SUMMARY
RNAi screens have implicated hundreds of host proteins as HIV-1 dependency factors (HDFs). While informative, these early studies overlap poorly due to false positives and false negatives. To ameliorate these issues, we combined information from the existing HDF screens together with new screens performed with multiple orthologous RNAi reagents (MORR). In addition to being traditionally validated, the MORR screens and the historical HDF screens were quantitatively integrated by the adaptation of an established analysis program, RIGER, for the collective interpretation of each gene’s phenotypic significance. False positives were addressed by the removal of poorly expressed candidates through gene expression filtering, as well as with GESS, which identifies off-target effects. This workflow produced a quantitatively integrated network of genes that modulate HIV-1 replication. We further investigated the roles of GOLGI49, SEC13, and COG in HIV-1 replication. Collectively, the MORR-RIGER method minimized the caveats of RNAi screening and improved our understanding of HIV-1–host cell interactions.
Graphical Abstract
INTRODUCTION
Each infectious HIV-1 virion contains a collection of 12 distinct viral proteins, including integrase (IN) and reverse transcriptase (RT), as well as two copies of a 9 kb genome. Any viral requirements not self-fulfilled by these contents must be satisfied by the infected host cell’s resources, a dynamic that has resulted in significant morbidity and mortality. HIV-1 infection of a host cell begins with the binding of the virus’ envelope spike (ENV) to the coreceptors CD4 and either CXCR4 or CCR5 (Goff, 2007). Once engaged, ENV fuses the viral and host membranes to create a pore through which the viral contents enter the cell. Upon entry, the virus uses the host’s dinucleotide triphosphates to reverse transcribe its RNA genome into DNA, forming a preintegration complex (PIC). The PIC courses along microtubules toward the nucleus, which it enters via the nuclear pore complex (NPC). Emerging amidst the chromatin, the PIC interacts with LEDGF/p75, leading to HIV-1’s preferential integration into an actively transcribed gene (Ciuffi et al., 2005). The integrated provirus next exploits the host’s transcriptional machinery to produce viral mRNAs, the most crucial being Tat, which together with the host complex, PTEF-b, ensures transcriptional elongation along the provirus. Once synthesized, the soluble viral components are packaged within a core of structural proteins: p24 capsid (CA), matrix, p7, and p6. ENV is translated on the endoplasmic reticulum (ER), modified in the Golgi, and trafficked to the surface to ultimately coat the viral envelope. Viral budding and abscission relies on the ESCRT proteins, which execute roles similar to those played during cytokinesis (Sundquist and Krausslich, 2012).
Elucidating host-viral interactions has been a longstanding pursuit of the scientific community, with the goal of using such knowledge to both treat and cure disease. Yet, although we know that HIV exploits multiple proteins, there remain many viral life-cycle processes that are at best partially defined. We and others have performed genetic screens to identify HIV-1 dependency factors (HDFs) and uncovered many host genes involved in HIV infection (Brass et al., 2008; König et al., 2008; Zhou et al., 2008). However, while this work has produced successes, it has also been hampered by low concordance across the screens due to false negatives, false positives, and a gradation of small interfering RNA (siRNA) efficacies resulting in variable hypomorphism.
With the goal of approaching a systems-level understanding of HIV-host factor interactions and to improve upon the initial HDF screens and address the lack of overlap between siRNA screens in general, we expanded our earlier efforts by using multiple orthologous RNAi reagents (MORR) coupled with integrative analysis tools. Our rationale in pursuing this strategy was 2-fold: first to take advantage of the strengths of several independent RNAi design approaches, and second to analyze the data sets using RNAi-focused informatics tools and screen-specific gene expression data. We analyzed the MORR screens by selecting candidates using a cutoff coupled with a reagent redundancy validation round. In addition, the screens were also assessed by quantitatively integrating all of the primary data sets using an established bioinformatics program to provide a global statistical evaluation of each gene’s role in HIV-1 replication. This comprehensive effort is validated by the discovery of known factors and the identification of multiple novel HDFs, demonstrating that using MORR can improve our understanding of HIV-1-host cell interactions.
RESULTS
MORR Screens Find Host Factors that Modulate HIV-1 Replication
One source of variability in the historical HDF screens is their being performed by different groups with distinct reagents. To control for this, we investigated HIV-1-host cell interactions by employing MORR with an established image-based assay where HeLa cells expressing CD4 and CXCR4 are transfected with siRNAs, then exposed to infectious HIV-1 (HIV-IIIB; Brass et al., 2008). Postinfection, the cells are immunostained for CA in conjunction with DNA staining to determine cell number. Due to several points, including infectivity, we used a different HeLa cell line, P4–P5 MAGI cells (MAGI), instead of the TZM-bl HeLa cell line used in the SMARTpool screen.
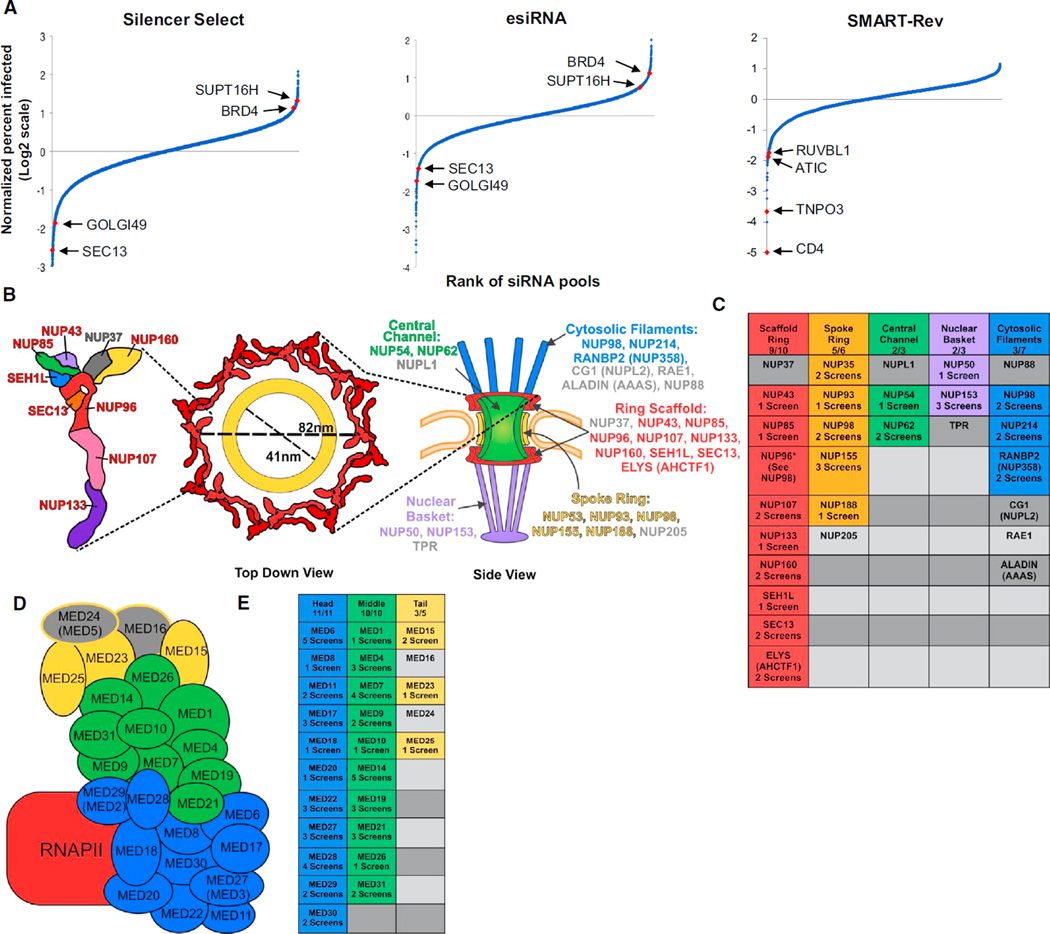
We screened three libraries: Silencer Select (21,584 pools, Ambion), esiRNA (15,300 pools, Sigma), and the SMARTpool RefSeq27 Reversion Human 5 (SMART-Rev, 4,506 pools, Dharmacon; Figure 1A; Table S1). The last library is a collection of replacement siRNAs produced because of RefSeq revisions. These libraries employ unique design strategies and/or target additional genes (SMART-Rev), making them complementary to one other and the earlier libraries. The esiRNAs are mixtures of overlapping siRNAs (18–25 bps) made by endoribonuclease digestion of longer RNAs (Kittler et al., 2007). Thus, less concentration-dependent off-target effects (OTEs) are expected versus conventional pools. Silencer Select siRNAs incorporate locked nucleic acids (LNAs) to increase the strength and specificity of binding to mRNA (Puri et al., 2008). An assumption with the MORR approach is that each of the libraries uses unique siRNA sequences for a given gene; this is likely because the libraries were designed with distinct algorithms. A random survey of 1017 Silencer Select and 1,356 matched SMARTpool siRNAs targeting the same 339 genes found <5% overlap (Table S1).
Figure 1. MORR Screens Identify Host Factors that Modulate HIV-1 Replication.
(A) The results of the screens with the siRNA pools ranked in order of their normalized percent infection (log2 scale).
(B) NPC genes required for HIV-1 replication. Schematic of the NPC (left), with scaffold ring (red), spoke ring (yellow), central channel (green), nuclear basket (purple), and cytosolic filaments (blue) components. Detail of the NPC’s spoke ring (far left) demonstrating the high percentage (90%) of components, including SEC13 (orange), that scored in one or more of the HDF screens (red lettered). The estimated diameter of the NPC lumen is provided. For reference the HIV-1 PIC is ~28 nm across (Miller et al., 1997).
(C) NPC genes that met selection criteria in one or more of the HDF screens are grouped and color-coded by their respective component in the table at right, with gray denoting a gene that did not score in any of the screens.
(D) MED genes required for HIV-1 replication. Schematic of MED (left), with the head (blue), middle (green), and tail (yellow) components shown as well as the complex’s association with RNA polymerase II (RNAPII, red).
(E) MED genes that met selection criteria in any of the screens are highlighted in their respective component colors in the table at right, with gray signifying a gene that did not meet selection criteria in any screen.
