Abstract
Most assassin bugs are predators that act as important natural enemies of insect pests. Mitochondrial (mt) genomes of these insects are double-strand circular DNAs that encode 37 genes. In the present study, we explore the duplication and rearrangement of tRNA genes in the mt genome of Reduvius tenebrosus, the first mt genome from the subfamily Reduviinae. The gene order rearranges from CR (control region)-trnI-trnQ-trnM-ND2 to CR-trnQ-trnI2-trnI1-trnM-ND2. We identified 23 tRNA genes, including 22 tRNAs commonly found in insects and an additional trnI (trnI2), which has high sequence similarity to trnM. We found several pseudo genes, such as pseudo-trnI, pseudo-CR, and pseudo-ND2, in the hotspot region of gene rearrangement (between the control region and ND2). These features provided evidence that this novel gene order could be explained by the tandem duplication/random loss (TDRL) model. The tRNA duplication/anticodon mutation mechanism further explains the presence of trnI2, which is remolded from a duplicated trnM in the TDRL process (through an anticodon mutation of CAT to GAT). Our study also raises new questions as to whether the two events proceed simultaneously and if the remolded tRNA gene is fully functional. Significantly, the duplicated tRNA gene in the mitochondrial genome has evolved independently at least two times within assassin bugs.
Keywords: mitochondrial genome, tRNA gene rearrangement, TDRL model, Reduviidae
1. Introduction
Mitochondria are not only organelles with their own genetic material that provide energy for cells, mitochondria are also involved in the control of the cell cycle and cell growth [1]. The typical mitochondrial (mt) genome of animals usually consists of a single circular chromosome with 37 genes (13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes) and a control region (CR) [2]; however, gene order is usually different between major groups [3]. With the rapid development of sequencing techniques, an increasing number of complete mt genome sequences have been determined, which has made mt genomes a popular molecular marker for inferring ancient evolutionary relationships. As much, mt has become the model for studying the mechanism of genome rearrangement [4,5,6,7,8].
Most insect mt genomes have the same gene arrangement as Drosophila yakuba which is acknowledged to be the ancestral pattern for insects [1,9]; however, there have been reports of gene rearrangements in many taxa, such as thrips [10,11], lice [12,13,14], wasps [15,16], and unique-headed bugs [17]. Within these groups, there are reports of rearrangements involving tRNA genes, protein-coding genes (PCGs) and even fragmented mt genomes. Among the several mechanisms proposed to explain gene order rearrangements [5,6,18], the tandem duplication/random loss (TDRL) model is most often used, and is generally considered the most recognized explanation for this occurrence in insects. According to the TDRL model, random deletion of redundant duplicated genes results in novel gene orders [18,19]. Which genes are lost depends on whether the random mutations affect the normal function of genes, and whether the derived pseudo gene will be eventually deleted from the genome [3,20]. In addition to the TDRL model, mechanisms including transposition [19], tandem duplication/nonrandom loss (TDNR) [20], inversion [21], tRNA mis-priming [22], tRNA duplication/anticodon mutations [22,23], and intramolecular recombination [24] have been proposed to account for mt gene rearrangements that we cannot easily or lucidly explain by relying only on the TDRL model.
Assassin bugs (Reduviidae), the second largest family of Heteroptera (true bugs), include close to 7000 species in almost 1000 genera and 25 subfamilies [25]. Presently, the mt genomes of 12 assassin bugs from seven subfamilies have been sequenced. Eleven of these 12 mt genomes contain the typical 37 genes and are arranged in the hypothesized ancestral gene order of insects; however, the mt genome of Brontostoma colossus has 38 coding genes, including the typical 37 genes and an extra trnR [26].
In this study, we report the complete mt genome sequence of Reduvius tenebrosus (R. tenebrosus), the first representative mt genome of the subfamily Reduviinae. The mt genome of R. tenebrosus has novel arrangements in the genomic region between the CR and ND2 (Q-I2-I1-M). The presence of pseudo genes, the positions of intergenic spacers, and a copy of tRNA genes provide evidence that this novel gene order can be explained by the TDRL model and the tRNA duplication/anticodon mutation mechanism. We also sequenced mt genome fragments between the CR and the ND2 of two other species (Reduvius gregorgi and Acanthaspis cincticrus) to further explore whether gene rearrangement is a common feature shared among the Reduviinae.
2. Results and Discussion
2.1. The Mitochondrial (mt) Genome of the Assassin Bug, Reduvius tenebrosus
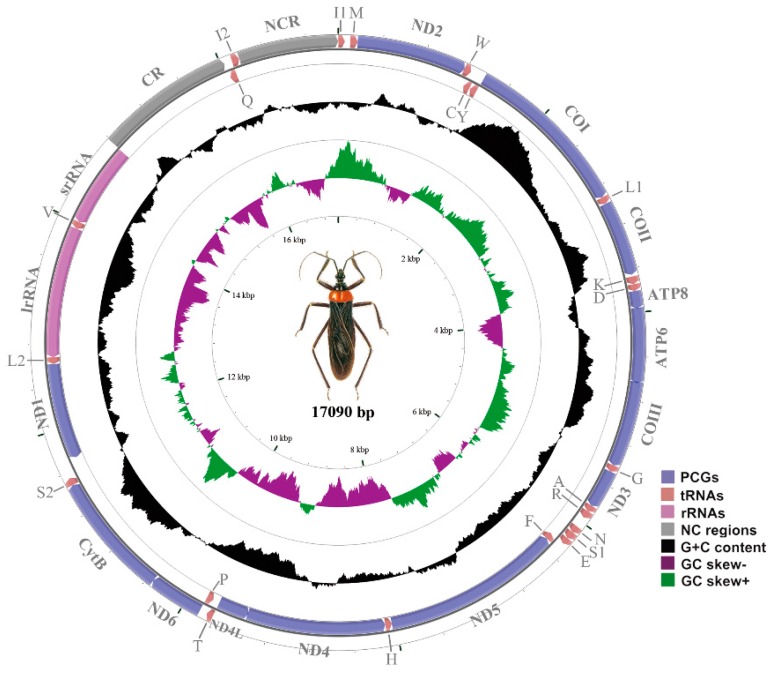
The mt genome of R. tenebrosus is a circular DNA molecule 17,090 bp in length. It contains 38 coding genes (13 PCGs, two rRNAs and 23 tRNAs) and a 1251-bp CR (Figure 1; Table S1). In addition to the typical 37 genes commonly found in arthropod mt genomes, an additional trnI (named trnI2) was identified in this genome. Twenty-four genes are encoded on the majority strand (J-strand) and the other 14 genes are annotated on the minority strand (N-strand). In addition to the CR, we observed 11 non-coding regions (NCR) ranging from 1 to 917 bp. We identified 11 gene overlaps, ranging from 1 to 14 bp in length (Table S1).
