Abstract Abstract
The Mediterranean Region is one of the world’s biodiversity hot-spots, which is also characterized by high level of endemism. Approximately 2100 species of leaf beetle (Coleoptera; Chrysomelidae) are known from this area, a number that increases year after year and represents 5/6% of the known species. These features, associated with the urgent need to develop a DNA-based species identification approach for a broad spectrum of leaf beetle species, prompted us to develop a database of nucleotide sequences, with a solid taxonomic background, for all the Chrysomelidae Latreille, 1802 sensu latu inhabiting the Mediterranean region. The Mediterranean Chrysomelidae Barcoding project, which has started in 2009, involves more than fifty entomologists and molecular biologists from different European countries. Numerous collecting campaigns have been organized during the first seven years of the project, which led to the collection of more than 5000 leaf beetle specimens. In addition, during these collecting campaigns two new allochthonous species for Europe, namely Ophraella communa LeSage, 1986 and Colasposoma dauricum Mannerheim, 1849, were intercepted and some species new to science were discovered (e.g., Pachybrachis sassii Montagna, 2011 and Pachybrachis holerorum Montagna et al., 2013). DNA was extracted from 1006 specimens (~13% of the species inhabiting the Mediterranean region) and a total of 910 cox1 gene sequences were obtained (PCR amplification efficiency of 93.8%). Here we report the list of the barcoded subfamilies, genera and the number of species for which cox1 gene sequences were obtained; the metadata associated with each specimen and a list of problematic species for which marker amplification failed. In addition, the nucleotide divergence within and between species and genera was estimated and values of intraspecific nucleotide divergence greater than the average have been discussed. Cryptocephalus quadripunctatus G. A. Olivier, 1808, Cryptocephalus rugicollis G. A. Olivier, 1791 and Exosoma lusitanicum Linnaeus, 1767) are representatives of these cases.
Keywords: Leaf beetles, molecular taxonomy, DNA barcoding, Cytochrome c oxidase subunit 1, C-bar project
Introduction
In the last decades we have witnessed what has been defined as the “taxonomy impediment” (Rodman and Cody 2003) indicating the crisis in taxonomic studies due primarily to a shortage of time and taxonomists (Wheeler 2004, Wheeler et al. 2004, Wilson 2004), a situation that is made even more critical due to the decrease in the funding of natural history studies. The causes of the taxonomy crisis are many and complex, and a comprehensive analysis of this situation is beyond our purpose (see as example Boero 2001, Tautz et al. 2003). In our view, the causes can be described by the sentence …a lack of prestige and resources that is crippling the continuing cataloguing of biodiversity (Godfray 2002). If we consider the increased rate of species extinction (Thomas et al. 2004) amplified by climate change and habitat erosion due to exploitation by human beings the situation is worsened. A DNA-based strategy, which plays a central role in modern taxonomic studies, has been proposed by different authors as a methodology to overcome the identified problems (Tautz et al. 2002, Tautz et al. 2003, Hebert et al. 2004, Goldstein and DeSalle 2010) whilst maintaining the importance of a traditional approach mainly based on morphology. Interestingly, in a survey conducted among Coleopteran taxonomists, taxonomic initiatives based on DNA have been regarded of potential utility in solving the “taxonomy impediment”, even if a few consider it absolutely useless (Löbl 2005). Currently, in the scientific world, an agreement on the correct approach to be adopted has not yet been reached. The “gold standard” for species identification studies based on molecular markers (e.g. mitochondrial cytochrome oxidase subunit I–cox1, or the nuclear small ribosomal subunit–SSU 18S rRNA) is to develop sequence databases used as a reference, beginning with DNA extracted from type