Abstract
In this study, we determined the host relationship of Agrotis ipsilon moths by identifying pollen species adhering them during their long-distance migration. Pollen carried by A. ipsilon moths was collected from 2012 to 2014 on a small island in the center of the Bohai Strait, which is a seasonal migration pathway of this pest species. Genomic DNA of single pollen grains was amplified by using whole genome amplification technology, and a portion of the chloroplast rbcL sequence was then amplified from this material. Pollen species were identified by a combination of DNA barcoding and pollen morphology. We found 28 species of pollen from 18 families on the tested moths, mainly from Angiosperm, Dicotyledoneae. From this, we were able to determine that these moths visit woody plants more than herbaceous plants that they carry more pollen in the early and late stages of the migration season, and that the amounts of pollen transportation were related to moth sex, moth body part, and plant species. In general, 31% of female and 26% of male moths were found to be carrying pollen. Amounts of pollen on the proboscis was higher for female than male moths, while the reverse was true for pollen loads on the antennae. This work provides a new approach to study the interactions between noctuid moth and their host plants. Identification of plant hosts for adult moths furthers understanding of the coevolution processes between moths and their host plants.
Keywords: Agrotis ipsilon, pollen, molecular identification, “insect-host plant” coevolution
1. Introduction
Plants and their insect herbivores represent more than 50% of all known species on earth [1]. The interaction between these two ecological partners is one of the most common and consequential of ecological associations [2]. Flowers and insects co-evolved [3]. Plants depending upon insects for pollination have evolved pollen that adheres readily to the insects’ exterior [4]. The yield of many crops usually increases with the number of pollinators, meanwhile, adults of numerous insect species feed on nectar, pollen and other plant exudates that are frequently associated with flowers [3]. The first step in understanding how these ecological associations function is to determine the range of host plants used by particular herbivorous insects.
Noctuidae is the largest family of Lepidoptera, containing over 40,000 currently recognized species [5]. Noctuids are a prominent feature of terrestrial insect faunas and food webs, including many species of economic importance [6]. Within the noctuids, larvae and adults usually differ considerably in their nutritional requirements and food ecology. Often, larvae feed on the structural tissue of their host plants while adults feed primarily or exclusively on plant-provided food supplements such as nectar and pollen [7]. Many studies focus on larval host records by direct observations of herbivory in the field and laboratory feeding trials [8,9,10,11]. Unfortunately, identifying the diet of adults using these methods is challenging. Direct observation of noctuid moths is limited by their nocturnal and flight habits. In laboratory-feeding trials, insects will often feed on host plants not normally consumed in nature, resulting in an overestimation of their diet breadths [8].
Nectar is a sugar-rich plant secretion that may contain low levels of amino acids, proteins, lipids, vitamins and secondary plant compounds, as well as other organic compounds and minerals [12]. Adults of many insect species feed on nectar to meet nutritional requirements for successful reproduction. As a result of this feeding activity, adults become contaminated with pollen, and the identification of pollen species can be used to determine the insect’s host plants [3]. Although conventional pollen morphology is widely used for plant identification, in some cases, pollen of closely related plants has highly similar morphology [13,14].
Alternatively, DNA bar coding has great potential for determining the dietary compositions of organisms [15,16,17,18]. For this purpose, a partial region of plant DNA sequence needs to be amplified, and compared to a reference database such as GenBank [15,16]. Genomic DNA of single pollen grains can be amplified by using the whole genome amplification technology for identification of the source plant species [19]. Previous studies have demonstrated that such molecular markers have the potential to identify insect herbivore diets at the plant family or genus level [15,16]. Accurate host identification is affected by the completeness of the database of reference DNA sequences, reference libraries are also used [15,16,18]. Moreover, the unambiguous identification of host plants at the species-level using this approach is not yet certain [18]. Therefore, in the present study, a combined approach of DNA barcoding and pollen morphology was used to identify pollen species.
The black cutworm, Agrotis ipsilon (Hufnagel) (Lepidoptera: Noctuidae), is a key pest of cotton, maize, potatoes, beans, and other crops [20]. Its long-distance migration behavior has allowed this species to disperse throughout Africa, Europe, Asia, and the Americas [21,22,23]. Wu and Guo (1997) and Jiang et al. (2000) found that adult feeding can significantly increase the flight capacity of moths [24,25], and that migratory A. ipsilon were often contaminated with pollen [26]. Therefore, in this study, the pollen found adhered to A. ipsilon moths was identified. We also investigated the quantities of pollen attached to various parts of bodies of A. ipsilon moths.
2. Results
2.1. Plant Hosts Inferred from Pollen
Twenty-eight pollen species from at least 18 families were detected on A. ipsilon moths. The combination of DNA data using rbcL, pollen morphology, and distribution data of plant allowed us to identify 12 samples to the species level, including Castanea mollissima Blume, Citrus sinensis Blanco, Melia azedarach Linn., Flueggea virosa (Roxb. Ex Woigt) Voigt, Olea europaea L., Amorpha fruticosa Linn., Alniphyllum fortunei (Hemsl.) Makino, Robinia pseudoacacia L., Pterocarya rhoifolia Siebold et Zucc., Castanopsis echinocarpa Miq., Elaeagnus umbellata Thunb. and Chenopodium album L., and 11 of the samples to the genus level, including Prunus, Ligustrum, Holboellia, Llex, Taraxacum, Pinus, Helianthus, Brassica, Adenophora, Smilax, and Aster. (Table 1, Figure 1, Supplementary Text S1). The success rates for a combination of pollen morphology, DNA barcoding, distribution data to the species-level and genus-level identification were 42.86% and 82.14%, respectively, while DNA barcoding were 21.43% and 78.57%, and pollen morphology were 7.14% and 71.43%, respectively.
