, other genes listed in Fig. 2 are affected more by copy number changes. Specifically, while hit by intragenic breakpoints in at least two cases, PARK2, DCC, CDKN2A, HUWE1, MRO, PTPRD, and TP73 are more commonly deleted, and GATA6, OSBPL1A, DPYS, and TRPS1 more commonly gained; through more distal events. As expected many of these genes cluster together by location, such as SMAD4, DCC, and MRO at 18q21.2, with co-gains/losses in cases. However, interestingly adjacent genes GATA6 and ZNF521, at 18q11.2, have quite distinct patterns of gain and loss. Similar numbers of both gains and losses are observed for many genes, potentially indicative of passenger events.
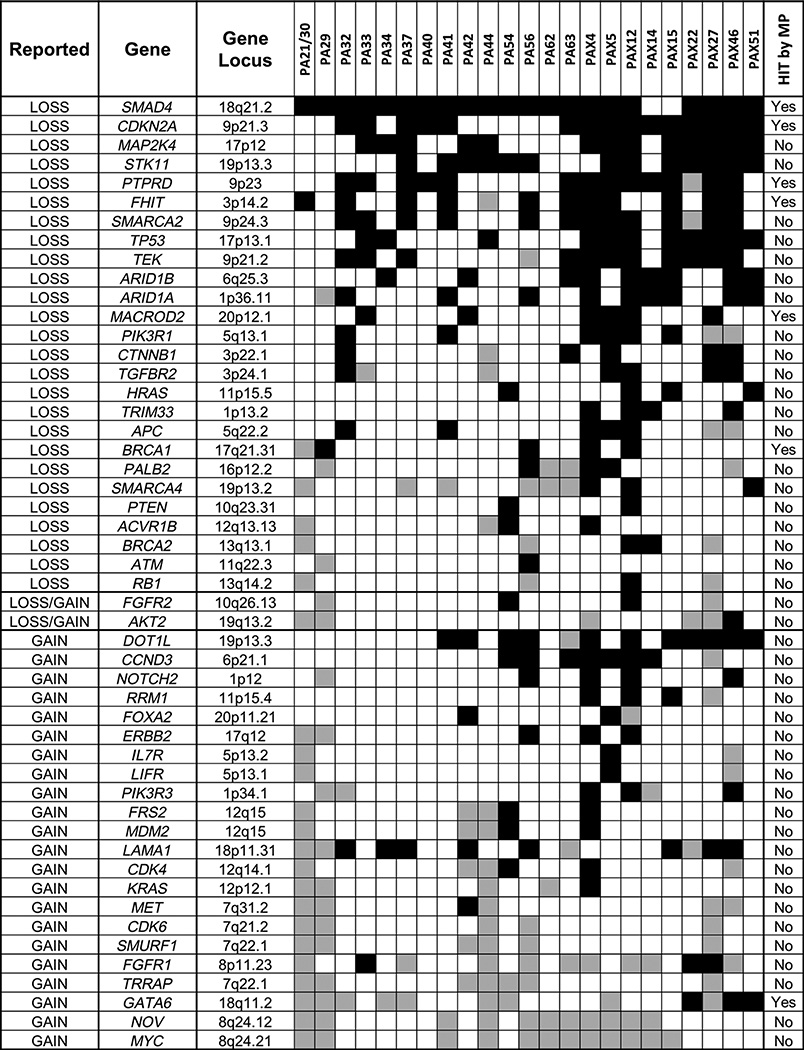
A number of commonly reported gained or lost genes in pancreatic cancers were not hit directly by intragenic breakpoints in this study. We therefore investigated the copy number levels of a panel of 50 commonly reported gained/lost genes in PDAC from literature (Fig. 4; refs. 6–7, 12–17, 21–23). In addition to the genes previously reported in Fig. 2, recurrent losses were observed for MAP2K4, STK11, SMARCA2, TEK, ARID1A and ARID1B. Conversely, MYC, NOV, TRRAP, and FGFR1 were commonly gained. As expected, the well-reported MYC oncogene (15–17) was consistently gained in 12 cases. NOV, adjacent to MYC at 8q24 was also cogained without intragenic breakpoints in 11 of these cases. TRRAP and SMURF1 on chromosome 7q22.1 were also consistently gained in seven and six cases, respectively. Reported gains of GATA6 and FGFR1 were also each prevalent in nine cases (15–17, 21); however, additional three cases of loss were observed for each gene. Significantly, loss of ARID1A and ARID1B, together with SMARCA2, document the reported impact on the SWI/SNF pathway (21–22). MAP2K4, STK11, SMARCA2, and TEK were also confirmed frequently lost without direct intragenic breakpoints, as was TP53 in this study. Mutations of TP53 in 79% of cases were complemented with additional copy losses, predicting homozygous loss of function in at least nine cases. Interestingly, loss of TP73 was observed in 12 cases, five of which presented with no evidence of TP53 loss.
