Abstract
Objective
To provide a more global view of adipocyte changes in human insulin resistance by proteomics analyses.
Methods
Baseline biopsies of abdominal subcutaneous adipose tissue were obtained from 23 non-diabetic subjects. Euglycemic clamps were used to divide subjects into an insulin-resistant group (IR, N=10) and an insulin-sensitive (IS, N=13) group, which were of similar age and gender but unequal adiposity (greater in IR). Proteins of isolated adipocytes were quantified by mass spectrometry using normalized spectral abundance factors.
Results
Of 1,245 proteins assigned, 30 were detected in at least 12 of the 23 subjects that differed significantly in abundance ≥1.5-fold between IR and IS. IR displayed a pattern of increased cytoskeletal proteins and decreased mitochondrial proteins and FABP4 and FABP5. In subgroup analyses of adiposity-matched subjects, several of these changes were less pronounced in IR but the abundance of proteins related to lipid metabolism and the unfolded/misfolded protein response were significantly and unfavorably altered.
Conclusions
These results confirm lower abundance of mitochondrial proteins and suggest increased cytoskeletal proteins and decreased FABP4 and FABP5 in subcutaneous adipocytes of typical insulin-resistant individuals. Changes in proteins related to lipid metabolism and the unfolded/misfolded protein may discriminate insulin-resistant from insulin-sensitive individuals of equal adiposity.
Keywords: Adipocyte, proteomics, insulin resistance
Introduction
A reduced ability of adipocytes to take up and retain free fatty acids, leading to ectopic lipid accumulation, and abnormalities in the release of adipokines by adipocytes are critical factors in insulin resistance and the development of type 2 diabetes mellitus (1). Amongst others, hitherto identified mechanisms underlying these abnormalities include impaired adipogenesis, activation of pro-inflammatory pathways, endoplasmatic reticulum (ER) stress, reduced mitochondrial mass or function, and lipid droplet dysfunction (2–4). However, the molecular changes in adipocytes that cause systemic insulin resistance in humans remain poorly understood, which may be due to a narrow research focus (such as on a cellular process, biochemical pathway or a limited number of proteins) of previous studies. Therefore, a more global examination of adipocyte alterations may help identifying additional important factors in insulin resistance.
Studies using global gene expression analysis of subcutaneous adipose tissue have found patterns of gene expression changes that suggest defects in adipocyte differentiation, cell-cycle control and up-regulation of extracellular matrix constituents in subjects with obesity (5); down-regulation of major metabolic pathways, including branched-chain amino acid, fatty acid, carbohydrate, and mitochondrial energy metabolism and up-regulation of immune response genes were found from lean subjects to subjects with obesity and the metabolic syndrome (6). Moreover, in adolescents with obesity, an increased ratio of visceral to subcutaneous adipose tissue was associated with down-regulation of lipogenesis/adipogenesis and insulin resistance (7). However, mRNA levels are not deterministic of protein abundance (8), while the latter is a closer reflection of the activity level of a biological pathway. Moreover, little is known about the mechanisms that explain why excess body fat is associated with insulin resistance in most but not all individuals (9), which is critical for identifying targets to dissociate obesity from its metabolic complications.
In the present study, we therefore used an unbiased proteomics approach previously developed by our group (10, 11) to identify molecular factors in adipocytes that may be important in systemic insulin resistance in typical insulin-resistant individuals and that may discriminate metabolically unhealthy insulin-resistant individuals from metabolically healthy insulin-sensitive individuals with similar excess body fat.
Methods
After informed written consent, recruited subjects underwent a medical history, physical examination, screening laboratory tests and a 75 g oral glucose tolerance test. Inclusion criteria were 21–70 years of age and a body mass index (BMI) of 20–35 kg/m2. Twenty-three healthy non-diabetic subjects (8 men, 15 women) completed the study. The subjects were instructed to not depart from their regular diet or exercise for two days before the study. Subjects arrived at the Clinical Research Unit of the Center of Metabolic Biology, Arizona State University (ASU) at 8:00 AM after an overnight fast and underwent body composition measurement by DEXA, an adipose tissue biopsy (5–7 g) lateral to the umbilicus through a 1.5–2.0 cm incision under local anesthesia and a 120-min euglycemic-hyperinsulinemic (80 mU·m−2·min−1) clamp (12). To avoid selection bias, subjects were divided into an insulin-resistant group (IR) and an insulin-sensitive group (IS) based on predefined criteria of insulin sensitivity by the euglycemic clamp. An average glucose infusion rate (GINF) <5 mg·kg−1·min−1 during the last 30 minutes of the clamp was considered IR. The protocol was approved by the ASU Institutional Review Board.
Adipocyte proteomics analyses were performed as previously described (11) by one-dimensional gel electrophoresis followed by HPLC-ESI-MS/MS using a hybrid linear ion trap (LTQ)-Fourier Transform Ion Cyclotron Resonance (FTICR) mass spectrometer (Thermo Fisher, San Jose, CA) fitted with a PicoView™ nanospray source (New Objective, Woburn, MA). Data analyses and bioinformatics were performed as in (11). Normalized spectral abundance factors (NSAF) were used to determine protein abundance (13) as previously described (10, 11) with a false discovery rate of 5.27% at the peptide level and 0.28% at the protein level. Only peptides with ≥95% probability were considered. Criteria for protein identification included detection of at least 2 unique identified peptides and a probability score of ≥99%, based on Scaffold analysis.
Since a large number of proteins were assigned in at least one of the 23 subjects, only proteins detected in ≥12 of the 23 subjects [i.e. >50%; a commonly used threshold (10, 14, 15)] and whose average abundance was either increased or decreased ≥1.5-fold between IR and IS were used for statistical comparisons. Unpaired Student’s t-tests and the non-parametric Mann Whitney U test were performed, since about half of these proteins did not meet the assumption of normality.
Because of imbalances in adiposity (but not age and gender) between IR and IS, subgroup analyses were performed comparing IR subjects and IS subjects matched for BMI and percent body fat (N=8/group) in order to explore the molecular basis of metabolically unhealthy excess body fat. For this analysis, only proteins detected in ≥8 of the 16 subjects were included using otherwise the statistical approach described above. All statistical analyses were performed by SPSS Statistics Version 20.
Pathway analysis was performed using Ingenuity Pathway Analysis (Ingenuity Systems, Inc., Redwood City, CA). Only proteins that were identified in ≥12 of the 23 subjects and differed in abundance ≥1.5-fold between IR and IS were used in this analysis. A pathway was considered significantly enriched if the p-value for the pathway was <0.01 and the pathway included ≥5 identified proteins in order to avoid excessive influence of differences in only one or two proteins.
For Western blot analysis, proteins (10–15 µg) from adipocyte lysates were separated by Tris-Tricine PAGE and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA). Membranes were immunoblotted with the goat anti-human antibody against fatty acid binding protein (FABP) 4 and FABP5 (R&D Systems, MN) at 1:2500 and 1:1500 dilution, respectively. Anti-beta actin antibody (Sigma-Aldrich, MO) was used as the control. Donkey-anti-goat and goat-anti-mouse secondary antibodies IRDye 680 (Li-COR Biosciences, Lincoln, NE) were used at 1:15000 dilution. Blots were quantified by densitometry (Li-COR Odyssey CLx system Lincoln, NE).