Table 1.
Molecular and morphological identification of plant species by identifying pollen.
| Number | Identified Plants | rbcL-Molecular Identification | Pollen Morphology Identification | Distributed in China |
|---|---|---|---|---|
| 1 | Castanea mollissima | Sister to Castanea mollissima/Castanea sativa |
Castanea Mill./ Castanopsis Spach./Lithocarpus Bl. |
Castanea mollissima |
| 2 |
Prunus yedoensis/ Prunus subhirtella/ Prunus serrulata |
Sister to Prunus yedoensis/ Prunus subhirtella/Prunus serrulata |
Prunus L. |
Prunus yedoensis/ Prunus subhirtella/ Prunus serrulata |
| 3 |
Ligustrum lucidum/ Ligustrum sempervirens |
Ligustrum lucidum/Ligustrum sempervirens | Oleaceae |
Ligustrum lucidum/ Ligustrum sempervirens |
| 4 | Citrus sinensis | Sister to Citrus maxima/Citrus sinensis | Citrus sinensis | Citrus maxima/Citrus sinensis |
| 5 | Melia azedarach | Sister to Melia azedarach | Melia L. | Melia azedarach |
| 6 | Flueggea virosa | Sister to Flueggea virosa/Flueggea neowawraea | Eucommiaceae | Flueggea virosa |
| 7 |
Hoboellia parviflora/ Hoboellia grandiflora |
Sister to Stauntonia duclouxii/ Hoboellia parviflora/Hoboellia grandiflora/ Parvatia brunoniana |
Holboellia Wall. |
Stauntonia duclouxii/ Hoboellia parviflora/ Hoboellia grandiflora/ Parvatia brunoniana |
| 8 | Olea europaea | Sister to Olea europaea | Oleaceae | Olea europaea |
| 9 | Amorpha fruticosa | Sister to Amorpha fruticosa/Amorpha canescens | Amorpha L. | Amorpha fruticosa |
| 10 | Alniphyllum fortunei | Sister to Alniphyllum fortunei | Alniphyllum fortunei | Alniphyllum fortunei |
| 11 | Robinia pseudoacacia | Sister to Robinia pseudoacacia | Robinia L. | Robinia pseudoacacia |
| 12 | Llex cornuta/Ilex corallina | Sister to Llex cornuta/Ilex corallina | Llex L. | Llex cornuta/Ilex corallina |
| 13 | Pterocarya rhoifolia | Sister to Pterocarya rhoifolia | Pterocarya Kunth. | Pterocarya rhoifolia |
| 14 | Castanopsis echinocarpa | Sister to Castanopsis echinocarpa |
Castanea Mill./Castanopsis Spach./ Lithocarpus Bl. |
Castanopsis echinocarpa |
| 15 |
Taraxacum officinale/ Taraxacum platypecidium |
Sister to Taraxacum alpinum/ Taraxacum officinale/Taraxacum platypecidium |
Taraxacum Weber |
Taraxacum officinale/ Taraxacum platypecidium |
| 16 |
Pinus tabuliformis/ Pinus thunbergii/Pinus nigra/ Pinus densiflora/Pinus kesiya/ Pinus sylvestris/Pinus tropicalis |
Sister to Pinus tabuliformis/Pinus thunbergii/ Pinus nigra/Pinus densiflora/Pinus densata/ Pinus hwangshanensis/Pinus kesiya/ Pinus sylvestris/Pinus tropicalis/ Pinus yunnanensis |
Pinus tabuliformis/Pinus thunbergii/ Pinus nigra/Pinus densiflora/Pinus kesiya/ Pinus sylvestris/Pinus tropicalis |
Pinus tabuliformis/ Pinus thunbergii/ Pinus nigra/Pinus densiflora/ Pinus kesiya/Pinus sylvestris/ Pinus tropicalis |
| 17 |
Brassica rapa/Brassica napus/ Brassica oleracea/Brassica juncea |
Sister to Brassica rapa/Brassica napus/ Brassica oleracea/Brassica juncea |