Figure 4.
Copy number levels of commonly gained and lost genes in PDAC. Fifty genes reported from literature (6–7, 12–17, 21–23) with recurrent gains or losses of copy numbers are presented detailing loss (black shading) or gain (gray shading) for each case in this study. The gene locus cytoband and whether the gene is additionally hit directly by a breakpoint in our data set also presented. Genes are ordered according to prevalence of gains or losses in cases.
Genes observed mutated in this dataset were also contrasted with mutations reported in other pancreatic cancer genomic studies, including The Cancer Genome Atlas (24), the International Cancer Genome Consortium (7) and the John Hopkins group (Supplementary Table S3; ref. 6). While this study confirms the high occurrence of somatic variation reported in literature for a subset of key driver genes involved in PDAC, it also highlights the heterogeneity and potential passenger or auxiliary functions of the majority of reported mutated genes.
Evaluation of SMAD4, FHIT, and ZNF521 RNA expression
The influence of somatic mutations in SMAD4, FHIT, and ZNF521 on gene expression levels were assessed using RNA-Seq data from nine of the tumors. All tumors presented with reduced SMAD4 expression, with an average 3.1-fold reduction compared with levels in matched normal ductal epithelial cells (Nd; Fig. 5A). The level of SMAD4 expression loss was consistent with the degree of SMAD4 deletion. FHIT expression levels were also reduced in every tumor, with an average 5.7-fold reduction (Fig. 5B). Reduction in FHIT expression in cases without predicted deletions and also in PA44 where a gain of FHIT gene sequence was predicted (Fig. 3C) suggests that other regulatory factors contributed to the reduced expression of these genes. For ZNF521, an overall trend of reduced expression in tumors was also observed (Fig. 5C). Overall 40%of the commonly hit genes had greater than 2-fold change in average expression compared with normal (Supplementary Fig. S3), supporting potential mechanistic roles in tumor progression.
Figure 5.
RNA expression of SMAD4, FHIT, and ZNF521. RNA-Seq coverage levels are presented for SMAD4 (A), FHIT (B), and ZNF521 (C) on the whole gene level for each normal duct (Nd; black bars) and tumor (T; gray bars) sample per case. The y-axis represent reads per kilobase per million. The median (med) values are also presented for each gene (hatched bars). Cases predicted with loss or gain of copy number by DNA mutation analysis are exemplified by * or Δ, respectively.
Pathways influenced by rearrangements and SNV/Indels
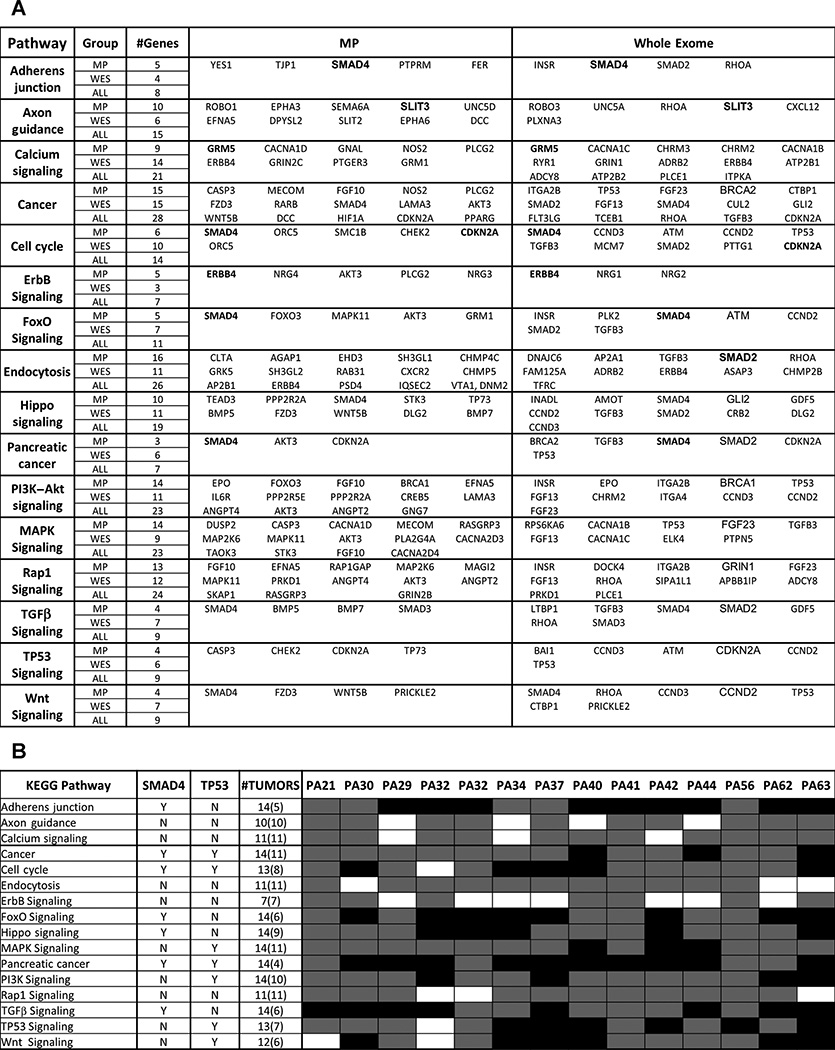