Figure 1.
The mitochondrial genome of Reduvius tenebrosus. Arrows indicate the orientation of gene transcription. Abbreviations of gene names are: ATP6 and ATP8 for adenosine triphosphate (ATP) synthase subunits 6 and 8; COI–III for cytochrome oxidase subunits 1–3; CytB for cytochrome b; ND1–6 and ND4L for nicotinamide adenine dinucleotide hydrogen (NADH) dehydrogenase subunits 1–6 and 4L; srRNA and lrRNA for large and small rRNA subunits; trnX (where X is replaced by one letter amino acid code of the corresponding amino acid), for transfer RNA (L1: CUN; L2: UUR; S1: AGN; S2: UCN; I1 and I2 indicate two trnI in different positions). The plotted GC content is the deviation from the average GC content of the entire sequence. The GC-skew was plotted as the deviation from the average GC-skew of the entire sequence. The inner cycle indicates the location of genes in the mitochondrial (mt) genome.
The nucleotide composition of the R. tenebrosus mt genome displays bias toward A and T (J-strand: A = 39.7%, T = 27.5%, G = 12.4%, C = 20.4%, see Table S2). In fact, the overall AT content is lower in comparison with the other 12 mt genomes of assassin bugs (Table S3). The mt genome exhibits obvious AT-skew (0.18) and GC-skew (−0.24), which is in accordance with the common strand bias of insect mtDNA [27]. Genome-wide patterns of base composition may be shaped by the highly asymmetric effects of transcription on mutagenesis, including unequal exposure of the strands to DNA damage and differential chance for repair [28]. The codon usage of PCGs also contributes to a genome-wide bias toward AT (see Table S4). At the third codon position, A or T are overwhelmingly overrepresented compared to G or C. Ultimately, the overrepresentation of A and T is attributable to nucleotide compositional bias. The six most prevalent codons are the AT-rich codons, ATT, ATA, TTA, TTT, AAT, and TAT.
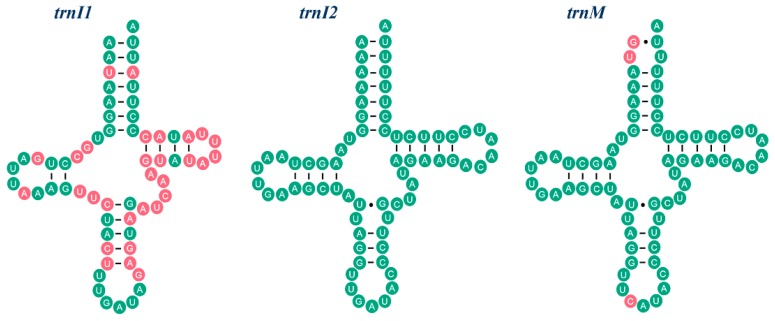
The typical 22 tRNAs commonly found in insects and the additional trnI (trnI2) are present in R. tenebrosus. The lengths vary between 60 and 71 bp. It is worth mentioning that the sequences of trnI2 and trnM only differ in three nucleotide positions, whereas the similarity between sequences of trnI1 and trnI2 is relatively low (Figure 2). The typical clover-leaf structure can be predicted in 22 tRNA genes, with the exception of trnS1 (in which the dihydrouridine, DHU, arm is reduced to a simple loop, as is true in most true bugs) [17,29,30]. The locations and lengths of rRNA genes are similar to those of other assassin bug mt genomes. For instance, lrRNA is located between trnL2 and trnV and is 1255 bp long. srRNA is flanked by trnV and the CR and is 784 bp long. The existence of the extra trnI raises the issue of whether both of the trnI function normally simultaneously. Both trnI1 and trnI2 may be functional genes for that both of them can form an intact secondary structure of standard mitochondrial tRNA genes [31,32] and have the identical anticodon sequence (GAU). Beyond that, the number of the ATC codon of R. tenebrosus tends to be higher than that of the other 12 species, and the amount of ATT codon is obviously lower compared to others. However, the total number of isoleucine codons (ATT + ATC) in R. tenebrosus (365) has no significant difference when compared to other sequenced assassin bugs, and the statistical test shows that the relative frequencies of ATT vs. ATC codons in both R. tenebrosus and Triatoma dimidiata mt PCGs strongly deviate (p < 0.001) from the average value of the other 13 assassin bugs (Table S3).
Figure 2.
The secondary stem-loop structure and sequence similarity of trnI1, trnM and trnI2. Inferred Watson-Crick bonds are illustrated by lines, whereas GU bonds are illustrated by dots. Sequences of trnI1 and trnM are compared with trnI2 respectively. The identical nucleotides with trnI2 are labeled with green circles. The variable nucleotides are highlighted in red.
Moreover, the AT contents of R. tenebrosus and T. dimidiate are the lowest two of the 13 sequenced species. To determine whether this codon usage deviation is due to the duplication of isoleucine tRNAs or the relatively high GC content, we calculated the ratios of codons ending in G or C vs. the total codons of each amino acid in the 13 assassin bugs (Table S5). The results indicate that the preference of codons ending in G or C than A or T is evident for almost all codon boxes in R. tenebrosus. Therefore, the strong deviation in the relative frequency of ATT vs. ATC codons in R. tenebrosus may be related to genome-wide bias in base composition. Ultimately, the functions of the two trnI require more verification, such as transcriptome analysis.