Analysis of the MORR Screens
The MORR screens were analyzed using a traditional reagent redundancy method as well as by quantitatively integrating all of the primary data sets. The latter approach considers the magnitude of the effects of all the independent RNAi reagents tested in order to address issues of partial penetrance for each gene. Each method has it strengths, as we discuss below. In the traditional method, genes were chosen for evaluation if the percentage of HIV-1 infected cells was ≤50% or ≥200% of the plate mean and cell number was ≥50% of the plate mean. Next, in the validation round, the siRNA pool components were tested individually (Table S1). For the esiRNA pools, we retested the candidates from the primary screen. In the validation data sets, genes with two or more siRNAs that confirmed were deemed high confidence based on the reagent redundancy principle (Echeverri et al., 2006).
The MORR screens identified several pathways and macromolecular complexes whose components were shared across the three high-confidence data sets (i.e., NPC; Figures 1B–1E; Tables S1 and S2) as well as the high-confidence data sets from the earlier screens that used three distinct libraries: one constructed by QIAGEN for the Genomics Institute of the Novartis Research Foundation (QIAGEN, König et al., 2008), one designed by Rosetta InPharmatics and synthesized by QIAGEN for Merck & Company (Rosetta, Zhou et al., 2008), and the commercially available Dharmacon siRNA library (SMARTpool; Brass et al., 2008). The QIAGEN library targets each of ~20,000 genes with up to three pools containing two oligos per pool. The Rosetta library targets 19,709 genes with 22,329 pools of three siRNAs per gene. The SMARTpool library targets 21,121 genes using pools of four siRNAs per gene. For the NPC, the strongest dependency was seen with the spoke ring and scaffold ring genes (Figures 1B and 1C; Table S2). Among the scaffold ring proteins, only NUP37 failed to score in one or more screens. Among the mediator (MED) components recovered across the screens, we detected a stronger viral reliance on proteins located in the head and middle portions as compared to the tail (Figures 1D and 1E; Table S2), suggesting that the tail plays a smaller role in long terminal repeat (LTR) activation.
The earlier screens identified three HDFs (RELA, MED6, and MED7) in all three screens and 34 in two or more screens (Figure S1A; Table S2). The MORR screens recovered 13 of these original 34 common HDFs and expanded the list of HDFs in two or more screens to 114, out of a total of 1,117 selected across all six screens (Figures S1B and S1C). Because RNAi produces variable hypomorphism, we assessed how the 114 HDFs scored in the Silencer Select screen and found their mean percent infection value was less than the screen as a whole (Figure S1D); this suggested that the phenotypic significance of genes whose depletion resulted in partial penetrance might be better appreciated by quantitatively integrating the screens’ data.
Low Overlap and High False-Positive Rates among All HDF Screens
A comparison of the high-confidence lists from the MORR screens and the three previous screens (Table S2) demonstrated a low percentage of exact gene overlap (Silencer Select 19.9%; SMARTpool 19.2%; SMART-Rev 14.0%; esiRNA 12.7%; Rosetta 10.7%; QIAGEN 10.2%). This likely arises from a combination of OTEs, false negatives, variable hypomorphism, and differences between methods; these assumptions are consistent with previous comparisons revealing that while similar pathways and complexes are likely to be detected across screens, the overlap of exact genes is much less (Hao et al., 2013; Meier et al., 2014). Comparing the primary data sets of the HDF screens for both MED and NPC revealed that the majority of subunits for each complex passed selection criteria (Figures 1B–1E and S2A–S2B; Table S2). For the QIAGEN library primary screen data set (König et al., 2008), we selected genes as HDFs if two or more of the pools decreased infection to <55%. Cell viability data were not available for this data set. For the Rosetta (Zhou et al., 2008) and SMARTpool (Brass et al., 2008) primary screen data sets screens, we selected pools that decreased infection to <50% without decreasing cell number to <40% of the negative control. Using these criteria, the screen data sets detected 92.3% of MED and 72.4% of NPC components, attesting to the comprehensiveness (saturation) of the collective HDF screens (Figure S2; Table S2). Interestingly, while some of the subunits scored in multiple screens, others scored in just one or two. While these results highlight the variable hypomorphism inherent in RNAi screening, they also demonstrate that the screens in aggregate are approaching saturation for these complexes.
The Silencer Select and SMART-Rev screens identified 33 previously unrecognized high-confidence HDFs that were validated with two or more individual siRNAs (Tables S1 and S2). Of note, siRNAs against TNPO3 were not in the esiRNA library; however, a TNPO3 esiRNA inhibited HIV-I (Figure S2C). Although we did find several common pathways and clusters across the MORR screens, the exact gene overlap was again low (12%–20%; Table S2); however, it was at least 2-fold higher than the largest seen earlier (SMARTpool and Rosetta, 18 genes or 6.4%). In contrast to the earlier screens, this occurred with the MORR screens being performed identically. We conclude that along with OTEs and screen design, varying siRNA efficacies result in false negatives and contribute to the low exact gene overlap between similar screens.
Quantitative Integration of the HDF Screens Using RIGER
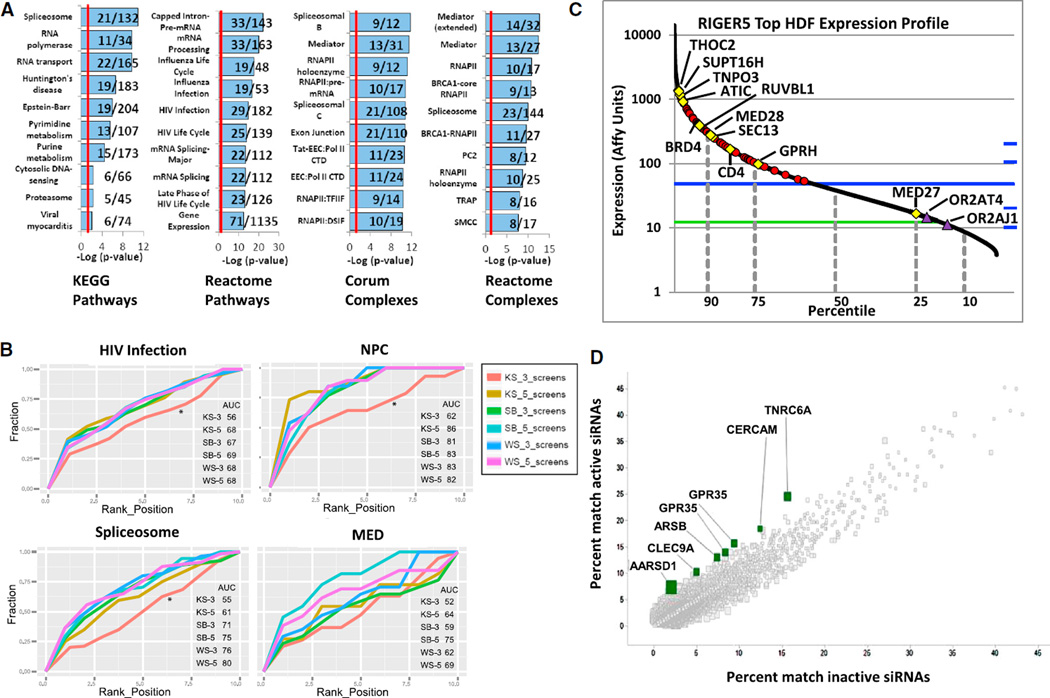
The Silencer Select screen’s mean percent infected cell value was greater than that of the 114 common HDFs even though only 54 of these genes met selection criteria, revealing that some siRNAs were partially inhibiting HIV-1 (Figure S1D). This suggested that one contribution to the lack of overlap is a gradation of hypomorphism produced by distinct siRNA reagents combined with absolute cutoffs in hit selection. We also used a second analysis method that produces an aggregate phenotypic significance score derived by quantitatively integrating multiple primary data sets. To this end, we sought an RNAi-focused method that calculates the cumulative significance of multiple independent RNAi reagents all targeting the same gene. Among several, we chose the RNAi gene enrichment ranking (RIGER) method, which uses a weighted likelihood ratio to calculate a gene-specific enrichment score based on the rank distribution of each individual RNAi reagent among all those screened (Luo et al., 2008). This enrichment score is represented by a p value that denotes the likelihood that the gene plays a role in the phenotype of interest. Although RIGER was originally designed to analyze similar screens using different cell lines and the same pooled RNAi library, we reasoned that it could also be used to evaluate the HDF screens that assess the same phenotype but use different RNAi libraries. Because the three screens (Silencer Select, esiRNA, and SMARTpool) performed by our group were most alike, we used RIGER to analyze these first (RIGER3; Table S3). To simplify this analysis, we replaced the results of the corresponding siRNAs in the original SMARTpool data set with the revised SMART-Rev screen values. We found that of the three RIGER integrative approaches, the second best (SB), weighted sum (WS), or Kolmogorov-Smirnov (KS), either the SB or WS method performed optimally (see below). A comparison of the highest ranking 355 RIGER3 HDFs to the top candidates from 2 of the earlier HDF screens performed by König et al. (QIAGEN; König et al., 2008) and Zhou et al. (Rosetta; Zhou et al., 2008) revealed a significant overlap in exact genes detected, with 321% more exact genes than expected seen for Konig et al. (22 genes, p value = 1.5 × 10−8, Fisher exact test) and 309% more for Zhou et al. (27 genes, p value = 6.4 × 10−10). Next, the analysis was extended to all five screens (RIGER5; Table S3; Zhou et al., 2008; König et al., 2008; Brass et al., 2008). For RIGER5, we employed all three approaches (SB, WS, and KS) and used the Liptak method to derive a combined p value and q-value (false discovery rate [FDR]; Liptak, 1958). Both the RIGER3 and RIGER5 analyses highly ranked multiple known HDFs: TNPO3 and MED28 (Table S3). Gene enrichment analyses of the top-ranked 528 genes from RIGER5 HDF (q-value < 0.2) was performed using ConsensusPathDB-human (http://cpdb.molgen.mpg.de) in conjunction with the REAC-TOME, CORUM, and KEGG databases (Figure 2A; Table S3). Consistent with RIGER identifying relevant factors, these analyses demonstrated the significant enrichment of known HDF pathways and complexes, including the spliceosome, RNA polymerase-associated factors, MED, NPC, and components of the Rev-associated mRNA export pathway. Gene enrichment analysis of the RIGER3 HDFs (top 500 genes, score < 0.127, p value < 0.03; Table S3) was similar. For the HRFs, we performed a RIGER analysis using the MORR screen data sets and the SB method (Table S3). Gene enrichment analysis was also performed using the top 260 RIGER3 HRFs (RIGER score < 0.1057, p value < 0.02; Table S3). Interestingly, this showed that both subunits of the facilitates-chromatin-transcription (FACT) complex, SUPT16H and SSRP1, ranked highly.