The limited overlap in genes containing rearrangements and point mutations/small indels raised the possibility that the different types of mutations were associated with independent tumor development pathways. However, analysis of protein–protein interactions (PPI) and KEGG pathways (25) influenced by WES and/or rearrangements identified 16 main pathways (Fig. 6A) with mutations evenly distributed between large genomic rearrangements and SNVs. While the high incidence of TP53 and SMAD4 mutations influenced the pathway analyses, many additional genes in the selected pathways were modified in these tumors. The number of tumors affected by each pathway is highlighted (Fig. 6B). Pathways in cancer, cell cycle, and TP53 signaling were all affected by rearrangements and SNVs. Adherens junction, tight junction, endocytosis and axon guidance as well a number of major cell signaling pathways including TGFβ, Wnt, Hippo, PI3K-Akt, MAPK, and ErbB signaling pathways were similarly affected by both mutation types across all tumors.
Figure 6.
Significant pathways in PDAC. A, sixteen pathways significantly affected in our data set. Columns indicate NGS group, number of genes mutated in each pathway in the mate pair (MPseq), whole exome (WES), or both (ALL) datasets. Specific genes in each pathway detected in MPseq or WES data are presented. B, distribution of affected cases for each pathway. The number of affected tumors and the involvement of TP53 and SMAD4 in a specific pathway are indicated. Gray shading indicates a case is affected through mutations in genes in addition to SMAD4 or TP53, while black shading indicates cases affected solely through SMAD4 or TP53.
Signaling networks in PDAC
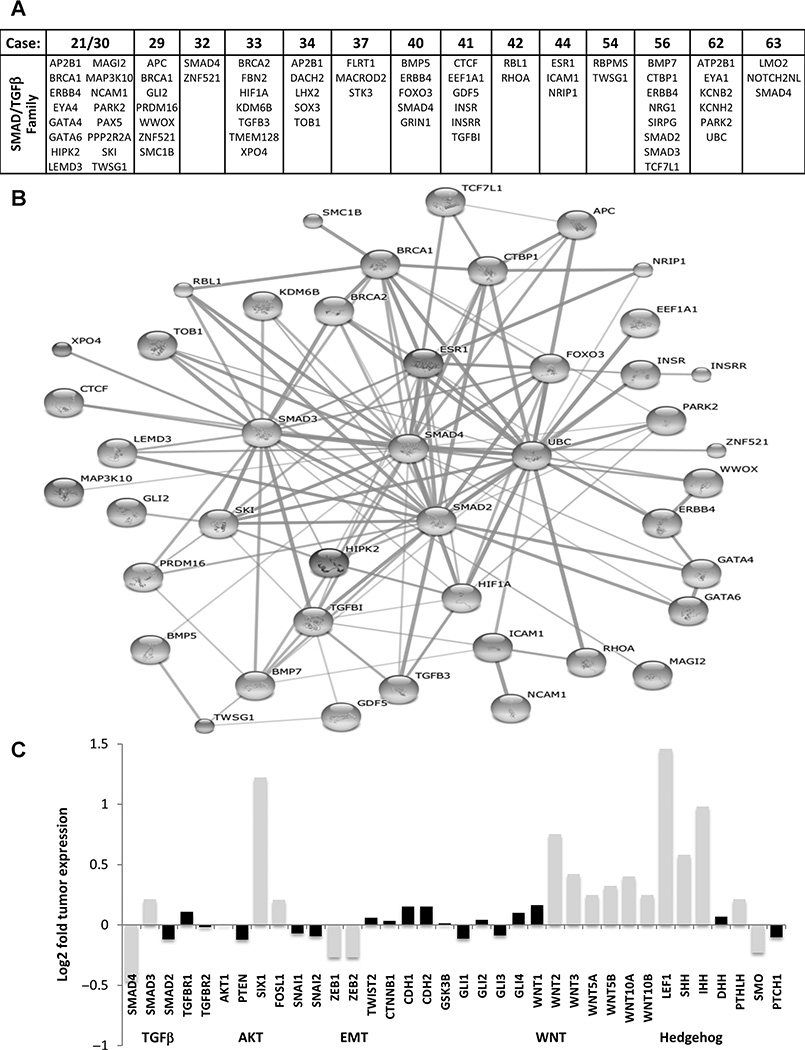
Next, we investigated whether other genes associated with SMAD4 function/binding and signaling pathways were also mutated in the PDACs. Significantly, all tumors had alterations in several genes implicated in SMAD function (Fig. 7A). Figure 7B illustrates a network of these genes visualizing predicted PPI using the STRING database v9.1 (25). In addition, the effect of SMAD4 deletion on TGFβ signaling was reflected in reduced mRNA levels of 27 known downstream target genes encompassing SMAD-response-elements (SRE; refs. 26–27). Approximately 67% of these 27 genes showed reduced levels of expression and just 15% retained significant increased expression (Supplementary Fig. S4). Interestingly, expression levels of TGFβ receptors and other SMAD genes did not change significantly (Fig. 7C), suggesting that signaling occurred through SMAD-independent pathways, such as PI3K/AKT (28–29). While PI3K/AKT levels did not change significantly, levels of FOSL1, a downstream effector of the PI3K/AKT signaling pathway were increased, indicating increased PI3K/AKT activity. In addition, Six1, linked to a SMAD-independent TGFβ activation pathways and EMT, was highly overexpressed (30). However, other markers of EMT including β-catenin, E-cadherin, N-cadherin, SNAIL1/2, and TWIST1/2 did not display EMT-associated altered expression in the PDACs.
Figure 7.
SMAD4 pathway genes and signaling networks. A, somatically mutated genes with binding or pathway related functions with SMAD4/TGFβ signaling are presented per case. B, a network of the listed genes visualizing predicted protein–protein interactions (PPI) using the STRING database v9.1. C, median mRNA expression levels for specific genes represented as Log2 of median tumor levels divided by median dN levels. Black shaded bars indicate those genes where expression was gained or decreased less than fifty percent and thus deemed an insignificant change in expression.