Unconventional start codons in the mt PCGs of insects have been reported in previous studies, such as the start codon GTG in ND5 [33] and ND1 [30], TTG in COI [29,30] and ND3 [34], and the tetra nucleotide start codon ATAA in ND4 [10] and COI [35]. In the mt genome of R. tenebrosus, GTG is found as start codon in both ND1 and ND4L. Other PCGs all initiate with ATN (Table S6). Furthermore, four PCGs (COI, COIII, ND5, ND4) have incomplete stop codons (a single T), which may be made into a completed and proper TAA stop codon by RNA polyadenylation during the transcript process [36]. The other PCGs terminate with either TAA or TAG (Table S6). The use of start and stop codons highlights the similar patterns in the 13 sequenced mt genomes of assassin bugs (Table S6).
2.2. NonCoding Regions, Pseudo Genes and a Novel Gene Order in the Reduvius tenebrosus mt Genome
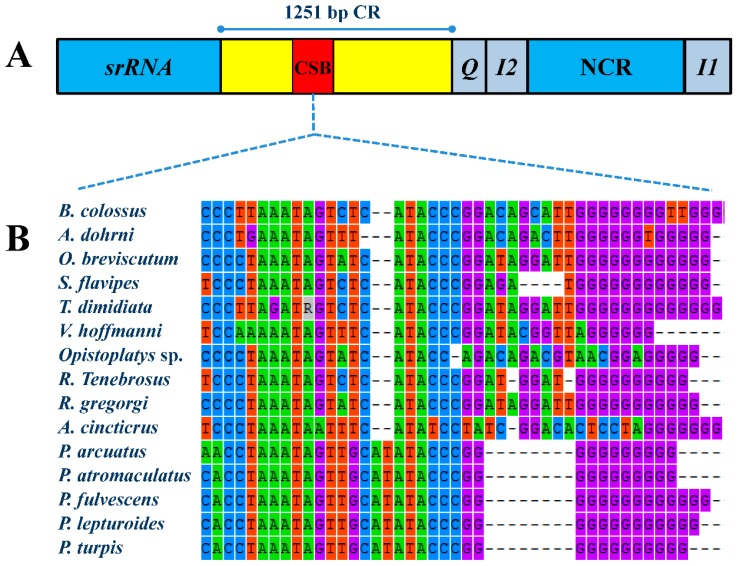
There are a total of 2441 bp NCRs (also referred as intergenic spacers, IGS) in the mt genome of R. tenebrosus. The 1251 bp of the CR is the longest NCR. It is located between the srRNA and the trnQ, and has 66.2% of the AT content. We failed to detect tandem repeat sequences, which is one of the common characteristics of assassin bug control regions [33,37]. However, the sequence alignments of 15 assassin bug control regions (two of which were sequenced fragments between CR and ND2, see below) indicate the presence of a conserved sequence block (CSB), including a G element of 10 bp (Figure 3B), which has been reported in most assassin bugs, as well as some dipterans (referred to as G islands) [38]. They are generally thought to play a role in the replication mechanism [39].
Figure 3.
The conserved sequence block in the control region of assassin bug mt genomes: (A) the organization of the control region in the R. tenebrosus mt genome; and (B) the conserved sequence block in the control region of the sequenced assassin bug mt genomes. CR is the abbreviation for control region and CSB is the abbreviation for conserved sequence block. Four bases are labeled in different colors: Adenine (A)—green, Guanine (G)—purple, Thymine (T)—red and Cytosine (C)—blue.
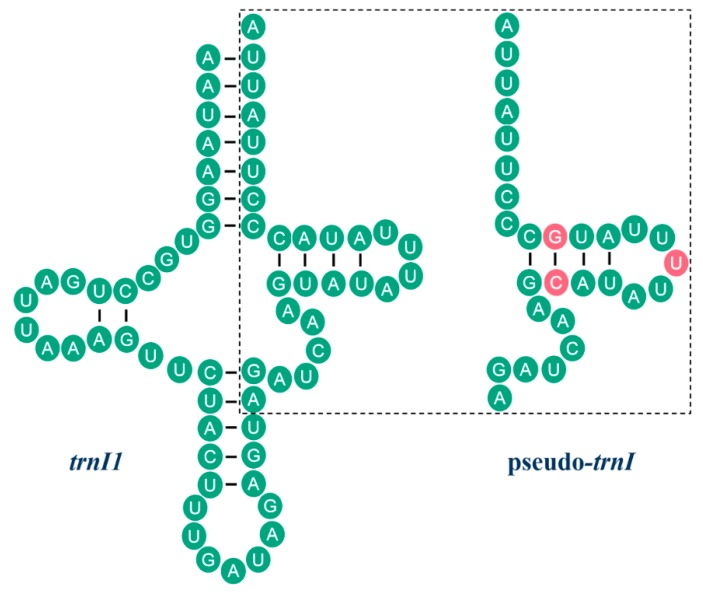
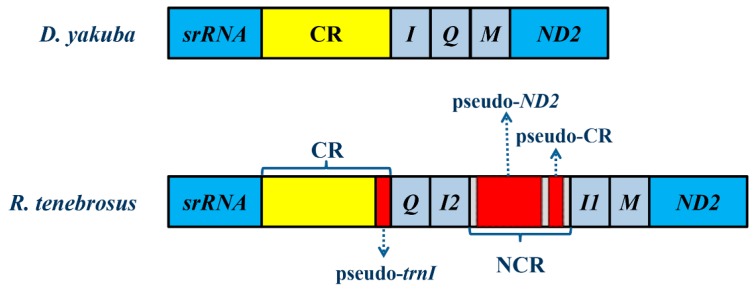
Apart from the CR, there is a relatively long NCR (917 bp) located between two trnI genes (Figure 3A), and 10 short NCRs ranging from 1 to 145 bp long (Table S1). In the long NCR, a 605 bp region shares significant similarity (82.26%) with the homologous sequences in ND2. Additionally, the 197 bp region is almost identical to its counterpart in the CR, except for five point mutations (Figure S1). We also identified a 28 bp region at the end of the CR which is similar to the sequences near the 5’ end of trnI (Figure 4). The significant sequence similarities between these regions indicate the presence of pseudo genes in the CR, ND2, and trnI. Therefore, we define these three regions as pseudo-CR, pseudo-ND2, and pseudo-trnI, respectively (Figure 5).