Cruciferae |
Brassica rapa/Brassica napus/ Brassica oleracea/ Brassica juncea |
| 18 | Elaeagnus umbellata | Sister to Elaeagnus umbellata | Elaeagnus L. | Elaeagnus umbellata |
| 19 | Helianthus L. | Sister to Helianthus L. | Compositae | Helianthus L. |
| 20 |
Adenophora trachelioides/ Adenophora remotiflora |
Sister to Adenophora trachelioides/ Adenophora remotiflora/Hanabusaya asiatica |
Adenophora Flash. |
Adenophora trachelioides/ Adenophora remotiflora |
| 21 | Smilax L. | Sister to Smilax L. | Smilax L. | Smilax L. |
| 22 | Chenopodium album | Sister to Chenopodium ficifolium/ Chenopodium album |
Chenopodium album |
Chenopodium ficifolium/ Chenopodium album |
| 23 | Rosaceae | Unidentifiable | Rosaceae | Rosaceae |
| 24 | Rosaceae | Unidentifiable | Rosaceae | Rosaceae |
| 25 | Violaceae | Unidentifiable | Violaceae (close to Viola) | Violaceae |
| 26 | Compositae | Unidentifiable | Compositae (close to Coreopsis, Sigebeckia) | Compositae |
| 27 | Compositae | Unidentifiable | Compositae (also similar to Cosmos, Kalimeris, Aster) | Compositae |
| 28 | Aster L. | Unidentifiable | Aster L. (also similar to Chrysanthemum, Matricaria) | Aster L. |
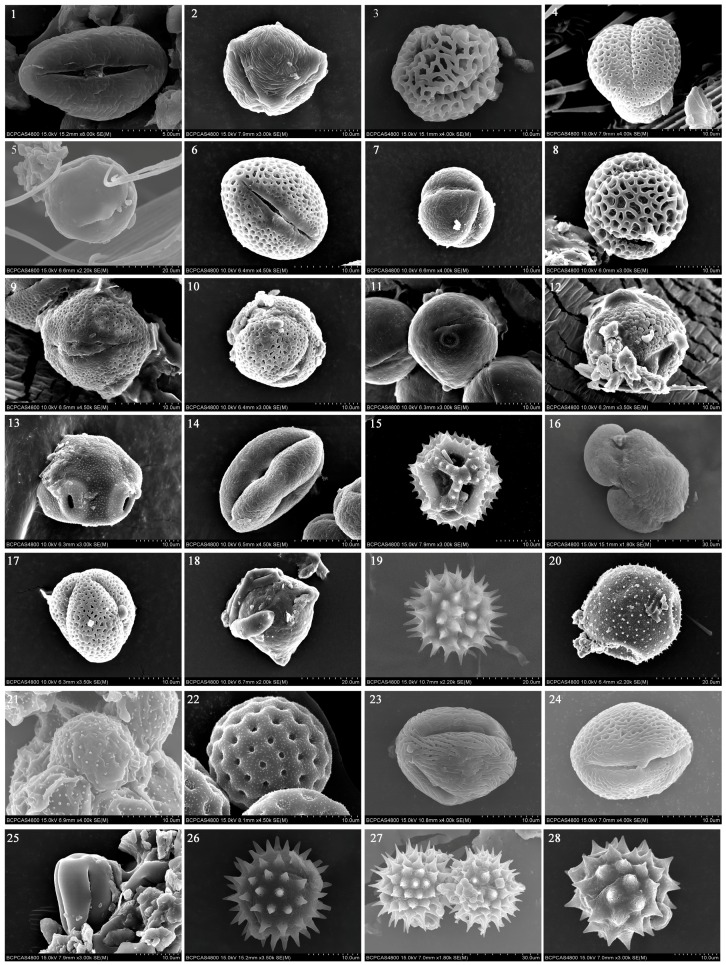
Figure 1.
Scanning electron microscopy (SEM) microphotographs of the examined pollen species. 1. Castanea mollissima; 2. Prunus yedoensis/Prunus subhirtella/Prunus serrulata; 3. Ligustrum lucidum/Ligustrum sempervirens; 4. Citrus sinensis; 5. Melia azedarach; 6. Flueggea virosa; 7. Hoboellia parviflora/Hoboellia grandiflora; 8. Olea europaea; 9. Amorpha fruticosa; 10. Alniphyllum fortunei; 11. Robinia pseudoacacia; 12. Llex cornuta/Ilex corallina; 13. Pterocarya rhoifolia; 14. Castanopsis echinocarpa; 15. Taraxacum officinale/Taraxacum platypecidium; 16. Pinus tabuliformis/Pinus thunbergii/Pinus nigra/Pinus densiflora/Pinus kesiya/Pinus sylvestris/Pinus tropicalis; 17. Brassica rapa/Brassica napus/Brassica oleracea/Brassica juncea; 18. Elaeagnus umbellata; 19. Helianthus annuus; 20. Adenophora trachelioides/Adenophora remotiflora; 21. Smilax L.; 22. Chenopodium album; 23. Rosaceae; 24. Rosaceae; 25. Violaceae (close to Viola); 26. Compositae (close to Coreopsis, Sigebeckia); 27. Compositae (also similar to Cosmos, Kalimeris, Aster); 28. Aster L. (also similar to Chrysanthemum, Matricaria).
2.2. Frequency of Pollen Detection Rates on Body Parts of A. ipsilon
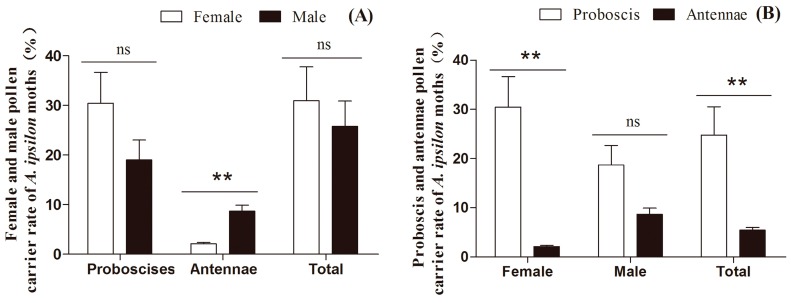
For whole body analyses, there was no significant sex-related difference in frequency of pollen occurrence on A. ipsilon moths, either for year groups (2012–2014) (Table 2, Figure 2), with 30.9% of female and 25.8% of male moths contaminated with plant pollens, or years analyzed individually: 2012, 2013 and 2014 (Table 2, Supplementary Figure S1C).
Table 2.
Chi-squared test and a Student’s t-test for tested frequencies of pollen deposition on A. ipsilon.
