Abstract
The androgen receptor (AR) is the dominant growth factor in prostate cancer (PCa). Therefore, understanding how ARs regulate the human transcriptome is of paramount importance. The early effects of castration on human PCa have not previously been studied 27 patients medically castrated with degarelix 7 d before radical prostatectomy. We used mass spectrometry, immunohistochemistry, and gene expression array (validated by reverse transcription-polymerase chain reaction) to compare resected tumour with matched, controlled, untreated PCa tissue. All patients had levels of serum androgen, with reduced levels of intraprostatic androgen at prostatectomy. We observed differential expression of known androgen-regulated genes (TMPRSS2, KLK3, CAMKK2, FKBP5). We identified 749 genes downregulated and 908 genes upregulated following castration. AR regulation of α-methylacyl-CoA racemase expression and three other genes (FAM129A, RAB27A, and KIAA0101) was confirmed. Upregulation of oestrogen receptor 1 (ESR1) expression was observed in malignant epithelia and was associated with differential expression of ESR1-regulated genes and correlated with proliferation (Ki-67 expression).
Patient summary
This first-in-man study defines the rapid gene expression changes taking place in prostate cancer (PCa) following castration. Expression levels of the genes that the androgen receptor regulates are predictive of treatment outcome. Upregulation of oestrogen receptor 1 is a mechanism by which PCa cells may survive despite castration.
Keywords: Prostate cancer, Androgen receptor, Castration, Clinical trial, Immunohistochemistry, Gene transcription
Take Home Message
This first-in-man study defines the rapid gene expression changes taking place in prostate cancer (PCa) following castration. Upregulation of ESR1 is a mechanism by which PCa cells may survive despite castration.
Prostate cancer (PCa) is the second most common cause of cancer death in men in the developed world [1]. The androgen receptor (AR) controls PCa growth. Previously, studies of the long-term effects (>3 mo) of medical castration have demonstrated differing transcriptional response following luteinising hormone-releasing hormone (LHRH) analogue and antiandrogens [2] and have implicated Wnt/β-catenin signalling in castration-resistant PCa [3]. We used a new drug called degarelix to rapidly decrease testosterone levels and inform the early in vivo response of human PCa to castration.
1. Patients and methods
1.1. Study approval
We obtained full ethics approval for all elements of studies NCT01852864 (REC ref:11/H0311/2) and NCT00967889 (REC ref:01/4/061).
1.2. Clinical sample collection
Twenty-seven patients with high-risk PCa (according to the D’Amico classification) [4] were administered 240 mg subcutaneous degarelix (donated by Ferring Pharmaceuticals Inc, Parsippany, NJ, USA) 7 d before radical prostatectomy (RP) (Supplementary Fig. 1a). These patients were matched for risk factors (age, serum prostate-specific antigen, tumour grade and stage) with 20 patients undergoing RP without neoadjuvant degarelix (Supplementary Table 1). Study and control patients underwent surgery contemporaneously using standard operative technique, with similar ischemic time before sampling as described [5]. Serum samples were obtained at 8 am prior to surgery.
1.3. Assay of androgen levels in body fluids and tissue
Fresh frozen prostate cores were homogenised, and steroids were extracted and quantified by liquid chromatography coupled with tandem mass spectrometry.
1.4. Gene expression profiling
We hybridised mRNA onto Illumina HumanHT-12 v4 Expression BeadChips (Illumina, San Diego, CA, USA).
1.5. Accession numbers
Study data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE72920. Expression data were validated for known AR-regulated genes (KLK3, FASN), genes found to be AR regulated in our expression data with known AR binding sites in promoter regions [6] (FAM129A, KIAA0101, RAB27A) and genes of biological importance (AMACR, ESR1, cyclin D1) by reverse transcription-polymerase chain reaction (RT-PCR) and immunohistochemistry (IHC).
1.6. Quantitative real-time polymerase chain reaction
We performed RT-PCR in triplicate using SYBR Green and the 7900HT Real-Time PCR System (Applied Biosystems, Foster City, CA, USA).