Figure 4.
The secondary stem-loop structure and sequence similarity of trnI1 and pseudo-trnI. Inferred Watson-Crick bonds are illustrated by lines, whereas GU bonds are illustrated by dots. The identical nucleotides between trnI1 and pseudo-trnI are labeled with green circles. The sequences in the dotted box represent the homologous sequences between trnI1 and pseudo-trnI. The variable sites are in red.
Figure 5.
Gene rearrangement and the locations of three pseudo genes in the region between the control region and ND2 in R. tenebrosus. Three pseudo genes are labeled with dotted arrows. CR is the abbreviation for control region and NCR is the abbreviation for non-coding region.
Heteroptera, also called true bugs, with the largest number of published complete mt genomes in the order Hemiptera, show only gene rearrangement events in a few taxa (Table 1), such as unique-headed bugs [29], pyrrhocoroid bugs [40] and flat bugs [41,42]. In the mt genome of R. tenebrosus, we found the presence of an extra trnI in the tRNA gene rearrangement between CR and ND2. This rearrangement changed the ancestral gene order of CR-I-Q-M-ND2 to the novel gene order of CR-Q-I2-I1-M-ND2 (Figure 5). Although tRNA rearrangement is a relatively common occurrence, the presence of extra tRNAs is a rare event. In Reduviidae, this phenomenon has only been reported in Brontostoma colossus, in which an additional copy of the trnR is brought out by the TDRL mechanism [26]. Interestingly, the two copies of trnR in B. colossus are absolutely identical, whereas in R. tenebrosus, trnI2 is not merely a copy of trnI1; instead, the extra trnI2 is highly similar to trnM (Figure 2).
Table 1.
Mitochondrial gene rearrangements in true bug mt genomes.
| Classification | Species | Rearrangement | GenBank Accession | Reference |
|---|---|---|---|---|
| Aradidae | Brachyrhynchus hsiaoi | I-Q→Q-I | HQ441232 | [41] |
| Aradidae | Aradacanthia heissi | I-Q→Q-I; W-C→C-W | HQ441233 | [42] |
| Aradidae | Neuroctenus parus | I-Q→Q-I | EU427340 | [40] |
| Pyrrhocoridae | Dysdercus Cingulatus | T-P→P-T | NC_012421 | [40] |
| Largidae | Physopelta gutta | T-P→P-T | NC_012432 | [40] |
| Enicocephalidae | Stenopirates sp. | T-P-ND6-CytB-S2-ND1-L2-lrRNA-V-srRNA-CR→CytB-S2-CR-lrRNA-V-srRNA-ND1-L2-P-T-ND6 | NC_016017 | [17] |
| Reduviidae | Brontostoma Colossus | A-R-N-S1→R-A-R-N-S1 | KM044501 | [26] |
| Reduviidae | Reduvius Tenebrosus | I-Q-M→Q-I2-I1-M | KC887529 | Present study |
In order to further probe whether the gene rearrangement of R. tenebrosus is a common feature in the subfamily Reduviinae, we amplified and sequenced two fragments between the CR and ND2 from two additional species of Reduviinae—Reduvius gregorgi and Acanthaspis cincticrus (GenBank accession numbers: KX241473 and KX241472). The results show that both fragments retain the ancestral arrangement (CR-I-Q-M-ND2). This suggests that genome rearrangement is not widely distributed within Reduviinae.
2.3. Mechanisms Account for the Gene Rearrangements in Reduvius tenebrosus mt Genome
According to the TDRL model, duplication of part of the mt genome is caused by slipped-strand mispairing or erroneous identification of the origin of light-strand replication. As part of this process, one gene from each of the duplicated gene pairs is randomly eliminated, at which point it becomes a pseudo gene. Pseudo genes then accumulate additional mutations and become NCRs. It is also possible that pseudo genes may become lost, which creates a novel gene order [3,18]. As mentioned above, the presence and location of pseudo-trnI, pseudo-ND2, pseudo-CR and trnI2 may shed light on whether a novel gene order can be explained by the TDRL model.
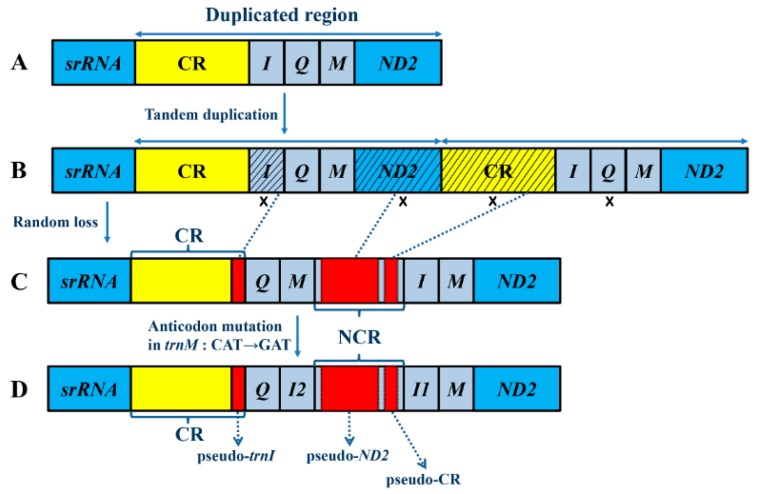
The steps of the TDRL are as follows: First, the gene CR-trnI-trnQ-trnM-ND2 (Figure 6A) is tandemly duplicated and generates two sets of the same gene cluster (CR-trnI-trnQ-trnM-ND2-CR-trnI-trnQ-trnM-ND2) (Figure 6B). Then, a new order CR-trnQ-trnM-trnI-trnM-ND2 is generated after one gene form each of the five duplicated gene pairs is randomly eliminated and becomes a pseudo gene. The pseudo genes then accumulate additional mutations and become NCRs or even be lost (see Figure 6C). The presence of pseudo-trnI in the CR, as well as the presence of pseudo-ND2 and pseudo-CR in the long NCR between trnI2 and trnI1, combined with the significant similarities between trnI2 and trnM, all support our speculated steps of gene duplication and loss in gene rearrangements. Meanwhile, the case that gene rearrangement between CR and ND2 only occurs in R. tenebrosus among the 13 sequenced assassin bug mt genomes and two fragments between CR and ND2 of two Reduviinae species also suggest a recent origin of duplication events as well.