| Female and Male Pollen Carrier Rate of A. ipsilon Moths | Proboscis and Antennae Pollen Carrier Rate of A. ipsilon Moths | ||||||
|---|---|---|---|---|---|---|---|
| Proboscis | 2012 | χ2 | 4.30 | Female | 2012 | χ2 | 52.46 |
| df | 1 | df | 1 | ||||
| p | 0.038 | p | <0.001 | ||||
| 2013 | χ2 | 8.56 | 2013 | χ2 | 34.09 | ||
| df | 1 | df | 1 | ||||
| p | 0.003 | p | <0.001 | ||||
| 2014 | χ2 | 10.57 | 2014 | χ2 | 97.75 | ||
| df | 1 | df | 1 | ||||
| p | 0.001 | p | <0.001 | ||||
| 2012–2014 | t | 1.57 | 2012–2014 | t | 9.25 | ||
| df | 4 | df | 4 | ||||
| p | 0.19 | p | 0.001 | ||||
| Antennae | 2012 | χ2 | 13.11 | Male | 2012 | χ2 | 8.52 |
| df | 1 | df | 1 | ||||
| p | <0.001 | p | 0.004 | ||||
| 2013 | χ2 | 4.39 | 2013 | χ2 | 4.84 | ||
| df | 1 | df | 1 | ||||
| p | 0.036 | p | 0.028 | ||||
| 2014 | χ2 | 8.89 | 2014 | χ2 | 11.34 | ||
| df | 1 | df | 1 | ||||
| p | 0.003 | p | 0.001 | ||||
| 2012–2014 | t | −6.73 | 2012–2014 | t | 2.62 | ||
| df | 4 | df | 4 | ||||
| p | 0.003 | p | 0.059 | ||||
| Proboscis and antennae | 2012 | χ2 | 0.049 | Female and male | 2012 | χ2 | 48.560 |
| df | 1 | df | 1 | ||||
| p | 0.82 | p | <0.001 | ||||
| 2013 | χ2 | 2.10 | 2013 | χ2 | 31.54 | ||
| df | 1 | df | 1 | ||||
| p | 0.15 | p | <0.001 | ||||
| 2014 | χ2 | 3.74 | 2014 | χ2 | 3.74 | ||
| df | 1 | df | 1 | ||||
| p | 0.053 | p | <0.001 | ||||
| 2012–2014 | t | 0.59 | 2012–2014 | t | 5.09 | ||
| df | 4 | df | 4 | ||||
| p | 0.59 | p | 0.007 | ||||
Figure 2.
(A) Frequencies of pollen deposition on female and male proboscises, antennae and total (proboscis and antennae) of A. ipsilon moths; (B) Frequencies of pollen deposition on proboscis and antennae of female, male and total (female and male) of A. ipsilon moths. Double asterisks (**) indicates there was significant difference at the 1% level as determined by a t-test, and ns indicates there was no significant difference.
For the proboscises only, pollen detection rates were higher on female moths (30.4%) than males (19.0%) for 2012–2014 as a group (Table 2, Figure 2), and significantly for the same years individually: 2012, 2013 and 2014 (Table 2, Supplementary Figure S1A).
However, the frequency of pollen occurrence on antennae was lower for females (2.1%) than for males (8.7%) during 2012–2014 as a group (Table 2, Figure 2), and for the same years individually: 2012, 2013 and 2014 (Table 2, Supplementary Figure S1B).
The frequency of pollen deposits was higher on the proboscis (24.7%) than the antennae (5.4%), for 2012–2014 as a group (Table 2, Figure 2), and for the same years individually: 2012, 2013 and 2014 (Table 2, Supplementary Figure S1F). This difference was true for both female moths (Table 2, Supplementary Figure S1D,E).
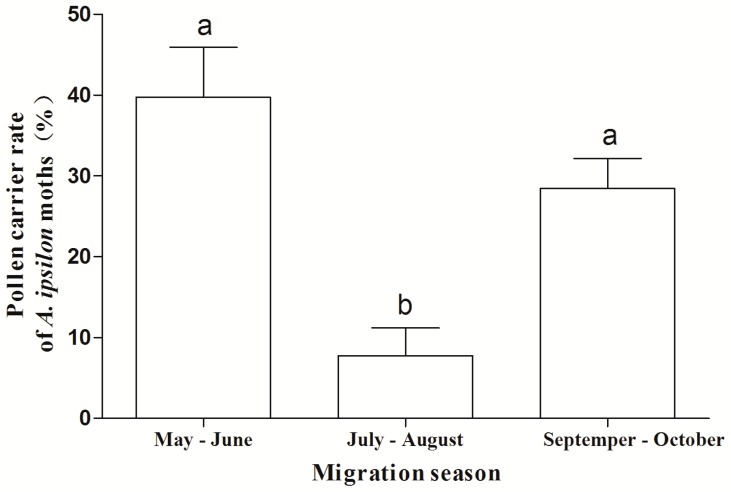
With respect to the phenology of migration, the frequency of pollen deposits on bodies of A. ipsilon in the early part of the migration period (39.7%) and in the late period (28.5%) were both significantly higher than that in the middle period of the migration season (7.8%) during 2012–2014 (F = 12.61, df1 = 2, df2 = 6, p = 0.007) (Figure 3).
Figure 3.
Frequencies of pollen deposition on migratory A. ipsilon near the Bohai Sea area in different migration stages during 2012–2014. Bars sharing the same letter mean there were no significant differences at the 5% level by Tukey’s honestly significant difference (HSD) tests.
2.3. Characteristic of Plants Whose Pollen Was Found on A. ipsilon
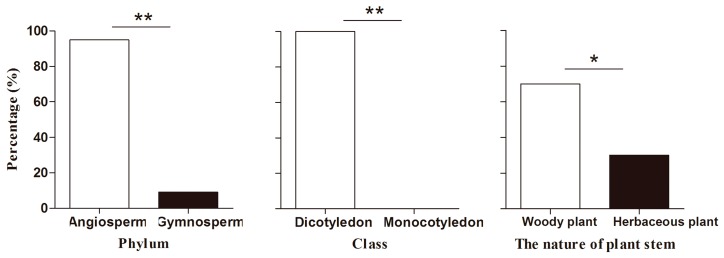
Chi-squared tests showed that significantly more species whose pollen was found on A. ipsilon moths were Angiosperm and Dicotyledon plants than Gymnosperms (χ2 = 32.40, df = 1, p < 0.001) or Monocotyledons (χ2 = 40.00, df = 1, p < 0.001). This led us to determine that moths visited woody plants more often than herbaceous plants (χ2 = 6.40, df = 1, p = 0.011) (Figure 4).