and type series specimens preserved in Museum dry collections. The main problem with this strategy is related to the conservation status of the old dry specimens; 18th and 19th century specimens have fragmented DNA (not easily amplified through standard PCR approaches targeting fragments of 500-700 bp) and are often infested by fungal hypha, which contaminate the insect’s genomic DNA. Even with the advent of high-throughput sequencing technologies to solve the problem of fragmented sequences, the contamination due to fungal DNA remains. Developing strategies for the acquisition and storage of molecular data to address molecular taxonomy purposes, we face another problem, which affects the DNA sequences deposited in publicly available databases, i.e. the accuracy of specimen identification. In light of these issues, an alternative strategy has been adopted in the Mediterranean Chrysomelidae Barcoding project (C-Bar). The aim of the C-Bar project is to develop a reference database of cox1 gene sequences for all the Chrysomelidae (excluding Bruchinae Latreille, 1802), the Megalopodidae Latreille, 1802 and the Orsodacnidae Thomson, 1859 (hereafter indicated as Chrysomelidae or leaf beetles sensu latu – s. l.) inhabiting the Mediterranean region. The study area of C-Bar includes all the states that possess coastline on the Mediterranean Sea or territories characterized by Mediterranean-type habitat plus Romania and Switzerland (Figure 1). Starting from the Catalogue of Palaearctic Coleoptera (Löbl and Smetana 2010), about 2100 species of Chrysomelidae s. l. (corresponding to an estimated 5/6% of all described species) are present in this area. The Mediterranean Region is one of the world’s biodiversity “hot-spots” (Myers et al. 2000, Cuttelod et al. 2008), which is characterized by exceptional concentrations of species with high levels of endemism that inhabit one of the most populated areas. The assumption of high levels of endemic species inhabiting the Mediterranean Region is also valid for leaf beetles (Biondi et al. 2013, Sassi 2006). Although the Mediterranean region has been the subject of investigation by generations of entomologists, knowledge of Chrysomelidae inhabiting this area is far from being fully known. The number of leaf beetle species new to science described from the Mediterranean region in the last decades, associated with the fact that they are widespread among different genera, confirms the need to increase the effort in biodiversity-based studies (e.g. Cryptocephalus O.F. Muller, 1764, Chrysolina Motschulsky, 1860, Gonioctena Motschulsky, 1860, Longitarsus Berthold, 1827, Psylliodes Berthold, 1827, Colaspidea Laporte de Castelnau, 1833; Bastazo 1997, Biondi 1997, Sassi 2001, Leonardi 2007, Daccordi and Ruffo 2005, Baviera 2007, Vela and Bastazo 2012, Zoia 2014).
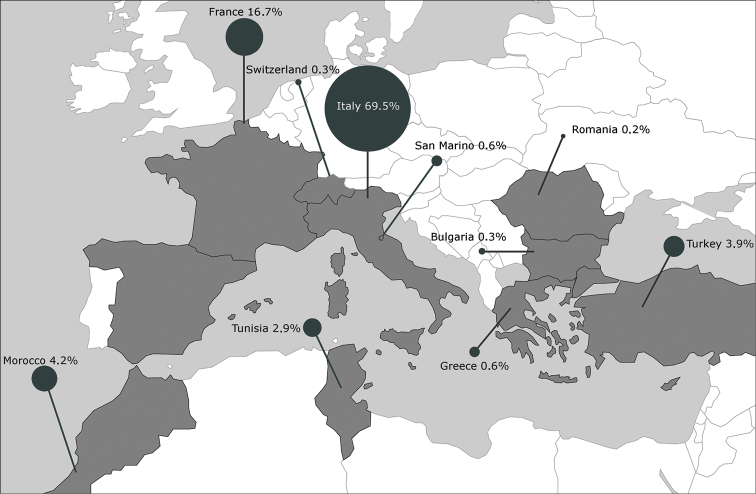
Figure 1.
Area investigated by the Chrysomelidae Barcoding project. The countries in which were performed the collecting campaigns are reported in dark grey. The percentage of the total processed specimens is reported for each country.
In this project are involved taxonomists, specialized in different leaf beetle clades, in order to guarantee the accurate specimen identification. In our view, the adoption of this strategy is a way to bring together traditional (intended as based on morphology) and molecular taxonomy in order to tentatively overcome the “taxonomy impediment” (Rodman and Cody 2003).
The purpose of this paper is to report the preliminary results achieved during the first seven years of the project in order to show the potential of a cooperation between molecular biologists and traditional taxonomists. In particular, we report: i) the method adopted and issues arisen in the development of the sequence dataset; ii) the list of subfamilies, genera and the number of species for which cox1 gene sequences were obtained; iii) the metadata associated with the processed organisms; iv) mean values of intraspecific and interspecific nucleotide divergence v) the new species described and the important faunistic findings.
