Abstract
The second messenger 3′,5′-cyclic adenosine monophosphate (cAMP) is increasingly recognized as having many different roles in plant responses to environmental stimuli. To gain further insights into these roles, Arabidopsis thaliana cell suspension culture was treated with 100 nM of cell permeant 8-bromo-cAMP for 5 or 10 min. Here, applying mass spectrometry and comparative proteomics, 20 proteins were identified as differentially expressed and we noted a specific bias in proteins with a role in abiotic stress, particularly cold and salinity, biotic stress as well as proteins with a role in glycolysis. These findings suggest that cAMP is sufficient to elicit specific stress responses that may in turn induce complex changes to cellular energy homeostasis.
Keywords: cAMP-dependent proteome, biotic stress, abiotic stress, glycolysis, TCA cycle
1. Introduction
Environmental factors such as biotic and abiotic stresses can cause constraints on the growth, development and productivity of plants. These stresses also disturb cellular homeostasis, and consequently, a rapid response is initiated to alleviate the impact of stress. Signaling molecules including second messengers such as cyclic nucleotides play an important role in this early phase. Cyclic nucleotides and 3′,5′-cyclic adenosine monophosphate (cAMP) in particular have long been established as important messengers in prokaryotes as well as in lower and higher eukaryotes [1]. In plants, cAMP has been reported to have direct and/or indirect roles in many developmental processes including pollen growth [2] and response to biotic stress [3,4,5].
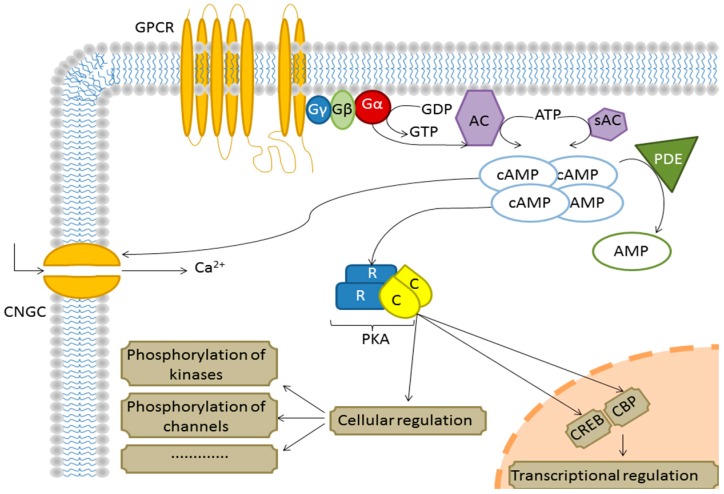
The levels of cAMP have been shown to increase in response to biotic stress and subsequently influence calcium (Ca2+) influx by targeting membrane cyclic nucleotide-gated channels (CNGCs) [6], thereby increasing cytosolic free Ca2+. Plant CNGCs are ligand and voltage-gated channels functioning in sensory signal transduction and have been proposed to regulate Ca2+ influx into the cytosol [7,8]. The CNGCs also have a role in plant development and plant responses to biotic and abiotic stress [9]. CNGCs open upon binding of either cAMP or 3′,5′-cyclic guanosine monophosphate (cGMP) which act as potential activating ligands [8] and close upon binding of Ca2+/calmodulin (Figure 1). The cyclic nucleotides and calmodulin competitively bind to overlapping binding sites at the C-terminus in the cytosolic part of the channel [10,11]. In turn, Ca2+ amplifies the signal as part of the cellular response [12]. In Arabidopsis, there are 20 annotated CNGCs [13,14] and CNGC2 in particular is an inward-rectifying potassium (K2+) channel that is blocked by Ca2+ [8]. Mutation of CNGC2 results in an impaired hypersensitive response to avirulent pathogens and cAMP-/cGMP-dependent cytosolic Ca2+ elevation [15].
Figure 1.
An illustration of some signal transduction pathways mediated by the G-protein coupled receptor based on the animal system. When adenylyl cyclase (AC) is activated by the Gα-subunit of the G-protein coupled receptor (GPCR), it catalyzes the formation of cAMP. Cyclic AMP then activates many substrates and kinases such as protein kinase A (PKA) which will regulate many biological processes. Cyclic nucleotide-gated channel (CNGC); soluble adenylyl cyclase (sAC); phosphodiesterase (PDE); cAMP response element-binding (CREB); CREB-binding protein (CBP); catalytic subunit of PKA (C); regulatory subunit of PKA (R).
At the structural level, voltage-independent channels (VICs) in the plasma membrane of Arabidopsis root cells have been reported to be highly sensitive to cAMP and these channels are also implicated in salt tolerance in Arabidopsis seedlings by reducing sodium (Na+) influx [16]. Furthermore, an increasing number of plant mononucleotide cyclases, including adenylate cyclases (enzymes that synthesis cAMP from ATP), have been predicted [17,18] and experimentally confirmed [2,19].
Mass spectrometry-based approaches have been employed previously to study cAMP in plants, firstly to determine the concentration of cAMP [20], secondly to look at the interactome network of cAMP [21], and finally to look at the cAMP-dependent responses [22]. In the latter study, systems-level analyses showed that upon treatment of Arabidopsis plants with 1 or 10 µM of cAMP, proteins involved in responses to temperature, light and photosynthesis were changing in abundance at 1 and 3 h after treatment. However, previous studies have used high concentrations of cAMP, which may not be physiologically relevant. Therefore, we have undertaken studying cAMP-dependent proteome changes at low cAMP concentration and monitoring early cellular responses with a view to gain further insight into the possible physiological implications at cellular levels.