Figure 4.
The characteristic of pollen source plants of migratory A. ipsilon during 2012–2014. Single asterisk (*) or double asterisks (**) indicates there was significant difference at the 5% or 1% level as determined by chi-squared test.
3. Discussion
We identified host plant used by A. ipsilon moths using DNA bar coding, pollen morphology, and known distributions of plants. Our results indicated that A. ipsilon moths fed on nectar from a very wide of plants, including 28 species from 18 families of mainly Dicotyledoneae in the Angiosperms. Kishimoto-Yamada et al. (2013) inferred host plant families of chrysomelid beetles including herbs, shrubs and vines [27]. However, a preference of A. ipsilon for visiting woody plants more than herbaceous plants was detected in our study. Different host plants can play important roles in the population increase of insects [28]. Both quality and quantity of food influence insect development and fecundity [29], and fitness effects may differ between different sources of nectar. Therefore, further research is needed to explore the preference of A. ipsilon moths for particular host plants.
Agrotis ipsilon is a long-distance migratory pest, often undertaking trans-regional migrations. The identification of pollen species can be used to determine the geographical origin of the insect since some plants grow only in certain ecological zones or geographic locations. For example, Hendrix and Showers (1992) found that A. ipsilon adults captured in Iowa and Missouri contained pollen from plants that only grow in south and southwest Texas [4]. Our results found pollen from Ligustrum lucidum Ait./Ligustrum sempervirens (Franch.) Lingelsh, C. sinensis, M. azedarach, Hoboellia parviflora (Hemsl.) Gagnepl./Hoboellia grandiflora Réaub., O. europaea, A. fruticosa, A. fortunei, R. pseudoacacia, llex cornuta Lindl. Et Paxton/Ilex coralline Franch. and C. echinocarpa on A. ipsilon adults captured in Beihuang from late May to June. Since these species grow in the Yangtze River basin and further south (over 800 km) [29], these adults must have foraged on these flowers there, then migrated northward to Beihuang, providing evidence for the northward spring migration of A. ipsilon [22].
Our use of DNA-based pollen analysis to provide species-level plant identifications validates this novel method, which has great potential for the study of plant-herbivore interactions [15]. The Consortium for the Barcode of Life-plant Working Group (CBOL) has recommended the two-locus combination of rbcL + matK as the best plant barcode due to its universality, sequence quality, and species discrimination [30]. Unfortunately, matK is difficult to amplify universally using currently available primer sets [31]. We chose the rbcL intron mainly because it has the highest level of coverage in GenBank among potential barcoding markers; the universality of PCR primers. A recent study also demonstrated that the rbcL sequences successfully identified the host plants families of rolled-leaf beetles [16]. rbcl is a good DNA barcoding region for plants at the family and genus levels, but for identification at the species level, a combination of pollen morphology and DNA barcoding is required. The efficiency of traditional pollen identification, which is highly dependent on human expertise and prohibitively time-consuming for large-scale studies, is improved by the use of DNA barcoding.
Our results showed that the frequency of pollen deposits on the proboscis was higher on female than male moths, which may be due to differences in the nutrient requirements of the sexes, with females generally having a higher nutrient demand than males [32]. The reason for higher frequencies of pollen deposits on antennae of male vs. female moths may be due to the different shape of the antennae between the sexes. The more branched antennae of male A. ipsilon moths are more favorable for pollen retention due to their greater surface area.
For the proboscis, when pollen was found, many grains were typically present, suggesting active contact through feeding rather than casual contact through wind-blown contamination. Pollen were rarely found on the antenna (5.4%), so future studies should focus on examining pollen found on A. ipsilon proboscises. Differences in the frequency of pollen deposits on A. ipsilon throughout the migratory period may be caused by differences in the abundance of nectar plants or the nutritional requirements of the moths, a topic that requires further investigation.
Flowers and insects have co-evolved [8], and the relationship between flowering plants and pollinators can be seen as mutually beneficial. Adult herbivores require nutrients or energy supplements from flowers for reproduction or flight, and flower-visiting herbivores are important pollinators [33]. The yield of many crops are dependent on the pollinators that visit their flowers, and yields are usually increased with the help of pollinators [4]. Our results showed A. ipsilon adults to be effective pollinators of C. mollissima, C. sinensis, M. azedarach, F. virosa, O. europaea and others. A. ipsilon is a long-distance disperser, undertaking regular migration across different agricultural areas. Therefore, the gene exchange of plants can be achieved across large regions with the help of A. ipsilon adults [34]. However, A. ipsilon larvae are pests of many crops, such as cotton, maize, potatoes, beans, and cruciferous vegetables [16]. Ultimately, the more knowledge we have about A. ipsilon, the better and more efficient the management practices that can be developed.
4. Experimental Section
4.1. Agrotis ipsilon Sampling
Agrotis ipsilon moths were collected using light traps. Moths were collected from sunset to sunrise, except during power outages or periods of heavy rain, using a vertical-pointing searchlight trap (model DK.Z.J1000B/t, 65.2 cm in diameter, 70.6 cm in height, and 30 in spread angle), equipped with a 1000-W metal halide-lamp (model JLZ1000BT; Shanghai Yaming Lighting Co., Ltd., Shanghai, China) on the top of a house (500 m elevation) on the 2.5 km2 island of Beihuang (BH, 38°24′ N; 120°55′ E) in the Bohai Gulf 40 km from the mainland to the north and 60 km from land to the south [34,35,36]. Collection were made every night from April to October during 2012–2014. Samples were collected with nylon net bags (60 mesh) which were changed manually every 2 h each night. Twenty moths (or all individuals if the total captured was <20) were removed from collection bags every morning. Each moth was killed by crushing its thorax, and it was placed into a 2 mL crytube and held at −20 °C in a freezer before microscopic examination.