For measurement of adipocyte size, isolated adipocytes were mixed well, placed on a siliconized glass slide in a silicone well and covered with a glass coverslip. Cell diameter was measured manually using phase contrast microscopy (BX51, Olympus Inc., Hicksville, NY) at 10× magnification and Image J. About 200–300 cells were measured for each subject.
Results
Subjects’ Characteristics
The division of subjects based on pre-defined insulin sensitivity criteria resulted in 10 IR subjects and 13 IS subjects, with the average GINF required during the euglycemic clamp differing nearly 2.5-fold. The demographic, physical and metabolic characteristics of both groups are shown in Table 1. Compared to the IS subjects, IR subjects were significantly more obese but of similar age and gender distribution. Moreover, IR subjects had significantly increased HbA1c, triglyceride and non-esterified fatty acid (NEFA) levels, and showed a trend for increased systolic and diastolic blood pressure and decreased HDL consistent with the metabolic syndrome. Fat cell diameter was also significantly greater (142±5 vs 104±6 µm, P<0.0001).
Table 1.
Subjects’ Demographic, Physical and Metabolic Characteristics
| IR | IS | P-value | |
|---|---|---|---|
| Male/female | 4/6 | 4/9 | NS |
| Age (years) | 49±4 | 50±4 | NS |
| BMI (kg/m2) | 30.4±1.2 | 25.8±0.9 | <0.007 |
| Weight (kg) | 89.8±4.8 | 73.1±3.9 | <0.015 |
| Fat mass (kg) | 34.9±2.2 | 25.5±1.8 | <0.004 |
| Lean mass (kg) | 50.4±3.4 | 44.4±3.0 | NS |
| Fasting plasma glucose (mmol/l) | 5.2±0.1 | 4.9±0.1 | 0.06 |
| 2-h plasma glucose OGTT (mmol/l) | 7.6±0.5 | 6.0±0.3 | <0.018 |
| Fasting plasma insulin (pmol/l) | 67.0±5.6 | 21.4±2.8 | <0.001 |
| HbA1c (%) | 5.9±0.1 | 5.5±0.1 | <0.012 |
| Plasma triglycerides (mmol/l) | 1.8±0.2 | 1.0±0.1 | <0.001 |
| Plasma total cholesterol (mmol/l) | 5.0±0.3 | 4.6±0.2 | NS |
| Plasma HDL cholesterol (mmol/l) | 1.2±0.1 | 1.5±0.1 | 0.085 |
| Plasma LDL cholesterol (mmol/l) | 3.1±0.3 | 2.7±0.2 | NS |
| Plasma NEFA (µmol/l) | 1,091±141 | 544±36 | <0.004 |
| Glucose infusion rate in clamp (mg·kg−1·min−1) | 3.7±0.3 | 9.0±0.5 | <0.001 |
| Systolic blood pressure (mmHg) | 128±4 | 118±3 | 0.087 |
| Diastolic blood pressure (mmHg) | 81±3 | 73±2 | 0.068 |
Data are presented as means ± SE
Proteomics Analysis of Adipocytes from IR and IS Subjects
When the results from all 23 subjects were combined and redundant results were collapsed, a total of 1,245 proteins were assigned in ≥1 of the 23 subjects (Table S1); 383 proteins were detected in ≥12 of the 23 subjects. Of these 383 proteins, 94 differed in abundance ≥1.5-fold between IR and IS, of which 30 were statistically significant by unpaired Student’s t-test or the Mann Whitney U test (P<0.05, Table 2). Nineteen proteins were less abundant in IR, 11 were more abundant. Of the 19 proteins with lower abundance, 9 were assigned to mitochondrion in subcellular location, including methylmalonate-semialdehyde dehydrogenase (gene name ALDH6A1), acetyl-CoA acetyltransferase (gene name ACAT1), LETM1 and EF-hand domain-containing protein 1 (gene name LETM1), succinyl-CoA ligase [GDP-forming] subunit beta (gene name SUCLG2), cytochrome c (gene name CYCS), succinyl-CoA ligase [GDP-forming] subunit alpha (gene name SUCLG1), tricarboxylate transport protein (gene name SLC25A1), medium-chain specific acyl-CoA dehydrogenase (gene name ACADM), and ADP/ATP translocase 2 (gene name SLC25A5). In contrast, amongst the proteins with increased abundance in IR, cytochrome b5 reductase 1 (gene name CYB5R1) was the only mitochondrial protein, which is involved in desaturation and elongation of fatty acids and cholesterol biosynthesis. The proportion of mitochondrial proteins amongst the proteins with decreased abundance (9/19) was significantly greater than that amongst the proteins with increased abundance (1/11) (P<0.05, Fisher Exact probability test). Three non-mitochondrial proteins with significantly decreased abundance were proteins involved in lipid metabolism including fatty acid binding protein 4 (gene name FABP4), fatty acid binding protein 5 (gene name FABP5), and abhydrolase domain containing 5 (gene name ABHD5), which functions in phosphatidic acid biosynthesis and regulates triacylglycerol storage. Of the 10 non-mitochondrial proteins with higher abundance in IR, 5 were involved in the cytoskeleton, including kinesin 1 heavy chain (gene name KIF5B), septin 7 (gene name SEPT7), filamin A (gene name FLNA), spectrin alpha chain, non-erythrocytic 1 (gene name SPTAN1), and tubulin beta-2A chain (gene name TUBB2A). Profilin (gene name PFN1) was the only cytoskeletal protein with lower abundance.
Table 2.