Materials and methods
Specimen collection and identification
More than 50 entomologists, from different European Countries, have joined the C-Bar project and have actively participated in samples collection. During the first seven years of the project (from 2009 to 2015) numerous collecting campaigns were organized from March to September of each year. The specimens were collected using different methods: from the vegetation by sweep net or by beating sheet, and directly by hand in specific habitats (e.g. under stones or digging the host plant roots). All the collected specimens were placed in 5 ml vials filled with absolute ethanol in order to preserve the genomic DNA. Within an hour of specimen collection, the mixture in the vials was replaced with fresh absolute ethanol in order to obtain better sample dehydration and preservation for long-term storage. Each vial was preserved at -20°C and was labeled by a unique identifier plus other metadata related to the sampling locality (i.e. Country, Province, Region, exact site, latitude, longitude and elevation), the date of collection, the collector/s and other ecological information related to the specimens.
Specimen manipulation and dissection (when necessary) were completed with the auxiliary use of a stereomicroscope. Images of the specimen habitus were acquired by a reflex camera (Canon EOS 450D, macro objective 60 mm or 100 mm with a set of macro extension tubes) or with Axiocam 506 mounted on Zeiss Axio Zoom V16. The specimens were morphologically identified by Italian taxonomists expert in different leaf beetle clades (most of them are listed among the authors of the present article). The nomenclature adopted in the C-bar project follows the work of Bouchard et al. (2011) at the levels of family and subfamily, while at the levels of genus and species was adopted the recently published Catalogue of Palaearctic Coleoptera–Chrysomeloidea (Löbl and Smetana 2010).
DNA extraction, PCRs and sequence quality control
DNA extraction was performed in two different ways since it took place in different laboratories (Biodiversity Institute of Ontario, University of Guelph and Department of Agricultural and Environmental Sciences, Università degli Studi di Milano): for 950 samples the DNA was extracted from one hind leg while for the 56 remaining samples the DNA was extracted from the whole specimen, after the removal of the abdomen. The latter procedure ensures to keep specimen morphology intact. In both cases, DNA was purified using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). Here we describe the adopted non-destructive procedure: the specimen was taken off from absolute ethanol and dried in single 1.5 ml vials for 45 minutes at 30°C; after the removal of the abdomen with the use of sterile pins and tweezers the specimen was placed in 180 µL of ATL lysis buffer (Qiagen) with 200 ng/mL proteinase K (Sigma Aldrich, St. Louis, MO, USA) at 56°C for 12 hours. The following steps of the DNA extraction were performed according to the manufacturer’s instructions of Qiagen DNeasy Blood and Tissue Kit. After DNA extraction, the specimens were dry mounted on pins together with genitalia and kept for future reference. A quote of the extracted DNA was preserved in the C-bar DNA library at -80°C for long term storage and a rate was preserved at -20° in order to perform the following amplifications. A fragment of 658 bp at the 5’-end of the mitochondrial (cox1) was amplified with primers LCO1490 5’-GGT CAA CAA ATC ATA AAG ATA TTG G / HCO2198 5’-TAA ACT TCA GGG TGA CCA AAA AAT CA (Folmer et al. 1994). When this pair of primers resulted in unsuccessful amplification of the target marker, other primers amplifying the same gene region were used, i.e. LepF1 5’-ATT CAA CCA ATC ATA AAG ATA TTG G / LepR1 5’-TAA ACT TCT GGA TGT CCA AAA AAT CA (Hebert et al. 2004). Successful amplifications were determined by gel electrophoresis. PCR products were directly sequenced on both strands using the marker-specific primers from ABI technology (Applied Biosystems, Foster City, CA, USA). The obtained sequences were edited using Geneious R8 (Biomatters Ltd., Auckland, New Zealand) and primers, pseudogenes and contaminations removed. Finally, they were deposited in the Bold Systems (Ratnasingham and Hebert 2007) and in the European Nucleotide Archive (Montagna et al. under revision).
Intraspecific and intrageneric nucleotide divergence
The obtained cox1 gene sequences were aligned at codon level using MUSCLE (Edgar 2004) with default parameters. A pairwise nucleotide distance matrix was estimated starting from the aligned sequences implementing the Kimura-two-parameter (K2P) model (Kimura 1980), considered as an adequate evolutionary nucleotide model when p-distances between sequences are low (Nei and Kumar 2000). The nucleotide distance matrix was used for the calculation of the mean intraspecific and interspecific nucleotide distances and for the calculation of mean intrageneric distance; these analyses were performed using the R package Spider (Brown et al. 2012). We also calculated nucleotide intraspecific distances for some species with a wide range of distribution.