4.2. Pollen Examination and Scanning Electron Microscopy (SEM) Preparation
Because Bryant et al. [37] found only 4% of pollen on Helicoverpa zea legs and eyes and suggested focusing examination efforts on the proboscis, in this study we examined the proboscis and antenna. A. ipsilon adult heads were removed from the body and examined at 200× magnification to make a preliminary identification of pollen adhering to their proboscis or antenna. To prevent contamination, a section of paper towel (9 × 9 cm) was placed on the microscope stage and changed after each sample, and forceps were also cleaned after each sample. Pollen grains found on a proboscis or antenna were mounted on aluminum stubs coated with gold in a sputter coater and immediately photographed under a Hitachi S-4800 cold field emission scanning electron microscope (Hitachi High-Technologies Co., Tokyo, Japan).
4.3. DNA Extraction from a Single Pollen Grain, PCR Amplification, Cloning and DNA Sequencing
Single pollen grains were isolated from the aluminum stubs and placed in individual PCR tubes containing 1 μL of lysis solution (NaOH with a concentration of 0.1 M, plus 2% Tween-20). NaOH and Tween-20 were purchased from the Beijing Chemical Reagent Co., Ltd. (Beijing, China). After the pollen grains were deposited, 5 μL of mineral oil was added. The samples were spun briefly (1 min at 1000× g) before incubation at 95 °C for 17 min 30 s to lyse the pollen grains. After lysis, equimolar aliquots of 1 μL Tris-EDTA (TE) buffer were added to neutralize the samples, which were then spun briefly [19]. The resulting solution was used as the template for Whole Genome Amplification (WGA) by illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare UK Ltd., London, UK). The WGA products of single pollen grains were used to amplify plant plastid DNA.
The partial region of chloroplast rbcL was amplified using PCR primers, namely rbcla forward (5′-ATGTCACCACAAACAGAAAC-3′), rbcLa reverse (5′-TCGCATGTACCTGCAGTAGC-3′) [38], rbclb forward (5′-ATGTCACCACAAACAGAAAC-3′) and rbcLb reverse (5′-GAAACGGTCTCTCCAACGCAT-3′) [39]. PCR amplification was performed using a thermal cycler (GeneAmp PCR System 9700, Applied Biosystems, Foster City, CA, USA) under the following conditions: initial activation at 94 °C for 4 min, 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 45 s, followed by final incubation at 72 °C for 10 min. Five sequencing replicates were taken for each pollen. The PCR reactions contained 100 ng of extracted DNA, 0.2 μM primer pairs, 2 mM dNTPs, 5 μL 10 × Long and Accurate (LA) Taq buffer, and 1 U LA Taq polymerase (TaKaRa, Beijing, China) for a total volume of 50 μL.
PCR products were purified with a Gel Extraction Kit (Tiangen, Beijing, China) and ligated directly into the pGEM-T Cloning Vector (Promega, Madison, WI, USA). Each DNA-containing plasmid was isolated from cultured E. coli cells by an alkaline miniprep method. The presence of inserts was verified by PCR with the M13 forward (5′-GTTTTCCCAGTCACGAC-3′) and reverse primers (5′-CAGGAAACAGCTATGAC-3′) and sequenced using Sanger sequencing by the company Biomed (Beijing, China).
4.4. Pollen Identification and the Characteristics of Pollen Source Plants
Identification of rbcL sequences was performed individually through similarity Basic Local Alignment Search Tool (BLAST) searches against GenBank [40]. The unknown sequence was considered a member of the best hit of the query sequences when it was completely consistent with them, and the unknown species sequence was considered to be the same genus as indicated by its top hits if there were differences between the sequences. Then the species were corrected according to their morphologic features and the known plant distributions. The pollen’s morphological features were identified using the published scanning electron microscopy (SEM) images in the atlas of pollen flora of China and pollen flora of China woody plants by SEM [41,42]. The pollen grains that could be classified to genus or species level were used to identify the source plants of pollen.
4.5. Data Analysis
Statistical analyses were conducted using SPSS 13.0 (SPSS Inc., Chicago, IL, USA) [43]. The frequency of pollen deposits on A. ipsilon during different migration phases was analyzed using one-way ANOVA, and the differences between the mean values were compared using Tukey’s HSD (honestly significant difference) test. The differences in the annual mean frequencies of pollen occurrence on female and male body parts (proboscis, antennae, or total (combined proboscis and antennae)) of A. ipsilon moths, and the differences in the annual mean frequency of pollen deposition on the proboscis or antennae of female, male, and total (female and male) A. ipsilon moths were analyzed by Student’s t-tests. The differences in frequency of pollen deposition on female and male body parts (proboscis, antennae or total (proboscis and antennae)) of A. ipsilon moths in each year, the differences of in such rates between proboscis and antennae of female, male and total (female and male) A. ipsilon moths in each year, and the characteristics of pollen source plants were all analyzed by a Chi-squared tests. All proportion data were logit transformed before being analyzed.
5. Conclusions
This study developed an effective approach for examining noctuid moth–host plant interactions. Unlike larvae that mainly feed on herbaceous plants, A. ipsilon moths obtain nutrition primarily from woody plants. We also found evidence that female A. ipsilon moths have a higher nutrient demand than males. Our results are helpful for understanding the coevolution processes between insects and their host plants, and for developing more efficient management practices of A. ipsilon.