Figure 2. MORR Screen Gene Enrichment Analyses and Gene Expression Filtering.
(A) Gene enrichment analyses of the RIGER5 HDFs. Highly enriched KEGG, REACTOME, and CORUM complexes and pathways among the top 550 RIGER5 HDFs are shown (Table S3). Overlying fractions to the right of each pathway’s or complex’s name indicate the number of HDFs identified in the analysis over the total number of genes in the set. The red line indicates a −log (p value) of 1.3 (p value of 0.05). Specific complex and pathways are abbreviated, with a key to their full names provided in Supplemental Experimental Procedures.
(B) RIGER analyses comparisons. The RIGER3 and RIGER5 HDF data sets produced using one of three methods (WS, SB, or KS) were compared by assessing their respective levels of enrichment for each of four expert-selected HDF gene sets (HIV infection, NPC, spliceosome, MED; http://www.reactome.org). An enrichment estimate for each derivation was calculated by determining the area under a curve (AUC) generated by plotting the percent fraction (fraction) of expert-selected genes encountered moving from the lowest to highest p value on the ranked gene lists (rank position).
(C) Microarray analysis was used to determine the levels of gene expression in the HeLa MAGI cells used in the MORR screens. The complete data set is arrayed with the red symbols representing the top 85 RIGER5 HDFs shown in the HDF cell model (Figure 3A). Specific HDFs and HRF/HCFs are noted (yellow). Two likely OTEs, the olfactory receptors (OR2AT4 and OR2AJ1), are shown in purple. The blue line represents the intron probe median. The green line represents the antisense probe mean. MED27, whose expression level falls below that of the intron probe median, is shown.
(D) OTE analysis using genome-wide enrichment of seed sequence matches (GESS). All siRNAs (313) that scored in the validation round for the Silencer Select library screen were analyzed. The significant enrichment of RefSeq CDSs that have a 7-mer seed match to these 313 siRNAs was determined by comparison to a set of inactive validation round siRNAs. Several potential OTEs are highlighted (green squares). A similar analysis was performed to look for on-viral effects (OVEs) using the HIV-1 genome sequence (Figure S2D).
The MORR-RIGER Method Enhances Enrichment
The RIGER3 and RIGER5 HDF data sets were compared by determining their respective levels of enrichment for each of four expert-selected gene sets (HIV infection, NPC, spliceosome, MED; http://www.reactome.org; Figure 2B; Table S3). Enrichment was calculated by determining the area under a curve (AUC) generated by plotting the percent fraction of expert-selected genes encountered moving from high to low on the ranking lists. A completely random set of genes would generate a line represented by Y = X and have an AUC of 50. The three RIGER methods performed similarly across the test sets, with the best overall AUCs seen with RIGER5 SB. For three of four gene sets, the KS method was significantly improved by including all five screens as compared to the analysis of only three screens. The greatest advantage of integrating all five screens was seen with the MED gene set. The analyses using the SB and WS methods did not show any significant differences between RIGER3 and RIGER5. Interestingly, the RIGER5 SB analysis showed an AUC advantage over the individual MORR screens (SMARTpool, Silencer Select, and esiRNA) using either the NPC or MED test sets, with this being most notable in the initial part of the curve that covered genes whose loss decreased HIV infection by >50% (Figure S3A). The RIGER method also demonstrated an AUC advantage over the individual MORR screens in highly ranking the 34 (three original screens) and 114 (all six screens) common HDFs (Figure S3B; Table S3). Overall, the increased enrichment among these sets suggests that the use of the MORR-RIGER method to quantitatively integrate the screen data sets produces a less biased and more robust HDF network.
Using Gene Expression Filtering to Decrease OTEs
The highest AUCs were seen for the NPC and the lowest in the HIV infection set; this may arise because several genes in the latter set (i.e., HLA; Table S3) are not expressed in the screening cells. Microarray analysis (Affymetrix GeneChip human 2.0 ST array) was used to determine the levels of gene expression in the MAGI cells. The values for the probe set in the microarray were matched to the genes present in the siRNA libraries based on searching several identifiers. Overall, we matched 17,205 (81.5%) and 18,168 (83.6%) genes with expression data in the SMARTpool and Silencer Select screens, respectively (Tables S1 and S4). The median of the negative control intron probe set was used as a cutoff for gene expression, producing a list of 12,115 common genes in the siRNA libraries that are likely to be expressed in the MAGI cells as well as the other HeLa cell line used by Zhou et al. (Rosetta; Zhou et al., 2008). We used this expression data as a filter (gene expression filtering) to remove RIGER5 HDFs whose mRNAs were present at levels less than the intron probe median because they most likely represented OTEs. Using this approach we found that among the top 150 RIGER5 SB genes only 26 fell below the intronistic median (17.3%), including two olfactory receptor genes (Figure 2C). There was no correlation between gene expression and RIGER ranking (Figure S4). However, while this approach is helpful, it identified MED27 as an OTE, which is unlikely. Therefore, with certain exceptions, we envision gene expression filtering to be useful for decreasing OTEs.
High False Negatives Impact Screen Comparisons
Comparison of the screens highlighted that while either distinct screens or those done by the same group using different siRNA libraries may detect functionally related genes, they are much less effective in detecting the same gene (Bushman et al., 2009). As we and others have noted, false negatives as well as false positives occur frequently in RNAi screening (Adamson et al., 2012; Hao et al., 2013). Earlier comparisons have had to qualify these observations due to potential variation in experimental design and reagents; however, in this instance, we have controlled for these variables between the esiRNA and Silencer Select screens and still find a low degree of overlap within two known HDF complexes, MED and NPC (Figures 1, 2, S1, and S2; Table S2). Extending this comparison made apparent that the degree of false negatives is both library and screen dependent (Table S2). For example, the Rosetta library performed well in the primary screen when identifying MED (8.3% estimated false-negative rate for MED), even though secondary screens removed these candidates from the published high-confidence list, considerably lowering the overlap between this and the other two early HDF screens. Similarly, with the NPC genes, the Rosetta screen was the most effective; however, in this instance, toxicity was a major factor in diminishing overlap, suggesting that the creation of hypomorphs with less penetrant phenotypes was optimal. Based on the average value seen with MED and NPC, the screens displayed 35.7%–66% false-negative rates (Rosetta 35.7%, Silencer Select 59.0%, QIAGEN 65.9%, SMARTpool 63.3%, and esiRNA 66.0%; Table S2).
RIGER ranks genes based partly on their similarly scoring across multiple screens and in some cases is improved by the integration of additional screen data sets. For example, genes that decreased infection across most of the screens (MED6 and MED8) had lower p values in RIGER5 versus RIGER3. In contrast, COG2-4, which scored only in the MORR screens, displayed larger p values in RIGER5 than in RIGER3. This is also seen with the greater number of MED and NPC subunits identified as HDFs using a simple cataloging of those that met selection criteria in any of the siRNA screens (ALL5) versus those that were ranked in the top 550 in RIGER5 (RIGER5; Figures S2A and S2B).
Use of GESS to Rule Out Prominent OTEs
Genome-wide enrichment of seed sequence matches (GESS) identifies OTEs (Sigoillot et al., 2012). OTEs result primarily from the binding of siRNA seed sequences to cognate mRNAs that they were not designed to deplete. GESS detects OTEs by searching for matches between the collective RefSeq mRNAs (coding sequences [CDSs], 5′ and 3′ UTRs) and the seed sequences of the individual siRNAs that elicit the phenotype of interest in the validation round (active siRNAs). The siRNAs that do not score in the validation round (inactive siRNAs), or a scrambled set of the active siRNA sequences, serve as negative controls. An mRNA that pairs more frequently with the validation round seed sequences compared to the scrambled or inactive sequences suggests an OTE. Using GESS, we found that several genes were recognized by the Silencer Select validation round seed sequences more frequently than the scrambled sequences (Figure 2D; Table S5). However, these results did not pass a statistical significance test (Fisher’s exact test with Benjamini-Hochberg correction, alpha = 0.05). None of the putative OTE target genes represent a known or putative HDF. Based on the GESS analysis, we estimate that <10% of the validation round siRNAs that confirmed in the Silencer Select screen are OTEs.
We used GESS to look for siRNAs that inhibit HIV-1 by promiscuously binding to viral mRNAs (on-viral effects [OVEs]) using the HIV-1 genome and detected no significant events (Figure S2D). Therefore, the Silencer Select screen was not dominated by one or more prominent OTEs, as with certain screens (Sigoillot et al., 2012). The likely OTEs identified by gene expression filtering but that were not noted by GESS as OTEs suggest that there are more complex interactions occurring, possibly involving less potent OTEs.