Results and discussion
Until now, C-Bar collecting campaigns have investigated some areas of Bulgaria, France, Greece, Italy, Morocco, Romania, Spain, Switzerland, Turkey and Tunisia (Figure 1). The sampling efforts that have been accomplished until now led to the collection of more than 5000 Chrysomelidae specimens. During the identification process, some specimens of previously unknown species were recognized, these samples were used for the description of the following species: Pachybrachis sassii (Montagna 2011) from the Giglio Island in the Tuscan Archipelago; Pachybrachis holerorum (Montagna et al. 2013) from the Northern Apennines and Oulema mauroi Bezděk & Baselga, 2015, from Northen Italy. Other samples collected during the C-Bar collecting campaigns were used in a revision of Colaspidea genus that led to the description of seven new species (Zoia 2014). All these new taxa were formally described by a traditional morphological approach, in some cases molecular data were added to confirm the existence of the new species. Besides the discovery of new taxa, two allochthonous species new to Europe, namely Ophraella communa (Boriani et al. 2013) and Colasposoma dauricum (Montagna et al. in press), were intercepted. Ophraella communa is a leaf beetle of Nearctic origin accidentally introduced in 1996 in Taiwan (Wang and Chiang 1998) and Japan (Takizawa et al. 1999); the species rapidly spread in East Asia and few years ago we intercepted it in the Northern part of Italy (Boriani et al. 2013). Colasposoma dauricum is a species originally present in the North and Central-East of Asia, it has never been observed out of its original range until our interception in 2011 in Piedmont (North of Italy).
Among the collected samples, the DNA was extracted from 1006 specimens and PCRs targeting a fragment of the cox1 gene performed. PCRs with the selected primer pairs lead to successful amplification in 93.8% of the cases (62 specimens failed the amplification). Among the specimens for which the amplification failed, 43 specimens belong to the subfamily Cryptocephalinae Gyllenhaal, 1813: 18 species of Cryptocephalus (40 specimens); interestingly cox1 sequences have never been obtained for Cryptocephalus therondi Franz, 1949, Cryptocephalus cantabricus Franz, 1958 and Cryptocephalus etruscus Sassi, 1995. We can hypothesize the presence of mutations in the annealing region of the used primers. Sequences obtained from Clytra laeviuscula Ratzeburg, 1837, Clytra quadripunctata Linnaeus, 1758, Cryptocephalus cristula Dufour, 1843, Cryptocephalus octoguttatus Linnaeus, 1767, Lachnaia tristigma Lacordaire, 1848 and Oomorphus concolor Sturm, 1807 did not possess an open reading frame and were thus considered as nuclear pseudogenes. Twenty-seven sequences were discarded because of contamination from exogenous DNA. A total of 910 cox1 sequences (267 species corresponding to ~13% of those inhabiting the Mediterranean region) were obtained, the size of the sequences was > 400 bp in ~99% of the cases.
We observed that only two species, namely Cryptocephalus violaceus Laicharting, 1781 and Cryptocephalus duplicatus Suffrian, 1845, sharing the same haplotype can not be discriminated through DNA barcoding. In this and in similar cases a barcoding failure can be confirmed only ensuring the correct identification of the samples by expert taxonomists. Therefore 99.3% of the species (265) for which we obtained cox1 sequences possessed unique haplotypes, allowing their molecular identification. The mean intraspecific nucleotide distance value is of 2%, while the mean interspecific and intrageneric distances result of, respectively, 25.2% and of 19.8%. The obtained intraspecific value are higher than that inferred in a previous study on Coleoptera (Pentinsaari et al. 2014). This results might be the effect of geographical distances among localities of collection of co-specific specimens; a possible alternative explanation is the presence of cryptic species. Among the species showing high intraspecific nucleotide distance noteworthy are the cases of Cryptocephalus rugicollis (2.8% [0%, 5.5%]), Exosoma lusitanicum (6.7% [0.2%, 9.2%]) and Cryptocephalus quadripunctatus that shows a mean intraspecific distance (3% [0%, 4.9%]). To test the formulated hypotheses further analyses, including the use of other mitochondrial and nuclear markers as well as a wider sample of specimens, are required.