Proteins that were identified in at least 12 of the 23 subjects and that differed significantly (P < 0.05) in abundance by a factor ≥1.5 between the averages of the IR group and the IS group
| Protein Name (Gene name) | Fold change in IR |
Student’s t-test |
Mann Whitney U test |
IR/IS with protein assigned |
Total unshared spectra per group |
|
|---|---|---|---|---|---|---|
| P-value | P-value | IR | IS | |||
| Fatty acid-binding protein, epidermal (FABP5) |
0.14±0.16 | <0.001 | <0.001 | 1/12 | 7 | 54 |
| Hemoglobin subunit beta (HBB) |
0.20±0.22 | <0.009 | <0.013 | 3/10 | 5 | 38 |
| 10-formyltetrahydrofolate dehydrogenase (ALDH1L1) |
0.23±0.15 | <0.001 | <0.001 | 5/13 | 18 | 124 |
| Succinyl-CoA ligase [GDP- forming] subunit beta, mitochondrial (SUCLG2) |
0.33±0.22 | <0.032 | <0.015 | 3/11 | 5 | 28 |
| Histone H3.2 (HIST2H3C; HIST2H3A; HIST2H3D) |
0.34±0.25 | <0.06 | <0.026 | 2/10 | 5 | 19 |
| Cytochrome c (CYCS) | 0.39±0.23 | <0.034 | <0.006 | 4/13 | 11 | 44 |
| Methylmalonate- semialdehyde dehydrogenase, mitochondrial (ALDH6A1) |
0.39±0.15 | <0.005 | <0.005 | 7/13 | 20 | 83 |
| Isoform 2 of
Leucyl-cystinyl aminopeptidase (LNPEP) |
0.39±0.24 | <0.05 | <0.067 | 6/10 | 8 | 31 |
| Abhydrolase domain- containing protein 5 (ABHD5) |
0.40±0.20 | <0.046 | <0.058 | 3/10 | 5 | 19 |
| LETM1 and EF-hand domain-containing protein 1, mitochondrial (LETM1) |
0.43±0.16 | <0.02 | <0.01 | 6/13 | 21 | 79 |
| Medium-chain specific acyl- CoA dehydrogenase, mitochondrial (ACADM) |
0.43±0.21 | <0.05 | <0.036 | 6/13 | 22 | 84 |
| Hemoglobin subunit alpha (HBA2; HBA1) |
0.45±0.22 | <0.047 | <0.022 | 10/12 | 46 | 148 |
| Profilin-1 (PFN1) | 0.46±0.20 | <0.02 | <0.036 | 5/12 | 45 | 140 |
| Succinyl-CoA ligase [GDP- forming] subunit alpha, mitochondrial (SUCLG1) |
0.47±0.18 | <0.038 | <0.022 | 6/12 | 20 | 62 |
| ADP/ATP translocase 2 (SLC25A5) |
0.47±0.22 | <0.076 | <0.026 | 6/10 | 8 | 31 |
| Isoform 1 of Cell division control protein 42 homolog (CDC42) |
0.48±0.17 | <0.049 | <0.058 | 5/11 | 8 | 26 |
| Fatty acid-binding protein, adipocyte (FABP4) |
0.49±0.16 | <0.026 | <0.036 | 9/13 | 528 | 1703 |
| Acetyl-CoA acetyltransferase, mitochondrial (ACAT1) |
0.50±0.12 | <0.01 | <0.006 | 9/13 | 43 | 131 |
| Tricarboxylate transport protein, mitochondrial (SLC25A1) |
0.61±0.15 | <0.047 | <0.026 | 9/13 | 26 | 69 |
| Kinesin-1 heavy chain (KIF5B) |
1.50±0.14 | <0.012 | <0.018 | 10/12 | 75 | 81 |
| Dihydropyridine receptor alpha 2 subunit (CACNA2D1) |
1.56±0.17 | <0.03 | <0.031 | 10/12 | 82 | 81 |
| Septin-7 (SEPT7) | 1.64±0.25 | <0.05 | <0.26 | 10/10 | 40 | 42 |
| Isoform 1 of Atlastin-3 (ATL3) |
1.76±0.31 | <0.04 | <0.036 | 10/12 | 68 | 66 |
| Carbonyl reductase [NADPH] 1 (CBR1) |
1.76±0.30 | <0.075 | <0.031 | 9/9 | 51 | 51 |
| Isoform 2 of Filamin-A (FLNA) |
1.78±0.33 | <0.038 | <0.077 | 10/11 | 197 | 183 |
| Isoform 1 of Spectrin alpha chain, brain (SPTAN1) |
2.05±0.41 | <0.024 | <0.05 | 9/12 | 99 | 95 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1) |
2.19±0.40 | <0.024 | <0.026 | 9/8 | 55 | 47 |
| NADH-cytochrome b5 reductase 1 (CYB5R1) |
2.50±0.62 | <0.038 | <0.058 | 7/7 | 12 | 8 |
| Isoform 1 of Liver carboxylesterase 1 (CES1) |
2.63±0.63 | <0.019 | <0.026 | 10/10 | 60 | 36 |
| Tubulin beta-2A chain (TUBB2A) |
2.94±0.80 | <0.017 | <0.05 | 8/9 | 18 | 10 |
Data are presented as means ± SE
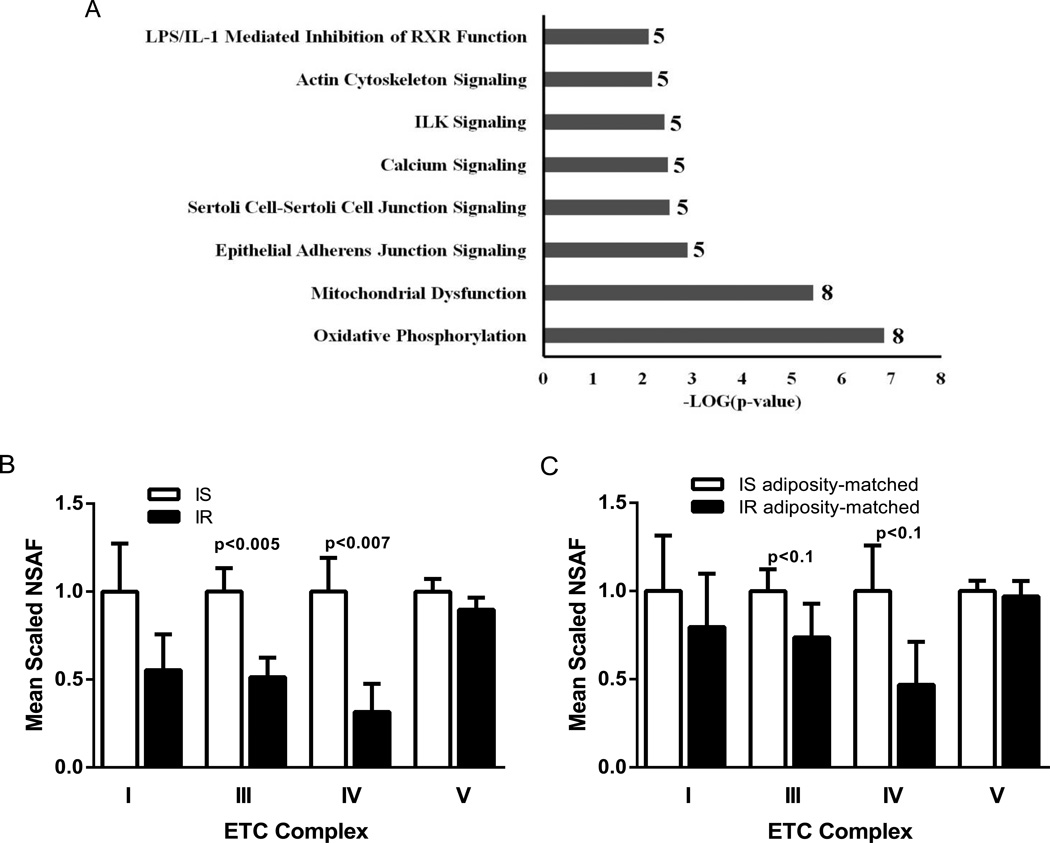
Eight pathways were significantly enriched in proteins with altered abundance in IR by Ingenuity Pathway Analysis (Figure 1). Of these, oxidative phosphorylation showed the highest significance (P<0.000001). Manually curated sets of the individual respiratory chain complexes indicated 50–70% reduced protein abundances of complexes I, III and IV in IR, of which complex III and complex IV reached significance (P<0.005 and P<0.007, respectively) (Figure 1). Complex II proteins were not detected and complex V proteins were similar between both groups. The protein set comprising adenine nucleotide translocators (ANT), which export ATP from the mitochondrial matrix and imports ADP into the matrix, was approximately 60% less abundant in IR (P<0.015).
Figure 1.