Several genes associated with FHIT function and DNA repair mechanisms were also mutated in all tumors (Supplementary Fig. S5). FHIT expression has been reported to suppress proliferation and promote apoptosis by blocking the PI3K–Akt pathway (31). Thus, loss of SMAD4 and FHIT could synergistically contribute to increased PI3K–AKT pathway activity.
As expected from the mutation-based pathway analysis, expression of WNT signaling genes was increased, with substantial overexpression of WNT 2, 3, 5, and 10 isoforms (Fig. 7C). LEF1, a major transcription factor of the Wnt pathway, driving cell migration, invasion, and proliferation, was very highly expressed in all cancers in the absence of increased β-catenin. Members of the Hedgehog gene family, Sonic and Indian Hedgehog, were also highly overexpressed as previously reported for pancreatic cancer (32–33). However, this signaling pathway was not highlighted by mutations or rearrangements (Fig. 6).
Discussion
MP sequencing of PDACs demonstrated that, in addition to point mutations, driver genes and pathways of PDAC are commonly affected by direct intragenic breakpoints during large genomic rearrangement events. The overlap in genes mutated by rearrangement and point mutation was limited, consistent with the well-reported intertumor heterogeneity in pancreatic cancer (6–8). As expected a high frequency of KRAS (100% cases) and TP53 (74%) mutations were observed and other than common KRAS-activating mutations, recurrent somatic mutations were not observed. Similar to studies of SNV in PDACs (6–8), few genes were frequently rearranged. In addition to rearrangements in the commonly reported SMAD4, FHIT, WWOX, and CDKN2A genes, rearrangements with intragenic breakpoints directly hitting a gene in greater than 10% of cases, included; ZNF521, PARK2, GATA6, DCC, CSMD3, IMPA2, CABLES1, RAB31, and HUWE1.
The majority of CNV studies in PDAC center on gains and losses from array-based techniques (13–17). In this study, MPseq allowed us to uniquely focus on genes hit with direct intragenic breakpoints through large genomic rearrangements, presenting strong evidence for altered or lost gene function. While the commonality in mutated genes was limited, basic KEGG pathway analysis performed independently on MPseq and WES datasets revealed extensive overlap in the affected pathways. Hence, while different genes may have different susceptibilities to point mutations or rearrangements, both mutational mechanisms have an impact on the same key regulatory pathways. This observation supports the hypothesis that altered expression of a selection of genes hit by intragenic breakpoints through large genomic rearrangements does contribute to PDAC progression similarly to SNV/Indels. In addition, while many rearrangements are predicted to be early events in tumor progression, our results confirm that rearrangements can affect key tumor genes late in the process of tumor development.