Among the nine subfamilies for which cox1 sequences were obtained (Table 1), Cryptocephalinae and Galerucinae Latreille, 1802 were better represented. In the first subfamily are listed 111 species (83 species of Cryptocephalini Gyllenhaal, 1813 and 28 of Clytrini Lacordaire, 1848, 426 specimens in total) while the second counts 88 species (24 species of Galerucini Latreille, 1802 and 64 of Alticini Spinola, 1844, 274 specimens in total). The unbalanced sampling towards Cryptocephalini, which in some way might affect the obtained results, could be explained by the fact that most of the C-bar specimens have been collected by Sassi and Montagna, which mainly work on this clade and are likely to have developed collecting strategies that increase their sampling (Figure 1).
Table 1.
List of the barcoded subfamilies and genera with the number of species and specimens belonging to each taxon.
| Subfamily | Genus | Nsa | bNspec | Nb |
|---|---|---|---|---|
| Zeugophorinae Böving & Craighead, 1931 | Zeugophora Kunze, 1818 | 1 | 1 | 1 |
| Orsodacninae Thomson, 1859 | Orsodacne Latreille, 1802 | 3 | 7 | 2.3 |
| Donacinae Kirby, 1837 | Donacia Fabricius, 1775 | 2 | 6 | 3 |
| Criocerinae Latreille, 1804 | Crioceris Muller, 1764 | 3 | 18 | 3 |
| Lilioceris Reitter, 1912 | 1 | |||
| Lema Fabricius, 1798 | 1 | |||
| Oulema Gozis, 1886 | 1 | |||
| Cassidinae Gyllenhal, 1813 | Cassida Linnaeus, 1758 | 14 | 61 | 3.4 |
| Hypocassida Weise, 1893 | 2 | |||
| Hispa Linnaeus, 1767 | 1 | |||
| Dicladispa Gestro, 1897 | 1 | |||
| Chrysomelinae Latreille, 1802 | Chrysolina Motschulsky, 1860 | 13 | 117 | 3.4 |
| Chrysomela Linnaeus, 1758 | 3 | |||
| Entomoscelis Chevrolat, 1836 | 1 | |||
| Gastrophysa Chevrolat, 1836 | 1 | |||
| Gonioctena Motschulsky, 1860 | 3 | |||
| Oreina Chevrolat, 1836 | 6 | |||
| Plagiosterna Motschulsky, 1860 | 1 | |||
| Phratora Chevrolat, 1836 | 1 | |||
| Plagiodera Chevrolat, 1836 | 1 | |||
| Prasocuris Latreille, 1802 | 1 | |||
| Timarcha Latreille, 1829 | 3 | |||
| Galerucinae Latreille, 1802 | Agelastica Chevrolat, 1836 | 1 | 274 | 3.1 |
| Arima Chapuis, 1875 | 1 | |||
| Calomicrus Stephens, 1831 | 3 | |||
| Exosoma Jacoby, 1903 | 2 | |||
| Diabrotica Chevrolat, 1836 | 1 | |||
| Galeruca Geoffroy, 1762 | 5 | |||
| Galerucella Crotch, 1873 | 3 | |||
| Lochmaea Weise, 1883 | 2 | |||
| Luperus Geoffroy, 1762 | 6 | |||
| Nymphius Weise, 1900 | 2 | |||
| Sermylassa Reitter, 1913 | 1 | |||
| Altica Muller, 1764 | 4 | |||
| Aphthona Chevrolat, 1842 | 6 | |||
| Argopus Fischer von Waldheim, 1824 | 1 | |||
| Arrhenocoela Foudras, 1860 | 1 | |||
| Chaetocnema Stephens, 1831 | 2 | |||
| Crepidodera Chevrolat, 1836 | 5 | |||
| Derocrepis Weise, 1886 | 2 | |||
| Dibolia Latreille, 1829 | 2 | |||
| Epitrix Foudras, 1860 | 1 | |||
| Hermaeophaga Foudras, 1860 | 1 | |||
| Hippuriphila Foudras, 1860 | 1 | |||
| Longitarsus Berthold, 1827 | 9 | |||
| Lythraria Bedel, 1897 | 1 | |||
| Neocrepidodera Heikertinger, 1911 | 6 | |||
| Phyllotreta Chevrolat, 1836 | 4 | |||
| Podagrica Chevrolat, 1836 | 1 | |||
| Psylliodes Berthold, 1827 | 12 | |||
| Sphaeroderma Stephens, 1831 | 2 | |||
| Cryptocephalinae Gyllenhal, 1813 | Cryptocephalus Geoffroy, 1762 | 73 | 426 | 3.8 |
| Pachybarchis Chevrolat, 1836 | 8 | |||
| Stylosomus Suffrian, 1848 | 2 | |||