A) Significantly enriched pathways in human adipocytes revealed by proteomics and Ingenuity Pathway Analysis (IPA). Only proteins that were identified in at least 12 of the 23 subjects and differed in abundance by a factor ≥1.5 between the averages of the insulin-resistant group and the insulin-sensitive group were used in this analysis. The total number of proteins for a given pathway in this study is denoted besides each bar. B+C) Mean scaled NSAF values of manually curated protein sets for complexes I, III, IV and V of the electron transport chain in all 10 insulin-resistant (IR) and 13 insulin-sensitive (IS) subjects (B), and in 8 IR and 8 IS subjects matched for adiposity (C). Values in the IS group were set at 1. Complex II proteins were not detected. Data are given as means ± SE. P-values by Mann Whitney U test.
Comparison of IR and IS Subjects Matched for Adiposity
IR subjects and IS subjects matched for adiposity had similar BMI (29.0±0.8 vs 28.0±0.3 kg/m2, P=0.32) and percent body fat (40±2 vs 38±2%), and were of similar age (46±4 vs 47±5 years) and gender distribution (3/5 vs 4/4 male/female). Despite matching for adiposity, IR continued having increased HbA1c (5.9±0.1 vs 5.4±0.1%, P<0.05), triglyceride (1.9±0.2 vs 1.0±0.1 mmol/l, P<0.01) and NEFA concentrations (1,047±156 vs 552±35 µmol/l, P<0.03), and by study design reduced GINF during the euglycemic clamp (4.0±0.2 vs 8.2±0.3 mg·kg−1·min−1, P<0.0001). Fat cell diameter also remained greater in IR than IS (137±5 vs 112±6 µm, P<0.018).
A total of 1,148 proteins were assigned in ≥1 of the 16 subjects (Table S2), of which 408 were detected in ≥8 subjects. One-hundred-fourteen proteins differed in abundance ≥1.5-fold between adiposity-matched IR and IS, of which 28 were statistically significant (P<0.05, Table 3). Eighteen of these 28 proteins were less abundant in IR, 10 were more abundant; 9 were common to those significantly different between all IR and IS subjects. Of the 18 proteins with lower abundance, 3 were assigned to mitochondrion in subcellular location, including methylmalonate-semialdehyde dehydrogenase (ALDH6A1), succinyl-CoA ligase [GDP-forming] subunit beta (SUCLG2) and isoform 1 and 2 of enoyl-CoA delta isomerase 1 (gene name ECI1). None of the 10 proteins with higher abundance were assigned to mitochondrion. Complex III and complex IV protein sets showed a trend for being approximately 30 and 50% less abundant in IR, respectively (P<0.1)(Figure 1), whereas the ANT set remained significantly less abundant by approximately 70% (P<0.019). Of the proteins with altered abundance in IR, four were directly involved in lipid metabolism, including FABP5, abhydrolase domain-containing protein 5 (gene name ABHD5), isoform 1 and 2 of enoyl-CoA delta isomerase 1 (decreased) and phospholipase C, delta 1 (gene name PLCD1) (increased); three proteins were involved in the misfolded/unfolded protein response, namely UDP-glucose:glycoprotein glucosyltransferase (gene name UGCGL1), ribophorin I (gene name RPN1) (increased) and protein disulfide-isomerase A1 (gene name P4HB) (decreased).
Table 3.
Changes in the abundance of proteins between IR and IS subjects matched for adiposity (8 IR and 8 IS) that were found to be significantly different between all IR and IS subjects (10 IR and 13 IS)
| Protein Name (Gene name) | Fold change in IR |
Student’s t-test |
Mann Whitney U test |
IR/IS with protein assigned |
Total unshared spectra per group |
|
|---|---|---|---|---|---|---|
| P-value | P-value | IR | IS | |||
| Fatty acid-binding protein, epidermal (FABP5) |
0.34±0.22 | <0.024 | <0.015 | 1/7 | 7 | 37 |
| Hemoglobin subunit beta (HBB) |
0.22±0.12 | <0.013 | <0.015 | 2/7 | 3 | 26 |
| 10-formyltetrahydrofolate dehydrogenase (ALDH1L1) |
0.24±0.09 | <0.001 | <0.002 | 4/8 | 8 | 78 |
| Succinyl-CoA ligase [GDP- forming] subunit beta, mitochondrial (SUCLG2) |
0.45±0.20 | <0.032 | <0.021 | 3/8 | 5 | 22 |
| Histone H3.2 (HIST2H3C; HIST2H3A; HIST2H3D) |
0.58±0.26 | <0.069 | <0.038 | 2/7 | 5 | 16 |
| Cytochrome c (CYCS) | 0.76±0.29 | =0.40 | <0.065 | 4/8 | 11 | 25 |
| Methylmalonate- semialdehyde dehydrogenase, mitochondrial (ALDH6A1) |
0.43±0.12 | <0.029 | <0.029 | 5/8 | 12 | 53 |
| Isoform 2 of Leucyl-cystinyl aminopeptidase (LNPEP) |
0.61±0.19 | =0.17 | =0.19 | 5/6 | 7 | 19 |
| Abhydrolase domain- containing protein 5 (ABHD5) |
0.49±0.19 | <0.004 | <0.011 | 2/8 | 3 | 15 |
| LETM1 and EF-hand domain-containing protein 1, mitochondrial (LETM1) |
0.77±0.18 | =0.33 | =0.23 | 6/8 | 21 | 45 |
| Medium-chain specific acyl- CoA dehydrogenase, mitochondrial (ACADM) |
0.62±0.21 | =0.29 | =0.33 | 4/8 | 18 | 45 |
| Hemoglobin subunit alpha (HBA2; HBA1) |
0.67±0.06 | <0.013 | <0.007 | 8/8 | 38 | 85 |
| Profilin-1 (PFN1) | 0.64±0.21 | =0.25 | =0.33 | 5/7 | 45 | 94 |
| Succinyl-CoA ligase [GDP- forming] subunit alpha, mitochondrial (SUCLG1) |
0.91±0.24 | =0.77 | =0.44 | 6/8 | 20 | 31 |
| ADP/ATP translocase 2 (SLC25A5) |
0.47±0.13 | =0.12 | =0.10 | 5/6 | 7 | 24 |
| Isoform 1 of Cell division control protein 42 homolog (CDC42) |
0.55±0.19 | <0.093 | <0.083 | 4/7 | 6 | 19 |
| Fatty acid-binding protein, adipocyte (FABP4) |
0.72±0.19 | =0.29 | =0.33 | 7/8 | 515 | 1111 |
| Acetyl-CoA acetyltransferase, mitochondrial (ACAT1) |
0.74±0.13 | =0.21 | =0.19 | 8/8 | 39 | 76 |
| Tricarboxylate transport protein, mitochondrial (SLC25A1) |
0.70±0.14 | =0.20 | =0.16 | 7/8 | 23 | 49 |
| Kinesin-1 heavy chain (KIF5B) |
1.43±0.09 | <0.015 | <0.021 | 8/8 | 60 | 59 |
| Dihydropyridine receptor alpha 2 subunit (CACNA2D1) |
1.69±0.16 | <0.019 | <0.021 | 8/8 | 62 | 52 |
| Septin-7 (SEPT7) | 1.16±0.19 | =0.32 | =0.96 | 8/7 | 30 | 34 |
| Isoform 1 of Atlastin-3 (ATL3) |
1.32±0.22 | =0.35 | =0.57 | 8/8 | 53 | 56 |
| Carbonyl reductase [NADPH] 1 (CBR1) |
1.09±0.19 | =0.99 | =0.57 | 7/8 | 36 | 49 |
| Isoform 2 of Filamin-A (FLNA) |
1.38±0.16 | =0.13 | =0.44 | 8/7 | 160 | 150 |
| Isoform 1 of Spectrin alpha chain, brain (SPTAN1) |
1.46±0.28 | =0.21 | =0.33 | 7/8 | 79 | 84 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1) |
1.64±0.28 | =0.20 | =0.19 | 7/7 | 44 | 43 |
| NADH-cytochrome b5 reductase 1 (CYB5R1) |
2.15±0.45 | <0.059 | <0.065 | 6/6 | 10 | 6 |
| Isoform 1 of Liver carboxylesterase 1 (CES1) |
2.05±0.40 | <0.063 | =0.13 | 8/7 | 41 | 26 |
| Tubulin beta-2A chain (TUBB2A) |
2.45±0.67 | <0.081 | =0.23 | 6/7 | 15 | 8 |
Data are presented as means ± SE
Out of the 30 proteins that were found to be ≥1.5-fold and significantly altered in abundance between all IR and IS subjects, 17 remained ≥1.5-fold different and 9 remained significantly different between the adiposity-matched IR and IS (Table 2).