Acknowledgments
This work was supported by the National Key Project of Scientific and Technical Supporting Programs (grant number 2012BAD19B05), and the National Natural Science Foundation (grant number 31321004).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/6/851/s1, Figure S1: Frequencies of pollen deposition on female and male proboscises (A), antennae (B) and total (proboscis and antennae) (C) of A. ipsilon moths; Frequencies of pollen deposition on proboscis and antennae of female (D), male (E) and total (female and male) (F) of A. ipsilon moths. Single asterisk (*) or double asterisks (**) indicates there was significant difference at the 5% or 1% level as determined by chi-squared test, and ns indicates there was no significant difference, Text S1: The rbcl sequences of the examined pollen species.
Author Contributions
Yongqiang Liu performed the experiments, analyzed the data, and participated in designing the study and drafting the manuscript; Xiaowei Fu, Limi Mao and Zhenlong Xing performed parts of the experiments and participated in analyzing the data; Kongming Wu designed the study, coordinated the work, and wrote and revised the manuscript; All authors contributed to and approved the final version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Futuyma D.J., Agrawal A.A. Macroevolution and the biological diversity of plants and herbivores. Proc. Natl. Acad. Sci. USA. 2009;106:18054–18061. doi: 10.1073/pnas.0904106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forister M.L., Novotny V., Panorska A.K., Baje L., Basset Y., Butterill P.T., Cizek L., Coley P.D., Dem F., Diniz I.R., et al. The global distribution of diet breadth in insect herbivores. Proc. Natl. Acad. Sci. USA. 2015;112:442–447. doi: 10.1073/pnas.1423042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones G.D., Jones S.D. The uses of pollen and its implication for entomology. Neotrop. Entomol. 2001;30:341–350. doi: 10.1590/S1519-566X2001000300001. [DOI] [Google Scholar]
- 4.Hendrix W.H., III, Showers W.B. Tracing black cutworm and armyworm (Lepidoptera: Noctuidae) northward migration using Pithecellobium and Calliandra pollen. Environ. Entomol. 1992;21:1092–1096. doi: 10.1093/ee/21.5.1092. [DOI] [Google Scholar]
- 5.Wagner D.L. Moths. In: Levin S.A., editor. Encyclopedia of Biodiversity. Volume 4. Academic Press; San Diego, CA, USA: 2001. pp. 249–270. [Google Scholar]
- 6.Wäckers F.L., Romeis J., Rijn P.V. Nectar and pollen feeding by insect herbivores and implications for multitrophic interactions. Annu. Rev. Entomol. 2007;52:301–323. doi: 10.1146/annurev.ento.52.110405.091352. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell A., Mitter C., Regier J.C. Systematics and evolution of the cutworm moths (Lepidoptera: Moctuidae): Evidence from two protein-coding nuclear genes. Syst. Entomol. 2006;31:21–46. doi: 10.1111/j.1365-3113.2005.00306.x. [DOI] [Google Scholar]
- 8.Barone J.A. Host-specificity of folivorous insects in a moist tropical forest. J. Anim. Ecol. 1998;67:400–409. doi: 10.1046/j.1365-2656.1998.00197.x. [DOI] [Google Scholar]
- 9.Wood T.K., Olmstead K.L. Latitudinal effects on treehopper species richness (Homoptera: Membracidae) Ecol. Entomol. 1984;9:109–115. doi: 10.1111/j.1365-2311.1984.tb00703.x. [DOI] [Google Scholar]
- 10.Janzen D.H. Ecological characterization of a Costa Rican dry forest caterpillar fauna. Biotropica. 1988;20:120–135. doi: 10.2307/2388184. [DOI] [Google Scholar]
- 11.Hodkinson I.D., Casson D. A lesser predilection for bugs: Hemiptera (Insecta) diversity in tropical rain forests. Biol. J. Linn. Soc. 1991;43:101–109. doi: 10.1111/j.1095-8312.1991.tb00587.x. [DOI] [Google Scholar]
- 12.Baker H.G., Baker I. A brief historical review of the chemistry of floral nectar. In: Bentley B., Elias T., editors. The Biology of Nectaries. Columbia University Press; New York, NY, USA: 1983. pp. 126–152. [Google Scholar]
- 13.Salmaki Y., Jamzad Z., Zarre S., Bräuchler C. Pollen morphology of Stachys (Lamiaceae) in Iran and its systematic implication. Flora. 2008;203:627–639. doi: 10.1016/j.flora.2007.10.005. [DOI] [Google Scholar]
- 14.Khansari E., Zarre S., Alizadeh K., Attara F., Aghabeigic F., Salmaki Y. Pollen morphology of Campanula (Campanulaceae) and allied genera in Iran with special focus on its systematic implication. Flora. 2012;207:203–211. doi: 10.1016/j.flora.2012.01.006. [DOI] [Google Scholar]
- 15.Jurado-Rivera J.A., Vogler A.P., Reid C.A., Petitpierre E., Gómez-Zurita J. DNA barcoding insect-host plant associations. Proc. R. Soc. Biol. Sci. Ser. B. 2009;276:639–648. doi: 10.1098/rspb.2008.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro S.P., Jurado-Rivera J.A., Gomez-Zurita J., Lyal C.H.C., Vogler A.P. DNA profiling of host-herbivore interactions in tropical forests. Ecol. Entomol. 2010;35:18–32. doi: 10.1111/j.1365-2311.2009.01145.x. [DOI] [Google Scholar]