2. Results and Discussion
In order to obtain a cAMP-dependent proteome, Arabidopsis ecotype Columbia-0 (Col-0) cell suspension cultures were treated with 100 nM of cell permeant 8-Br-cAMP and samples were collected at 5 and 10 min after treatment. Extracted proteins from three biological replicates were digested with trypsin and the peptides were labeled with tandem mass tag (TMT) six-plex for quantitative analysis. A total of 1023 quantifiable proteins were identified (false discovery rate: 0.8%). Proteins present in at least two biological replicates from each treatment time point were compared to their corresponding controls. Twenty proteins were detected as differentially expressed, with at least a ±1.5-fold change (±0.6 in log2 transformation) and a statistically significant p-value of ≤0.05 (Table 1). After 5 min of cAMP treatment, three proteins increased in abundance and four proteins decreased in abundance, while after 10 min of cAMP treatment, 10 proteins increased in abundance and four proteins decreased in abundance. In this study, only a few of the proteins fulfilled the set thresholds and it is of particular note that in addition to each protein being detected in at least two out of three biological replicates, proteins from cAMP-treated cells were compared with mock-treated cells collected at the same time point.
Table 1.
3′,5′-Cyclic adenosine monophosphate (cAMP)-responsive proteins after 5 and 10 min treatment.
| Accession Number | Protein Name | Fold Change (Log2) | p-Value | GO Term |
|---|---|---|---|---|
| Proteins identified changing after 5 min of cAMP treatment | ||||
| At3g16460 | Jacalin-related lectin 34 | 4.495 | 0.01358 | A |
| At2g01140 | Fructose-bisphosphate aldolase 3 | 3.532 | 0.01473 | B, D, E |
| At1g20450 | Early response to dehydration 10 (ERD10) | 0.971 | 0.04245 | A, B, E |
| At2g37220 | RNA-binding protein | −0.607 | 0.00180 | A, C |
| At1g08110 | Glyoxalase I | −0.651 | 0.00912 | B, D, E |
| At1g14980 | Chaperonin 10 | −0.733 | 0.02203 | |
| At2g27710 | 60S acidic ribosomal protein family | −1.476 | 0.03411 | A |
| Proteins identified changing after 10 min of cAMP treatment | ||||
| At1g23100 | GroES-like family protein | 39.566 | 0.02049 | |
| At1g24360 | 3-Oxoacyl-[acyl-carrier-protein] reductase | 9.238 | 0.01110 | A |
| At1g28200 | FH interacting protein 1 | 2.979 | 0.04999 | |
| At1g14980 | Chaperonin 10 | 1.771 | 0.04747 | |
| At1g48920 | Nucleolin like 1 | 1.434 | 0.03283 | |
| At1g53240 | Mitochondrial malate dehydrogenase | 0.846 | 0.03931 | A, B, C, D, E |
| At2g38540 | Lipid transfer protein 1 | 0.844 | 0.04096 | |
| At2g41430 | Early response to dehydration 15 (ERD15) | 0.756 | 0.03081 | B, C, D |
| At2g47730 | Glutathione S-transferase Φ 8 | 0.726 | 0.04412 | A, B, C |
| At3g16450 | Jacalin-related lectin 33 | 0.628 | 0.02041 | A |
| At1g11580 | Methylesterase PCR A | −0.606 | 0.03673 | C |
| At4g21860 | Methionine sulfoxide reductase B2 | −0.697 | 0.04911 | |
| At4g38740 | Rotamase cyclophilin 1 (ROC1) | −0.706 | 0.00733 | D |
| At5g47200 | RAB GTPase homolog 1A | −0.856 | 0.03223 | |
GO, gene ontology; A, response to cold (GO:0009409); B, response to salt stress (GO:0009651); C, response to bacterium (GO:0009617); D, response to cadmium ion (GO:0046686); E, glycolytic process (GO:0006096).
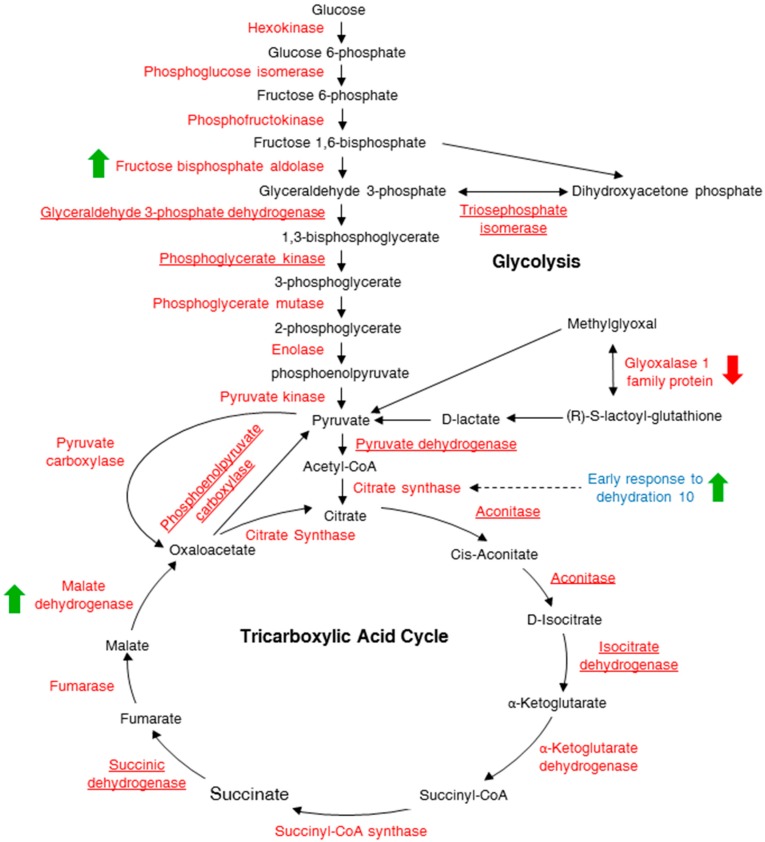
Gene ontology (GO) analysis using FatiGO+ allowed classification of identified proteins in each group based on their enrichments as compared to the normal distribution [23,24]. Eight of the 20 differentially expressed proteins are in the category “response to cold”, six proteins are in the category “response to salt stress”, five proteins are in both “response to bacterium” and “response to cadmium ion” and four proteins are enriched in the category “glycolytic process” (Table 1). Although 24 proteins that are involved in the glycolytic process were identified (Table S1), only four were significantly changing in abundance at either 5 or 10 min after cAMP treatment. The four proteins include fructose-bisphosphate aldolase 3 (FBA3; At2g01140), mitochondrial malate dehydrogenase (MDH; At1g53240) and early response to dehydration 10 (ERD10; At1g20450) which increased in abundance while glyoxalase I (At1g08110) decreased (Figure 2).
