Abstract
Objective (1) To develop an automated algorithm to predict a patient’s response (ie, if the patient agrees or declines) before he/she is approached for a clinical trial invitation; (2) to assess the algorithm performance and the predictors on real-world patient recruitment data for a diverse set of clinical trials in a pediatric emergency department; and (3) to identify directions for future studies in predicting patients’ participation response.
Materials and Methods We collected 3345 patients’ response to trial invitations on 18 clinical trials at one center that were actively enrolling patients between January 1, 2010 and December 31, 2012. In parallel, we retrospectively extracted demographic, socioeconomic, and clinical predictors from multiple sources to represent the patients’ profiles. Leveraging machine learning methodology, the automated algorithms predicted participation response for individual patients and identified influential features associated with their decision-making. The performance was validated on the collection of actual patient response, where precision, recall, F-measure, and area under the ROC curve were assessed.
Results Compared to the random response predictor that simulated the current practice, the machine learning algorithms achieved significantly better performance (Precision/Recall/F-measure/area under the ROC curve: 70.82%/92.02%/80.04%/72.78% on 10-fold cross validation and 71.52%/92.68%/80.74%/75.74% on the test set). By analyzing the significant features output by the algorithms, the study confirmed several literature findings and identified challenges that could be mitigated to optimize recruitment.
Conclusion By exploiting predictive variables from multiple sources, we demonstrated that machine learning algorithms have great potential in improving the effectiveness of the recruitment process by automatically predicting patients’ participation response to trial invitations.
Keywords: patient-directed precision recruitment, socioeconomic status, predictive modeling, machine learning
INTRODUCTION
Challenges with patient recruitment for clinical trials represent major barriers to the timely and efficacious conduct of translational research.1 Despite a long-term effort made by the National Institutes of Health to enhance clinical trial accrual, trial enrollment rates are not improving, and even lower participation rates are reported in minority and underserved populations.2–7 Previous research suggested that a remarkable number of clinical trials were extended or closed prematurely because of recruitment problems.8–10 This can lead to a significant waste of financial resources or underpowered studies that report on clinically relevant research questions with insufficient statistical power. The potential consequences and costs of failed clinical trials due to poor recruitment highlight the urgent need to identify strategies that could optimize and improve patient enrollment.
Studies have reported various predictors that impact the successful recruitment of patients for clinical trials.4,11–34 At the patient level, demographic characteristics such as age, race, and gender have been commonly recognized as influential factors on patients’ participation.4,19,21,23,33,35,36 Patients’ financial and socioeconomic status (SES), measured through factors such as insurance payer and education level, have also been shown to correlate with decision-making.11,12,19,29,32 Besides these objective factors, the impact of patients’ subjective attitudes towards research is thought to be considerable – attitudes are also potentially influenced by family members and care providers, particularly in a pediatric setting.4,16,18,20,24,25,27,32,36,37 Additionally, each clinical trial has unique characteristics that could impact a patient’s willingness to participate, including time demands and scheduling, trial type (eg, randomized trial), and financial incentive.4,25,27,33 Some of these characteristics could confound a patient’s clinical status when influencing their participation decisions. For instance, deterioration of a patient’s health could motivate the family to participate in a disease-specific trial, while lower severity of illness could potentially have the opposite effect.19,38 Other factors, such as seasonality and clinical environment have also proven to be influential in recruitment success.14,36
Despite these efforts, barriers remain in the application of such findings toward interventions aimed at facilitating patient recruitment. Since the majority of the work has focused on a handful of clinical trials,2,5,11,12,19,29,32,35,36,38 small subgroups of the general population (eg, race, gender, and ethic group),4,32,34 or with a specific type of trial design,16,24,27,37 generalizability of findings, and of the predictors studied, remain of unclear significance to a broad range of clinical trials. Although the use of such findings to tailor trial invitations for individual patients was widely accepted as a future direction,13,24,29 few have actually trialed implementation, possibly due to the labor-intensive process of collecting and reviewing pertinent information in the busy clinical care setting.13 Therefore, there is a critical need for automated methods to analyze influential factors on patients’ decision making to support patient-directed precision recruitment.
Machine learning is a field of computer science that employs mathematical algorithms to learn the relation between, and make prediction on, sets of data. The algorithms operate by formulating a model from example inputs (ie, training data) to make data-driven predictions on unseen samples (ie, test data). Machine learning has been widely used on a variety of clinical decision support tasks, including patient clinical status detection, sign and symptom identification for specific diseases, and phenotype discovery.39–43 In particular, earlier studies have provided evidence for the effectiveness of machine learning in predicting human behaviors such as suicide attempt and conflict.44–46 Nevertheless, few studies attempted to explicitly predict human attitude towards clinical trial participation.36 Additional study is therefore required to fill this gap in the body of knowledge.
To take the next step, we investigated different predictors from multiple sources and developed a machine learning-based algorithm to support patient recruitment. Our specific aims are: (1) to develop an automated algorithm to predict a patient’s response (ie, if the patient agrees or declines) before he/she is approached for a clinical trial invitation, (2) to assess the algorithm performance and the predictors on real-world patient recruitment data for a diverse set of clinical trials in an urban tertiary care pediatric emergency department (ED), and (3) to identify directions for future studies in predicting patients’ participation response. The study is the first, known to us, investigation of influential factors from multiple sources to predict patients’ participation preference. Our long-term objective is to develop an automated approach to patient recruitment that will achieve a more effective, patient-directed paradigm in clinical trial enrollment.
METHODS
We included all clinical trials for patients occurring at the Cincinnati Children’s Hospital Medical Center (CCHMC) ED between January 1, 2010 and December 31, 2012. Approval of the study was given by the CCHMC institutional review board and a waiver of consent was authorized.
The pediatric ED at CCHMC is an urban, level one trauma center with 6 triage rooms, 42 beds, and 3 trauma bays. Its challenging clinical environment offers a unique opportunity for implementing and evaluating the proposed algorithm: the ED has a busy clinical care setting with approximately 70 000 patient visits annually and treating illness takes precedence over patient recruitment. In addition, families are often less likely to support ED research because of fears that it could delay treatment or distract from care provided by their physician in an emergency situation.47,48 As such, automatically identifying the likelihood of patients’ participation preference before approaching the patient or family promises benefits for clinical trial enrollment.