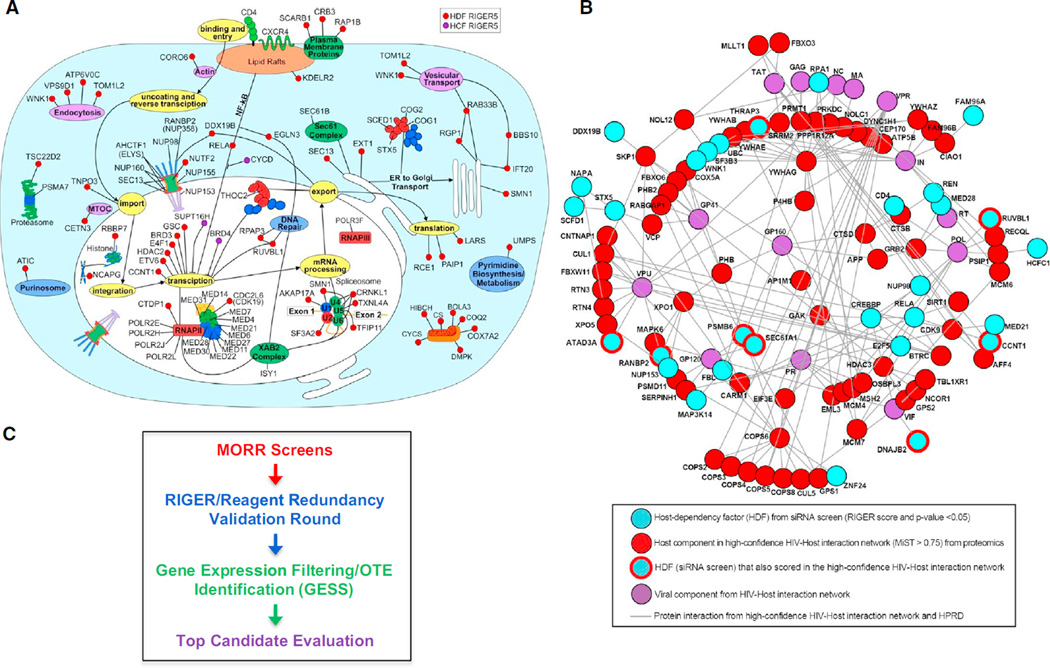
We next combined the RIGER5 analysis and gene expression filtering to generate a top 150 HDF list (Table S6). Using this list, we constructed an updated hypothetical model cell representing key steps in the HIV-1 life cycle as well as where 86 of the RIGER5 HDFs might function based on the literature (Figure 3A; Table S6; Brass et al., 2008). Inclusion criteria were the HDF possessing a gene expression value greater than the intron control as well as literature reporting a function. Genes in the RIGER top 150 that did not meet these criteria are not pictured (Table S6). We have also created a table containing RIGER rankings and the normalized primary screen data from all of the screens for these RIGER top 85, as well as the common 34 HDFs and the HDFs that were further evaluated below (Table S7). Given the near saturation of multiple major complexes in the RIGER5 data set, as well as their overlap with the events of the HIV-1 life cycle, this model likely approximates the majority of the virus’ requirements in HeLa cells.
Figure 3. MORR Screen HDF Model and HIV-Host Proteome Analyses.
(A) Using the RIGER5 list as a starting point, we constructed an updated hypothetical model cell depicting key steps in the HIV-1 life cycle as well as where 85 of the HDFs might function based on the literature (Table S6).
(B) Network showing connections between HDFs (top-ranked 123 from RIGER3 SB analysis) and host proteins that interacted with HIV viral components (Jäger et al., 2012). Human protein reference database (HPRD).
(C) MORR screening is followed by RIGER analysis and traditional reagent redundancy validation. Gene expression filtering and the GESS program are next used to remove likely OTEs. Based on these results, candidates are selected for further evaluation.
Network Analysis of HDFs and HIV-Human Protein Complexes
A proteomics study identified >430 host factors that interact with HIV-1 proteins using a quantitative ranking system (MiST; Jager et al., 2012). To find connections between this resource and HDFs, we performed a network analysis of the RIGER3 (p values < 0.05; Table S3) and proteomics data sets (MiST score > 0.75) using a heuristic approach starting with a central scaffold of first-order (direct) HDF-HDF interactions. We next extended this to direct interactions between a HDF and a host factor that interacted with HIV [viral component (Figure 3B), first-order (direct interactions with enrichment p ≤ 0.002)]. This analysis identified several HDFs that directly interact with an HIV protein (HDF: viral protein, ATAD3A: Vpu, RANBP2: Vpu, PSMB6:gp160, THRAP3: Gag, RUVBL1: POL, CCNT1: Tat, DNAJB2: Vif, SEC61A1: ENV). Among these HDF-HIV interactors was RUVBL1, a NuA4 histone acetyltransferase complex member that modulates transcriptional activity via histone modification (Jha et al., 2008). In line with this broad-acting role, RUVBL1’s depletion with either of three siRNAs inhibited the replication of all the retroviruses tested, including MLV-GFP and pPHAGE-GFP, the latter being dependent on a cytomegalovirus (CMV) cis element for expression (Figures S5A–S5C).
To this point, we have outlined a workflow for RNAi screening starting with MORR screens followed by a reagent-redundancy-based validation round and quantitative integration of screen data sets using RIGER. We have also used the GESS program and gene expression filtering in an attempt to decrease OTEs (Figure 3C). In this next section, we further evaluate several top-candidate HDFs identified above.
HIV-1 Requires Specific Host Cell Nucleotide Synthesis Enzymes
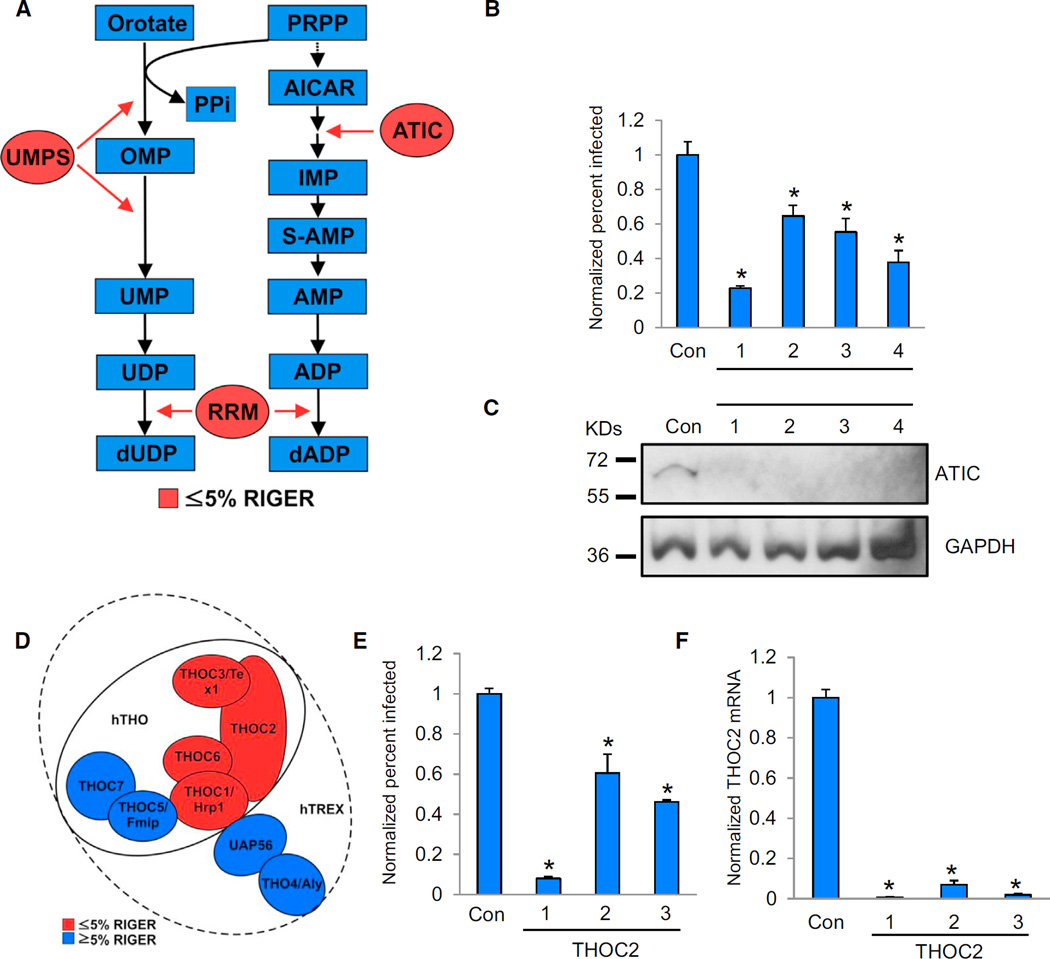
The MORR screens detected multiple factors involved in nucleotide metabolism. While wholly expected that HIV-1 requires such pathways, these results nonetheless identify specific enzymes as HDFs. For example, the MORR screens identified HDFs involved in pyrimidine (UMPS) and purine metabolism (ATIC; ADSS; Cheong et al., 2004), as well as both subunits (RRM1 and RRM2) of a holoenzyme necessary for the formation of deoxyribonucleotides from ribonucleotides (RNR; Figure 4A). To validate the role of ATIC in viral replication, we transfected cells with siRNAs against ATIC or a control nontargeting siRNA (Con). After 72 hr, the transfected cells were challenged with HIV-IIIB for 48 hr and the percent infection determined. In parallel, we also assessed siRNA-mediated depletion (Figures 4B and 4C). All the siRNAs depleted ATIC and inhibited HIV-1, consistent with ATIC being an HDF.
Figure 4. HIV-1 Requires Specific Nucleotide Synthesis Enzymes and the THOC/TREX Complex.
(A) Multiple nucleotide synthesis enzymes (ATIC, UMPS, and RRM) were detected in the screens and RIGER analyses (≤5% RIGER = lowest 5% q-values).
(B) MAGI HeLa cells were transfected with either nontargeting control siRNA (Con) or siRNAs targeting ATIC (1–4). At 72 hr posttransfection, cells were infected with HIV-1 IIIB. After 48 hr, cells were immunostained with an anti-HIV-1 p24 CA antibody (anti-CA). The percentage of infected cells was measured using image analysis software and normalized to the control siRNA transfected cells.
(C) Whole-cell lysates from the indicated cells in (B) were subjected to immunoblotting using the noted antibodies. kDa, kilodaltons; GAPDH, loading control.
(D) Schematic model of the human THO/TREX complexes (Viphakone et al., 2012). Subunits detected in the MORR screens and RIGER5 SB analysis are shown in red (≤5% RIGER = lowest 5% q-values).
(E) HeLa cells were transfected with the indicated siRNAs and assessed as in (B).
(F) siRNA-mediated depletion of THOC2 was evaluated by quantitative PCR (qPCR) measurement of mRNA levels from the cells in (E). mRNA levels were normalized to those of the control siRNA transfected cells.