Figure 2.
A schematic diagram of the glycolysis and tricarboxylic acid (TCA) cycle pathways showing proteins differentially expressed post cAMP treatment. The glycolysis and TCA metabolic products are shown in black, the enzymes are in red and a protein associated with the TCA cycle is shown in blue. The green arrows indicate proteins that accumulate in response to cAMP while the red arrow indicates a protein that is reduced in quantity. All the other proteins underlined are some of the proteins identified in the current study that are not changing in abundance after cAMP treatment.
Fructose-bisphosphate aldolase catalyzes two reversible reactions. The first involves the enzymatic conversion of fructose 1,6-bisphosphate into the triose phosphates, dihydroxyacetone phosphate and glyceraldehyde 3-phosphate in gluconeogenesis. The second reaction involves the condensation reaction of fructose-1,6-biphosphate and sedoheptulose-1,7-biphosphate in the Calvin cycle [25]. The mRNA levels of FBA3 have been shown to increase in response to abscisic acid (ABA) and salicylic acid (SA) and to various abiotic stresses such as salinity, drought, cold and heat, but the expression levels were noted to decrease in response to cadmium (Cd2+) ions [26]. Increased abundance of FBA3 was also observed at 5 min after cAMP treatment (Table 1) and this implicates cAMP in abiotic stress responses.
Malate dehydrogenase reversibly catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD+ to NADH in the tricarboxylic acid (TCA) cycle [27]. The control of the TCA cycle activity is dependent on several enzymes, including aconitase, fumarase, succinate dehydrogenase, 2-oxoglutarate dehydrogenase and MDH, which all have different flux control coefficients [28]. A flux control coefficient is a quantifiable parameter measuring the contribution or effect of enzymes on the overall steady-state flux of a metabolic pathway. Mitochondrial MDH has the highest flux control coefficient (1.76) and is considered one of the rate-limiting steps [28]. Plant mitochondrial MDH is also important in oxidizing NADH in the TCA cycle and in the malate/aspartate shuttle in photorespiration [29]. Malate dehydrogenase has been shown to increase in abundance in response to oxidative stress [30] and flooding [31]. In tomato (Solanum lycopersicum), mutations in mitochondrial MDH affect growth and fruit yield [32], while in Arabidopsis, single and double knockouts of the two mitochondrial MDH genes appear to have little effect on growth and development [33]. In addition to MDH increasing in abundance in response to cAMP, ERD10 also increased in abundance in response to cAMP treatment (Table 1). An increase in abundance of ERD10 has previously been shown to have a protective role by slowing the heat-induced aggregation and/or inactivation rate of various substrates such as alcohol dehydrogenase and citrate synthase, the enzyme that catalyses the first step of the TCA cycle [34]. Given that increases in both MDH and ERD10 abundance have been observed to increase the respiration rate [29,34], an increase in MDH and ERD10 in response to cAMP implicates cAMP directly or indirectly in the modulation of the TCA cycle and, with it, energy metabolism.
In contrast, glyoxalase I, a protein that detoxifies methylglyoxal, a cytotoxic by-product of glycolysis [35], decreases in abundance in response to cAMP. In Arabidopsis, glyoxalase 1 (At1g08110) has been shown to decrease in abundance under abiotic stress conditions including salinity, drought, osmotic and cold [36]. Given that proteins ERD10, mitochondrial MDH and FBA3 increase and glyoxalase I decreases, we hypothesize that cAMP has a regulatory function in abiotic stress responses in general and the modulation of the TCA cycle in particular.
Of the 20 differentially expressed proteins identified in this study, five proteins, 3-oxoacyl-[acyl-carrier-protein] reductase (At1g24360), rotamase cyclophilin 1 (ROC1; At4g38740), RNA-binding protein (At2g37220), early response to dehydration 15 (ERD15; At2g41430) and mitochondrial MDH, have been associated with the response to ABA. Abscisic acid controls several aspects of development and adaptation to stress [37,38,39]. The 3-oxoacyl-[acyl-carrier-protein] reductase that catalyzes the first reduction step in fatty acid biosynthesis [40] is repressed by ABA in guard cells of Arabidopsis [41], but it increases in abundance in response to cAMP treatment in Arabidopsis cell suspension culture (Table 1). On the other hand, ROC1, a signaling protein component controlling plant responses to light, is an important link between phytochrome signaling and brassinosteroid sensitivity [42]. Rotamase cyclophilin 1 has been shown to decrease in abundance in response to abiotic stress and after ABA treatment [43], and this is consistent with the response to cAMP (Table 1).