Immunoblot Analysis
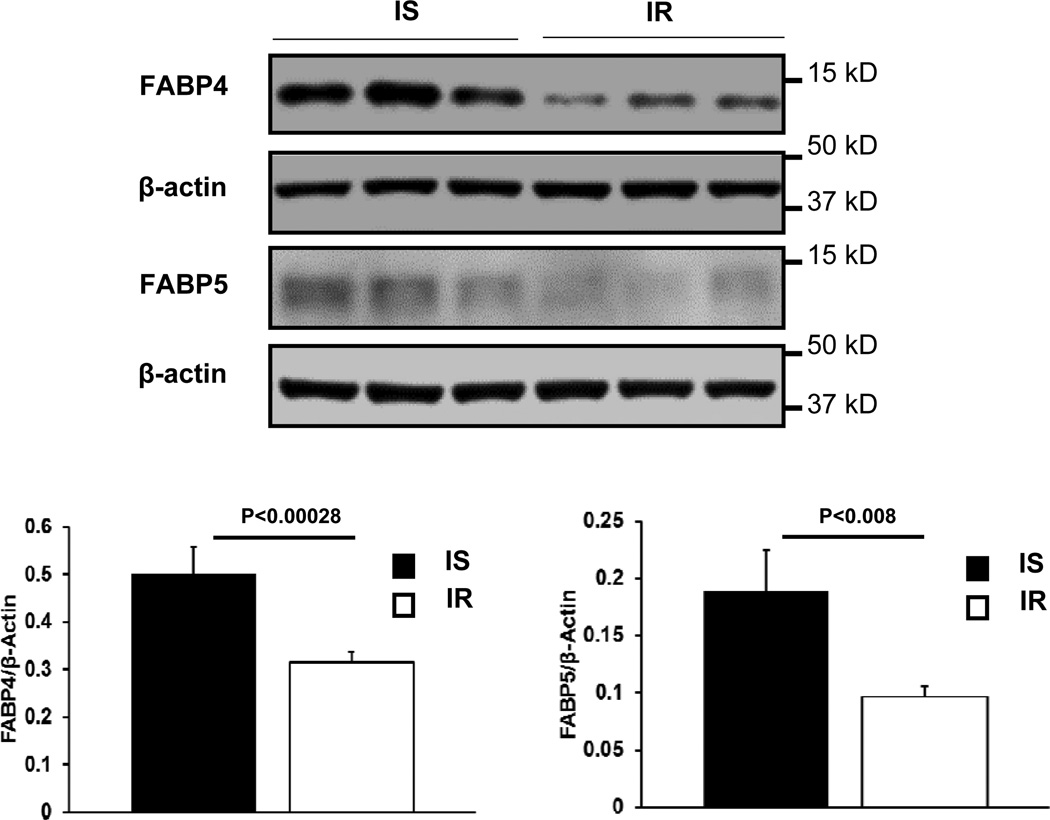
Contrary to the proteomics findings, previous animal studies have found that deficiency of FABP4 and FABP5 protects from insulin resistance whereas increased levels of FABP5 in adipose tissue induce insulin resistance (16, 17). Therefore, we performed immunoblot analyses of FABP4 and FABP5 in 19 subjects (7 IR, BMI 30.6±1.6 kg/m2; 12 IS, BMI 26.0±0.9 kg/m2) from whom we had sufficient remaining specimens. Immunoblots confirmed reduced expression of FABP4 and FABP5 in IR by approximately 30 (P<0.0003) and 50% (P<0.008), respectively (Figure 2).
Figure 2.
Immunoblot analyses comparing the abundance of FABP4 (A) and FABP5 (B) in subcutaneous adipocytes from insulin-resistant (IR; N=7) and insulin-sensitive subjects (IS; N=12). Density values from blots were expressed relative to β-actin determined on the same blots. Representative blots from the IR and the IS subjects (three per group) are shown. Data are shown as means ± SE. P-values by Mann Whitney U test.
Discussion
A wide array of adipose tissue abnormalities have been found to be associated with and implicated in systemic insulin resistance (2–4). These result from investigations that often have focused on a particular protein, cellular process or biochemical pathway, making it challenging to discern larger patterns of adipose tissue abnormalities in the insulin-resistant state. In the present study, we therefore used an unbiased global proteomics approach to further explore the molecular mechanisms that underlie systemic insulin resistance in humans.
Previous studies indicate that mitochondrial mass is reduced in white adipocytes or white adipose tissue of insulin-resistant ob/ob and high fat fed C57BL/6J mice (18, 19). Furthermore, in white adipose tissue of diabetic mice, mitochondrial number, electron transport chain enzymatic activity, oxidative phosphorylation and β-oxidation were reduced and mitochondrial morphology was altered (20). In individuals with obesity and T2DM compared to younger, leaner, healthy individuals the mitochondrial number and the expression of key genes related to mitochondrial function were found to be significantly reduced in white adipose tissue and improved by pioglitazone treatment (21). The present study extends these findings by showing significantly decreased abundance of mitochondrial proteins in non-diabetic insulin-resistant individuals compared to insulin-sensitive individuals matched for age and gender distribution. Of the 19 proteins with lower abundance in the IR group, 9 were assigned to mitochondrion, whereas of the proteins with increased abundance, only one was assigned to mitochondrion (P<0.05 for difference in distribution). Of the mitochondrial proteins with decreased abundance, several are involved in fatty acid, triacylglycerol and ketone body metabolism, branched-chain amino acid degradation, and other intermediary metabolism including the citric acid cycle. In addition, in analyses of protein sets, complex I, III and IV were 50–70% reduced with the reductions in complex III and IV being highly significant (both P<0.013). These changes may not only contribute to dysregulated lipid metabolism and ectopic lipid accumulation but also the increased circulating BCAA levels that are typically seen and implicated in systemic insulin resistance in humans (22). However, in the IR subjects compared to the IS subjects matched for adiposity, fewer proteins remained significantly less abundant suggesting that abnormal adipocyte mitochondria may be less important in explaining metabolic healthy versus metabolic unhealthy obesity but be largely secondary to acquired excess body fat. This notion is consistent with studies in identical twin pairs discordant for obesity and physical fitness that have demonstrated a coordinated reduction in adipose tissue transcript levels of genes involved in mitochondrial oxidative phosphorylation and the cellular mitochondrial copy number in the co-twins with obesity compared with the co-twins without obesity (23, 24).