Figure 6.
The hypothetical process of gene rearrangement in the model of tandem duplication/random loss and duplication/anticodon mutation: (A) the ancestral gene order in insects; (B) the tandem duplication of CR/trnI/trnQ/trnM/ND2; (C) the random loss of the duplicated trnI, ND2, CR and trnQ; and (D) the anticodon mutation in trnM which changes trnM (CAT) to trnI2 (GAT). “x” indicates the random loss of the duplicated genes. Shaded areas indicate the genes or regions that are partially lost. Dotted arrows connect the original genes to the rearranged products. Random lost genes and their corresponding pseudo genes are indicated by blue dotted lines. CR is the abbreviation for control region and NCR is the abbreviation for non-coding region.
Obviously, the novel gene order CR-trnQ-trnI2-trnI1-trnM-ND2 cannot be entirely explained by the TDRL mechanism, because this model only generates the order CR-trnQ-trnM-trnI-trnM-ND2. As such, we use the duplication/anticodon mutation mechanism [23] to further explain the transformation from trnM to trnI2 (Figure 6D), in which trnI2 remolds from the duplicated trnM through an anticodon switch (CAT to GAT) and two other site mutations. We also use the tRNA duplication/anticodon mutation model to explain the occurrence of the duplicated trnW in Bothriometopus which is derived from trnE via anticodon mutations (UUC to UCA) [43]. This duplication of a tRNA gene, followed by an anticodon change, provides an example of the types of events that could underlie shifts in the genetic code (which occur often during mtDNA evolution [22,44]).
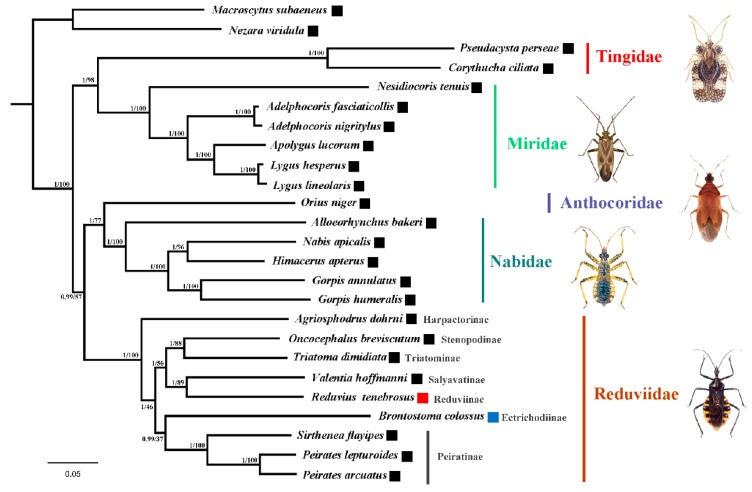
The presence of extra tRNAs is a rare event in the family Reduviidae. Extra tRNAs have only been found in B. colossus (the subfamily Ectrichodiinae) with a duplicated trnR derived from the TDRL mechanism [26] and in R. tenebrosus (the subfamily Reduviinae) with an extra trnI2 generated by the tRNA duplication/anticodon mutation model. Phylogenetic analyses showed almost identical topologies for the relationships among the five cimicomorphan families (Figure 7). Within Reduviidae, the sister-group relationship between Ectrichodiinae and Piratinae and a close relationship between Reduviinae and Salyavatinae is supported. This is consistent with previous analyses based on mitochondrial and nuclear ribosomal genes [45]. These results indicate that the duplicated tRNA gene in the mitochondrial genome has evolved independently at least two times within Reduviidae.
Figure 7.
The phylogenetic relationships among the seven reduviid subfamilies and five cimicomorphan families inferred from the mt genome sequences. Numbers close to the branching points are Bayesian posterior probabilities and maximum likelihood (ML) bootstrap support values. The black boxes refer to species without duplicated tRNAs. Red boxes indicate duplicate tRNA derived from the tRNA duplication/anticodon mutation model. Blue boxes represent duplicated tRNA caused by the tandem duplication/random loss model.
Mitochondrial (mt) genome rearrangements in many other taxa occur frequently in the region between the CR and ND2, such as the rearrangement from trnI-trnQ-trnM to trnM-trnI-trnQ in Formicidae (Hymenoptera) [46] and Ditrysia (Lepidoptera) [47], and from trnI-trnQ-trnM to trnQ-trnI-trnM in Aradidae [41,42]. In Hymenoptera and Lepidoptera, identical rearrangement occurs independently, with numerous possible combinations of duplications and deletions. Many studies have noted that tandem duplications and gene deletions may be subject to mechanistic constraints, so that genes flanking the origins of strand replication (e.g., the CR) are more likely to be duplicated. This forms a “hot spot” for gene rearrangement that makes convergent gene order rearrangement more probable [3]. For the sake of “hot spot”, local or even fully homoplastic rearrangements may be more common than previously thought, therefore, when genome orders are employed as important molecular signatures, we should make sure the data are considered in a phylogenetic and systematic context. Moreover, we should make comparisons of complete gene orders rather than focusing solely on local arrangements [48].
3. Materials and Methods
3.1. Specimen Collection and DNA Extraction
We collected specimens of R. tenebrosus from the Maolan National Nature Reserve, Guizhou, China. The specimens were stored in 100% ethanol at −20 °C at the Entomological Museum of China Agricultural University (Beijing, China). We extracted total DNA from the thoracic muscle of one adult specimen using the DNeasy DNA Extraction kit (Qiagen, Hilden, Germany).