In Arabidopsis, RNA-binding proteins have also been shown to play a role in ABA signaling during germination and drought tolerance [44], and ABA affects tyrosine dephosphorylation of a chloroplast-localized RNA-binding protein (At2g37220) in particular, and may modify its RNA-binding activity and thereby regulate gene expression [45]. The RNA-binding protein is related to the maize glycine-rich RNA-binding protein A, and the encoding gene has been shown to be induced by ABA and this is also a target for phosphorylation [46,47]. The protein has also been observed to increase in abundance at least two-fold in response to cold treatment [48]. However, in response to cAMP, the RNA-binding protein decreased in abundance (Table 1). It is therefore important to follow up on the phosphorylation status of this RNA-binding protein to see whether cAMP affects tyrosine dephosphorylation much like in the response to ABA.
Another protein that is involved in ABA responses is ERD15. In addition to being a negative regulator of ABA responses, ERD15 has a role in the defense against pathogens [38]. Besides the central role of ABA in controlling responses to abiotic stress stimuli, ABA also influences biotic stress responses and may interfere with signaling that is regulated by other hormones including SA [49]. ABA treatment prior to infection can increase the susceptibility of Arabidopsis to Pseudomonas syringae pv. tomato, while decreased ABA levels can improve SA-dependent defenses, suggesting that ABA modulates SA-dependent defense responses [50]. Expression of ERD15 was observed to increase at least 50 times more in Arabidopsis inoculated with Paenibacillus polymyxa than in untreated plants [51]. Given that ERD15 is induced by biotic stress, rapidly but transiently induced in response to ABA [52] and increased in abundance in response to cAMP treatment, this again is consistent with the hypothesis that cAMP is part of the biotic stress response.
Glutathione S-transferase Φ8 (GSTF8, At2g47730), a marker for early stress and defense responses, also increases in abundance in response to cAMP (Table 1). The expression of GSTF8 can be induced by a range of biotic and abiotic stresses, hydrogen peroxide and in response to SA [53,54,55,56,57]. Overall this increase in abundance of GSTF8 upon induction by various stresses is also an indirect support for a role of cAMP in both biotic and abiotic stress responses.
The response to cAMP also leads to enrichment in the GO categories “response to cold” and “response to salt stress” (Table 1) and the proteins in these categories include the mitochondrial MDH, GSTF8, RNA-binding protein, 3-oxoacyl-(acyl-carrier-protein) reductase and ERD10. Interestingly, ERD10 has been reported to be a general anti-stress protein that is up-regulated in response to a broad range of abiotic stresses [58] and was previously shown to increase in abundance after treatment of Arabidopsis leaves with 1 or 10 µM cAMP [22]. Furthermore, it is not uncommon for salinity stress-responsive proteins to also be responsive to Cd2+ ion stress and for both responses to confer a degree of cross-protection [59]. It is therefore not surprising to see that five out of the six proteins enriched in the category “response to salt stress” are also enriched in the category “response to cadmium ion” (Table 1).
Treatment of Arabidopsis roots with Cd2+ ions (10 µM) has been shown to alter the root proteome, e.g., an increase in the accumulation of proteins involved in the synthesis of glutathione-derived metal-binding proteins such as ATP sulfurylase, glutathione S-transferase, latex allergen-like proteins [60]. Interestingly, treatment of Arabidopsis seedlings with Cd2+ ions (50 µM) has been observed to increase the cellular cAMP concentration [61], and this in turn can induce the expression of enzymes involved in the phenylpropanoid pathway [62]. The phenylpropanoid pathway synthesizes precursors and metabolites protecting against abiotic stress including Cd2+ [63,64]. Taken together, the changing abundance of at least some proteins in response to Cd2+ may indeed require a direct or indirect interaction with cAMP.
What is the link between cAMP, Cd2+ and salinity? In Arabidopsis, an increase in cAMP levels significantly reduced Na+ influx in roots and it was shown that VIC-mediated Na+ currents are down-regulated by cAMP [16]. Further, treatment of Arabidopsis roots with 100 mM NaCl causes a decrease in cellular levels of cAMP, leading to a deactivation of protein kinase A. Thus, during salinity stress, cAMP suppresses expression of the Na+ efflux pump [16]. In addition, five of the differentially expressed proteins enriched in the category “response to salt stress” (Table 1) have been previously observed to undergo differential expression in response to low Cd2+ stress [65]. Most of the proteins enriched in “response to cadmium ion” are also enriched in the category “response to salt stress”, suggesting that cAMP has a role in both salinity and Cd2+ stress responses.
Twelve of the differentially expressed proteins identified (Table 1) have previously been linked to cabbage leaf curl virus infection [66]. Of these, seven show a similar differential accumulation pattern to cAMP (Table 2) consistent with a generalized role in plant defense against pathogens.
Table 2.
Comparison between proteins responsive to cAMP and proteins involved in pathogen response.
| Accession Number | Protein Name | cAMP Treatment | Pathogen Response * | ||
|---|---|---|---|---|---|
| Fold Change (Log2) | p-Value | Fold Change (Log2) | p-Value | ||
| AT3G16460 | Jacalin-related lectin 34 | 4.495 | 0.01358 | 0.274 | 4.90 × 10−5 |
| AT2G01140 | Fructose-bisphosphate aldolase 3 | 3.532 | 0.01473 | 0.372 | 1.48 × 10−5 |
| AT1G20450 | Early response to dehydration 10 (ERD10) | 0.971 | 0.04245 | −0.756 | 7.00 × 10−6 |
| AT2G37220 | RNA-binding protein | −0.607 | 0.00180 | −0.539 | 2.35 × 10−5 |
| AT2G27710 | 60S acidic ribosomal protein family | −1.476 | 0.03411 | −0.692 | 2.25 × 10−7 |
| AT1G24360 | 3-Oxoacyl-[acyl-carrier-protein] reductase | 9.238 | 0.01110 | −0.265 | 3.97 × 10−4 |
| AT1G53240 | Mitochondrial malate dehydrogenase | 0.846 | 0.03931 | −0.599 | 1.11 × 10−5 |
| AT2G38540 | Lipid transfer protein 1 | 0.844 | 0.04096 | −2.701 | 5.04 × 10−25 |
| AT1G11580 | Methylesterase PCR A | −0.606 | 0.03673 | −0.194 | 1.86 × 10−3 |
| AT4G21860 | Methionine sulfoxide reductase B2 | −0.697 | 0.04911 | −0.766 | 2.55 × 10−8 |
| AT4G38740 | Rotamase cyclophilin 1 (ROC1) | −0.706 | 0.00733 | −0.827 | 1.40 × 10−7 |
| AT5G47200 | RAB GTPase homolog 1A | −0.856 | 0.03223 | 0.498 | 2.11 × 10−7 |
* Proteins reported in [66].