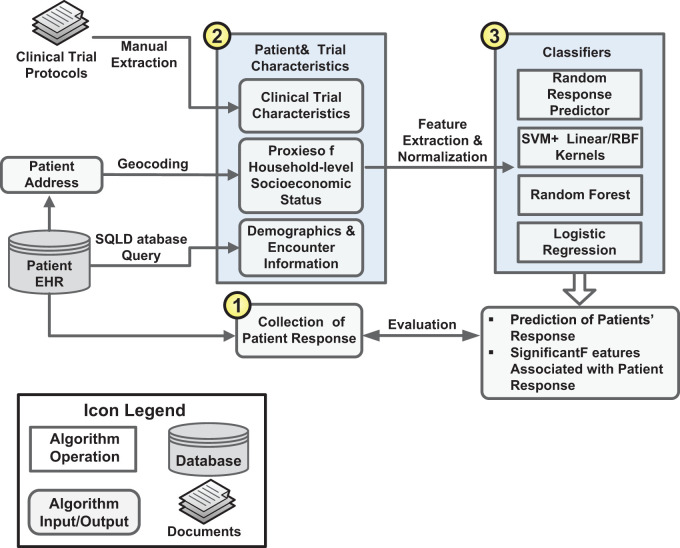
Figure 1 diagrams the overall processes of the study and the details of each process are provided below.
Figure 1:
The overall processes of the study.
Clinical Trials and Gold Standard Patient Response
All 18 clinical trials that recruited patients in the ED and required patients’ (and/or parents’) consent/assent before enrollment were included in the study. The trials covered a variety of clinical areas, interventions, observations, randomized trials, and trials requiring long-term follow-ups. Trial descriptions are presented in Table A1, Supplementary Appendix.
In current practice, patient recruitment in the ED is performed on a per visit basis. A clinical research coordinator (CRC) matches patients with the actively enrolling trials open on the patients’ date of visit and approaches the eligible patients if consent is required. Therefore, in this study we treated each patient visit (referred to as “encounter”) as the unit of analysis. During the study period, patients in 3444 encounters were eligible for at least one of the trials and were approached for enrollment. Patients in 99 encounters (2.87%) were excluded due to lack of documentation in the Electronic Health Record (EHR) (ie, no patient response documented or unidentified home addresses), resulting in a set of 3345 encounters for the study. We retrospectively collected the patients’ actual response (ie, agree or decline) to the trial invitations from the CRC study database, which served as a gold standard set to train and evaluate the predictive models. To use these data, we labeled the consent and decline response as {+1, −1}, respectively.
Patient and Clinical Trial Characteristics
Based on the literature, we collected a list of demographic characteristics, measures of socioeconomic status, and clinical factors from multiple sources. The list consists of three categories of variables (Table 1). First, encounter information (denoted by EI) that was documented during the patients’ visits, including the demographics, visit data, and clinical status. Clinical status was represented by number of arrival complaints, category of chief complaint, priority in triage, and acuity of clinical problem. Second, we identified proxies of the patients’ socioeconomic status (denoted by SES). To estimate SES, the patients’ addresses were geocoded or mapped using ArcGIS software (Redlands, California).49 This allowed for the identification of the census tract, or neighborhood, in which each patient lived. Ten socioeconomic variables, available at the census tract level, were then extracted from the 2008 to 2012 US Census American Community Survey to be used as proxies for household-level SES.50 Third, clinical trial characteristics (denoted by CTC) that could influence patients’ participation decisions were identified.33 Two CRCs who recruited patients for the 18 trials manually reviewed the trial protocols and abstracted the corresponding characteristics (Table A2, Supplementary Appendix).
Table 1:
The List of Variables Collected from Multiple Sources
| Variable Name | Variable Description |
|---|---|
| Age (EI) | Patient’s age |
| Gender (EI) | Patient’s gender |
| Race (EI) | Patient’s race |
| Ethnicity (EI) | Patient’s ethnicity |
| Insurance type (EI) | Patient’s insurance type (eg, commercial, Medicare, and self-pay) |
| Arrival means (EI) | The arrival means of the patient (eg, walk in, by private car and other) |
| Arrival time (EI) | The time interval in which the patient arrived (at 2-h increment) |
| Arrival season (EI) | The season of the patient visit (spring, summer, autumn, winter) |
| Guardian presence (EI) | Is the patient escorted by his/her legal guardian? (yes/no) |
| Arrival complaint (EI) | Number of arrival complaints |
| Chief complaint (EI) | The category of the patient’s chief complaint (eg, abdominal pain) |
| Pain score (EI) | The pain score evaluated by the clinicians (normalized from 0 to 10) |
| Triage priority (EI) | Is this patient a triage priority? (yes/no) |
| Acuity (EI) | The acuity of the patient’s chief complaint (from 1 to 5: 1 indicating urgent complaint and 5 nonurgent complaint) |
| Length of stay (EI) | Patient’s length of stay (at 30-min increment) |
| Disposition (EI) | The disposition of the patient (admit, discharge, transfer, other) |
| Poor (SES) | Percentage of persons within a census tract at <100% poverty line |
| Extreme poor (SES) | Percentage of persons within a census tract at <50% poverty line |
| Unemployment (SES) | Unemployment rate within a census tract for persons ≥16 years in the workforce |
| Income (SES) | Median household income within a census tract |
| Occupied house (SES) | Percentage of housing units that are occupied within a census tract |
| House value (SES) | Median value of owner-occupied houses within a census tract |
| Crowded house (SES) | Percentage of households with ≥1 person per room within a census tract |
| Rent house (SES) | Percentage of households who rent their home within a census tract |
| Own car (SES) | Percentage of households who do not own a car within a census tract |
| Marriage (SES) | Percentage of persons aged ≥15 years who have never married within a census tract |
| Education (SES) | Percentage of persons aged ≥25 years with less than 12th grade education within a census tract |
| Complexity (CTC) | Amount of information provided to the patient (simple, moderate, complex) |
| Time required (CTC) | Length of time required for the trial (brief, moderate, extensive) |
| Invasiveness (CTC) | Level of invasiveness of the trial (from 1 to 5: 1 indicating noninvasive and 5 highly invasive) |
| Incentive (CTC) | Amount of compensation |
| Conductor (CTC) | Conductor of the clinical trial (patient, parent, CRC, nurse, and physician) |
| Trial type (CTC) | Type of the clinical trial (observation, intervention, other) |
| Randomization (CTC) | Is the trial a randomized trial? (yes/no) |
| Disease specific (CTC) | Is the trial a disease specific trial? (yes/no) |
| Multi-center (CTC) | Is the trial a multi-center trial? (yes/no) |
| Sample required (CTC) | Does the trial require samples (eg, blood sample)? (yes/no) |
| Follow-up visit (CTC) | Does the trial require follow-up visits? (yes/no) |
| Follow-up call (CTC) | Does the trial require follow-up calls? (yes/no) |
| Insurance restriction (CTC) | Does the trial only enroll Medicare or self-pay patients? (yes/no) |
| Sensitive topic (CTC) | Does the trial involve sensitive topics? (yes/no) |
“EI” in “Variable Name” indicates an “Encounter Information” variable, “SES” a socioeconomic status variable and “CTC” a clinical trial characteristics variable.