A novel and perhaps unexpected finding in the present study was the marked reduction of FABP5 (also known as mal1) and to a lesser degree of FABP4 (also known as aP2) in adipocytes from the IR individuals. Although we had previously validated our NASF method for protein quantification (10), we confirmed these findings by immunoblot analyses. Significant differences in FABP5 but not FABP4 persisted in comparisons of IR and IS subjects matched for adiposity. FABPs are a family of 14–15-kDa proteins that bind with high affinity to hydrophobic ligands, such as saturated and unsaturated long-chain fatty acids and eicosanoids. FABP4 regulates transport NEFA, increases the hydrolytic activity of hormone-sensitive lipase and functions as an intercompartmental shuttle from the cytosol to the nucleus for PPARγ agonists (25). Global deficiency of FABP4 partially protects mice against the development of insulin resistance associated with genetic or diet-induced obesity (26, 27). Further, FABP5−/− mice and double FABP4−/− and FABP5−/− mice exhibit strong protection from diet-induced obesity, insulin resistance and type 2 diabetes (16, 17), whereas mice expressing high levels of FABP5 in adipose tissue display decreased systemic insulin sensitivity (16). Species differences may explain the apparent differences between these animal data and the present human data. However, an alternative reconciling explanation is that the inverse relationship between FABP4 and/or FABP5 levels and systemic insulin sensitivity may be mediated through adipose tissue other than abdominal subcutaneous adipose tissue. Therefore, further research is needed to determine the relation of FABPs and insulin sensitivity in humans.
Another novel change observed in the present studies was the increased abundance of several cytoskeletal proteins in the IR subjects. Profilin-1 was the only cytoskeletal protein with lower abundance, which binds to actin and prevents its polymerization at high concentrations but enhances it at low concentrations (28). Interactions between the cytoskeletal network and mitochondria play critical roles in mitochondrial fusion and fission, morphology, localization as well as function (29). Interestingly, we had previously observed increased microtubule-related proteins but decreased mitochondrial proteins also in insulin-resistant skeletal muscle by proteomics analysis (10). These observations not only suggest the existence of such reciprocal changes across several tissues but also a possible connection between cytoskeletal and mitochondrial abnormalities in insulin resistance.
It is well-established that insulin resistance is generally associated with adipose tissue inflammation. It may hence be surprising that we did not observe changes in the abundance of adipokines/cytokines, consistent with inflammation, in adipocytes of the IR group. However, this is probably due to the fact that adipokines/cytokines are secreted and that many are also produced by adipose tissue-resident immune cells (30).
In addition to comparing typical insulin-resistant individuals to insulin-sensitive individuals, we performed subgroup analysis of adiposity-matched IR and IS subjects in order to explore the molecular basis for metabolically unhealthy excess body fat. In this analysis we found significantly increased adipocyte levels of UDP-glucose:glycoprotein glucosyltransferase 1 and ribophorin I but significantly decreased levels of protein disulfide-isomerase A1 in the IR group. UDP-glucose:glycoprotein glucosyltransferase 1 is a central gatekeeper for ER quality control for glycoproteins. It recognizes and re-glucosylates glycoproteins with minor folding defects, leading to their retention in the ER by the chaperones calreticulin and calnexin (31). Ribophorin I has more recently been identified to have similar functions, leading to the retention of misfolded proteins by interaction with malnectin (32). Protein disulfide-isomerase A1, in addition to its foldase function, plays an important role as a molecular chaperone by inhibiting the aggregation of unfolded/misfolded proteins at the ER (33). Abnormal protein disulfide-isomerase A1 can lead to accumulation of misfolded proteins and ER stress with multiple adverse cellular consequences, including various neurodegenerative diseases, such as Alzheimer’s and Parkinson’s disease (33). Altered protein levels indicative of ER stress has previously been found in adipose tissue of insulin-resistant individuals with obesity compared to lean insulin-sensitive individuals (34). However, the present study is the first to observe that such abnormalities may differentiate metabolically unhealthy insulin-resistant individuals from metabolically healthy insulin-sensitive individuals of similar excess body fat.
Another interesting finding is that the IR subjects compared to the IS subjects matched for adiposity had significantly altered levels of several proteins related to lipid metabolism, including FABP5, abhydrolase domain-containing protein 5, enoyl-CoA delta isomerase 1 (decreased) and phospholipase C, delta 1 (increased). ABHD5 increases the activity of adipose triglyceride lipase, which catalyzes the first step of triacylglycerol hydrolysis. Interestingly, mutations in ABHD5 result in Chanarin-Dorfman syndrome, where triacylglycerol accumulates in various tissues, including the liver and muscle (35). Enoyl-CoA delta isomerase 1 is a mitochondrial enzyme critical for beta-oxidation of unsaturated fatty acids. Conversely, phospholipase C, which was increased in IR, catalyzes the hydrolysis of the membrane phospholipid phosphatidylinositol 4,5-bisphosphate (PIP2) to produce inositol 1,4,5-trisphosphate (IP3) and 1,2-diacylglycerol (DAG). Together with reduced succinyl-CoA ligase and a tendency for reduced levels of proteins involved in the TCA cycle and oxidative phosphorylation, this constellation suggests a condition that favors the accumulation of lipids and lipid intermediates, such as DAG, that can trigger problematic cellular events, including impaired insulin signaling (36).
We recognize that the present study has several limitations. First, there are significant differences between various fat depots (37) but we only examined abdominal subcutaneous adipocytes. Second, the descriptive nature of the data does not allow determination of causality. Third, despite applying filters, we used 94 proteins for statistical comparisons and found 30 proteins to be different between all IR and IS at a p-value of <0.05. Accordingly, there may have been 5 false discoveries (94 × 0.05 = 4.7) leading to a false discovery rate (false positive discoveries/number of significant discoveries) of approximately 16% (=4.7/30) similar to previous omics studies (38–40). Therefore, caution should be applied in interpreting the data and extrapolating the data to adipocytes in general.
In conclusion, novel findings that emerged from our studies include lower abundances of FABP4 and FABP5, and higher abundance of cytoskeletal proteins in abdominal subcutaneous adipocytes from typical insulin-resistant individuals; moreover, we confirm reductions in adipocyte mitochondrial proteins. Comparison of adiposity-matched IR and IS subjects suggest that changes in the abundance of proteins related to lipid metabolism and the unfolded/misfolded protein response discriminate metabolically unhealthy from metabolically healthy individuals of similar excess body fat. These findings may be used to generate new hypotheses to be tested by more focused studies.
Supplementary Material
Table 4.