Finally, a previous study using 1 or 10 µM cAMP treatments of Arabidopsis leaves for 1 or 3 h implicated cAMP in light and temperature responses [22]. Despite the much higher concentration and later time points used in the earlier study, three proteins, methylesterase PCR A (At1g11580), 60S acidic ribosomal protein family (At2g27710) and RNA-binding protein (At2g37220), were common and decreased in abundance in response to cAMP, adding to the growing evidence for a key role of cAMP in the transduction and/or modulation of environmental stimuli.
3. Materials and Methods
3.1. Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) cell suspension culture was grown in 250 mL Erlenmeyer flasks containing 100 mL of Gamborg’s B5 medium with vitamins (Sigma-Aldrich, St. Louis, MO, USA) [67] supplemented with 3% (w/v) sucrose, 0.05 µg·mL−1 (v/v) kinetin, 1 mg·mL−1 2,4-dichlorophenoxyacetic acid and 0.05% (w/v) MES. Cells were grown in a growth chamber (Innova® 43, New Brunswick Scientific Co., Edison, NJ, USA) under photosynthetic light with 12 h light/12 h dark cycles at 23 °C and orbital agitation at 120 rpm and sub-cultured every 1-days.
3.2. cAMP Treatment and Protein Extraction
At 10 days post-subculturing, three biological replicate flasks were treated with 100 nM of 8-bromo-cAMP and cells of each mock (equal volume of water) or cAMP treated were collected at 0, 5 and 10 min post-treatment. Media were drained off using Stericup® filter unit (Millipore, Billerica, MA, USA), and the cells were immediately snap frozen in liquid nitrogen and stored at −80 °C until use. Approximately 1 g of cells was ground to a fine powder with mortar and pestle in liquid nitrogen and proteins were precipitated in trichloroacetic acid in acetone, vortexed and incubated overnight. Precipitated proteins were pelleted, washed and re-suspended in urea lysis buffer (7 M urea, 2 M thiourea, 4% (w/v) 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate). Approximately 100 µg of total soluble protein extract was reduced, alkylated, digested with trypsin and purified using Sep-Pak Vac tC18 100 mg cartridge (Waters, Milford, MA, USA), as described previously [68], prior to drying in a Speed Vac concentrator (Thermo Scientific, Bremen, Germany).
3.3. Peptide Labeling Using Tandem Mass Tag and Peptide Fractionation by OFFGEL Fractionator
Purified and dried tryptic peptides were labeled with tandem mass tag (TMT™) six-plex (Thermo Scientific, Bremen, Germany) according to manufacturer’s instructions. Each biological replicate was labeled separately, pooled together and then fractionated using a 3100 OFFGEL fractionator (Agilent Technologies, Santa Clara, CA, USA) using 24-well high resolution immobilized pH gradient strips, pH 3–10, as described previously [68].
3.4. Protein Identification by LTQ Orbitrap and Quantification of Differentially Expressed Proteins
Dried peptide fractions were re-suspended in a solution containing 5% (v/v) acetonitrile and 0.1% (v/v) formic acid and analyzed by an LTQ-Orbitrap Velos (Thermo Scientific, Bremen, Germany) coupled with a nanoelectrospray ion source (Proxeon Biosystems, Odense, Denmark) for nano-LC–MS/MS analyses. The MS scan range was 350 to 1600 m/z with the normalized collision-induced dissociation at 35.0 V. The top 10 precursor ions were selected in the MS scan with resolution R = 60,000 for fragmentation in the linear ion trap. All spectra were submitted for protein identification to MASCOT search engine (Matrix Science, London, UK) as described previously [22], except that TMT labeling was added as a fixed modification. Identified proteins were evaluated and quantitated using Scaffold Q+ software, version 4.0.4 (Proteome Software, Portland, OR, USA). Proteins were considered as positive identifications if they were identified with a minimum of two unique peptides, a MASCOT ion score ≥26, a peptide probability of 95% and a protein threshold of 99%. Abundance levels of positively identified proteins from cAMP-treated cells that were present in at least two technical replicate were compared with mock-treated cells collected at the same time point. Differential expression of a protein was considered significant if the fold change was at least ±1.5-fold change (±0.6 in log2 transformation) and statistical significance p-value of ≤0.05.
3.5. Computational Analysis of Functional Enrichment
The gene ontology (GO) and functional categorization analyses of the differentially expressed proteins were performed using FatiGO+ tool in Babelomics version 5 suite [69,70].
4. Conclusions
cAMP treatment causes changes in the proteome that are diagnostic for biotic and abiotic responsive proteins. Furthermore, cAMP also affects abundance levels of enzymes in the glycolytic pathway and the TCA cycle and this is likely to have direct implications for the energy-transducing pathways and ATP generation. Finally, cAMP may conceivably link biotic and abiotic stress responses in stress-dependent changes of energy metabolism.