3.2. Polymerase Chain Reaction (PCR) Amplification and Sequencing
The complete mt genome was amplified by polymerase chain reaction (PCR) in overlapping fragments with 13 universal insect mt primers [49], and eight species-specific primers designed from sequenced fragments. We list all the primers used in this study in Table S7. We conducted PCR and sequencing reactions following the guidelines set forth by Li et al. [29,50].
3.3. Genome Annotation and Sequence Analysis
We assembled raw sequences into contigs using BioEdit version 7.0.5.3 [51]. We annotated the complete mt genome sequence using the MITOS webserver with the invertebrate mitochondrial code [52]. We re-confirmed the tRNA genes by using tRNAscan-SE Search Server version 1.21 [53]. tRNA genes that could not be identified by using the MITOS webserver [52] and tRNAscan-SE were determined by using sequence similarity comparison with tRNA genes of other true bugs. Finally, we manually checked all automatic annotations. We deposited the complete mt genome of R. tenebrosus in the GenBank database under the accession number KC887529.
We computed nucleotide composition and codon usage by using MEGA 6.0 program [54]. We used AT-skew [(A − T)/(A + T)] and GC-skew [(G − C)/(G + C)] to measure nucleotide compositional differences between genes [55]. Synonymous codon composition is represented by relative synonymous codon usage (RSCU) [56]. We used the chi-square test (with 5% significance level) to evaluate the statistical significance of our results. We assumed the relative frequency of ATT vs. ATC to be equal from species to species as a null hypothesis. We evaluated the deviation of the relative frequency in a species (from the average value among the 13 assassin bug species) with the chi-square test [32].
3.4. Phylogenetic Analysis
We used sequences from 13 PCGs and two rRNAs from 23 cimicomorphan insects, as well as two outgroup species from Pentatomomorpha for phylogenetic analyses. Details of species we used in this study are shown in Table S8.
We individually aligned each PCG based on codon-based multiple alignments using the MAFFT algorithm within the TranslatorX online platform (with the L-INS-i strategy and default setting) [57]. We aligned the sequences of rRNAs using the MAFFT 7.0 online server (with the G-INS-i strategy) [58]. We concentrated the alignments of the individual genes into a large dataset by removing the third codon position of the PCGs. We analyzed the dataset under Bayesian (BI) and maximum likelihood (ML) frameworks using MrBayes 3.2.3 (with the GTR + I + G model) [59] and RAxML-HPC2 8.1.11 (with the GTR + I + G model) [60] respectively. We created separate partitions for each gene in the dataset. Bootstrap ML analyses with 1000 replicates were performed with the fast ML method implemented in RAxML using the GTRGAMMA model [61]. For MrBayes, two simultaneous runs of 10 million generations were conducted for the datasets and trees were sampled every 1000 generations, with the first 25% discarded as burn-in.
4. Conclusions
In this study, we discovered and described the duplication, remolding and rearrangement of tRNA genes in the complete mt genome of the assassin bug R. tenebrosus. Pseudo genes and rearranged tRNA genes in the hotspot region of gene rearrangement (between the control region and ND2) indicate that the main mechanism underlying the rearrangement of the R. tenebrosus mt genome is the TDRL model. Additionally, the tRNA duplication/anticodon mutation mechanism further explains the presence of trnI2, which is remolded from a duplicated trnM in the TDRL process (through an anticodon mutation of CAT to GAT). However, it is unclear whether the two events proceed simultaneously, and if the remolded tRNAs are functional. The remolding of duplicated tRNA genes is a rare event in insect mt genomes and this novel mt gene rearrangement in assassin bugs improves our understanding of the evolution of mt gene arrangements in insects.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Nos. 31372229, 31420103902, and 31401991), the National Basic Research Program of China (No. 2013CB127600), the Beijing Natural Science Foundation (Nos. 6144027, 6152016), the National Key Technology R & D Program of the Ministry of Science and Technology (2012BAD19B00), the Special Fund for Scientific Research (No. 2012FY111100), and the Chinese Universities Scientific Fund (Nos. 2016QC025, 2016QC072, and 2016ZB001).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/6/951/s1.
Author Contributions
Hu Li and Wanzhi Cai conceived and designed the experiments; Pei Jiang, Hu Li, Fan Song and Yao Cai performed the experiments; Pei Jiang, Fan Song, Hu Li and Jinpeng Liu analyzed the data; Pei Jiang, Hu Li, Fan Song, Yao Cai and Wanzhi Cai contributed reagents/materials/analysis tools; and Pei Jiang and Hu Li wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.McBride H.M., Neuspiel M., Wasiak S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 2.Boore J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.San Mauro D., Gower D.J., Zardoya R., Wilkinson M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol. Biol. Evol. 2006;23:227–234. doi: 10.1093/molbev/msj025. [DOI] [PubMed] [Google Scholar]
- 4.Boore J.L., Collins T.M., Stanton D., Daehler L.L., Brown W.M. Deducing the pattern of arthropod phylogeny from mitochondrial DNA rearrangements. Nature. 1995;376:163–165. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- 5.Bensch S., Härlid A. Mitochondrial genomic rearrangements in songbirds. Mol. Biol. Evol. 2000;17:107–113. doi: 10.1093/oxfordjournals.molbev.a026223. [DOI] [PubMed] [Google Scholar]
- 6.Mueller R.L., Boore J.L. Molecular mechanisms of extensive mitochondrial gene rearrangement in plethodontid salamanders. Mol. Biol. Evol. 2005;22:2104–2112. doi: 10.1093/molbev/msi204. [DOI] [PubMed] [Google Scholar]