Significantly, many well-reported key regulatory genes with altered expressions by gains and losses, did not involve intragenic breakpoints. As expected, the well-reported MYC oncogene was confirmed consistently gained (15–17), being observed in 12 cases, but no intragenic MYC breakpoints were observed in tumors. The absence of intra-genic breakpoints may emphasize the need to retain oncogenic functions, which can be predicted to be more often lost by rearrangements directly hitting a gene, as is the case for tumor suppressor genes; SMAD4, CDKN2A, and FHIT. Thus, direct intragenic hits on genes with oncogenic functions in cancer may often negatively impact tumor growth and thus are not selected for. Interestingly selective patterns of gains or losses, evident of more driver functions, were only observed for a limited number of genes, further emphasizing the inherent heterogeneity, with many affected genes passengers in PDAC progression. Similar to other studies GATA6 gains were prevalent in this study (15–17, 21). However, additional cases with GATA6 loss were evident, as were cases with direct intragenic breakpoints in combination with gains. Loss of SWI/SNF pathway genes were also common in this dataset (21–22); however, interestingly, no direct intragenic breakpoints were detected in ARID1A, ARID1B, or SMARCA2. Loss of TP73, a tumor suppressor gene in the TP53 family of transcription factors was also prevalent in this dataset, being observed in 12 cases, five of which had no evidence of loss of TP53 function. While not directly linked to PDAC, the TP73 pathway has been linked to therapy for TP53-deficient PDACs (34). Further implicating the TP53 family in PDAC, one case also presented with a potentially damaging D157N mutation in TP63.
The previously reported role of TGFβ signaling in PDAC (21) was well defined, with all tumors presenting with direct somatic mutations in genes in the pathway and concomitant reduced expression of SMAD-targeted genes. Functionally, reduced SMAD signaling may result in SMAD-independent induction of PI3K/AKT pathway activity similarly to induction of AKT signaling in the absence of the PTEN tumor suppressor (28–29, 35). High levels of SIX1 (Fig. 5C) were interesting considering a reported role for SIX1 in SMAD-independent TGFβ activation through induction of cyclin D1 and EMT (30, 36–37). However, upregulation ofCCND1and induction of EMT expression profiles were not observed. Joint activation of WNT and HH signaling by TGFβ is a conserved feature of TGFβ signaling in EMT (38). In line with current clinical targeting strategies towards WNT/HH pathways for PDAC (39–40), both pathways were highly active nodes in PDAC in the current study. However, in contrast to WNT pathway genes, few HH pathway genes were directly mutated in our dataset.
SMAD4 loss was observed at a level similar to KRAS-activating mutations. In addition, to underscoring the high incidence of SMAD4 loss in PDAC, this integrated study enabled a detailed picture of the varied structural rearrangements that drive this loss. The diverse nature of deletions predicted at least a single copy loss of SMAD4 in 92% of cases, with six cases predicting progressive homozygous loss-of-function. Mutation of SMAD4 has been commonly reported in 50% of PDAC (41). However, without complete integrated datasets of somatic breakpoints, SNV/Indel, and CNV, alterations may have been overlooked in many prior studies assessing the contribution of SMAD4 to pancreatic cancer. Loss of SMAD4 was paralleled with reduced SMAD4 expression in each case studied. Importantly, a number of studies have reported that SMAD4 hemozygosity is sufficient to significantly inhibit both TGFβ and BMP signal transduction and result in the differential expression of a broad subset of target genes likely to underlie tumor formation (42–44). Thus, the loss of at least one SMAD4 allele in 92% of tumors and the associated reduced RNA-Seq expression of SMAD4 in all PDAC tumors studied; suggest that SMAD4 loss is an essential driver of PDAC progression. Additional hits in the TGFβ signaling pathways may synergize with SMAD4 alterations and alleviate the need for substantially reduced SMAD4 expression during tumor progression. ZNF521, encoding a zinc finger protein known to interact with SMAD4 for activation of BMP target genes (45) had aberrations in multiple cases. It is interesting to speculate that disruption of ZNF521 may have a similar effect as SMAD4 disruption on PDAC development. However, no pattern of mutual exclusion between the two events occurring in cases was observed (Fig. 3C). Significantly, ZNF521 lies proximal to the gene for transcription factor GATA6 on 18q11.2, previously reported gained in PDAC with a function on WNT signaling pathways (17). While the two genes had quite distinct patterns of gain and loss within tumors (Fig. 2), the potential for ZNF521 variations as passenger events in the GATA6 rearrangements remains to be determined.