Results throughout are the mean of three independent experiments ± SD. *p ≤ 0.05 (Student’s t test).
THOC2, the COG Complex, and SUPT16H Play Roles in HIV-1 Replication
The THO/TREX complex couples transcription to the export of poly(A) RNA and mRNAs derived from intronless genes (Strässer et al., 2002). Multiple THO/TREX components were highly ranked by RIGER5, including THOC1/Hpr1 (p value < 0.05), THOC2 (p value < 0.001), and THOC3/Tex1 (p value < 0.05; Figure 4D). Three independent siRNAs targeting THOC2 validated for both their targeting efficacy and their effects on HIV-1 infection (Figures 4E and 4F). Additional subunits (THOC4, THOC5, and THOC7) were ranked lower by RIGER, perhaps due to the existence of different complexes with varying significance for HIV-1 replication. In support of this idea, and in accordance with an existing model (Figure 4D; Viphakone et al., 2012), THOC1/Hpr1, THOC3/Tex1, and THOC6 may directly associate with THOC2, suggesting these proteins may form a subcomplex that is important for viral replication or that silencing any one of them may decrease the stability of the others, thereby enhancing penetrance.
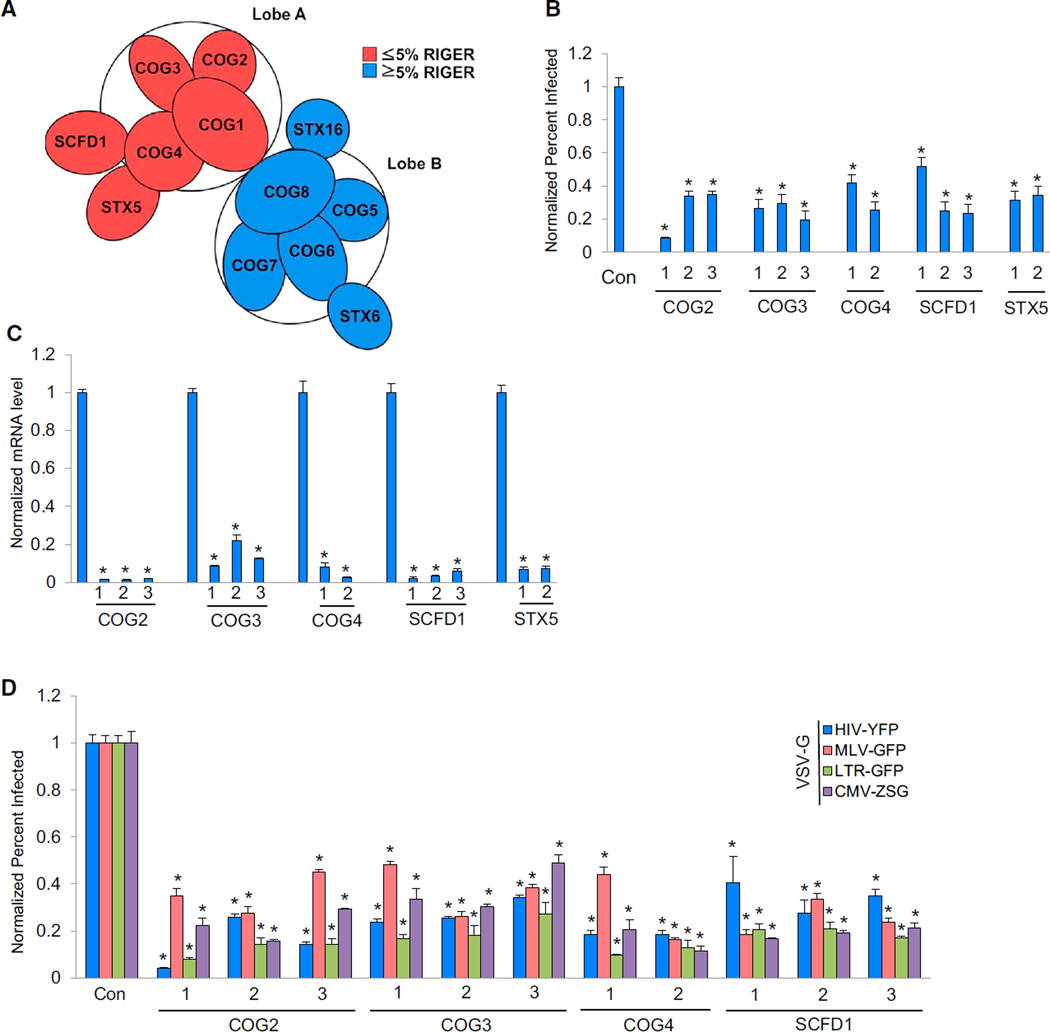
COG functions in retrograde vesicular transport within the Golgi and is critical for the recycling of glycosyltransferases (Miller and Ungar, 2012). In mammalian cells, COG consists of eight subunits (COG1–8) arranged into two lobes: lobe A (COG-LA, COG1–4) and lobe B (COG-LB, COG5–8). Our earlier screen had identified COG-LA as being required for HIV-1 replication along with the COG-interacting proteins STX5 and SCFD1, and this was seen again in both the Silencer Select and esiRNA screens, with RIGER3 highly ranking COG-LA along with STX5 and SCFD1 (Figure 5A; Table S3). Coupled RNAi and phenotype assays showed that multiple siRNAs targeting each of the COG-LA subunits, or STX5 and SCFD1, decreased viral replication and the mRNA levels of their respective targets (Figures 5B and 5C). We next used several vesicular stomatitis virus-G (VSV-G) pseudotyped viruses to investigate the role of these factors in replication: The HIV-yellow fluorescence protein (HIV-YFP), HIV-1 LTR-green fluorescence protein (LTR-GFP), and CMV Zoanthus species green (CMV-ZSG) viruses were used to evaluate involvement in ENV-mediated entry and/or Tat-dependent LTR transactivation, while the gamma retrovirus, Moloney leukemia virus-GFP (MLV-GFP) virus, was used to test for lentiviral specificity. The depletion of any three COG-LA members or SCFD1 reduced infection by all of these viruses (Figure 5D). These results indicate that COG-LA and SCDF1 modulate the infection of viruses expressing either VSV-G or HIV-1 ENV potentially via their regulation of glycosylation.
Figure 5. COG Plays a Role in HIV-1 Replication.
(A) Schematic of the COG complex. Subunits of COG lobe A (COG-LA) detected in the MORR HDF screens and RIGER5 analysis (≤5% RIGER = lowest 5% q-values) are shown in red. Genes in blue represent those with q values in the highest 95%.
(B) HeLa cells were transfected with the indicated siRNAs. At 72 hr after transfection, cells were infected with HIV-1 IIIB. After 48 hr, cells were immunostained with anti-CA. The percentage of infected cells was normalized to the control siRNA-treated cells.
(C) siRNA-mediated depletion was evaluated by qPCR measurement of the endogenous mRNA levels from (B). mRNA levels were normalized to those of the control siRNA-transfected cells.
(D) HeLa cells were transfected with either nontargeting control siRNA or the indicated siRNAs. At 72 hr posttransfection, cells were infected with one of the following VSV-G-pseudotyped viruses: HIV-YFP, MLV-GFP, LTR-GFP, or pPHAGE-CMV-ZSG. After incubation for 48 hr, the percentage of positive cells was determined and normalized to controls.
Results throughout are the mean of three independent experiments ± SD. *p ≤ 0.05 (Student’s t test).
SUPT16H and SSRP1 comprise the FACT complex, which regulates nucleosome formation and transcription elongation (Belotserkovskaya et al., 2003). Multiple siRNAs were confirmed in the validation round for SUPT16H as increasing viral infection, with the greatest impact seen with the transfer of viral supernatant (part two, Figures S6A and S6B). These data are consistent with experiments in yeast showing that FACT regulates the basal transcription of the 5′ HIV-LTR. Loss of SUPT16H helps alleviate repression of the cryptic promoters of latent HIV proviruses (Gallastegui et al., 2011). To test the significance of these events in a more physiologic setting, we stably transduced primary human CD4+ T cells with three independent small hairpin RNAs (shRNAs) against SUPT16H. When these cells were infected with HIV-1 IIIB, the depletion of SUPT16H produced a 3-fold increase in Gag mRNA levels (Figures S6C and S6D). Similar results were obtained with VSV-G-pseudotyped HIV-1 NL4-3-GFP, but not with MLV-GFP (Figure S6E). We next evaluated SUPT16H’s effect on the LTR promoter in HeLa cells. Using the lentiviral vectors described above, we demonstrated that SUPT16H levels modulated the activity of the HIV LTR, but not the CMV promoter (Figure S6F). We also used an LTR-driven luciferase reporter assay to show that the expression of HIV-1 Tat and the depletion of SUPT16H led to further enhanced reporter activity and that this required the presence of the TAR sequence, which recruits Tat to the LTR, consistent with previous work suggesting that SUPT16H is a HIV-1 competitive factor (HCF; Figure S6G).