1.7. Human tissue microarray construction and immunohistochemistry
PCa samples (index lesion) from 27 patients were identified on the haematoxylin and eosin–stained sections from formalin-fixed paraffin–embedded RP specimens and marked (A.W.). Triplicate 2-mm tumour cores were taken from these areas and re-embedded in human tissue microarray (TMA) paraffin blocks.
We used a BOND-III automated stainer (Leica Biosystems, Buffalo Grove, IL, USA) and Bond Polymer Refine Detection Kit (Leica Biosystems) for IHC. Scoring was by two blinded observers (including A.W.) until consensus was reached. Staining intensity was classified as 0 (none), 1 (weak), 2 (moderate), and 3 (high). For nuclear staining (ESR1, Ki-67, and CCND1), we used the following modified scoring systems to quantify the number of moderately or intensely staining nuclei as a proportion of all the malignant epithelial cells: ESR1 and CCND1 staining (0, no intensely staining positive nuclei; 1, 0.1–10% intensely stained nuclei per core; 2, 10–20% intensely staining nuclei per core; 3, >20% stained nuclei per core) and Ki-67 staining (0, no positive nuclei; 1, 1–5% positive nuclei; 2, 5–10% positive nuclei; 3, >10% positive nuclei).
1.8. Statistical analysis
All data are quoted as median plus or minus interquartile range, and p values <0.05 were considered significant. We use the paired t test for paired data; for unpaired data, we used an unpaired t test for parametric data and the Mann-Whitney test for nonparametric data. For TMA IHC scoring analysis, we computed the mean intensities for each sample and performed a Mann-Whitney test.
1.9. Gene set enrichment analysis
We used Gene Set Enrichment Analysis (GSEA) version 2.0.14 (The Broad Institute, Cambridge, MA, USA). We used the Reactome version 3.0 database to analyse differentially expressed genes (DEGs) following castration for enrichment of predefined gene sets. Using the JASPAR database in the Pscan Web interface (version 1.3), we analysed 500 bases upstream of the top 500 DEGs for enriched transcription factor binding sites (Fig. 2C).
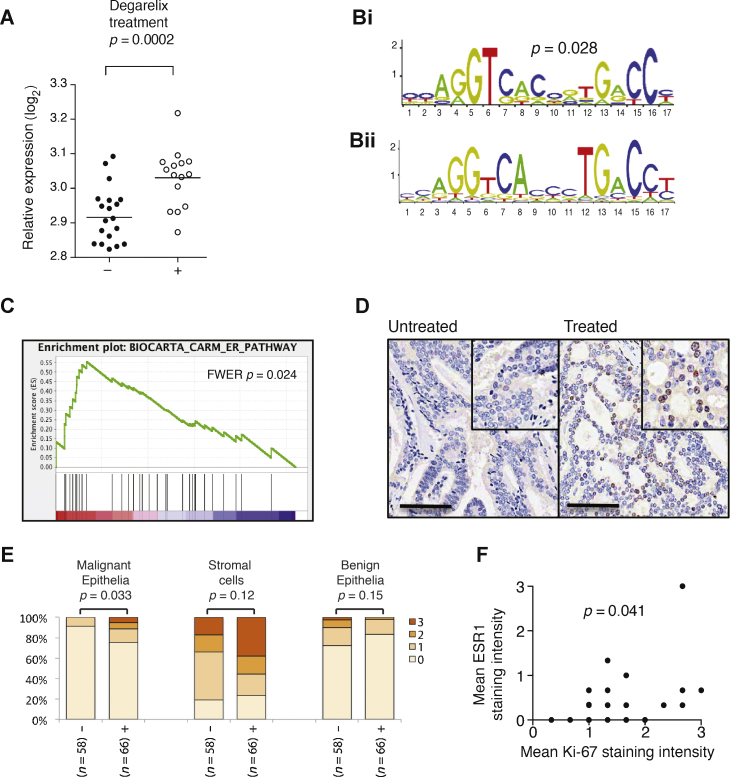
Fig. 2.