Proteins that differed significantly (P < 0.05) in abundance by a factor ≥1.5 between the averages of the IR group and the IS group matched for adiposity (N = 8/group) and that were assigned in at least 8 of the 16 subjects
| Protein Name (Gene name) | Fold change in IR |
Student’s t-test |
Mann Whitney U test |
IR/IS with protein assigned |
Total unshared spectra per group |
|
|---|---|---|---|---|---|---|
| P-value | P-value | IR | IS | |||
| Hemoglobin subunit beta (HBB) |
0.16±0.12 | <0.013 | <0.015 | 2/7 | 3 | 26 |
| 10-formyltetrahydrofolate dehydrogenase (ALDH1L1) |
0.18±0.09 | <0.001 | <0.002 | 4/8 | 8 | 78 |
| 26S proteasome non- ATPase regulatory subunit 7 (PSMD7) |
0.19±0.19 | <0.015 | <0.011 | 1/7 | 3 | 17 |
| Fatty acid-binding protein, epidermal (FABP5) |
0.19±0.19 | <0.024 | <0.015 | 1/7 | 7 | 37 |
| Abhydrolase domain- containing protein 5 (ABHD5) |
0.26±0.17 | <0.004 | <0.007 | 2/8 | 3 | 15 |
| Cystatin-B (CSTB) | 0.27±0.19 | <0.02 | <0.029 | 2/7 | 9 | 39 |
| 59 kDa protein (CCT8) | 0.28±0.10 | <0.011 | <0.007 | 5/8 | 8 | 39 |
| Histone H3.2 (HIST2H3C; HIST2H3A; HIST2H3D) |
0.33±0.24 | <0.069 | <0.038 | 2/7 | 5 | 16 |
| Methylmalonate- semialdehyde dehydrogenase, mitochondrial (ALDH6A1) |
0.33±0.11 | <0.029 | <0.021 | 5/8 | 12 | 53 |
| Succinyl-CoA ligase [GDP- forming] subunit beta, mitochondrial (SUCLG2) |
0.35±0.23 | <0.033 | <0.021 | 3/8 | 5 | 22 |
| Isoform 1 of
Abhydrolase domain-containing protein 14B (ABHD14B) |
0.36±0.16 | <0.024 | <0.029 | 4/8 | 6 | 21 |
| Isoform 1 and 2 of Enoyl- CoA delta isomerase 1, mitochondrial (gene name ECI1) |
0.36±0.19 | <0.028 | <0.029 | 3/7 | 7 | 28 |
| D-3-phosphoglycerate dehydrogenase (PHGDH) |
0.41±0.13 | <0.011 | <0.029 | 5/8 | 15 | 49 |
| Membrane-associated progesterone receptor component 1 (PGRMC1) |
0.41±0.19 | <0.066 | <0.05 | 4/8 | 8 | 24 |
| Protein disulfide-isomerase A1 (P4HB) |
0.45±0.19 | <0.036 | <0.065 | 4/8 | 18 | 50 |
| Isoform 2 of Heme-binding protein 2 (HEBP2) |
0.48±0.19 | <0.042 | <0.065 | 4/8 | 16 | 44 |
| Isoform 1 of Proteasome subunit alpha type-7 (PSMA7) |
0.62±0.22 | =0.164 | <0.038 | 7/8 | 14 | 36 |
| HBA2; HBA1 Hemoglobin subunit alpha |
0.63±0.06 | <0.013 | <0.007 | 8/8 | 38 | 85 |
| 1,4-alpha-glucan-branching enzyme (GBE1) |
1.50±0.17 | <0.044 | <0.05 | 8/8 | 96 | 93 |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) |
1.55±0.23 | <0.12 | <0.05 | 8/7 | 44 | 43 |
| Serum deprivation-response protein (SDPR) |
1.59±0.22 | <0.039 | <0.065 | 8/8 | 198 | 192 |
| Isoform 1 of
UDP- glucose:glycoprotein glucosyltransferase 1 (UGCGL1) |
1.64±0.19 | <0.036 | <0.029 | 8/8 | 65 | 57 |
| Dihydropyridine receptor alpha 2 subunit (CACNA2D1) |
1.72±0.20 | <0.018 | <0.029 | 8/8 | 62 | 52 |
| Ribophorin II isoform 2 precursor (RPN2) |
1.76±0.29 | <0.057 | <0.05 | 7/6 | 7 | 6 |
| Ribophorin I (RPN1) | 1.79±0.21 | <0.009 | <0.007 | 8/8 | 66 | 55 |
| NAD(P)H Dehydrogenase, Quinone (NQO2) |
3.98±1.31 | <0.05 | =0.105 | 6/4 | 14 | 6 |
| Protein NOXP20 (FAM114A1) |
5.51±1.48 | <0.015 | <0.021 | 6/2 | 17 | 4 |
| Phospholipase C delta-1 (PLCD1) |
9.40±3.16 | <0.021 | <0.003 | 7/2 | 11 | 2 |
Data are presented as means ± SE
Answers to Study Importance Questions.
Abnormal function of adipocytes plays an important role in insulin resistance and the development of type 2 diabetes mellitus, but the molecular changes in adipocytes that cause systemic insulin resistance in humans remain poorly understood.
Global gene expression has been used to further elucidate the potentially underlying mechanisms, but gene expression is a relatively poor reflection of the activity level of a biological pathway.
Protein expression is a closer reflection of the activity level of a biological pathway, but large scale unbiased proteomics analyses have hitherto net been undertaken.
Using such proteomics analyses, our findings suggest increased cytoskeletal proteins and decreased FABP4 and FABP5, and confirm lower abundance of mitochondrial proteins in subcutaneous adipocytes of typical insulin-resistant individuals.
Changes in proteins related to lipid metabolism and the unfolded/misfolded protein response may separate insulin-resistant from insulin-sensitive individuals of comparable adiposity.
These findings provide new information on potential factors or pathways that may individually or in concert induce insulin resistance in humans and discriminate metabolically unhealthy insulin-resistant from metabolically healthy insulin-sensitive individuals with similar excess body fat.
Acknowledgments
The authors gratefully acknowledge the assistance of the CRU staff and especially thank the study volunteers.
This study was supported in part by NIH grants R01DK47936 (LM), R01DK66483 (LM), R01DK081750 (ZY), R21DK082820 (CM) and the Translational Science Award 1-13-TS-27 (ZY) and the Clinical Research Grant 1-09-CR-39 (CM) from the American Diabetes Association.
Footnotes
None of the authors have any conflict of interest.