Acknowledgments
We thank Ludivine Thomas for her intellectual input during the project.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/6/852/s1.
Author Contributions
Chris Gehring, Claudius Marondedze and May Alqurashi designed the research. May Alqurashi performed the experiments and data analysis. All authors participated in writing and revising the manuscript and approved the final version.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gancedo J.M. Biological roles of cAMP: Variations on a theme in the different kingdoms of life. Biol. Rev. 2013;88:645–668. doi: 10.1111/brv.12020. [DOI] [PubMed] [Google Scholar]
- 2.Moutinho A., Hussey P., Trewavas A., Malhó R. Cyclic AMP acts as a second messenger in pollen tube growth and reorientation. Proc. Natl. Acad. Sci. USA. 2001;98:10481–10486. doi: 10.1073/pnas.171104598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooke C.J., Smith C.J., Walton T.J., Newton R.P. Evidence that cyclic AMP is involved in the hypersensitive response of Medicago sativa to a fungal elicitor. Phytochemistry. 1994;35:889–895. doi: 10.1016/S0031-9422(00)90633-2. [DOI] [Google Scholar]
- 4.Kim H.-S., Park S.-Y., Lee S., Adams E.L., Czymmek K., Kang S. Loss of cAMP-dependent protein kinase A affects multiple traits important for root pathogenesis by Fusarium oxysporum. Mol. Plant Microbe Interact. 2011;24:719–732. doi: 10.1094/MPMI-11-10-0267. [DOI] [PubMed] [Google Scholar]
- 5.Lemtiri-Chlieh F., Thomas L., Marondedze C., Irving H., Gehring C. Cyclic nucleotides and nucleotide cyclases in Plant stress responses. In: Shanker A., Venkateswarlu B., editors. Abiotic Stress Response in Plants—Physiological, Biochemical and Genetic Perspectives. InTech; Vienna, Austria: 2011. p. 346p. [Google Scholar]
- 6.Ma W., Qi Z., Smigel A., Walker R., Verma R., Berkowitz G. Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc. Natl. Acad. Sci. USA. 2009;106:20995–21000. doi: 10.1073/pnas.0905831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemtiri-Chlieh F., Berkowitz G.A. Cyclic adenosine monophosphate regulates calcium channels in the plasma membrane of Arabidopsis leaf guard and mesophyll cells. J. Biol. Chem. 2004;279:35306–35312. doi: 10.1074/jbc.M400311200. [DOI] [PubMed] [Google Scholar]
- 8.Qi Z., Verma R., Gehring C., Yamaguchi Y., Zhao Y., Ryan C., Berkowitz G. Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. USA. 2010;107:21193–21198. doi: 10.1073/pnas.1000191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kugler A., Köhler B., Palme K., Wolff P., Dietrich P. Salt-dependent regulation of a CNG channel subfamily in Arabidopsis. BMC Plant Biol. 2009;9:1–11. doi: 10.1186/1471-2229-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan B., Sherman T., Fromm H. Cyclic nucleotide-gated channels in plants. FEBS Lett. 2007;581:2237–2246. doi: 10.1016/j.febslet.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Ladwig F., Dahlke R.I., Stührwohldt N., Hartmann J., Harter K., Sauter M. Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell. 2015;27:1718–1729. doi: 10.1105/tpc.15.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeWald D.B., Torabinejad J., Jones C.A., Shope J.C., Cangelosi A.R., Thompson J.E., Prestwich G.D., Hama H. Rapid Accumulation of Phosphatidylinositol 4,5-Bisphosphate and Inositol 1,4,5-Trisphosphate Correlates with Calcium Mobilization in Salt-Stressed Arabidopsis. Plant Physiol. 2001;126:759–769. doi: 10.1104/pp.126.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frietsch S., Wang Y.-F., Sladek C., Poulsen L.R., Romanowsky S.M., Schroeder J.I., Harper J.F. A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc. Natl. Acad. Sci. USA. 2007;104:14531–14536. doi: 10.1073/pnas.0701781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konrad K.R., Hedrich R. The use of voltage-sensitive dyes to monitor signal-induced changes in membrane potential–ABA triggered membrane depolarization in guard cells. Plant J. 2008;55:161–173. doi: 10.1111/j.1365-313X.2008.03498.x. [DOI] [PubMed] [Google Scholar]
- 15.Ma W., Berkowitz G.A. Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol. 2011;190:566–572. doi: 10.1111/j.1469-8137.2010.03577.x. [DOI] [PubMed] [Google Scholar]
- 16.Maathuis F.J.M., Sanders D. Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 2001;127:1617–1625. doi: 10.1104/pp.010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gehring C. Adenyl cyclases and cAMP in plant signaling—Past and present. Cell Commun. Signal. 2010;8:15–19. doi: 10.1186/1478-811X-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marondedze C., Wong A., Thomas L., Irving H., Gehring C. Cyclic Nucleotide Monophosphates in Plants and Plant Signaling. In: Barrett J.E., Flockerzi V., Frohman M.A., Geppetti P., Hofmann F.B., Michel M.C., Page C.P., Thorburn A.M., Wang K., editors. Handbook of Experimental Pharmacology. Springer; Berlin, Germany; Heidelberg, Germany: 2016. pp. 1–17. [DOI] [PubMed] [Google Scholar]
- 19.Al-Younis I., Wong A., Gehring C. The Arabidopsis thaliana K+-uptake permease 7 (AtKUP7) contains a functional cytosolic adenylate cyclase catalytic centre. FEBS Lett. 2015;589:3848–3852. doi: 10.1016/j.febslet.2015.11.038. [DOI] [PubMed] [Google Scholar]