ESR1 is upregulated in response to degarelix treatment. (A) Dot plots to show mRNA expression levels of ESR1 in untreated and degarelix-treated prostate cancer (PCa) samples by expression array. (Bi) Using the Gene Set Enrichment Analysis (GSEA) to examine the distribution of known genes with the ESR1 binding motif, shown within their promoter regions, we found that these genes were enriched among genes that degarelix treatment upregulated when they were ranked by their statistical significance. The ESR1 binding motif analysed was described previously [7]. (Bii) This ESR1 binding motif closely matches the validated, experimentally derived ESR1 binding motif [10] shown here. (C) GSEA demonstrates enrichment of factors known to be involved with ESR1 signalling (from the National Cancer Institute BIOCARTA curated database) among genes differentially expressed in response to degarelix ranked by statistical significance. For this analysis, the degree to which the genes were enriched is defined by the running sum statistic called the normalised enrichment score, which was 2.036 (false discovery rate q-value of 0.027; p = 0.024). (D) Representative images of PCa samples stained by immunohistochemistry (IHC) for ESR1 in untreated and degarelix-treated patients with intense nuclear staining seen in the malignant epithelia of the treated but not the untreated samples. Scale bars = 250 μm. (E) IHC ESR1 staining was increased in treated (+) compared with untreated (−) PCa samples in malignant epithelia but not cancer-associated stroma or benign epithelia (samples in triplicate; sample sizes [n] indicate the number of subjects [20 or 27] times 3 minus missing or damaged samples on the human tissue microarray; staining intensity 0 [none] to 3 [strong]). Mean intensities from replicate samples for each patient were used to calculate statistical significance when comparing treated and untreated groups (Mann-Whitney test). (F) Graph shows correlation between intensity of staining by IHC for ESR1 and Ki-67. Spearman's ρ correlation coefficient of 0.338 (p = 0.041).
2. Results
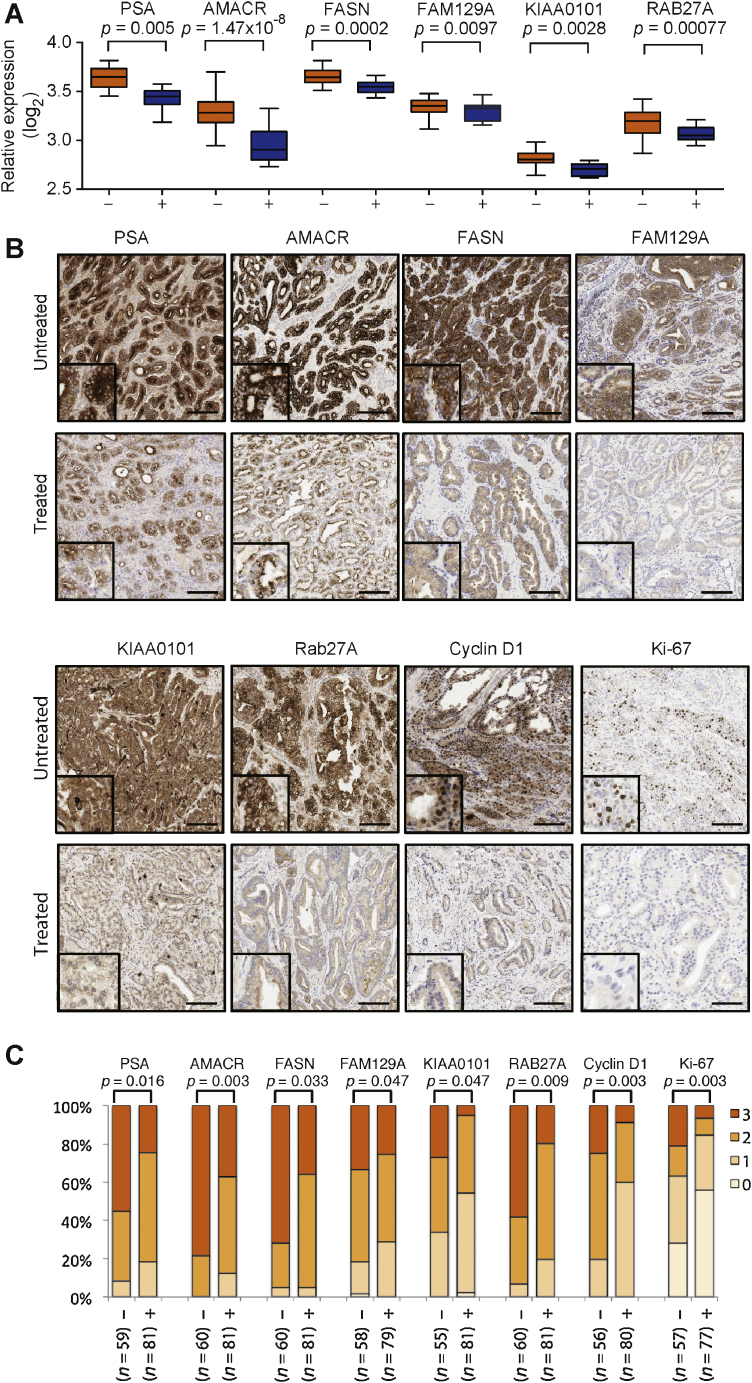
We saw no differences in the tumour or patient characteristics of the study and control cohorts (Supplementary Table 1). We saw a rapid decrease in the serum testosterone at 7 d after degarelix administration in all treated patients (11.73 ± 5.08 vs 1.19 ± 0.63 nmol/l) (Supplementary Fig. 1B), paralleled by a decrease in intratumoural androgens (Supplementary Fig. 1C). We identified 749 genes downregulated and 908 genes upregulated in response to castration. Expression levels of known AR-regulated genes—including TMPRSS2, FKBP5, KLK3, and FASN—were among those most strongly affected by treatment (Supplementary Table 2 and 3). We validated differential expression for eight genes by RT-PCR and IHC (Fig. 1B and 1C).