FHIT was commonly deleted in 50% of tumors, with associated loss in expression. While the loss of FHIT has been predicted as an early event in PDAC pathogenesis (46–47), PCR validation of alterations in tumor associated PanINs in this study predicted a late presentation, similar to SMAD4. FHIT is believed to function as a tumor suppressor and was previously found to be downregulated in PDAC promoting tumor growth, but the precise mechanism of action remains unclear (46–50). Reduced FHIT levels in tumors with no detected genomic alterations, suggests additional regulatory pathways or epigenetic regulation (50). FHIT is located at one of the most common fragile sites (FRA3B; ref. 49). In addition to FHIT, other common fragile site genes including WWOX, PARK2, IMMP2L, MACROD2, and NBEA, were also affected. At least one fragile site was hit in each tumor, emphasizing chromosome instability, but with no evident fragile site mutation profile within cases.
In conclusion, a wide spectrum of genes was influenced by genomic rearrangements in this PDAC study, with many key cancer genes hit directly by intragenic breakpoints. While minimal overlap was observed in genes mutated by rearrangements and point mutations, multiple commonly targeted pathways were identified, indicating the significance of both mutation types in driving PDAC progression. Overall, these results emphasize the needs for integrated data analyses including breakpoint, CNV and SNV analysis, which together with transcriptome data enable better inference of mechanisms of PDAC progression.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by the Mayo Biomarker Discovery Program, Center for Individualized Medicine, the Minnesota Partnership for Biotechnology and Medical Genomics, the Mayo Clinic Laboratory Medicine and Pathology Collaborative Research Funds, and an NIH Specialized Program of Research Excellence (SPORE) in Pancreatic Cancer (CA102701).
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: S.J. Murphy, S.N. Hart, S. Subramaniam, R.R. McWilliams, F.J. Couch, G. Vasmatzis
Development of methodology: S.J. Murphy, F.J. Couch, G. Vasmatzis
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S.J. Murphy, G. Halling, J. Felipe Lima, F. Rakhshan Rohakhtar, F.R. Harris, G.M. Petersen, T.D. Wiltshire, M.J. Truty, F.J. Couch, G. Vasmatzis
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S.J. Murphy, S.N. Hart, G. Halling, S.H. Johnson, J.B. Smadbeck, T. Drucker, F.R. Harris, F. Kosari, B.R. Kipp, M.J. Truty, R.R. McWilliams, F.J. Couch, G. Vasmatzis
Writing, review, and/or revision of the manuscript: S.J. Murphy, S.N. Hart, S.H. Johnson, T. Drucker, F.R. Harris, F. Kosari, S. Subramanian, B.R. Kipp, R.R. McWilliams, F.J. Couch, G. Vasmatzis
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): F. Rakhshan Rohakhtar, F.J. Couch, G. Vasmatzis Study supervision: R.R. McWilliams, F.J. Couch, G. Vasmatzis
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald TL, Hickner ZJ, Schmitz M, Kort EJ. Changing incidence of pancreatic neoplasms: a 16-year review of statewide tumor registry. Pancreas. 2008;37:134–138. doi: 10.1097/MPA.0b013e318163a329. [DOI] [PubMed] [Google Scholar]
- 3.Smeenk HG, Tran TC, Erdmann J, van Eijck CH, Jeekel J. Survival after surgical management of pancreatic adenocarcinoma: does curative and radical surgery truly exist? Langenbecks Arch Surg. 2005;390:94–103. doi: 10.1007/s00423-004-0476-9. [DOI] [PubMed] [Google Scholar]
- 4.Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. 2014;111:817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider G, Schmid RM. Genetic alterations in pancreatic carcinoma. Mol Cancer. 2013;2:15. doi: 10.1186/1476-4598-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy SJ, Hart SN, Lima JF, Kipp BR, Klebig M, Winters JL, et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology. 2013;145:1098–1109. doi: 10.1053/j.gastro.2013.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy SJ, Cheville JC, Zarei S, Johnson SH, Sikkink RA, Kosari F, et al. Mate-Pair sequencing of WGA DNA following LCM of prostate cancer. DNA Res. 2012;19:395–406. doi: 10.1093/dnares/dss021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy SJ, Aubry MC, Harris FR, Halling GC, Johnson SH, Terra S, et al. Identification of independent primary tumors and intrapulmonary metastases using DNA rearrangements in non-small-cell lung cancer. J Clin Oncol. 