- 20.Newton R., Gibbs N., Moyse C., Wiebers J., Brown E. Mass spectrometric identification of adenosine 3′:5′-cyclic monophosphate isolated from a higher plant tissue. Phytochemistry. 1980;19:1909–1911. doi: 10.1016/0031-9422(80)83002-0. [DOI] [Google Scholar]
- 21.Donaldson L., Meier S., Gehring C. The Arabidopsis cyclic nucleotide interactome. Cell Commun. Signal. 2016;14:10–27. doi: 10.1186/s12964-016-0133-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas L., Marondedze C., Ederli L., Pasqualini S., Gehring C. Proteomic signatures implicate cAMP in light and temperature responses in Arabidopsis thaliana. J. Proteom. 2013;83:47–59. doi: 10.1016/j.jprot.2013.02.032. [DOI] [PubMed] [Google Scholar]
- 23.Meier S., Gehring C. A guide to the integrated application of on-line data mining tools for the inference of gene functions at the systems level. Biotechnol. J. 2008;3:1375–1387. doi: 10.1002/biot.200800142. [DOI] [PubMed] [Google Scholar]
- 24.Alqurashi M., Meier S. Inferring biological functions of guanylyl cyclases with computational methods. In: Gehring C., editor. Cyclic Nucleotide Signaling in Plants. Volume 1016. Humana Press; Totowa, NJ, USA: 2013. pp. 225–234. [DOI] [PubMed] [Google Scholar]
- 25.Flechner A., Gross W., Martin W.F., Schnarrenberger C. Chloroplast class I and class II aldolases are bifunctional for fructose-1,6-biphosphate and sedoheptulose-1,7-biphosphate cleavage in the Calvin cycle. FEBS Lett. 1999;447:200–202. doi: 10.1016/S0014-5793(99)00285-9. [DOI] [PubMed] [Google Scholar]
- 26.Lu W., Tang X., Huo Y., Xu R., Qi S., Huang J., Zheng C., Wu C. Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene. 2012;503:65–74. doi: 10.1016/j.gene.2012.04.042. [DOI] [PubMed] [Google Scholar]
- 27.Nunes-Nesi A., Carrari F., Lytovchenko A., Smith A.M.O., Ehlers Loureiro M., Ratcliffe R.G., Sweetlove L.J., Fernie A.R. Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 2005;137:611–622. doi: 10.1104/pp.104.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araújo W.L., Nunes-Nesi A., Nikoloski Z., Sweetlove L.J., Fernie A.R. Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 2012;35:1–21. doi: 10.1111/j.1365-3040.2011.02332.x. [DOI] [PubMed] [Google Scholar]
- 29.Scheibe R. Malate valves to balance cellular energy supply. Physiol. Plant. 2004;120:21–26. doi: 10.1111/j.0031-9317.2004.0222.x. [DOI] [PubMed] [Google Scholar]
- 30.Taylor N.L., Heazlewood J.L., Day D.A., Millar A.H. Differential impact of environmental stresses on the pea mitochondrial proteome. Mol. Cell. Proteom. 2005;4:1122–1133. doi: 10.1074/mcp.M400210-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Komatsu S., Yamamoto A., Nakamura T., Nouri M.-Z., Nanjo Y., Nishizawa K., Furukawa K. Comprehensive analysis of mitochondria in roots and hypocotyls of soybean under flooding stress using proteomics and metabolomics techniques. J. Proteome Res. 2011;10:3993–4004. doi: 10.1021/pr2001918. [DOI] [PubMed] [Google Scholar]
- 32.Nunes-Nesi A., Araújo W.L., Fernie A.R. Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol. 2011;155:101–107. doi: 10.1104/pp.110.163816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tronconi M.A., Fahnenstich H., Gerrard Weehler M.C., Andreo C.S., Flügge U.-I., Drincovich M.F., Maurino V.G. Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism. Plant Physiol. 2008;146:1540–1552. doi: 10.1104/pp.107.114975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kovacs D., Kalmar E., Torok Z., Tompa P. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 2008;147:381–390. doi: 10.1104/pp.108.118208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thornalley P.J. Glyoxalase I—Structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 2003;31:1343–1348. doi: 10.1042/bst0311343. [DOI] [PubMed] [Google Scholar]
- 36.Mustafiz A., Singh A., Pareek A., Sopory S., Singla-Pareek S. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genom. 2011;11:293–305. doi: 10.1007/s10142-010-0203-2. [DOI] [PubMed] [Google Scholar]
- 37.Finkelstein R.R., Rock C.D. Abscisic acid biosynthesis and response. Arab. Book. 2002;1:852. doi: 10.1199/tab.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harwood J.L. Fatty acid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988;39:101–138. doi: 10.1146/annurev.pp.39.060188.000533. [DOI] [Google Scholar]
- 41.Leonhardt N., Kwak J.M., Robert N., Waner D., Leonhardt G., Schroeder J.I. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trupkin S.A., Mora-García S., Casal J.J. The cyclophilin ROC1 links phytochrome and cryptochrome to brassinosteroid sensitivity. Plant J. 2012;71:712–723. doi: 10.1111/j.1365-313X.2012.05013.x. [DOI] [PubMed] [Google Scholar]
- 43.Matsui A., Ishida J., Morosawa T., Mochizuki Y., Kaminuma E., Endo T.A., Okamoto M., Nambara E., Nakajima M., Kawashima M., et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- 44.Kuhn J.M., Schroeder J.I. Impacts of altered RNA metabolism on abscisic acid signaling. Curr. Opin. Plant Biol. 2003;6:463–469. doi: 10.1016/S1369-5266(03)00084-0. [DOI] [PubMed] [Google Scholar]
- 45.Ghelis T., Bolbach G., Clodic G., Habricot Y., Miginiac E., Sotta B., Jeannette E. Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol. 2008;148:1668–1680. doi: 10.1104/pp.108.124594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez J., Sanchez-Martinez D., Stiefel V., Rigau J., Puigdomenech P., Pages M. A gene induced by the plant hormone abscisic acid in response to water stress encodes a glycine-rich protein. Nature. 1988;334:262–264. doi: 10.1038/334262a0. [DOI] [PubMed] [Google Scholar]