GOLGI49 Is Required for HIV-1 Entry
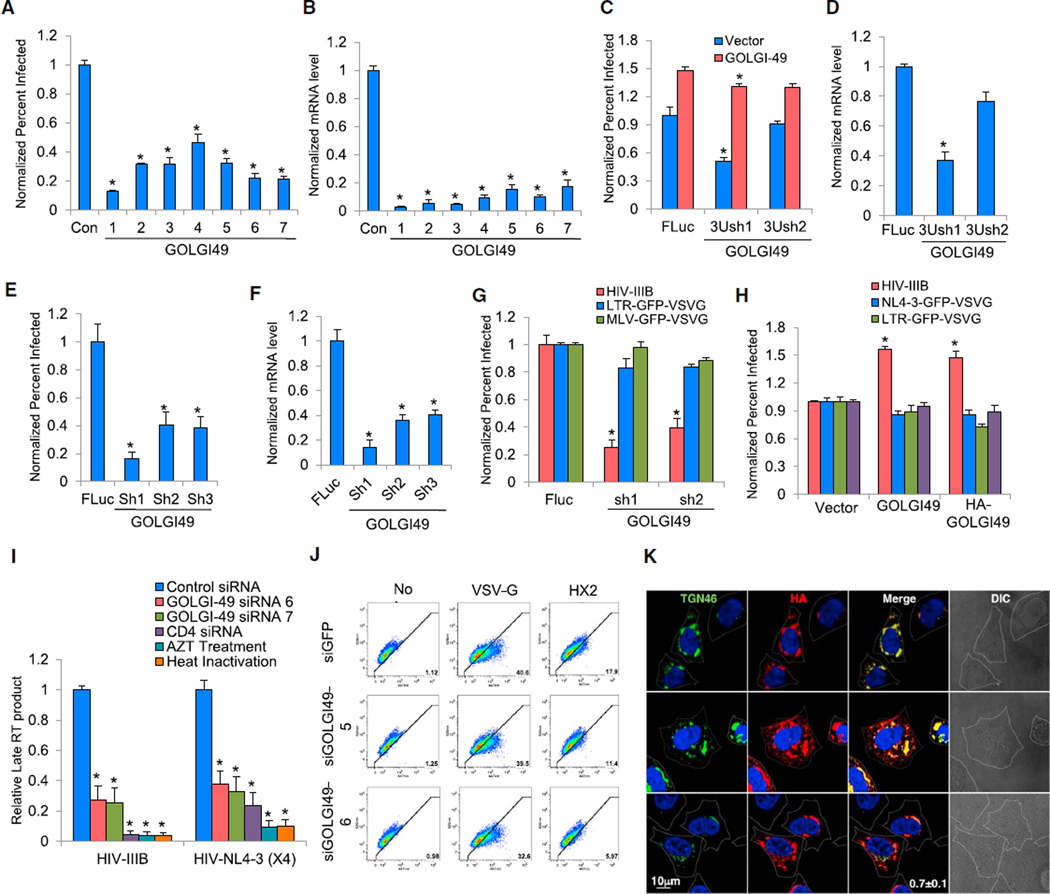
C3orf58 is a Golgi protein of unknown function that was a top candidate in the MORR screens (Takatalo et al., 2008). Based on this and the data below, we have named this gene GOLGI49. In validation experiments, the transfection of any of seven GOLGI49 siRNAs depleted mRNA levels and reduced the replication of both HIV-1 IIIB (X4-tropic) and BaL (R5-tropic) viruses (Figures 6A, 6B, and S7A). We then performed a rescue experiment by restoring HIV-I replication with a stably expressed GOLGI49 cDNA in conjunction with an shRNA targeting the endogenous mRNA’s 3′ UTR (Figures 6C and 6D). Expression of three distinct shRNAs in primary human CD4+ T cells reduced the infection of HIV-1 NL4-3-GFP, and these events correlated with efficient mRNA depletion (Figures 6E and 6F). In both gain- and loss-of-function experiments, we determined that GOLGI49 was specifically required for replication of HIV-1 IIIB, but not two VSV-G-pseudotyped HIV-1 viruses, leading us to postulate that GOLGI49 may be required during the ENV-dependent phase of infection (Figures 6G and 6H). Consistent with this notion, GOLGI49 depletion decreased late RT levels (Figure 6I). A β-lactamase-Vpr (BlaM-Vpr) fusion assay revealed that GOLGI49 depletion reduced the fusion of pseudotyped viruses expressing the ENV of HIV-IIIB (HX2B), but not viruses expressing VSV-G (Figure 6J). Surface expression of neither CD4 nor CXCR4 was altered from controls in multiple cell lines expressing shRNAs targeting GOLGI49 (Figure S7B). In keeping with a previous report, confocal imaging showed colocalization between stably expressed GOLGI49 containing a FLAG epitope tag and the known Golgi protein, TGN46 (Figure 6K) (Takatalo et al., 2008). Using MORR, we identified a Golgi-resident HDF, GOLGI49, whose loss results in an early block in the viral life cycle, most likely at the level of either viral binding or viral fusion to the host’s plasma membrane.
Figure 6. Depletion of GOLGI49 Prevents HIV-1 Entry.
(A) HeLa cells were transfected with the indicated siRNAs. At 72 hr posttransfection, cells were infected with HIV-1 IIIB. After 48 hr, cells were stained with anti-CA. The percentage of infected cells was measured and normalized to the control siRNA-treated cells.
(B) siRNA-mediated depletion was evaluated by qPCR measurement of mRNA levels from (A). mRNA levels were normalized to those of the control siRNA-transfected cells.
(C) TZM-bl HeLa cells stably expressing the indicated short hairpin RNAs (shRNA) against the 3′ UTR of the GOLGI49 mRNA (3Ush) or a control shRNA against firefly luciferase (FLuc) were either mock transfected (mock) or transiently transfected with a vector expressing the GOLGI49 cDNA. After 48 hr, the cells were infected with HIV-IIIB. After an additional 48 hr, the cells were stained with anti-CA. The percentage of infected cells was measured and normalized to the FLuc control cells.
(D) GOLGI49 3′ UTR shRNA-mediated depletion was confirmed by measuring mRNA from (C). mRNA levels were normalized to those of the FLuc control cells. 3Ush, 3′UTR shRNA.
(E) Primary human CD4+ T cells stably expressing either the control FLuc shRNA or one of the indicated shRNAs against GOLGI49 (Sh1-3) were infected with HIV-1 NL4-3-GFP. The percentage of GFP-positive cells was measured at 48 hr postinfection and normalized to the control cells.
(F) GOLGI49 shRNA-mediated depletion in primary CD4+ T cells in (E) was confirmed by qPCR. mRNA levels were normalized to those of the FLuc control cells.
(G) TZM-bl HeLa cells stably expressing the indicated shRNAs were infected with HIV-1 IIIB, LTR-GFP-VSV-G, or MLV-GFP-VSV-G. After 48 hr, the percentage of infected cells was measured and normalized to the levels found in the FLuc control cells.
(H) TZM-bl HeLa cells stably transduced with viruses containing an empty vector (pQCXIP, Vector), a GOLGI49 cDNA (GOLGI49), or an HA-tagged GOLGI49 cDNA (HA-GOLGI49) were infected with the indicated fluorescence protein reporter viruses. The percentage of infected fluorescent cells was measured at 48 hr postinfection and normalized to the control cells.
(I) HeLa cells were transfected with the indicated siRNAs. At 72 hr posttransfection, cells were infected with HIV-1 IIIB or HIV-1 NL4-3-GFP (X4). As negative controls, infections were performed in the presence of either azidothymidine (AZT) (2.5 uM) or using heat-inactivated virus. Cells were harvested at 18 hr postinfection and subjected to DNA extraction. Late RT product was measured by qPCR using described primer and probe set and normalized to the control siRNA-treated cells.
(J) HeLa cells were transfected with the indicated siRNAs (siGFP is a negative control siRNA targeting GFP). Cells were infected 72 hr later with an HIV-1 virus containing a BlaM-Vpr fusion protein pseudotyped with either VSV-G or an HIV-1 ENV (HX2, X4-tropic). Infected cells were incubated with the CCF2-AM substrate and fluorescence was measured. Cells at the lower-right corner of the diagonal distribution of the no-infection control (No) were gated as a positive population, and numbers in that region represent percent positive cells.
(K) Confocal imaging of HeLa cells stably expressing HA-GOLGI49 (HA, red) and the endogenous Golgi protein TGN46 (green). Cell outlines were rendered based on differential interference contrast (DIC) images (far right). Percent colocalization ± SD is provided in the bottom merged panel.
Results throughout are the mean of three independent experiments ± SD. *p ≤ 0.05 (Student’s t test).
The NPC Protein SEC13 Is Necessary for HIV-1 Replication after PIC Nuclear Entry but prior to Viral Integration
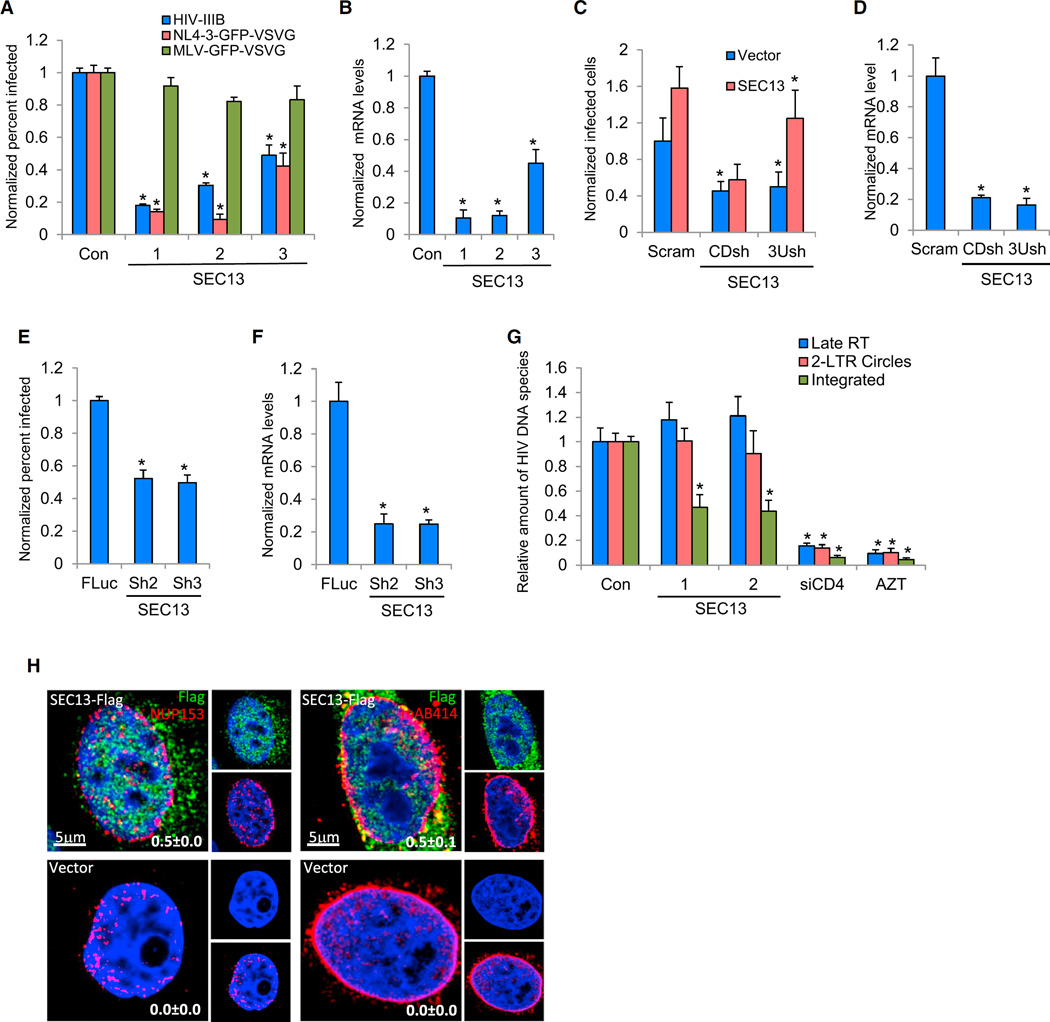
Depletion of SEC13 was found to inhibit HIV-1 infection. A WD-repeat family protein, SEC13, resides in both the ER, where it is involved in COPII vesicle formation and trafficking (Hsia et al., 2007), and the NPC’s cytosolic and nuclear portions. SEC13’s distinct residences, together with the evolving role of the NPC in HIV-1 replication, made it an interesting candidate for further evaluation. siRNAs against SEC13 each inhibited either HIV-1 IIIB or VSV-G-pseudotyped HIV-1 NL4-3-GFP (Figures 7A and 7B). This effect was lentiviral specific because SEC13’s loss did not alter infection by VSV-G-pseudotyped MLV-GFP. SEC13 depletion also lowered the expression of GFP from proviruses containing either an LTR or CMV promoter, indicating it was not specifically required for LTR activity (Figure S7C). We next rescued HIV-1 infection using an shRNA-resistant SEC13 cDNA (Figures 7C and 7D). SEC13 was also required for HIV-1 replication in primary human CD4+ T cells (Figures 7E and 7F) with similar results seen with the Jurkat T cell line (Figures S7D and S7E). While depletion of SEC13 did not alter the levels of either late RT products or 2-LTR circles, it did reduce HIV-1 integration, suggesting that it functions in the intranuclear portion of the HIV-1 life cycle (Figure 7G). In keeping with its reported locations, confocal imaging showed that SEC13 localized to both the ER and the nucleus, with the latter population partially colocalizing with other NPC proteins (Figure 7H).