Since the goal of the automated algorithm was to predict patients’ decisions during the encounter to assist CRCs’ patient prioritization, all the information used in the study was either available in the EHR, or could be imputed from information in the EHR (eg, home address and SES data), before patient discharge. Some variables discussed in the literature (eg, patients’ treatment preference) were not available before patient approach, so they were not included in the developed algorithm. In addition, the current practice allows the CRCs to approach stable patients for enrollment without need for notifying ED physicians. Consequently, the physicians’ attitude towards the patients’ decisions on trial enrollment were not available in the study database. Therefore, we did not investigate physicians’ influence on patients’ decision making, although “deference to physician opinion” was mentioned as an influential factor in the literature.16,37
Predictive Modeling of Patients’ Participation Response
Predictive modeling was applied to capture the mathematical relationship between a patient’s response and the patient and trial variables with the goal of weighing and identifying influential features for the patient’s decision-making. The process consisted of two steps: (1) features were extracted from the multiple-sourced sets of data and were normalized and (2) different machine learning techniques were leveraged to build the predictive models.
Feature Extraction and Normalization.
Since we collected variables from multiple sources, most of them required pre-processing and normalization before use in the predictive models. Following the methodology from our previous studies, the nominal variables (eg, gender and insurance type) were converted to binary features.51 The numerical variables (eg, age, length of stay and SES data) were first discretized into bins. The supervised discretization method, “ChiMerge,” was then applied to merge bins using the Chi-square test to reduce feature dimensions.52,53 Finally, the ordinal features generated by ChiMerge were converted to binary features. In the experiments the ChiMerge method was always trained on the data that was never part of the test set.
Predictive Modeling.
We leveraged machine learning methodology to build models for predicting patients’ participation response. The baseline approach (denoted by BASELINE) simulated the current practice in which the CRCs randomly approached eligible patients without prioritization. It was implemented as a random response predictor using a binomial probability model, where the probabilities of agreeing/declining a trial invitation were optimized using maximum likelihood estimation.54 The algorithm randomly generated a response to a trial invitation based on the consent/decline probabilities learned from the training data of the trial. We then compared the baseline with two typical machine learning classifiers: (1) logistic regression (LR), a direct probability model that measures the linear relationship between the features and the patients’ response and (2) a support vector machine (SVM) with linear kernel, a nonprobabilistic model that constructs a hyperplane in the feature space to separate the patients’ agree and decline response.54 We chose this sample of classifiers on purpose because they allowed us to analyze coefficients on individual features. The coefficients implied importance of the features in making predictions, and they were useful for identifying predictive factors associated with the patients’ participation decisions.
To take into account the possibility of presence of correlated features, we also validated the results with two additional machine learning algorithms: (1) SVM with a radial basis function (RBF) kernel that captures the nonlinearity of the feature space55 and (2) a random forest that constructs a multitude of decision trees each of which learns a highly irregular combination of the features.56
The classifiers output predictive values ranged between −∞ and +∞ to represent a patient’s response to a trial invitation. We used a default threshold to place the predictions in binary form: if a predictive value was greater than 0, we assigned +1 to the output suggesting that the patient agreed to participate. Otherwise, we assigned −1, suggesting that the patient declined.
Experiments
Evaluation Metrics
To assess algorithm performance, we adopted three evaluation metrics that are customary in biomedical science: (1) Precision = True Positives/(True Positives+False Positives) (denoted by P); (2) Recall=True Positives/(True Positives + False Negatives) (denoted by R); and (3) F-measure = 2P × R/(P + R) (denoted by F), which is the harmonic mean of precision and recall.57,58 We also generated receiver operating characteristics curves and measured the area under the curve (denoted by AUC) to assess balance between sensitivity and specificity.59 Since the goal of this study was to identify patients willing to participate in clinical trials, we adopted the F-measure as the primary metric to evaluate the algorithms.
Experiment setup
We performed a stratified random sampling based on numbers of patients approached for each trial to split the data into two sets, 90% for training and development and 10% for testing and error analysis. Ten-fold cross-validation was applied on the training and development set to tune the hyper-parameters for the predictive models. The cost parameters (C) of the LR and the SVM models were optimized using grid search parameterization (screened at 2 increments from 2−20 to 220). The same strategy was applied to find the optimal parameter for the RBF kernel (screened at 2 increments from 2−15 to 25) and the optimal number of trees for random forest (screened at 2 increments from 2 to 211). The models with optimal parameters were then applied to the test data for performance comparison and error analysis.
The above experimental setup (denoted by in-domain setup) assumed that data from all trials were available for model development. To assess the generalizability of the models on new trials that might not be available for training, we also developed an out-of-domain test simulation. In the simulation a model was trained on samples from all clinical trials except a test trial (denoted by X). The best model with optimal parameters was then evaluated on the samples from X as its out-of-domain performance. In each experiment the predictive model was selected from LR, SVM with linear and RBF kernels, and random forest, which generated the best F-measure in parameter optimization. The experiments were repeated until we tested all trials.
In addition to algorithm comparison, we tested the three variable sets (EI, SES, and CTC data) individually and in combination to validate their respective contribution to the predictive models. The experiments were conducted using the in-domain setup and LR was used as the predictive model.
Finally, to identify predictive factors associated with patients’ decisions, the features from the multivariable LR model that were associated with patients’ response at P ≤ .1 level were exported for feature analysis. To increase the interpretability of the feature coefficients, we used binary features derived from the nominal variables and ordinal features from the numerical variables. Our experiments showed that there was no significant difference between the performances of LR using ordinal and binary features derived from the numerical variables.