- 47.Testi M.G., Croce R., Laureto P.P.-D., Bassi R. A CK2 site is reversibly phosphorylated in the photosystem II subunit CP29. FEBS Lett. 1996;399:245–250. doi: 10.1016/S0014-5793(96)01333-6. [DOI] [PubMed] [Google Scholar]
- 48.Amme S., Matros A., Schlesier B., Mock H.-P. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J. Exp. Bot. 2006;57:1537–1546. doi: 10.1093/jxb/erj129. [DOI] [PubMed] [Google Scholar]
- 49.Mauch-Mani B., Mauch F. The role of abscisic acid in plant–pathogen interactions. Curr. Opin. Plant Biol. 2005;8:409–414. doi: 10.1016/j.pbi.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 50.Mohr P.G., Cahill D.M. Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct. Plant Biol. 2003;30:461–469. doi: 10.1071/FP02231. [DOI] [PubMed] [Google Scholar]
- 51.Timmusk S., Wagner E.G.H. The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: A possible connection between biotic and abiotic stress responses. Mol. Plant-Microbe Interact. 1999;12:951–959. doi: 10.1094/MPMI.1999.12.11.951. [DOI] [PubMed] [Google Scholar]
- 52.Kariola T., Brader G., Helenius E., Li J., Heino P., Palva E.T. Early Response to Dehydration 15. A negative Regulator of ABA-responses in Arabidopsis. Plant Physiol. 2006;142:1559–1573. doi: 10.1104/pp.106.086223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen W., Chao G., Singh K.B. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 1996;10:955–966. doi: 10.1046/j.1365-313X.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- 54.Chen W., Singh K.B. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 1999;19:667–677. doi: 10.1046/j.1365-313x.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 55.Wagner U., Edwards R., Dixon D., Mauch F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 2002;49:515–532. doi: 10.1023/A:1015557300450. [DOI] [PubMed] [Google Scholar]
- 56.Uquillas C., Letelier I., Blanco F., Jordana X., Holuigue L. NPR1-Independent Activation of Immediate Early Salicylic Acid-Responsive Genes in Arabidopsis. Mol. Plant Microbe Interact. 2004;17:34–42. doi: 10.1094/MPMI.2004.17.1.34. [DOI] [PubMed] [Google Scholar]
- 57.Perl-Treves R., Foley R.C., Chen W., Singh K.B. Early Induction of the Arabidopsis GSTF8 Promoter by Specific Strains of the Fungal Pathogen Rhizoctonia solani. Mol. Plant Microbe Interact. 2004;17:70–80. doi: 10.1094/MPMI.2004.17.1.70. [DOI] [PubMed] [Google Scholar]
- 58.Kimura M., Yamamoto Y.Y., Seki M., Sakurai T., Sato M., Abe T., Yoshida S., Manabe K., Shinozaki K., Matsui M. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem. Photobiol. 2003;77:226–233. doi: 10.1562/0031-8655(2003)077<0226:IOAGRB>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 59.Smýkalová I., Zámečníková B. The relationship between salinity and cadmium stress in barley. Biol. Plant. 2003;46:269–273. doi: 10.1023/A:1022815013951. [DOI] [Google Scholar]
- 60.Roth U., von Roepenack-Lahaye E., Clemens S. Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+ J. Exp. Bot. 2006;57:4003–4013. doi: 10.1093/jxb/erl170. [DOI] [PubMed] [Google Scholar]
- 61.Pietrowska-Borek M., Nuc K., Chadzinikolau T., Lechowska K. Cadmium stress causes accumulation of cAMP and this signal molecule induces phenylpropanoid pathway in Arabidopsis thaliana seedlings; Proceedings of the First Polish-German Biochemical Societies Joint Meeting; Poznon, Poland. 11–14 September 2012; poster 4.30. [Google Scholar]
- 62.Pietrowska-Borek M., Nuc K. Both cyclic-AMP and cyclic-GMP can act as regulators of the phenylpropanoid pathway in Arabidopsis thaliana seedlings. Plant Physiol. Biochem. 2013;70:142–149. doi: 10.1016/j.plaphy.2013.05.029. [DOI] [PubMed] [Google Scholar]
- 63.Dixon R.A., Paiva N.L. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferrer J.-L., Austin M.B., Stewart C., Jr., Noel J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008;46:356–370. doi: 10.1016/j.plaphy.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skórzyńska-Polit E., Drażkiewicz M., Krupa Z. The Activity of the Antioxidative System in Cadmium-Treated Arabidopsis thaliana. Biol. Plant. 2003;47:71–78. doi: 10.1023/A:1027332915500. [DOI] [Google Scholar]
- 66.Ascencio-Ibáñez J.T., Sozzani R., Lee T.J., Chu T.M., Wolfinger R.D., Cella R., Hanley-Bowdoin L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008;148:436–454. doi: 10.1104/pp.108.121038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gamborg O.L., Miller R.A., Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- 68.Marondedze C., Wong A., Groen A., Serrano N., Jankovic B., Lilley K., Gehring C., Thomas L. Exploring the Arabidopsis proteome: Influence of protein solubilization buffers on proteome coverage. Int. J. Mol. Sci. 2015;16:857–870. doi: 10.3390/ijms16010857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Al-Shahrour F., Díaz-Uriarte R., Dopazo J. FatiGO: A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 70.BabelomicsV5. [(accessed on 1 September 2015)]. Available online: http://babelomics.bioinfo.cipf.es/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.