Figure 7. SEC13 Is Needed for HIV-1 Replication after Nuclear Entry but prior to Integration.
(A) MAGI HeLa cells were transfected with the indicated siRNAs and infected 72 hr later with the noted viruses. The percentage of infected cells was measured at 48 hr postinfection and normalized to the control siRNA-treated cells.
(B) siRNA-mediated depletion was tested by measuring SEC13 mRNA from the cells in (A). mRNA levels are normalized to those of the negative control cells.
(C) TZM-bl HeLa cells stably expressing an shRNA against the coding sequence of the SEC13 mRNA (CDsh), the 3′ UTR of the SEC13 mRNA (3Ush), or a scrambled control shRNA (Scram) were transfected with either an empty expression vector (vector) or a vector expressing the SEC13 cDNA. After 48 hr, the cells were infected with HIV-IIIB. After an additional 48 hr, the cells were stained with anti-CA. The percentage of infected cells was measured and normalized to the control siRNA-transfected cells. The exogenously expressed SEC13 cDNA rescued HIV replication in the 3Ush cells, but not the CDsh cells.
(D) SEC13 shRNA targeting efficacy was confirmed by measuring mRNA from cells in (C). Values were normalized to the mRNA level of the scramble shRNA control cells.
(E) Primary CD4+ T cells stably expressing either FLuc shRNA or two different SEC13 shRNAs were infected with NL4-3-GFP-VSV-G. The percentage of infected cells was measured 48 hr later and normalized to the values determined for the FLuc shRNA-transduced cells.
(F) SEC13 shRNA-mediated depletion efficacy in cells in (E) was determined. mRNA levels were normalized to those of FLuc shRNA-transduced cells.
(G) HeLa-T4 cells were transfected with the indicated siRNAs for 72 hr. Cells were then infected with HIV-1 IIIB. As a control, nontransfected cells were pretreated with AZT. DNA was harvested and subjected to qPCR assays measuring late RT (blue), 2-LTR circles (red), or integrated proviral DNA species (integrated, green). The relative level of viral DNAs was normalized to cells transfected with the control siRNA.
(H) Confocal imaging of HeLa cells stably expressing SEC13-FLAG (green) or the empty vector control (Vector) immunostained using either an antibody that recognizes the NPC protein, NUP153 (red), or one that recognizes the NPC complex (mAB414, red). Nuclear DNA is shown in blue. Percent colocalizations of either NUP153 or mAB414 with SEC13 ± SD from multiple cells from multiple fields are provided in the bottom right corner of the merged panel.
Results throughout are the mean of three independent experiments ± SD. *p ≤ 0.05 (Student’s t test).
DISCUSSION
RNAi screens have revealed the dependencies of multiple pathogens and in the case of HIV-1 enabled the discovery of numerous HDFs (Brass et al., 2008; König et al., 2008; Zhou et al., 2008). However, the overlap of candidates identified across these efforts has been low (Bushman et al., 2009; Goff, 2008). To address this issue and attempt to saturate the HDF screen, we set out to generate a high-confidence list of host proteins required for viral replication. To do this, we used three large-scale siRNA resources and integrated these primary data sets with those from earlier studies by adapting an established program, RIGER. This MORR-RIGER strategy produced a quantitatively integrated data set that highly ranked both known HDFs and previously unappreciated ones. A comparison of the RIGER3 and RIGER 5 HDF data sets with a set of expertselected HDFs (Reactome) showed that both analyses enriched for these test genes and that integrating all five screens improved this metric in several instances.
Identifying OTEs
To further improve the HDF data set, we determined expressed genes in the cells used for the screens and used this data to remove poorly expressed genes, which likely represent OTEs. A recently created OTE identification program, GESS, was used to rule out prominent OTEs. Therefore, by using RIGER to integrate all the HDF screens, and combining this with gene expression filtering, we produced a rigorous genome-wide data set that quantifies each gene’s role in HIV-1 replication.
Individual Screens Are Dominated by False Negatives
As different experimental approaches may contribute to interscreen variability, we kept these factors constant within the MORR screens. Nonetheless, a comparison of the data between the MORR screens demonstrates a low level of exact gene overlap. Thus, while the field has become accustomed to OTEs, these data also reveal the false negatives encountered using these resources, a concern recently hypothesized (Adamson et al., 2012; Hao et al., 2013). Having a data set with known positives allowed the direct testing of this possibility. A useful illustration of false negatives is the MED complex, of which only 4 out of 26 total components scored in four or more individual screens: MED6, 7, 14, and 28 (Table S2). Similar results are seen when comparing the NPC, THOC, and NF-κB pathway components across the screen data sets.
RIGER Reduces False Negatives
False negatives arise because of the variability in the penetrance of RNAi and the inability to easily take advantage of hypomorphs. We suggest that the near saturation of several protein complexes using the MORR-RIGER method may arise because while each siRNA library is limited by intrinsic false negatives, the use of a combined approach offsets this limitation by taking advantage of the best that each of the libraries has to offer. All of the previously published HIV screens provide lists with strict thresholds for siRNAs that score. However, siRNA effects are incomplete and produce a range of hypomorphic phenotypes that still provide information even if falling short of the selection cutoffs. Using RIGER to assess all five data sets minimizes the loss of data regarding potential HDFs whose phenotypic penetrance did not surpass a strict cutoff. We propose that a quantitatively integrated analysis of similar screens using orthologous reagents permits additional information to be extracted from each separate effort and strengthens the conclusions of this work. These analyses can also be used to evaluate the quality of a given screening effort or reagent. For example, if we know that MED components should be found in an HDF screen, we can determine how many of the total expected MED subunits were recovered. To evaluate this quality metric, we have surveyed each of the five HDF screens for the percentage of the total MED and NUPs detected. This analysis revealed that, overall, increasing the number of independent screens in a combined analysis improves the detection of relevant genes (KS method). These results also further clarify the origins of the lack of overlap seen among the earlier screens and empirically indicate that false negatives and hypomorphs play a significant role in screen-to-screen discordance.
HDFs
While the MORR screens confirmed many known host factors, they also identified several novel HDFs. Although more remains to be done to define the mechanism underlying these many host-viral interactions, we focused on validating a subset of genes that function at various stages of the HIV-1 life cycle: entry (GOLGI49), reverse transcription (ATIC), integration (SEC13), and transcription (inhibitory, SUPT16H). A growing number of HDFs play roles in the post-nuclear-entry HIV-1 life cycle (Di Nunzio, 2013). Depletion of SEC13 inhibited HIV-1 after nuclear entry but before proviral integration. SEC13 is a nucleoporin that localizes to both the nuclear and the cytoplasmic faces of the NPC. Nuclear SEC13 associates with chromatin and may be an activator of early transcription (Capelson et al., 2010). Together with its requirement for HIV integration, these data suggest that SEC13 may play a role in helping the HIV-1 PIC interact effectively with the host’s chromatin.
In carrying out this study, our goal was to improve our understanding of HIV-1’s dependencies by (1) minimizing false negatives by taking advantage of the best that each siRNA library offers, (2) using gene expression filtering to decrease OTEs, and (3) avoiding the pitfalls of hypomorphism and absolute cutoffs in candidate selection by integrating multiple screens (RIGER; Figure 3C). By integrating all of the data generated with the orthologous libraries, RIGER in effect performs a large-scale reagent redundancy-based evaluation of the entire genome’s role in HIV replication. However, since RIGER may bias against specific HDFs that possess wide phenotypic variation between screens, we favor using both RIGER and a traditional reagent redundancy-based validation method. This strategy may prove useful to integrate data sets from multiple screens for other phenotypes performed by separate groups that are currently hampered by OTEs, hypomorphs, and false negatives. Additional screening methods, i.e., haploid cells and CRISPR, can also be integrated together with siRNA efforts using RIGER. We will continue to improve these efforts by adding such capabilities as well as those integrating bioinformatic data sets to generate rankings based on screening results together with functional and physical gene associations.