RESULTS
Descriptive Statistics of the Data Set
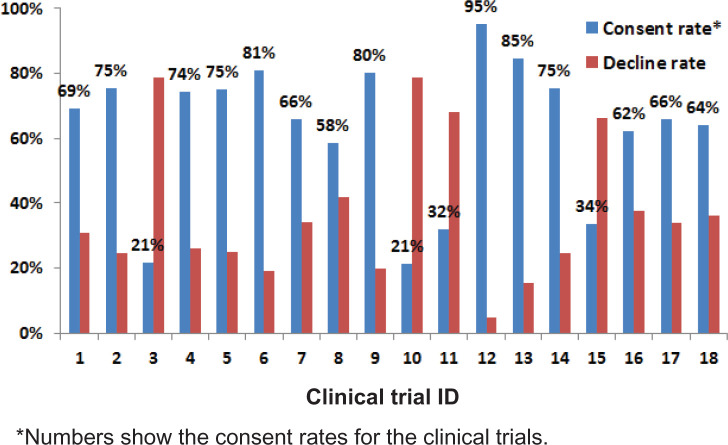
For the gold standard set, patients in 2039 encounters agreed to participate in the trials, which yielded an overall consent rate of 61%. Figure 2 shows the consent and decline rates for each clinical trial, suggesting a large variation in participation decisions across included trials. After stratified sampling and feature processing, the training set contained 3010 samples (1834/1176 agreed/declined) with 150 unique features. The test set had 335 samples (205/130 agreed/declined) with 142 features. In total there were 150 unique features in the data set.
Figure 2:
The consent and decline rates of the clinical trials.
Performance of Patient Response Prediction
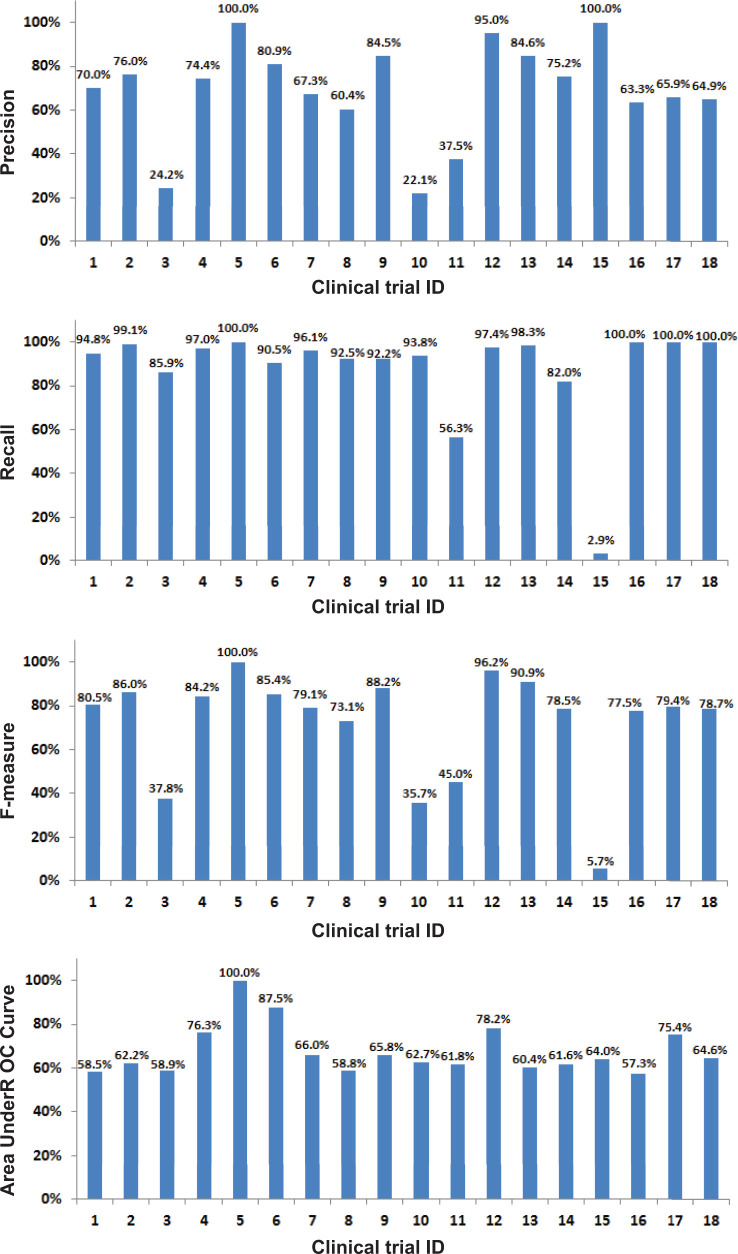
Table 2 shows the performance of different classification algorithms with all variables. All machine learning algorithms performed significantly better than the random predictor baseline (P < .001 on F-measure under paired t-test). On the 10-fold cross validation set, the SVM with RBF kernel achieved the best F-measure (80.15%), but the performance was not significantly better than that of LR (80.04%) and SVM with linear kernel (79.65%). On the test set, the three algorithms also achieved similar performance on F-measure, while the random forest algorithm performed approximately 4% lower. Figure 3 shows the performance on individual trials in the out-of-domain test simulation. The average performance (P/R/F/AUC: 69.2%/87.7%/72.3%/67.8%) was lower than that of the in-domain setup (Table 2), and the individual performances varied across the trials.
Table 2:
Performance of Different Classification Algorithms with all Variables.
| Classifier | Ten-fold cross validation performance (%) |
||||
|---|---|---|---|---|---|
| P | R | F | AUC | P-value* | |
| BASELINE | 61.68 | 61.58 | 61.54 | 50.64 | 1.06E-9 |
| Logistic Regression | 70.82 | 92.02 | 80.04 | 72.78 | 2.85E-1 |
| SVM + Linear Kernel | 70.22 | 92.02 | 79.65 | 69.91 | 2.83E-1 |
| SVM + RBF Kernel | 70.35 | 93.12 | 80.15 | 69.46 | N/A |
| Random Forest | 72.52 | 79.31 | 75.76 | 72.13 | 5.56E-6 |
| Classifier | Test set performance (%) |
||||
|---|---|---|---|---|---|
| P | R | F | AUC | ||
| BASELINE | 60.70 | 59.51 | 60.10 | 50.65 | |
| Logistic Regression | 71.52 | 92.68 | 80.74 | 75.47 | |
| SVM + Linear kernel | 70.52 | 92.20 | 79.92 | 68.07 | |
| SVM + RBF kernel | 69.46 | 93.17 | 79.58 | 70.58 | |
| Random Forest | 72.25 | 80.00 | 75.93 | 72.96 | |
Bold numbers indicate the best results.