- 7.Cameron S.L. Insect mitochondrial genomics implications for evolution and phylogeny. Annu. Rev. Entomol. 2014;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 8.Li H., Shao R., Song N., Song F., Jiang P., Li Z.H., Cai W.Z. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci. Rep. 2015;5:8527. doi: 10.1038/srep08527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clary D.O., Wolstenholme D.R. The mitochondrial DNA molecular of Drosophila yakuba: Nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- 10.Shao R., Barker S.C. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): Convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol. Biol. Evol. 2003;20:362–370. doi: 10.1093/molbev/msg045. [DOI] [PubMed] [Google Scholar]
- 11.Perlman S.J., Hodson C.N., Hamilton P.T., Opit G.P., Gowen B.E. Maternal transmission, sex ratio distortion, and mitochondria. Proc. Natl. Acad. Sci. USA. 2015;112:10162–10168. doi: 10.1073/pnas.1421391112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., Shao R., Song F., Zhou X.G., Yang Q.Q., Li Z.H., Cai W.Z. Mitochondrial genomes of two barklice, Psococerastis albimaculata and Longivalvus hyalospilus (Psocoptera: Psocomorpha): Contrasting rates in mitochondrial gene rearrangement between major lineages of Psocodea. PLoS ONE. 2013;8:951. doi: 10.1371/journal.pone.0061685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao R., Kirkness E.F., Barker S.C. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Res. 2009;19:904–912. doi: 10.1101/gr.083188.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao R., Zhu X.Q., Barker S.C., Herd K. Evolution of extensively fragmented mitochondrial genomes in the lice of humans. Genome Biol. Evol. 2012;4:1088–1101. doi: 10.1093/gbe/evs088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao M., Austin A.D., Johnson N.F., Dowton M. Coexistence of minicircular and a highly rearranged mtDNA molecule suggests that recombination shapes mitochondrial genome organization. Mol. Biol. Evol. 2014;31:636–644. doi: 10.1093/molbev/mst255. [DOI] [PubMed] [Google Scholar]
- 16.Wei S.J., Li Q., van Achterberg K., Chen X.X. Two mitochondrial genomes from the families Bethylidae and Mutillidae: Independent rearrangement of protein-coding genes and higher-level phylogeny of the Hymenoptera. Mol. Phylogenet. Evol. 2014;77:1–10. doi: 10.1016/j.ympev.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 17.Li H., Liu H., Shi A.M., Štys P., Zhou X.G., Cai W.Z. The complete mitochondrial genome and novel gene arrangement of the unique-headed bug Stenopirates sp. (Hemiptera: Enicocephalidae) PLoS ONE. 2012;7:951. doi: 10.1371/journal.pone.0029419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boore J.L. The duplication random-loss model for gene rearrangement exemplified by mitochondrial genomes of deuterosome animals. In: Sankoff D., Nadeau J.H., editors. Comparative Genomics: Empirical and Analytical Approaches to Gene Order Dynamics, Map Alignment and the Evolution of Gene Families. 2nd ed. Volume 1. Springer Netherlands; Dordrecht, The Netherlands: 2000. pp. 133–147. [Google Scholar]
- 19.Macey J.R., Larson A., Ananjeva N.B., Fang Z., Papenfuss T.J. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 1997;14:91–104. doi: 10.1093/oxfordjournals.molbev.a025706. [DOI] [PubMed] [Google Scholar]
- 20.Lavrov D.V., Boore J.L., Brown W.M. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: Duplication and non-random loss. Mol. Biol. Evol. 2002;19:163–169. doi: 10.1093/oxfordjournals.molbev.a004068. [DOI] [PubMed] [Google Scholar]
- 21.Smith M.J., Banfield D.K., Doteval K., Gorski S., Kowbel D.J. Gene arrangement in sea star mitochondrial DNA demonstrates a major inversion event during echinoderm evolution. Gene. 1989;76:181–185. doi: 10.1016/0378-1119(89)90022-X. [DOI] [PubMed] [Google Scholar]
- 22.Cantatore P., Gadaleta M.N., Roberti M., Saccone C., Wilson A.C. Duplication and remoulding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes. Nature. 1987;329:853–855. doi: 10.1038/329853a0. [DOI] [PubMed] [Google Scholar]
- 23.Rawlings T.A., Collins T.M., Bieler R. Changing identities: tRNA duplication and remolding within animal mitochondrial genomes. Proc. Natl. Acad. Sci. USA. 2003;100:15700–15705. doi: 10.1073/pnas.2535036100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lunt D.H., Hyman B.C. Animal mitochondrial DNA recombination. Nature. 1997;387:247. doi: 10.1038/387247a0. [DOI] [PubMed] [Google Scholar]
- 25.Weirauch C., Bérenger J.M., Berniker L., Forero D., Forthman M., Frankenberg S., Freedman A., Gordon E., Hoey-Chamberlain R., Hwang W.S., et al. An illustrated identification key to assassin bug subfamilies and tribes (Hemiptera: Reduviidae) Can. J. Arthropod Identif. 2014;26:1–115. [Google Scholar]
- 26.Kocher A., Kamilari M., Lhuillier E., Coissac E., Péneau J., Chave J., Murienne J. Shotgun assembly of the assassin bug Brontostoma colossus mitochondrial genome (Heteroptera, Reduviidae) Gene. 2014;552:184–194. doi: 10.1016/j.gene.2014.09.033. [DOI] [PubMed] [Google Scholar]
- 27.Wei S.J., Shi M., Chen X.X., Sharkey M.J., van Achterberg C., Ye G.Y., He J.H. New views on strand asymmetry in insect mitochondrial genomes. PLoS ONE. 2010;5:951. doi: 10.1371/journal.pone.0012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francino M.P., Ochman H. Strand asymmetries in DNA evolution. Trends Genet. 1997;13:240–245. doi: 10.1016/S0168-9525(97)01118-9. [DOI] [PubMed] [Google Scholar]
- 29.Li H., Liu H.Y., Song F., Shi A.M., Zhou X.G., Cai W.Z. Comparative mitogenomic analysis of damsel bugs representing three tribes in the family Nabidae (Insecta: Hemiptera) PLoS ONE. 2012;7:951. doi: 10.1371/journal.pone.0045925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hua J.M., Li M., Dong P.Z., Cui Y., Xie Q., Bu W.J. Phylogenetic analysis of the true water bugs (Insecta: Hemiptera: Heteroptera: Nepomorpha): Evidence from mitochondrial genomes. BMC Evol. Biol. 2009;9:951. doi: 10.1186/1471-2148-9-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumazawa Y., Nishida M. Sequence evolution of mitochondrial tRNA genes and deep-branch animal phylogenetics. J. Mol. Evol. 1993;37:380–398. doi: 10.1007/BF00178868. [DOI] [PubMed] [Google Scholar]