2014;32:4050–4058. doi: 10.1200/JCO.2014.56.7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy SJ, Wigle DA, Lima JF, Harris FR, Johnson SH, Halling G, et al. Genomic rearrangements define lineage relationships between adjacent lepidic and invasive components in lung adenocarcinoma. Cancer Res. 2014;74:3157–3167. doi: 10.1158/0008-5472.CAN-13-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willis JA, Mukherjee S, Orlow I, Viale A, Offit K, Kurtz RC, et al. Genomewide analysis of the role of copy-number variation in pancreatic cancer risk. Front Genet. 2014;13:5–29. doi: 10.3389/fgene.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattie M, Christensen A, Chang MS, Yeh W, Said S, Shostak Y, et al. Molecular characterization of patient-derived human pancreatic tumor xenograft models for preclinical and translational development of cancer therapeutics. Neoplasia. 2013;15:1138–1150. doi: 10.1593/neo.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birnbaum DJ, Adélaïde J, Mamessier E, Finetti P, Lagarde A, Monges G, et al. Genome profiling of pancreatic adenocarcinoma. Genes Chromosomes Cancer. 2011;50:456–465. doi: 10.1002/gcc.20870. [DOI] [PubMed] [Google Scholar]
- 16.Liang WS, Craig DW, Carpten J, Borad MJ, Demeure MJ, Weiss GJ, et al. Genome-wide characterization of pancreatic adenocarcinoma patients using next generation sequencing. PLoS One. 2012;7:e43192. doi: 10.1371/journal.pone.0043192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwei KA, Bashyam MD, Kao J, Ratheesh R, Reddy EC, Kim YH, et al. Genomic profiling identifies GATA6 as a candidate oncogene amplified in pancreatobiliary cancer. PLoS Genet. 2008;4:e1000081. doi: 10.1371/journal.pgen.1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y, Zhang R, Suzuki R, Li SQ, Roife D, Truty MJ, et al. Two-dimensional culture of human pancreatic adenocarcinoma cells results in an irreversible transition from epithelial to mesenchymal phenotype. Lab Invest. 2015;95:207–222. doi: 10.1038/labinvest.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drucker TM, Johnson SH, Murphy SJ, Cradic KW, Therneau TM, Vasmatzis G. BIMA V3: an aligner customized for mate pair library sequencing. Bioinformatics. 2014;30:1627–1629. doi: 10.1093/bioinformatics/btu078. [DOI] [PubMed] [Google Scholar]
- 20.Anders S, Pyl PT, Huber W. HTSeq: Analysing high-throughput sequencing data with Python. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birnbaum DJ, Mamessier E, Birnbaum D. The emerging role of the TGFβ tumor suppressor pathway in pancreatic cancer. Cell Cycle. 2012;11:683–686. doi: 10.4161/cc.11.4.19130. [DOI] [PubMed] [Google Scholar]
- 22.Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci USA. 2012;109:E252–E259. doi: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yachida S, White CM, Naito Y, Zhong Y, Brosnan JA, Macgregor-Das AM, et al. Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clin Cancer Res. 2012;18:6339–6347. doi: 10.1158/1078-0432.CCR-12-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin H, Chan MW, Liyanarachchi S, Balch C, Potter D, Souriraj IJ, et al. An integrative ChIP-chip and gene expression profiling to model SMAD regulatory modules. BMC Syst Biol. 2009;3:73. doi: 10.1186/1752-0509-3-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomis RR, Alarcón C, He W, Wang Q, Seoane J, Lash A, Massagué J. A FoxO-Smad synexpression group in human keratinocytes. Proc Natl Acad Sci USA. 2006;103:12747–12752. doi: 10.1073/pnas.0605333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniels CE, Wilkes MC, Edens M, Kottom TJ, Murphy SJ, Limper AH, Leof EB. Imatinib mesylate inhibits the profibrogenic activity of TGF-beta and prevents bleomycin-mediated lung fibrosis. J Clin Invest. 2004;114:1308–1316. doi: 10.1172/JCI19603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkes MC, Murphy SJ, Garamszegi N, Leof EB. Cell-type-specific activation of PAK2 by transforming growth factor -beta independent of Smad2 and Smad3. Mol Cell Biol. 2003;23:8878–8889. doi: 10.1128/MCB.23.23.8878-8889.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H, Zhang Y, Altomare D, Peña MM, Wan F, Pirisi L, Creek KE. Six1 promotes epithelial–mesenchymal transition and malignant conversion in human papillomavirus type 16-immortalized human keratinocytes. Carcinogenesis. 2014;35:1379–1388. doi: 10.1093/carcin/bgu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Q, Liu Z, Xie F, Liu C, Shao F, Zhu CL, Hu S. Fragile histidine triad (FHIT) suppresses proliferation and promotes apoptosis in cholangiocarcinoma cells by blocking PI3K-Akt pathway. Sci World J. 2014;2014:179698. doi: 10.1155/2014/179698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cengel KA. Targeting SonicHedgehog: a new way to mow down pancreatic cancer? Cancer Biol Ther. 2004;3:165–166. doi: 10.4161/cbt.3.2.780. [DOI] [PubMed] [Google Scholar]