*The P-value was calculated by comparing the F-measure between the best algorithm (SVM + RBF kernel) and the other algorithms using the paired t-test in 10-fold cross-validation.
N/A indicates that the performances between the two algorithms are identical and no P-value is returned.
Figure 3:
Performance of the predictive models on individual clinical trials under the out-of-domain test simulation.
Table 3 presents the performance of LR with different sets of variables. The LR with all variables achieved the best F-measure (set 7). Improvements were statistically significant over the LRs using individual sets, and the combination of EI and SES data (sets 1–4). Among the three variable sets, the CTC (set 3) achieved the best F-measure. The EI set performed worse, but combining it with CTC (set 5) significantly improved the F-measure (P = 2.66E-2 under paired t-test). The same improvement was also observed when comparing the combination of SES and CTC data (set 6) with the individual CTC data (P = 3.00E-3 under paired t-test).
Table 3:
Performance of Logistic Regression with Different Variable Sets
| Variable Set | Ten-fold cross validation performance (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Set | EI | SES | CTC | P | R | F | AUC | P-value* | |||
| 1 | √ | × | × | 65.33 | 81.95 | 72.69 | 61.25 | 3.39E-10 | |||
| 2 | × | √ | × | 61.61 | 91.80 | 73.72 | 52.15 | 1.05E-7 | |||
| 3 | × | × | √ | 70.64 | 90.12 | 79.20 | 72.22 | 9.60E-3 | |||
| 4 | √ | √ | × | 65.06 | 82.25 | 72.65 | 61.94 | 1.99E-8 | |||
| 5 | √ | × | √ | 70.70 | 90.50 | 79.38 | 72.23 | 5.28E-2 | |||
| 6 | × | √ | √ | 70.01 | 92.24 | 79.60 | 71.41 | 3.30E-1 | |||
| 7 | √ | √ | √ | 70.82 | 92.02 | 80.04 | 72.78 | N/A | |||
| Variable Set | Test set performance (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Set | EI | SES | CTC | P | R | F | AUC | ||||
| 1 | √ | × | × | 66.53 | 79.51 | 72.44 | 62.50 | ||||
| 2 | × | √ | × | 61.76 | 92.31 | 74.01 | 52.07 | ||||
| 3 | × | × | √ | 70.27 | 90.45 | 79.09 | 71.86 | ||||
| 4 | √ | √ | × | 66.67 | 79.14 | 72.37 | 61.99 | ||||
| 5 | √ | × | √ | 72.03 | 91.71 | 80.68 | 73.76 | ||||
| 6 | × | √ | √ | 70.41 | 91.71 | 79.66 | 70.20 | ||||
| 7 | √ | √ | √ | 71.86 | 92.20 | 80.77 | 75.47 | ||||
√ variable set used; × otherwise.
Bold numbers indicate the best results.
*The P-value was calculated by comparing the F-measure between the best algorithm (set 7) and the other algorithms using the paired t-test in 10-fold cross-validation.
N/A indicates that the performances between the two algorithms are identical and no P-value is returned.
Impact of Variables on Patients’ Participation Response
Table 4 presents features from the multivariate LR model that were associated with patients’ response at the P ≤ .1 level. Seventeen features were found to be statistically significant, 10 from EI, 6 from CTC, and 1 from SES data. Eight features had a positive effect on patients’ decision-making, meaning that they were predictive of patients’ (or parents’) agreeing to participate in clinical trials. The other nine features had a negative effect.
Table 4:
Variables Output by Logistic Regression That Were Significant at the P ≤ .1 Level (Ordered by Odds Ratio).
| Variable Index | Variable category | Variable description | OR (95% CI) |
|---|---|---|---|
| 1 | CTC | Disease specific trial: yes vs no+ | 5.29 (0.91, 30.89) |
| 2 | EI | Guardian presence: yes vs no* | 2.22 (1.03, 4.78) |
| 3 | EI | Arrival means: other means vs by car* | 1.56 (1.00, 2.44) |
| 4 | EI | Arrival season 1: winter vs summer* | 1.54 (1.14, 2.08) |
| 5 | EI | Arrival season 2: autumn vs summer* | 1.33 (1.00, 1.77) |
| 6 | EI | Race: White vs African American* | 1.31 (1.02, 1.69) |
| 7 | EI | Disposition: discharge vs admission+ | 1.29 (0.96, 1.74) |
| 8 | EI | Length of stay: every 30-min increment* | 1.06 (1.03, 1.09) |
| 9 | EI | Pain score: every 1-point increment* | 0.95 (0.92, 0.98) |
| 10 | SES | Extreme poor: every 3% increment* | 0.94 (0.88, 0.99) |
| 11 | CTC | Randomization: yes vs no+ | 0.43 (0.16, 1.14) |
| 12 | CTC | Multi-center: yes vs no* | 0.37 (0.13, 0.99) |
| 13 | CTC | Complexity 1: complex vs simple* | 0.24 (0.06, 0.96) |
| 14 | CTC | Complexity 2: moderate vs simple* | 0.16 (0.03, 0.80) |
| 15 | CTC | Follow-up visit: yes vs no* | 0.10 (0.02, 0.43) |
| 16 | EI | Chief complaint: swollen lymph nodes+ | 0.08 (0.004, 1.52) |
| 17 | EI | Chief complaint: pain+ | 0.06 (0.002, 1.86) |
*Variable significant at P ≤ .05 level, + variable significant at P ≤ .1 level. OR: odds ratio, CI: confidence interval.