- 17.Staudacher K., Wallinger C., Schallhart N., Traugott M. Detecting ingested plant DNA in soil-living insect larvae. Soil Biol. Biochem. 2011;43:346–350. doi: 10.1016/j.soilbio.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Robledo C., Erickson D.L., Staines C.L., Erwin T.L., Kress W.J. Tropical plant-herbivore networks: Reconstructing species interactions using DNA barcodes. PLoS ONE. 2013;8:851. doi: 10.1371/journal.pone.0052967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen P.H., Pan Y.B., Chen R.K. High-throughput procedure for single pollen grain collection and polymerase chain reaction in plants. J. Integr. Plant Biol. 2008;50:375–383. doi: 10.1111/j.1744-7909.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- 20.EI-Salamouny S., Lange M., Jutzi M., Huber J., Jehle J.A. Comparative study on the susceptibility of cutworms (Lepidoptera: Noctuidae) to Agrotis segetum nucleopolyhedrovirus and Agrotis ipsilon nucleopolyhedrovirus. J. Invert. Pathol. 2003;84:75–82. doi: 10.1016/j.jip.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Spitzer K. Seasonal adult activity of Scotia ipsilon Hfn. (Lepidoptera, Noctuidae) in Bohemia. Acta Entomol. Bohemoslov. 1972;69:396–400. [Google Scholar]
- 22.Jia P.H. A brief report on long distance migration and mark-recapture of cutworm, Agrotis ypsilon Rott. in China. Plant Prot. 1985;11:20. [Google Scholar]
- 23.Showers W.B., Smelser R.B., Keaster A.J., Whitford F., Robinson J.F., Lopez J.D., Taylor S.E. Recapture of marked black cutworm (Lepidoptera: Noctuidae) males after long-range transport. Environ. Entomol. 1989;18:447–458. doi: 10.1093/ee/18.3.447. [DOI] [Google Scholar]
- 24.Wu K.M., Guo Y.Y. Effects of food quality and larval density on flight capacity of cotton bollworm. Acta Entomol. Sin. 1997;40:51–57. [Google Scholar]
- 25.Jiang X.F., Luo L.Z., Hu Y. The effect of compensatory nutrition condition on flight ability of beet armyworm Spodoptera exigua (Hübner) Acta Phytophylacica Sin. 2000;27:327–332. [Google Scholar]
- 26.Singh O.P., Parihar S.B.B. Effect of different hosts on the development of Heliothis armigera Hub. Bull. Entomol. Res. 1988;29:2168–2172. [Google Scholar]
- 27.Kishimoto-Yamada K., Kamiya K., Meleng P., Diway B., Kaliang H., Chong L., Itioka T., Sakai S., Ito M. Wide host ranges of herbivorous beetles? Insights from DNA bar coding. PLoS ONE. 2013;8:851. doi: 10.1371/journal.pone.0074426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kehat M., Wyndham M. The effect of food and water on development, longevity, and fecundity in the Rutherglen bug, Nysius vinitor (Hemiptera: Lygaeidae) Aust. J. Zool. 1972;20:119–130. doi: 10.1071/ZO9720119. [DOI] [Google Scholar]
- 29.Fang J.Y., Wang Z.H., Tang Z.Y. Atlas of Woody Plant in China: Distribution and Climate. Springer; Berlin, Germany: 2009. [Google Scholar]
- 30.CBOL Plant Working Group A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X.W., Yang Y., Henry R.J., Rossetto M., Wang Y.T., Chen S.L. Plant DNA barcoding: From gene to genome. Biol. Rev. 2015;90:157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 32.Wheeler D. The role of nourishment in oogenesis. Annu. Rev. Entomol. 1996;41:407–431. doi: 10.1146/annurev.en.41.010196.002203. [DOI] [PubMed] [Google Scholar]
- 33.Faegri K., van der Pijl L. The Principles of Pollination Ecology. Pergamon; Oxford, UK: 1979. [Google Scholar]
- 34.Liu Y.Q., Fu X.W., Feng H.Q., Liu Z.F., Wu K.M. Trans-regional migration of Agrotis ipsilon (Lepidoptera: Noctuidae) in north-east Asia. Ann. Entomol. Soc. Am. 2015;108:519–527. doi: 10.1093/aesa/sav050. [DOI] [Google Scholar]
- 35.Feng H.Q., Wu K.M., Cheng D.F., Guo Y.Y. Radar observations of the autumn migration of the beet armyworm Spodoptera exigua (Lepidoptera: Noctuidae) and other moths in northern China. Bull. Entomol. Res. 2003;93:115–124. doi: 10.1079/BER2002221. [DOI] [PubMed] [Google Scholar]
- 36.Cheng D.F., Feng H.Q., Wu K.M. Scanning Entomological Radar and Radar Observation for Insect Migration. Science Press; Beijing, China: 2005. [Google Scholar]
- 37.Bryant V.M., Pendleton M., Murry R.E., Lingren P.D., Raulston J.R. Techniques for studying pollen adhering to nectar-feeding corn earworm (Lepidoptera: Noctuidae) moths using scanning electron microscopy. J. Econ. Entomol. 1991;84:237–240. doi: 10.1093/jee/84.1.237. [DOI] [Google Scholar]
- 38.Fay M.F., Swensen S.M., Chase M.W. Taxonomic affinities of Medusagyne oppositifolia (Medusagynaceae) Kew Bull. 1997;52:111–120. doi: 10.2307/4117844. [DOI] [Google Scholar]
- 39.Fazekas A.J., Burgess K.S., Kesanakurti P.R., Graham S.W., Newmaster S.G., Husband B.C., Percy D.M., Hajibabaei M., Barrett S.C.H. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE. 2008;3:851. doi: 10.1371/journal.pone.0002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Wang F.X., Qian N.F., Zhang Y.L., Yang H.Q. Pollen Flora of China. Science Press; Beijing, China: 1995. pp. 1–461. [Google Scholar]
- 42.Li T.Q., Cao H.J., Kang M.S., Zhang Z.X., Zhao N., Zhang H. Pollen Flora of China Woody Plants by SEM. Science Press; Beijing, China: 2010. pp. 1–1233. [Google Scholar]
- 43.SPSS Incorporation . SPSS 13.0 for the Windows. SPSS Inc.; Chicago, IL, USA: 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.