- 33.Petrova E, Matevossian A, Resh MD. Hedgehog acyltransferase as a target in pancreatic ductal adenocarcinoma. Oncogene. 2015;34:263–268. doi: 10.1038/onc.2013.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rödicker F, Stiewe T, Zimmermann S, Pützer BM. Therapeutic efficacy of E2F1 in pancreatic cancer correlates with TP73 induction. Cancer Res. 2001;61:7052–7055. [PubMed] [Google Scholar]
- 35.Fisher KW, Montironi R, Lopez Beltran A, Moch H, Wang L, Scarpelli M, et al. Molecular foundations for personalized therapy in prostate cancer. Curr Drug Targets. 2015;16:103–114. doi: 10.2174/1389450115666141229154500. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Tian T, Lv F, Chang Y, Wang X, Zhang L, et al. Six1 promotes proliferation of pancreatic cancer cells via upregulation of cyclin D1 expression. PLoS One. 2013;8:e59203. doi: 10.1371/journal.pone.0059203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin A, Xu Y, Liu S, Jin T, Li Z, Jin H, et al. Sineoculis homeobox homolog 1 protein overexpression as an independent biomarker for pancreatic ductal adenocarcinoma. Exp Mol Pathol. 2014;96:54–60. doi: 10.1016/j.yexmp.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Mill P, Mo R, Fu H, Grachtchouk M, Kim PC, Dlugosz AA, Hui CC. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 2003;17:282–294. doi: 10.1101/gad.1038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCleary-Wheeler AL, McWilliams R, Fernandez-Zapico ME. Aberrant signaling pathways in pancreatic cancer: a two compartment view. Mol Carcinog. 2012;51:25–39. doi: 10.1002/mc.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wall I, Schmidt-Wolf IG. Effect of Wnt inhibitors in pancreatic cancer. Anticancer Res. 2014;34:5375–5380. [PubMed] [Google Scholar]
- 41.Malkoski SP, Wang XJ. Two sides of the story? Smad4 loss in pancreatic cancer versus head-and-neck cancer. FEBS Lett. 2012;586:1984–1992. doi: 10.1016/j.febslet.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izeradjene K, Combs C, Best M, Gopinathan A, Wagner A, Grady WM, et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229–243. doi: 10.1016/j.ccr.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 43.Alberici P, Jagmohan-Changur S, De Pater E, Van Der Valk M, Smits R, Hohenstein P, et al. Smad4 haploinsufficiency in mouse models for intestinal cancer. Oncogene. 2006;25:1841–1851. doi: 10.1038/sj.onc.1209226. [DOI] [PubMed] [Google Scholar]
- 44.Alberici P, Gaspar C, Franken P, Gorski MM, de Vries I, Scott RJ, et al. Smad4 haploinsufficiency: a matter of dosage. Pathogenetics. 2008;1:2. doi: 10.1186/1755-8417-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsubara E, Sakai I, Yamanouchi J, Fujiwara H, Yakushijin Y, Hato T, et al. The role of zinc finger protein 521/early hematopoietic zinc finger protein in erythroid cell differentiation. J Biol Chem. 2009;284:3480–3487. doi: 10.1074/jbc.M805874200. [DOI] [PubMed] [Google Scholar]
- 46.Bloomston M, Kneile J, Butterfield M, Dillhoff M, Muscarella P, Ellison EC, et al. Coordinate loss of fragile gene expression in pancreatobiliary cancers: correlations among markers and clinical features. Ann Surg Oncol. 2009;16:2331–2238. doi: 10.1245/s10434-009-0507-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Birnbaum D, Adélaïde J, Popovici C, Charafe-Jauffret E, Mozziconacci MJ, Chaffanet M. Chromosome arm 8p and cancer: a fragile hypothesis. Lancet Oncol. 2003;4:639–642. doi: 10.1016/s1470-2045(03)01225-7. [DOI] [PubMed] [Google Scholar]
- 48.Saldivar JC, Bene J, Hosseini SA, Miuma S, Horton S, Heerema NA, et al. Characterization of the role of FHIT in suppression of DNA damage. Adv Biol Regul. 2013;53:77–85. doi: 10.1016/j.jbior.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao J, Chen XP, Li WL, Xia J, Du H, Tang WB, et al. Decreased fragile histidine triad expression in colorectal cancer and its association with apoptosis inhibition. World J Gastroenterol. 2007;13:1018–1026. doi: 10.3748/wjg.v13.i7.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuroki T, Trapasso F, Yendamuri S, Matsuyama A, Aqeilan RI, Alder H, et al. Allelic loss on chromosome 3p21.3 and promoter hypermethylation of semaphorin 3B in non-small cell lung cancer. Cancer Res. 2003;63:3352–3355. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.