DISCUSSION
Compared with the baseline random response predictor, all machine learning algorithms achieved significantly better performance in identifying patients’ participation decisions (Table 2). In addition, most of the algorithms achieved similar performance on F-measure, suggesting that the results are reliable. The lower performance made by the random forest approach was possibly due to the fact that the algorithm tends to learn a highly irregular pattern in the feature space and consequently overfits, if the sample size is not sufficiently large. The promising results suggest the potential of utilizing machine learning algorithms in improving the effectiveness of patient recruitment into clinical trials. Nevertheless, training only with data from out-of-domain trials decreased the predictive performance. Similar observations have been discussed in our earlier study.60 The generalizability of the predictive models varied across the clinical trials, suggesting that some trials could have unique patterns in influencing patients’ decision-making (which could be a combination of trial characteristics and studied patient characteristics) that cannot be learned from the others. As such, one should apply the predictive models more cautiously on new clinical trials. For instance, one could train a model on out-of-domain data and incrementally customize the model using online machine learning when data from new trials become available.61 Investigating such domain adaption methods to improve the machine learning-based algorithms is an interesting direction of our future work.
Among the influential variables, the CTC set was shown to be more predictive than the others (Table 3). The EI and SES data were less predictive. However, they did contribute unique information such that including them in the algorithm significantly improved the performance. By analyzing the significant features output by the LR algorithm (Table 4), on a more diverse set of clinical trials and patient data, we confirmed several findings reported in the literature. For clinical trial characteristics, we observed that patients were more likely to participate in disease-specific trials (var.1). They were less likely to participate in randomized and multi-center trials, more complex trials, and trials that required follow-up visits (vars.11–15).4,25,27,33,38 Regarding patients’ characteristics, White patients were more likely to participate than African Americans (var.6 in Table 4).4,33 In addition, patients from extremely poor areas were less likely to participate (var.10). During the patients’ visits, medical factors could affect their response to a trial invitation. A discharge disposition suggested a better clinical status and had a positive effect on the patients’ decision-making (var.7). In contrast, increasing pain and certain chief complaints were physical barriers to trial participation (vars. 9, 16, 17).32 In addition to medical factors, guardians’ presence usually motivated the pediatric patients to participate (var. 2). Longer length of stay in the ED also implied a greater chance for the CRCs to approach patients and recommend clinical trials (var. 8). Arrival by car might suggest longer travel time, and it could decrease the patients’ enthusiasm for contributing to a clinical trial (var. 3).12 Finally, we found that the families that visited the hospital during winter and autumn were more likely to participate compared with the families who visited during the summer (vars. 4, 5), such seasonal variability has been reported in earlier research.14
The developed algorithm and the findings could have the potential for a significant impact on the planning and patient prioritization in the recruitment process. The algorithm could facilitate recommendations of trials to patients in ways that maximize the chance of participation. For instance, the CRCs could recommend non-disease-specific trials (eg, trials 1 and 12 in Table A1, Supplementary Appendix) to patients who are going to be discharged rather than patients with increasing pain. We could also recommend the patient for trials that have more need for recruitment and are an acceptable match with a possibility of enrollment. In practice, the acceptable possibility of enrollment into a trial for a patient could be enumerated with an empirical value that balances trial need and enrollment likelihood. If a patient has an acceptable possibility of participating in multiple trials (eg, trial 1 and 2), the CRCs could recommend the trial that has fewer eligible participants (trial 2). They could also recommend the trial that is near the end of its recruitment but has not met the enrollment goals.
Error Analysis, Challenges, and Future Work
To identify challenges with predicting patients’ participation decisions, we performed error analysis for the LR algorithm on the test set. The algorithm made 90 errors, of which 16 were false negatives and 74 false positives. All false negatives came from the trials with very low consent rates (trials 3, 10, 11, and 15) and the majority of the patients (62.5%) were African Americans. These patient and trial factors could be overweighed on the samples and made the predictions bias towards a decision to decline. To alleviate this problem, we will develop advanced multi-layer classifiers in our future work to balance weights between different variable sets before aggregating them for prediction.51
To analyze the false positives, we manually reviewed the CRC’s notes documented during the recruitment process. The errors were grouped into four categories in Table 5. We observed that 37.8% of the errors were due to participants’ attitude towards research as was reported in earlier studies.4,36 Project planning is in progress to conduct surveys of patients and families to identify and integrate potential factors associated with patient, parent, and family attitudes and beliefs. Another category of error was time restraint as several families expressed interest in studies initially but the recruitment process was interrupted later on. This observation illustrates the challenge with integrating recruitment processes into busy clinical care settings. Although it is out of the scope of this study, we plan to implement real-time notification of patients’ activities in the ED to see if it helps streamline the workflow of clinical trial enrollment. Finally, concerns about study procedures and patients’ clinical status caused an additional 32.5% of the errors. Although potentially influential factors such as pain evaluation (in EI data), a proxy of education level (in SES data), and invasiveness (in CTC data) have been included in the algorithms, they might not be sufficiently informative and, hence, may have been outweighed by the other variables. Besides leveraging the multi-layer classifiers as described above, we will collect and investigate additional predictors to more accurately model the patients’ participation preference.
Table 5:
False Positive Errors Made by the LR Algorithm
| Category and percentage | Subcategory | Frequency |
|---|---|---|
| Participant Attitude (37.8%) | Generally not interested in research study | 28 |
| Time Restraints (29.7%) | Enrollment process interrupted by patient treatment | 5 |
| Parent(s) occupied by other activities (eg, taking care of the patient, working on insurance issue) | 11 | |
| Being discharged, not willing to stay | 6 | |
| Study Procedures (17.6%) | Could not complete the enrollment (eg, could not use computer or understand the protocol) | 2 |
| Concerns about additional invasive techniques (mainly blood draw) | 6 | |
| Concerns about privacy (access of patient EHR) | 2 | |
| Unspecified concerns | 3 | |
| Patient Status (14.9%) | Patient too tired (eg, sleeping or tired due to long stay in the ED) | 3 |
| Patient too ill (eg, headache and pain) | 8 |
One limitation of our study is that the SES data used is ecological, referring to the status of the geographic areas and not to individual households. The proxies might cause an inaccurate estimate of individuals’ SES and decrease the power of the SES variables (evidenced by the fact that only one was significant in the experiments). In the future, we plan to collect more accurate SES data from the patient and family via an innovative and privacy-compliant screening program (under development in the ED), which will make the predictive model more powerful. Another limitation of the study is that its evaluation is restricted to the ED clinical trials, where the physicians’ influence was not documented. To study its generalizability, we also plan to test the algorithms on more diversified clinical trials including oncology clinical trials, where the physicians could play more important roles in patients’ decision-making.16 Finally, whether using such predictive analytics to prioritize patients causes any impact on selection bias warrants further investigation in a prospective study in the future.