EXPERIMENTAL PROCEDURES
Cell Lines
Cells were from the NIH AIDS Reagent Repository: HeLa MAGI, HeLa T4, TZMbl, HeLa-Tat-III/CMV/d1EGFP (#11062), HeLa-Tat-III/LTR/d1EGFP (#11063, both from Parent et al., 2005), and Jurkat Clone E6-1 (# 177; Weiss et al., 1984).
Viruses
The HIV-1 IIIB strain and the NL4-3 plasmid were from the NIH AIDS repository. VSV-G pseudotyped viruses were created by cotransfecting pCG-VSV-G and pCG-Gag-Pol vectors using the viral constructs (HIV-YFP, HIV-1 LTR-GFP [LTR-GFP], CMV-ZSG [pPHAGE-CMV-ZSG], and MLV-GFP).
Antibodies
For the MORR screens, we used the primary antibody, anti-HIV-1 p24 mouse monoclonal antibody mab-183 (NIH AIDS Reagent Repository), and the secondary antibody, goat anti-mouse Alex Flour 488 (Invitrogen), for immunofluorescence detection of HIV-1 infection following a published protocol. SUPT16H (sc-165987), GAPDH (sc-25778), and actin (sc-7210) antibodies from Santa Cruz, and a mouse ATIC antibody (A8480) from Sigma, were used for immunoblots. Flow cytometry used CD4-FITC (Immunotech PN IM0448) and CXCR4-PE (BD Pharmigen 555974).
Plasmids
pQCXIP-HA-GOLGI49 and pQCXIP-GOLGI49 were constructed by subcloning GOLGI49/C3orf58 with or without an N-terminal hemagglutinin (HA) tag into the pQCXIP retroviral vector (Clontech) using AgeI and BamHI sites. pQCXIP-FLAG-SEC13 was constructed by subcloning SEC13 with N-terminal FLAG tag into the pQCXIP vector using NotI and BamHI sites. Pseudotyped viruses were packaged using pCG-VSV-G and pCG-Gag-Pol vectors. pDEST40-GOLGI49 and pDEST40-SEC13 were constructed through Gateway LR reactions of pDONR223-GOLGI49 or pDONR223-SEC13 (Open Biosystems) with a pcDNA-DESR40 vector (Invitrogen) using LR Clonase enzyme mix (Invitrogen).
siRNA Screening
The RNAi screens were done in triplicate using three siRNA libraries: Silencer Select, Ambion (21,584 siRNA pools, three oligos per pool); esiRNA, Sigma (15,300 siRNA pools, complex pools); and Dharmacon RefSeq27 Reversion Pools (4,506 siRNA pools, four oligos per pool) following a previous established protocol (Brass et al., 2008; Zhu et al., 2012). Briefly, HeLa MAGI cells (approximately 600 per well) were reverse transfected with 50 nM final concentration siRNA using Oligofectamine (Invitrogen) in 384-well plates. The cells were infected 72 hr later with HIV-IIIB in 30 µl fresh Dulbecco’s modified Eagle’s medium. Cells were fixed 48 hr later (4% paraformaldehyde; Sigma) in Dulbecco’s phosphate-buffered saline (D-PBS) (Sigma), permeabilized (0.2% Triton X-100 in D-PBS), and stained for intracellular HIV-1 p24 expression using the anti-p24 antibody mAB-183 (NIH AIDS Reagent Repository) and goat anti-mouse Alexa 488. Cellular DNA was stained using Hoechst 33342 (Invitrogen). Immunostained cells were imaged using an Image Xpress Micro microscope (Molecular Devices) and the images analyzed to determine percent infection and cell number (cell scoring module; Metamorph, Molecular Devices). The Qiagen (König et al., 2008) primary screen data set was kindly provided by S. Chanda (Burnham Institute). The Rosetta (Zhou et al., 2008) primary screen data set was kindly provided by A. Espeseth (Merck & Co.).
Luciferase Assay
293T cells were transfected with the indicated firefly luciferase reporters, along with pRL-TK (Promega) as a control. The cells were processed using the Dual-Glo Luciferase Assay System (Promega), and the plates were read using a Luminoskan Luminometer (Thermo Fisher Scientific). The HIV-1 LTR luciferase reporter construct was obtained through the AIDS Research and Reference Reagent Program. The TAR-deleted (dTAR) LTR luciferase reporter construct was a kind gift from Dr. Andrew Badley (Mayo Clinic). The HIV-1 Tat expression plasmid was a kind gift from Dr. Richard Mulligan (Harvard Medical School).
Supplementary Material
Highlights.
Multiple orthologous RNAi reagents were used to elucidate HIV-host interactions
Screens were traditionally validated and quantitatively phenotyped
All screen data sets were integrated to quantify each gene’s role in HIV-1 replication
Roles of GOLGI49, SEC13, COG, and THOC in HIV-1 replication were investigated
Acknowledgments
We thank S. Chanda (Burnham Institute) and A. Espeseth (Merck & Co.) for generously sharing their primary screen data, the ICCB-L (C. Shamu, S. Rudnicki, S. Johnston, K. Rudnicki, and D. Wrobel), UMass Medical School (P. Spatrick), the UMMS Genomics Core Facility (R. Fish, B. Hobbs, L. Benson, T. Brailey, and J. Barrett), and the Ragon Institute (M. Boyarina, K. Donnelly, and P. Richtmeyer). This research was funded by a grant from the Bill and Melinda Gates Foundation to A.L.B and S.J.E. A.L.B. is grateful to the Bill and Melinda Gates Foundation, the Burroughs Wellcome Fund, the UMass CFAR, and the NIH (1R01AI091786) for their generous support; S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.09.031.
REFERENCES
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNAbinding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, König R, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong CG, Wolan DW, Greasley SE, Horton PA, Beardsley GP, Wilson IA. Crystal structures of human bifunctional enzyme aminoimidazole- 4-carboxamide ribonucleotide transformylase/IMP cyclohydrolase in complex with potent sulfonyl-containing antifolates. J. Biol. Chem. 2004;279:18034–18045. doi: 10.1074/jbc.M313691200. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Di Nunzio F. New insights in the role of nucleoporins: a bridge leading to concerted steps from HIV-1 nuclear entry until integration. Virus Res. 2013;178:187–196. doi: 10.1016/j.virusres.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- Gallastegui E, Millán-Zambrano G, Terme JM, Chávez S, Jordan A. Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. J. Virol. 2011;85:3187–3202. doi: 10.1128/JVI.01920-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SP. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 2007;5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- Goff SP. Knockdown screens to knockout HIV-1. Cell. 2008;135:417–420. doi: 10.1016/j.cell.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, He Q, Wang Z, Craven M, Newton MA, Ahlquist P. Limited agreement of independent RNAi screens for virus-required host genes owes more to false-negative than false-positive factors. PLoS Comput. Biol. 2013;9:e1003235. doi: 10.1371/journal.pcbi.1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell. Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Surendranath V, Heninger AK, Slabicki M, Theis M, Putz G, Franke K, Caldarelli A, Grabner H, Kozak K, et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat. Methods. 2007;4:337–344. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- König R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liptak T. On the combination of independent tests. Magyar Tud Akad Mat Kutato Int Kozl. 1958;3:171–197. [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. Highly parallel identification of essential genes in cancer cells. Proc. Natl. Acad. Sci. USA. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R, Franceschini A, Horvath P, Tetard M, Mancini R, von Mering C, Helenius A, Lozach PY. Genome-wide siRNA screens reveal VAMP3 as a novel host factor required for Uukuniemi virus late penetration. J. Virol. 2014;88:8565–8578. doi: 10.1128/JVI.00388-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VJ, Ungar D. Re’COG’nition at the Golgi. Traffic. 2012;13:891–897. doi: 10.1111/j.1600-0854.2012.01338.x. [DOI] [PubMed] [Google Scholar]
- Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent M, Yung TM, Rancourt A, Ho EL, Vispé S, Suzuki-Matsuda F, Uehara A, Wada T, Handa H, Satoh MS. Poly(ADP-ribose) polymerase-1 is a negative regulator of HIV-1 transcription through competitive binding to TAR RNA with Tat.positive transcription elongation factor b (p-TEFb) complex. J. Biol. Chem. 2005;280:448–457. doi: 10.1074/jbc.M408435200. [DOI] [PubMed] [Google Scholar]
- Puri N, Wang X, Varma R, Burnett C, Beauchamp L, Batten DM, Young M, Sule V, Latham K, Sendera T, et al. LNA incorporated siRNAs exhibit lower off-target effects compared to 20-OMethoxy in cell phenotypic assays and microarray analysis. Nucleic Acids Symp. Ser. (Oxf.) 2008;52:25–26. doi: 10.1093/nass/nrn013. [DOI] [PubMed] [Google Scholar]
- Sigoillot FD, Lyman S, Huckins JF, Adamson B, Chung E, Quattrochi B, King RW. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Methods. 2012;9:363–366. doi: 10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, Hurt E. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Krausslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatalo M, Järvinen E, Laitinen S, Thesleff I, Rönnholm R. Expression of the novel Golgi protein GoPro49 is developmentally regulated during mesenchymal differentiation. Dev. Dyn. 2008;237:2243–2255. doi: 10.1002/dvdy.21646. [DOI] [PubMed] [Google Scholar]
- Viphakone N, Hautbergue GM, Walsh M, Chang CT, Holland A, Folco EG, Reed R, Wilson SA. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 2012;3:1006. doi: 10.1038/ncomms2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Wiskocil RL, Stobo JD. The role of T3 surface molecules in the activation of human T cells: a two-stimulus requirement for IL 2 production reflects events occurring at a pre-translational level. J. Immunol. 1984;133:123–128. [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Zhu J, Gaiha GD, John SP, Pertel T, Chin CR, Gao G, Qu H, Walker BD, Elledge SJ, Brass AL. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Reports. 2012;2:807–816. doi: 10.1016/j.celrep.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.