Fig. 1.
Degarelix treatment regulates expression of known androgen receptor (AR)–regulated genes and genes not previously identified as AR regulated. (A) Messenger RNA expression levels of known AR-regulated genes PSA and FASN as well as putative AR-regulated genes AMACR, FAM129A, RAB27A, and KIAA0101 were decreased in degarelix-treated (+) prostate cancer (PCa) samples compared with untreated (−) samples. (B) Representative immunohistochemistry (IHC) images of PCa samples from degarelix-treated and untreated patients. Inlay = ×2 magnification. Scale bars = 250 μm. (C) Bar chart to show the distribution of staining intensity of a tissue microarray made up of PCa samples from untreated (−) and degarelix-treated (+) patients stained by IHC (0 [no staining] to 3 [intense staining]; samples in triplicate; sample sizes [n] evaluated in each analysis include the number of subjects [20 or 27] times 3 minus missing or damaged samples; staining intensity 0 [none] to 3 [strong]). Mean intensities from replicate samples for each patient were used to calculate statistical significance when comparing treated and untreated groups (Mann-Whitney test).
Degarelix treatment decreased nuclear expression of proliferation marker Ki-67 protein and cell cycle progression (expression of nuclear CCND1) (Fig. 1B and 1C). We observed and validated decreased expression of three genes with limited evidence of AR regulation but with AR binding sites in their promoter regions (RAB27A, KIAA0101, and FAM129A) following degarelix treatment (Fig. 1A–1C)
2.1. ESR1 expression is upregulated in malignant epithelia, is pro-proliferative, and is associated with transcription of known ESR1 target genes
Degarelix treatment upregulated ESR1 mRNA (Fig. 2A). Using GSEA, we demonstrated enrichment of genes that had an ESR1 binding motif (Fig. 2B) [7] within the promoter region and genes known to be involved in ESR1 signalling (Fig. 2C) among those genes whose expression was upregulated with degarelix treatment (Fig. 2C).
IHC demonstrated expression of ESR1 by stromal cells but not epithelial cells (benign or malignant) in untreated samples, but 24% of degarelix-treated cancers stained positive for ESR1 in malignant epithelia compared with 8% of untreated samples (Fig. 2E). Despite an overall decrease in the expression of the proliferation marker Ki-67 in malignant glands following degarelix treatment (Fig. 1B and 1C); in those glands with increased nuclear ESR1 staining, proliferation was upregulated (Fig. 2F).