CONCLUSION
Our ultimate goal is to improve the effectiveness of the recruitment process by developing algorithms capable of predicting whether patients would agree to enroll in clinical trials when invited. In this study, by leveraging potentially predictive factors from multiple sources, we demonstrated that machine learning algorithms could achieve promising performance on the prediction of patients’ decisions. In a gold standard based evaluation of real-world clinical data and trials, the LR algorithm achieved 70.82%/92.02%/80.04%/72.78% (Precision/Recall/F-measure/AUC) on 10-fold cross validation and 71.52%/92.68%/80.74%/75.74% (Precision/Recall/F-measure/AUC) on the test set, significantly better than the baseline predictor that simulated the current practice. By analyzing the significant features identified through the algorithm, we also confirmed several findings that have been previously reported and identified challenges that could be mitigated to optimize recruitment. Further refinements are still required to improve algorithm accuracy, including the development of advanced multi-layer classifiers and the exploration of a broader and more precise set of predictors, including the collected socioeconomic data. If successful, the developed algorithm will pave the way to a more effective, patient-directed paradigm in clinical trial enrollment.
ACKNOWLEDGEMENTS
The authors thank Olga Semenova for her support in providing the clinical data.
CONTRIBUTORS
Y.N. conducted and coordinated the extraction of patient EHR data, ran the experiments, analyzed the results, created the tables and figures, and wrote the manuscript. A.F.B. provided the SES data, consulted on data quality and cleaning, and contributed to the manuscript. R.T. and J.D. extracted the clinical trial characteristics from the trial protocols, provided suggestions in collecting the predictors and contribute to the manuscript. I.S. conceptualized the work, provided suggestions in analyzing the results, and contributed to the manuscript. J.G.P. provided the patient EHR and clinical trial data, and contributed to the manuscript. J.W.D. provided suggestions in collecting the predictors, supervised the data extraction and the experiments, and contribute to the manuscript. All authors read and approved the final manuscript.
FUNDING
This work was partially supported by 1 R01 LM012230-01, 1U01HG008666-01, and 1 K23 AI112916 from the National Institutes of Health. Y.N. was also supported by internal funds from Cincinnati Children's Hospital Medical Center.
COMPETING INTERESTS
None.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1.Hunninghake DB, Darby CA, Probstfield JL. Recruitment experience in clinical trials: literature summary and annotated bibliography. Control Clin Trials. 1987;8(4 Suppl):6S–30S. [DOI] [PubMed] [Google Scholar]
- 2.Osann K, Wenzel L, Dogan A, et al. Recruitment and retention results for a population-based cervical cancer biobehavioral clinical trial. Gynecol Oncol. 2011;121(3):558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sibai T, Carlisle H, Tornetta P, 3rd. The darker side of randomized trials: recruitment challenges. J Bone Joint Surg Am. 2012;94 (Suppl 1):49–55. [DOI] [PubMed] [Google Scholar]
- 4.Penberthy L, Brown R, Wilson-Genderson M, et al. Barriers to therapeutic clinical trials enrollment: differences between African-American and white cancer patients identified at the time of eligibility assessment. Clin Trials. 2012;9(6):788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crutzen R, Bosma H, Havas J, et al. What can we learn from a failed trial: insight into non-participation in a chat-based intervention trial for adolescents with psychosocial problems. BMC Res Notes. 2014;7:824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams M, Caffrey L, McKevitt C. Barriers and opportunities for enhancing patient recruitment and retention in clinical research: findings from an interview study in an NHS academic health science centre. Health Res Policy Syst. 2015;13(8):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tincello DG, Kenyon S, Slack M, et al. Colposuspension or TVT with anterior repair for urinary incontinence and prolapse: results of and lessons from a pilot randomised patient-preference study (CARPET 1). BJOG. 2009;116(13):1809–1814. [DOI] [PubMed] [Google Scholar]
- 8.Foy R, Parry J, Duggan A, et al. How evidence based are recruitment strategies for randomized controlled trials in primary care? Experience from seven studies. Fam Pract. 2003;20(1):83–92. [DOI] [PubMed] [Google Scholar]
- 9.McDonald AM, Knight RC, Campbell MK, et al. What influences recruitment to randomized controlled trials? A review of trials funded by two UK funding agencies. Trials. 2006;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Treweek S, Mitchell E, Pitkethly M, et al. Strategies to improve recruitment to randomized controlled trials. Cochrane Database Syst Rev. 2010;14:MR000013. [DOI] [PubMed] [Google Scholar]
- 11.Lerman C, Rimer BK, Daly M, et al. Recruiting High-Risk Women into a Breast-Cancer Health Promotion Trial. Cancer Epidemiol Biomarkers Prevent. 1994;3(3):271–276. [PubMed] [Google Scholar]
- 12.Hurley SF, Huggins RM, Jolley DJ, et al. Recruitment Activities and Sociodemographic Factors That Predict Attendance at a Mammographic Screening-Program. Am J Public Health. 1994;84(10):1655–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross S, Grant A, Counsell C, et al. Barriers to participation in randomised controlled trials: a systematic review. J Clin Epidemiol. 1999;52(12):1143–1156. [DOI] [PubMed] [Google Scholar]
- 14.Haidich AB, Ioannidis JP. Determinants of patient recruitment in a multicenter clinical trials group: trends, seasonality and the effect of large studies. BMC Med Res Methodol. 2001;1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ettinger RL, Qian F, Xie XJ, et al. Evaluation and characteristics of “dropouts” in a longitudinal clinical study. Clin Oral Investig. 2004;8(1):18–24. [DOI] [PubMed] [Google Scholar]
- 16.Wright JR, Whelan TJ, Schiff S, et al. Why cancer patients enter randomized clinical trials: Exploring the factors that influence their decision. J Clin Oncol. 2004;22(21):4312–4318. [DOI] [PubMed] [Google Scholar]
- 17.Eng M, Taylor L, Verhoef M, et al. Understanding participation in a trial comparing cryotherapy and radiation treatment. Can J Urol. 2005;12(2):2607–2613. [PubMed] [Google Scholar]
- 18.King M, Nazareth I, Lampe F, et al. Impact of participant and physician intervention preferences on randomized trials: a systematic review. JAMA. 2005;293(9):1089–1099. [DOI] [PubMed] [Google Scholar]