| Clytra Laicharting, 1781 | 4 | |||
| Coptocephala Chevrolat, 1836 | 3 | |||
| Labidostomis Chevrolat, 1836 | 10 | |||
| Lachnaia Chevrolat, 1836 | 3 | |||
| Macrolenes Chevrolat, 1836 | 1 | |||
| Smaragdina Chevrolat, 1836 | 7 | |||
| Tituboea Lacordaire, 1848 | 1 | |||
| Eumolpinae Hope, 1840 | Chrysochus Chevrolat, 1836 | 1 | 5 | 1.7 |
| Colaspidea Laporte de Castelnau, 1833 | 1 | |||
| Macrocoma Chapuis, 1874 | 1 |
Ns indicates the number of barcoded species
Nspec and N indicates respectively the total number and the average number of barcoded specimens belonging to each subfamily
The metadata related to the specimens (i.e., specimen identification, collection identifier, collecting date, state, province, exact site of collection, latitude, longitude, elevation and collector/s) from which cox1 gene sequences were obtained, are available in a web site dedicated to the project (http://www.c-bar.org). Regarding the specimens collected within Italian administrative boundaries the metadata associated with the specimens are also available in the Biodiversity Database and GIS platform of the Italian National Network of Biodiversity. These faunistic data are useful because increase the awareness of species presence and distribution in the sampled area.
Conclusion
In this paper, we report that C-Bar project, besides having produced useful data for molecular taxonomy (cox1 sequences were obtained for about 13% of the species inhabiting the investigated area), has obtained important results also from the viewpoint of the classical taxonomy leading to the morphological description of same new species of Chrysomelidae. A further important achievement has been the interception of allochthonous species. These results have been obtained only thanks to the cooperation amongst the taxonomists specialized in different leaf beetle clades, which have ensured the correct identification of samples, the people involved in the extensive collecting campaigns and the molecular biologists.
The promising preliminary results that have been obtained encourage us to continue with this project since they strongly confirm the urgent need to increase the efforts in faunistic studies to uncover the real biodiversity of leaf beetles inhabiting the Mediterranean region. For these reasons, we are confident that the aim of C-bar project of developing a repository of cox1 sequences for the majority of the species of Chrysomelidae s. l. inhabiting the Mediterranean region may be achieved in the near future.
In conclusion, as demonstrated by the relevant results obtained during the first years of the project, we believe that DNA barcoding projects, when developed with the participation of taxonomists and molecular biologists, represent an opportunity to bring together two different worlds and may be considered the driving force able to revive interest in what can be regarded as the milestone of biological studies that is a-taxonomy, helping to fill the “taxonomy impediment”.
Acknowledgements
The Authors would like to thank Dr. Stefano Zoia for the work performed and precious suggestions. In addition, special thanks go to all those involved in the initiative and have helped in sample collection across the Mediterranean region (http://www.c-bar.org/about/people-actively-involved/).