References
- 1.Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004;116:337–350. doi: 10.1016/s0092-8674(03)01081-x. [DOI] [PubMed] [Google Scholar]
- 2.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kusminski CM, Scherer PE. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol Metab. 2012;23:435–443. doi: 10.1016/j.tem.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 5.Henegar C, Tordjman J, Achard V, Lacasa D, Cremer I, Guerre-Millo M, Poitou C, Basdevant A, Stich V, Viguerie N, et al. Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome Biol. 2008;9:R14. doi: 10.1186/gb-2008-9-1-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klimcakova E, Roussel B, Marquez-Quinones A, Kovacova Z, Kovacikova M, Combes M, et al. Worsening of obesity and metabolic status yields similar molecular adaptations in human subcutaneous and visceral adipose tissue: decreased metabolism and increased immune response. J Clin Endocrinol Metab. 2011;96:E73–E82. doi: 10.1210/jc.2010-1575. [DOI] [PubMed] [Google Scholar]
- 7.Kursawe R, Eszlinger M, Narayan D, Liu T, Bazuine M, Cali AM, et al. Cellularity and adipogenic profile of the abdominal subcutaneous adipose tissue from obese adolescents: association with insulin resistance and hepatic steatosis. Diabetes. 2010;59:2288–2296. doi: 10.2337/db10-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 9.Durward CM, Hartman TJ, Nickols-Richardson SM. All-cause mortality risk of metabolically healthy obese individuals in NHANES III. J Obes. 2012;2012:460321. doi: 10.1155/2012/460321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang H, Bowen BP, Lefort N, Flynn CR, De Filippis EA, Roberts C, et al. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes. 2010;59:33–42. doi: 10.2337/db09-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie X, Yi Z, Bowen B, Wolf C, Flynn CR, Sinha S, et al. Characterization of the Human Adipocyte Proteome and Reproducibility of Protein Abundance by One-Dimensional Gel Electrophoresis and HPLC-ESI-MS/MS. J Proteome Res. 2010;9:4521–4534. doi: 10.1021/pr100268f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol. 1979;237:E214–E223. doi: 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- 13.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, et al. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefort N, Glancy B, Bowen B, Willis WT, Bailowitz Z, De Filippis EA, et al. Increased reactive oxygen species production and lower abundance of complex I subunits and carnitine palmitoyltransferase 1B protein despite normal mitochondrial respiration in insulin-resistant human skeletal muscle. Diabetes. 2010;59:2444–2452. doi: 10.2337/db10-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caruso M, Ma D, Msallaty Z, Lewis M, Seyoum B, Al-janabi W, et al. Increased interaction with insulin receptor substrate 1, a novel abnormality in insulin resistance and type 2 diabetes. Diabetes. 2014;63:1933–1947. doi: 10.2337/db13-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda K, Uysal KT, Makowski L, Gorgun CZ, Atsumi G, Parker RA, et al. Role of the fatty acid binding protein mal1 in obesity and insulin resistance. Diabetes. 2003;52:300–307. doi: 10.2337/diabetes.52.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda K, Cao H, Kono K, Gorgun CZ, Furuhashi M, Uysal KT, et al. Adipocyte/macrophage fatty acid binding proteins control integrated metabolic responses in obesity and diabetes. Cell Metab. 2005;1:107–119. doi: 10.1016/j.cmet.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Wilson-Fritch L, Nicoloro S, Chouinard M, Lazar MA, Chui PC, Leszyk J, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rong JX, Qiu Y, Hansen MK, Zhu L, Zhang V, Xie M, et al. Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone. Diabetes. 2007;56:1751–1760. doi: 10.2337/db06-1135. [DOI] [PubMed] [Google Scholar]
- 20.Choo HJ, Kim JH, Kwon OB, Lee CS, Mun JY, Han SS, et al. Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia. 2006;49:784–791. doi: 10.1007/s00125-006-0170-2. [DOI] [PubMed] [Google Scholar]
- 21.Bogacka I, Xie H, Bray GA, Smith SR. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes. 2005;54:1392–1399. doi: 10.2337/diabetes.54.5.1392. [DOI] [PubMed] [Google Scholar]
- 22.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15:606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mustelin L, Pietilainen KH, Rissanen A, Sovijarvi AR, Piirila P, Naukkarinen J, et al. Acquired obesity and poor physical fitness impair expression of genes of mitochondrial oxidative phosphorylation in monozygotic twins discordant for obesity. Am J Physiol. 2008;295:E148–E154. doi: 10.1152/ajpendo.00580.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietilainen KH, Naukkarinen J, Rissanen A, Saharinen J, Ellonen P, Keranen H, et al. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS medicine. 2008;5:e51. doi: 10.1371/journal.pmed.0050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kralisch S, Fasshauer M. Adipocyte fatty acid binding protein: a novel adipokine involved in the pathogenesis of metabolic and vascular disease? Diabetologia. 2013;56:10–21. doi: 10.1007/s00125-012-2737-4. [DOI] [PubMed] [Google Scholar]
- 26.Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM. Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science. 1996;274:1377–1379. doi: 10.1126/science.274.5291.1377. [DOI] [PubMed] [Google Scholar]
- 27.Uysal KT, Scheja L, Wiesbrock SM, Bonner-Weir S, Hotamisligil GS. Improved glucose and lipid metabolism in genetically obese mice lacking aP2. Endocrinology. 2000;141:3388–3396. doi: 10.1210/endo.141.9.7637. [DOI] [PubMed] [Google Scholar]
- 28.Vidali L, Perez HE, Valdes Lopez V, Noguez R, Zamudio F, Sanchez F. Purification, characterization, and cDNA cloning of profilin from Phaseolus vulgaris. Plant Physiol. 1995;108:115–123. doi: 10.1104/pp.108.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anesti V, Scorrano L. The relationship between mitochondrial shape and function and the cytoskeleton. Biochim Biophys Acta. 2006;1757:692–699. doi: 10.1016/j.bbabio.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 30.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maattanen P, Gehring K, Bergeron JJ, Thomas DY. Protein quality control in the ER: the recognition of misfolded proteins. Semin Cell Dev Biol. 2010;21:500–511. doi: 10.1016/j.semcdb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Qin SY, Hu D, Matsumoto K, Takeda K, Matsumoto N, Yamaguchi Y, et al. Malectin forms a complex with ribophorin I for enhanced association with misfolded glycoproteins. J Biol Chem. 2012;287:38080–38089. doi: 10.1074/jbc.M112.394288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreu CI, Woehlbier U, Torres M, Hetz C. Protein disulfide isomerases in neurodegeneration: from disease mechanisms to biomedical applications. FEBS Lett. 2012;586:2826–2834. doi: 10.1016/j.febslet.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 34.Boden G, Duan X, Homko C, Molina EJ, Song W, Perez O, et al. Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals. Diabetes. 2008;57:2438–2444. doi: 10.2337/db08-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R. Neutral lipid storage disease: genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol. 2009;297:E289–E296. doi: 10.1152/ajpendo.00099.2009. [DOI] [PubMed] [Google Scholar]
- 36.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: unravelling the mechanism. Lancet. 2010;375:2267–2277. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee MJ, Wu Y, Fried SK. Adipose tissue heterogeneity: implication of depot differences in adipose tissue for obesity complications. Mol Aspects Med. 2013;34:1–11. doi: 10.1016/j.mam.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng D, Kille P, Feeney GP, Cunningham P, Handy RD, Hogstrand C. Dynamic transcriptomic profiles of zebrafish gills in response to zinc supplementation. BMC Genomics. 2010;11:553. doi: 10.1186/1471-2164-11-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahimov F, King OD, Leung DG, Bibat GM, Emerson CP, Jr, Kunkel LM, et al. Transcriptional profiling in facioscapulohumeral muscular dystrophy to identify candidate biomarkers. Proc Natl Acad Sci U S A. 2012;109:16234–16239. doi: 10.1073/pnas.1209508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geetha T, Langlais P, Luo M, Mapes R, Lefort N, Chen SC, et al. Label-free proteomic identification of endogenous, insulin-stimulated interaction partners of insulin receptor substrate-1. J Am Soc Mass Spectr. 2011;22:457–466. doi: 10.1007/s13361-010-0051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.