- 32.Kumazawa Y., Miura S., Yamada C., Hashiguchi Y. Gene rearrangements in gekkonid mitochondrial genomes with shuffling, loss, and reassignment of tRNA genes. BMC Genom. 2014;15:951. doi: 10.1186/1471-2164-15-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dotson E., Beard C.B. Sequence and organization of the mitochondrial genome of the Chagas disease vector, Triatoma dimidiata. Insect Mol. Biol. 2001;10:205–215. doi: 10.1046/j.1365-2583.2001.00258.x. [DOI] [PubMed] [Google Scholar]
- 34.Stewart J.B., Beckenbach A.T. Insect mitochondrial genomics: The complete mitochondrial genome sequence of the meadow spittlebug Philaenus spumarius (Hemiptera: Auchenorrhyncha: Cercopoidae) Genome. 2005;48:46–54. doi: 10.1139/g04-090. [DOI] [PubMed] [Google Scholar]
- 35.De Bruijn M.H.L. Drosophila melanogaster mitochondrial DNA, a novel organization and genetic code. Nature. 1983;304:234–241. doi: 10.1038/304234a0. [DOI] [PubMed] [Google Scholar]
- 36.Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 37.Zhao G.Y., Li H., Zhao P., Cai W.Z. Comparative mitogenomics of the assassin bug genus Peirates (Hemiptera: Reduviidae: Peiratinae) reveal conserved mitochondrial genome organization of P. atromaculatus, P. fulvescens and P. turpis. PLoS ONE. 2015;10:951. doi: 10.1371/journal.pone.0117862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira M.T., Barau J.G., Junqueira A.C., Feijão P.C., Rosa A.C., Abreu C.F., Azeredo-Espin A.M., Lessinger A.C. Structure and evolution of the mitochondrial genomes of Haematobia irritans and Stomoxys calcitrans: The Muscidae (Diptera: Calyptratae) perspective. Mol. Phylogenet. Evol. 2008;48:850–857. doi: 10.1016/j.ympev.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Saito S., Tamura K., Aotsuka T. Replication origin of mitochondrial DNA in insects. Genetics. 2005;171:1695–1705. doi: 10.1534/genetics.105.046243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hua J.M., Li M., Dong P.Z., Cui Y., Xie Q., Bu W.J. Comparative and phylogenomic studies on the mitochondrial genomes of Pentatomomorpha (Insecta: Hemiptera: Heteroptera) BMC Genom. 2008;9:951. doi: 10.1186/1471-2164-9-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., Shi A.M., Song F., Cai W.Z. Complete mitochondrial genome of the flat bug Brachyrhynchus hsiaoi (Hemiptera: Aradidae) Mitochondrial DNA. 2016;27:14–15. doi: 10.3109/19401736.2013.867437. [DOI] [PubMed] [Google Scholar]
- 42.Shi A.M., Li H., Bai X.S., Dai X., Chang J., Guilbert E., Cai W.Z. The complete mitochondrial genome of the flat bug Aradacanthia heissi (Hemiptera: Aradidae) Zootaxa. 2012;3238:23–38. [Google Scholar]
- 43.Cameron S.L., Johnson K.P., Whiting M.F. The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): Effects of extensive gene rearrangements on the evolution of the genome. J. Mol. Evol. 2007;65:589–604. doi: 10.1007/s00239-007-9042-8. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Li H., Wang P., Song F., Cai W.Z. Comparative mitogenomics of plant bugs (Hemiptera: Miridae): Identifying the AGG codon reassignments between serine and lysine. PLoS ONE. 2014;9:951. doi: 10.1371/journal.pone.0101375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weirauch C., Munro J.B. Molecular phylogeny of the assassin bugs (Hemiptera: Reduviidae), based on mitochondrial and nuclear ribosomal genes. Mol. Phylogenet. Evol. 2009;53:287–299. doi: 10.1016/j.ympev.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 46.Gotzek D., Clarke J., Shoemaker D. Mitochondrial genome evolution in fire ants (Hymenoptera: Formicidae) BMC Evol. Biol. 2010;10:951. doi: 10.1186/1471-2148-10-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y.Q., Ma C., Chen J.Y., Yang D.R. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: The ancestral gene arrangement in Lepidoptera. BMC Genom. 2012;13:951. doi: 10.1186/1471-2164-13-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Babbucci M., Basso A., Scupola A., Patarnello T., Negrisolo E. Is it an ant or a butterfly? Convergent evolution in the mitochondrial gene order of Hymenoptera and Lepidoptera. Genome Biol. Evol. 2014;6:3326–3343. doi: 10.1093/gbe/evu265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon C., Buckley T.R., Frati F., Stewart J.B., Beckenbach A.T. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2006;37:545–579. doi: 10.1146/annurev.ecolsys.37.091305.110018. [DOI] [Google Scholar]
- 50.Li H., Gao J.Y., Liu H.Y., Liu H., Liang A.P., Zhou X.G., Cai W.Z. The architecture and complete sequence of mitochondrial genome of an assassin bug Agriosphodrus dohrni (Hemiptera: Reduviidae) Int. J. Biol. Sci. 2011;7:792–804. doi: 10.7150/ijbs.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 52.Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., Pütz J., Middendorf M., Stadler P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 53.Lowe T.M., Eddy S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura K. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perna N.T., Kocher T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995;41:353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- 56.Sharp P.M., Li W.H. The codon adaptation index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1986;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abascal F., Zardoya R., Telford M.J. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 61.Abascal F., Posada D., Zardoya R. MtArt: A new model of amino acid replacement for Arthropoda. Mol. Biol. Evol. 2007;24:1–5. doi: 10.1093/molbev/msl136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.