Citation
Magoga G, Sassi D, Daccordi M, Leonardi C, Mirzaei M, Regalin R, Lozzia G, Montagna M (2016) Barcoding Chrysomelidae: a resource for taxonomy and biodiversity conservation in the Mediterranean Region. In: Jolivet P, Santiago-Blay J, Schmitt M (Eds) Research on Chrysomelidae 6. ZooKeys 597: 27–38. doi: 10.3897/zookeys.597.7241
References
- Bastazo G. (1997) Two new species of Longitarsus Berthold, 1827 from Southern Spain (Coleoptera: Chrysomelidae: Alticinae). Elytron 11: 31–38. [Google Scholar]
- Baviera C. (2007) First record of Gonioctena (Coleoptera Chrysomelidae, Chrysomelinae) in Sicily, with the description of Gonioctena theae n. sp. Italian Journal of Zoology 74: 389–393. doi: 10.1080/11250000701650719 [Google Scholar]
- Bezděk J, Baselga A. (2015) Revision of western Palaearctic species of the Oulema melanopus group, with description of two new species from Europe (Coleoptera: Chrysomelidae: Criocerinae). Acta Entomologica Musei Nationalis Pragae 55: 273–304 [Google Scholar]
- Biondi M. (1997) Longitarsus aubozaorum and Psylliodes littoralis, two new flea beetle species from Turkey. Fragmenta Entomologica 29: 383–390. [Google Scholar]
- Biondi M, Urbani F, D’Alessandro P. (2013) Endemism patterns in the Italian leaf beetle fauna (Coleoptera, Chrysomelidae). ZooKeys 332: 177–205. doi: 10.3897/zookeys.332.5339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boero F. (2001) Light after dark: the partnership for enhancing expertise in taxonomy. Trends in Ecology & Evolution 16: . doi: 10.1016/S0169-5347(01)02133-4 [DOI] [PubMed] [Google Scholar]
- Boriani M, Calvi M, Taddei A, Tantardini A, Cavagna B, Spadoni Andreani F, Montagna M, Bonini M, Lommen S, Müller-Schärer H. (2013) Ophraella communa segnalata in Italia su Ambrosia. L’informatore agrario 34: . [Google Scholar]
- Bouchard P, Bousquet Y, Davies AE, Alonso-Zarazaga MA, Lawrence JF, Lyal CH, Newton AF, Reid CA, Schmitt M, Slipiński SA, Smith AB. (2011) Family-group names in Coleoptera (Insecta). Zookeys 88: 1–972. doi: 10.3897/zookeys.88.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SDJ, Collins RA, Boyer S, Lefort MC, Olarte JM, Vink CJ, Cruickshank RH. (2012) SPIDER: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Molecular Ecology Resources 12: 562–565 doi: 10.1111/j.1755-0998.2011.03108.x [DOI] [PubMed] [Google Scholar]
- Cuttelod A, García N, Abdul Malak D, Temple H, Katariya V. (2008) The Mediterranean: a biodiversity hotspot under threat. In: Vié JC, Hilton-Taylor C, Stuart SN. (Eds) The 2008 Review of The IUCN Red List of Threatened Species. IUCN Gland, Switzerland. [Google Scholar]
- Daccordi M, Ruffo S. (2005) Considerazioni biogeografiche sulle Chrysolina delle province appenninica e sicula con descrizione di Chrysolina (Stichoptera) bourdonnei n. sp. Studi Trentini Scienze Naturali. Acta Biologica 81: 113–127. [Google Scholar]
- Edgar RC. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: . doi: 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. (1994) DNA primers for amplification of mitochondrial cytrochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299. [PubMed] [Google Scholar]
- Godfray HCJ. (2002) Challenges for taxonomy. Nature 417: 17–19. doi: 10.1038/417017a [DOI] [PubMed] [Google Scholar]
- Goldstein PZ, DeSalle R. (2010) Integrating DNA barcode data and taxonomic practice: determination, discovery, and description. Bioessays 33: 135–147. doi: 10.1002/bies.201000036 [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Penton EH, Burns J, Janzen DH, Hallwachs W. (2004) Ten species in one: DNA barcoding reveals cryptic species in neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences 101: 14812–14817. doi: 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. doi: 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Leonardi C. (2007) Dati inediti sul genere Psylliodes Latreille, con descrizione di quattro nuove specie mediterranee (Coleoptera Chrysomelidae). Atti della Società Italiana di Scienze Naturali e del Museo Civico di Storia Naturale di Milano 148: 161–240. [Google Scholar]
- Löbl I. (2005) Demography of coleopterist and their thoughts on DNA Barcoding and the Phylocode, with commentary. The Coleopterists Bulletin 59: 284–292. doi: 10.1649/850.1 [Google Scholar]