3. Discussion
In addition to identifying a large number of AR-regulated genes, we have confirmed AR regulation of three genes (RAB27A, FAM129, and KIAA0101) not proven to be AR regulated in vivo as well as AMACR, for which data were conflicting. ESR1 is essential for prostate carcinogenesis and implicated in PCa growth control [8]. Polymorphism of ESR1 is associated with PCa prognosis [9].
We observed rapid upregulation of ESR1 expression with castration (Fig. 2A). Increased ESR1 mRNA expression following prolonged castration using LHRH analogues has been observed [2], [3]. Upregulation of ESR1 expression may represent an intrinsic mechanism by which some malignant prostate epithelial cells proliferate despite castration.
Author contributions: Greg L. Shaw had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Shaw, Whitaker, Corcoran, Mills, Neal, Massie, Lynch, Nelson.
Acquisition of data: Shaw, Corcoran, Luxton, Kay, Miller, Warren, Arlt, Taylor, Shah, Ross-Adams.
Analysis and interpretation of data: Shaw, Dunning, Lamb, Eldridge, Russell, Ramos-Montoya.
Drafting of the manuscript: Shaw, Whitaker, Lamb, Nelson, Russell.
Critical revision of the manuscript for important intellectual content: Shaw, Mills, Neal, Massie.
Statistical analysis: None.
Obtaining funding: Shaw, Neal.
Administrative, technical, or material support: None.
Supervision: Mills, Neal.
Other (specify): None.
Financial disclosures: Greg L. Shaw certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: The Academy of Medical Sciences, the National Institute for Health Research, Cancer Research UK, and National Cancer Research provided funding for the design and conduct of the study as well as the collection, analysis, and interpretation of data.
Acknowledgments: The authors thank CRUK; the NIHR; the Academy of Medical Sciences (RG:63397); the National Cancer Research Prostate Cancer: Mechanisms of Progression and Treatment (ProMPT) collaborative (G0500966/75466); Hutchison Whampoa Limited; the Human Research Tissue Bank (Addenbrooke's Hospital, supported by the NIHR Cambridge BRC); and Cancer Research UK. We would like to thank those men with prostate cancer and the subjects who have donated their time and their samples to the Cambridge Biorepository that were used in this research. We also acknowledge the support of the research staff in S4, who so carefully curated the samples and the follow-up data (Jo Burge, Marie Corcoran, Anne George, and Sara Stearn).
Associate Editor: James Catto
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.eururo.2015.10.042.
Appendix A. Supplementary data
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Lehmusvaara S., Erkkila T., Urbanucci A. Chemical castration and anti-androgens induce differential gene expression in prostate cancer. J Pathol. 2012;227:336–345. doi: 10.1002/path.4027. [DOI] [PubMed] [Google Scholar]
- 3.Rajan P., Sudbery I.M., Villasevil M.E. Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. Eur Urol. 2014;66:32–39. doi: 10.1016/j.eururo.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Amico A.V., Whittington R., Malkowicz S.B. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 5.Warren A.Y., Whitaker H.C., Haynes B. Method for sampling tissue for research which preserves pathological data in radical prostatectomy. Prostate. 2013;73:194–202. doi: 10.1002/pros.22556. [DOI] [PubMed] [Google Scholar]
- 6.Sharma N.L., Massie C.E., Ramos-Montoya A. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23:35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Sandelin A., Wasserman W.W. Prediction of nuclear hormone receptor response elements. Mol Endocrinol. 2005;19:595–606. doi: 10.1210/me.2004-0101. [DOI] [PubMed] [Google Scholar]
- 8.Ricke W.A., McPherson S.J., Bianco J.J., Cunha G.R., Wang Y., Risbridger G.P. Prostatic hormonal carcinogenesis is mediated by in situ estrogen production and estrogen receptor alpha signaling. FASEB J. 2008;22:1512–1520. doi: 10.1096/fj.07-9526com. [DOI] [PubMed] [Google Scholar]
- 9.Gu Z., Wang G., Chen W. Estrogen receptor alpha gene polymorphisms and risk of prostate cancer: a meta-analysis involving 18 studies. Tumour Biol. 2014;35:5921–5930. doi: 10.1007/s13277-014-1785-4. [DOI] [PubMed] [Google Scholar]
- 10.Welboren W.J., van Driel M.A., Janssen-Megens E.M. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.