- Löbl I, Smetana A. (2010) Catalogue of Palaearctic Coleoptera, Vol. 6. Chrysomeloidea. Apollo Books, Stenstrup, 606–617. [Google Scholar]
- Montagna M. (2011) Pachybrachis sassii, a new species from the Mediterranean Giglio Island (Italy) (Coleoptera, Chrysomelidae, Cryptocephalinae). ZooKeys 155: 51–60. doi: 10.3897/zookeys.155.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna M, Sassi D, Giorgi A. (2013) Pachybrachis holerorum (Coleoptera: Chrysomelidae: Cryptocephalinae), a new species from the Apennines, Italy, identified by integration of morphological and molecular data. Zootaxa 3741: 243–253. doi: 10.11646/zootaxa.3741.2.3 [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. (2000) Biodiversity hotspots for conservation priorities. Nature 403: 853–858. doi: 10.1038/35002501 [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. (2000) Molecular Evolution and Phylogenetics. Oxford University Press, New York, 333 pp. [Google Scholar]
- Pentinsaari M, Hebert PDN, Mutanen M. (2014) Barcoding Beetles: A Regional Survey of 1872 Species Reveals High Identification Success and Unusually Deep Interspecific Divergences. PLoS ONE 9(9): . doi: 10.1371/journal.pone.0108651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PDN. (2007) BOLD: The Barcode of Life Data System (www.barcodinglife.org). Molecular Ecology Notes 7: 355–364. doi: 10.1111/j.1471-8286.2007.01678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodman JE, Cody JH. (2003) The taxonomic impediment overcome: NSF’s Partnerships for Enhancing Expertise in Taxonomy (PEET) as a model. Systematic Biology 52: 428–435. doi: 10.1080/10635150309326 [DOI] [PubMed] [Google Scholar]
- Sassi D. (2001) Nuove specie del genere Cryptocephalus vicine a Cryptocephalus marginellus (Coleoptera Chrysomelidae). Memorie della Società entomologica italiana 80: 107–138. [Google Scholar]
- Sassi D. (2006) Insecta Coleoptera Chrysomelidae Cryptocephalinae. In: Ruffo S, Stoch F. (Eds) Checklist and distribution of the Italian fauna. Memorie del Museo Civico di Storia Naturale di Verona, 221–223.
- Takizawa H, Saito A, Saito K, Hirano Y, Ohno M. (1999) Invading insect, Ophraella communa LeSage, 1986: range expansion and life history in Kanto district, Japan. Gekkan-Mushi 338: 26–31. [Google Scholar]
- Tautz D, Arctander P, Minelli A, Thomas RH, Vogler AF. (2002) DNA points the way ahead in taxonomy. Nature 418: . doi: 10.1038/418479a [DOI] [PubMed] [Google Scholar]
- Tautz D, Arctander P, Minelli A, Thomas RH, Vogler AF. (2003) A plea for DNA taxonomy. Trends in Ecology & Evolution 18: 70–74. doi: 10.1016/S0169-5347(02)00041-1 [Google Scholar]
- Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC, Erasmus BF, De Siqueira MF, Grainger A, Hannah L, Hughes L, Huntley B, Van Jaarsveld AS, Midgley GF, Miles L, Ortega-Huerta MA, Peterson AT, Phillips OL, Williams SE. (2004) Extinction risk from climate change. Nature 427: 145–148. doi: 10.1038/nature02121 [DOI] [PubMed] [Google Scholar]
- Vela JM, Bastazo G. (2012) A new Cryptocephalus of the C. curvilinea – group (Coleoptera, Chrysomelidae) from the Atlantic coastal marshes of Southern Spain and Portugal. Boletín de la Asociación Española de Entomología 36: 31–41. [Google Scholar]
- Wang CL, Chiang MY. (1998) New record of a fastidious chrysomelid, Ophraella communa LeSage (Coleoptera: Chrysomelidae), in Taiwan. Plant Protection Bulletin 40: 185–188. [Google Scholar]
- Wheeler QD. (2004) Taxonomic triage and the poverty of phylogeny. Philosophical Transactions of the Royal Society B 359: 571–583. doi: 10.1098/rstb.2003.1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler QD, Raven PH, Wilson EO. (2004) Taxonomy: Impediment or expedient? Science 303: 285. doi: 10.1126/science.303.5656.285 [DOI] [PubMed] [Google Scholar]
- Wilson EO. (2004) Taxonomy as a fundamental discipline. Philosophical Transactions of the Royal Society B 359: 739. doi: 10.1098/rstb.2003.1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoia S. (2014) The Mediterranean Colaspidea (Coleoptera Chrysomelidae Eumolpinae). Entomologia 2. doi: 10.4081/entomologia.2014.159 [Google Scholar]