- 19.Kim YJ, Peragallo N, DeForge B. Predictors of participation in an HIV risk reduction intervention for socially deprived Latino women: a cross sectional cohort study. Int J Nurs Stud. 2006;43(5):527–534. [DOI] [PubMed] [Google Scholar]
- 20.Sharp L, Cotton SC, Alexander L, et al. Reasons for participation and non-participation in a randomized controlled trial: postal questionnaire surveys of women eligible for TOMBOLA (Trial Of Management of Borderline and Other Low-Grade Abnormal smears). Clin Trials. 2006;3(5):431–442. [DOI] [PubMed] [Google Scholar]
- 21.Fletcher K, Mant J, Holder R, et al. An analysis of factors that predict patient consent to take part in a randomized controlled trial. Family Practice. 2007;24(4):388–394. [DOI] [PubMed] [Google Scholar]
- 22.Calamaro C. Cultural competence in research: research design and subject recruitment. J Pediatr Health Care. 2008;22(5):329–332. [DOI] [PubMed] [Google Scholar]
- 23.Klosky JL, Tyc VL, Lawford J, et al. Predictors of non-participation in a randomized intervention trial to reduce environmental tobacco smoke (ETS) exposure in pediatric cancer patients. Pediatr Blood Cancer. 2009;52(5):644–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mills N, Donovan JL, Wade J, et al. Exploring treatment preferences facilitated recruitment to randomized controlled trials. J Clin Epidemiol. 2011;64(10):1127–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murthy V, Awatagiri KR, Tike PK, et al. Prospective analysis of reasons for non-enrollment in a phase III randomized controlled trial. J Cancer Res Ther. 2012;8 (Suppl 1):S94–S99. [DOI] [PubMed] [Google Scholar]
- 26.Shah A, Efstathiou JA, Paly JJ, et al. Prospective preference assessment of patients' willingness to participate in a randomized controlled trial of intensity-modulated radiotherapy versus proton therapy for localized prostate cancer. Int J Radiat Oncol Biol Phys. 2012;83(1):e13–e19. [DOI] [PubMed] [Google Scholar]
- 27.Kaur G, Hutchison I, Mehanna H, et al. Barriers to recruitment for surgical trials in head and neck oncology: a survey of trial investigators. BMJ Open. 2013;3(4) pii:e002625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christie KM, Meyerowitz BE, Stanton AL, et al. Characteristics of breast cancer survivors that predict partners' participation in research. Ann Behav Med. 2013;46(1):107–113. [DOI] [PubMed] [Google Scholar]
- 29.Williams C, Maher C, Hancock M, et al. Recruiting rate for a clinical trial was associated with particular operational procedures and clinician characteristics . J Clin Epidemiol. 2014;67(2):169–175. [DOI] [PubMed] [Google Scholar]
- 30.Mills N, Blazeby JM, Hamdy FC, et al. Training recruiters to randomized trials to facilitate recruitment and informed consent by exploring patients' treatment preferences. Trials. 2014;15:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bucci S, Butcher I, Hartley S, et al. Barriers and facilitators to recruitment in mental health services: care coordinators' expectations and experience of referring to a psychosis research trial. Psychol Psychother. 2015;88(3):335–50. [DOI] [PubMed] [Google Scholar]
- 32.Hubacher D, Spector H, Monteith C, et al. Rationale and enrollment results for a partially randomized patient preference trial to compare continuation rates of short-acting and long-acting reversible contraception. Contraception. 2015;91(3):185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor RG, Hounchell M, Ho M, et al. Factors associated with participation in research conducted in a pediatric emergency department. Pediatr Emerg Care. 2015;31(5):348–352. [DOI] [PubMed] [Google Scholar]
- 34.Aponte-Rivera V, Dunlop BW, Ramirez C, et al. Enhancing Hispanic participation in mental health clinical research: development of a Spanish-speaking depression research site. Depress Anxiety. 2014;31(3):258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buis LR, Janney AW, Hess ML, et al. Barriers encountered during enrollment in an internet-mediated randomized controlled trial. Trials. 2009;10:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeomans Kinney A, Vernon SW, Shui W, et al. Validation of a model predicting enrollment status in a chemoprevention trial for breast cancer. Cancer Epidemiol Biomarkers Prev. 1998;7(7):591–595. [PubMed] [Google Scholar]
- 37.Jacobs SR, Weiner BJ, Reeve BB, et al. Organizational and physician factors associated with patient enrollment in cancer clinical trials. Clin Trials. 2014;11(5):565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dreyzin A, Barnato AE, Soltys KA, et al. Parent perspectives on decisions to participate in a phase I hepatocyte transplant trial. Pediatr Transplant. 2014;18(1):112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roque FS, Jensen PB, Schmock H, et al. Using electronic patient records to discover disease correlations and stratify patient cohorts. Plos Computat Biol. 2011;7(8):e1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deleger L, Brodzinski H, Zhai H, et al. Developing and evaluating an automated appendicitis risk stratification algorithm for pediatric patients in the emergency department. J Am Med Inform Assoc. 2013;20(e2):e212–e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connolly B, Matykiewicz P, Bretonnel Cohen K, et al. Assessing the similarity of surface linguistic features related to epilepsy across pediatric hospitals. J Am Med Inform Assoc. 2014;21(5):866–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doshi-Velez F, Ge Y, Kohane I. Comorbidity clusters in autism spectrum disorders: an electronic health record time-series analysis. Pediatrics. 2014;133(1):e54–e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhai H, Brady P, Li Q, et al. Developing and evaluating a machine learning based algorithm to predict the need of pediatric intensive care unit transfer for newly hospitalized children. Resuscitation. 2014;85(8):1065–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pestian JP. Using natural language processing to classify suicide notes. in AMIA Annu Symp Proc. 2008;2008:1091. [DOI] [PubMed] [Google Scholar]
- 45.Pestian JP, Matykiewicz P, Grupp-Phelan J, et al. Suicide note classification using natural language processing: a content analysis. Biomed Inform Insights. 2010;2010(3):19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perry C. Machine learning and conflict prediction: a use case. Stability. 2013;2(3):1–18. [Google Scholar]
- 47.Morris M, Besner D, Vazquez H, et al. Parental opinions about clinical research. J Pediatr. 2007;151(5):532–537. [DOI] [PubMed] [Google Scholar]
- 48.Cofield S, Conwit R, Barsan W, et al. Recruitment and retention of patients into emergency medicine clinical trials. Acad Emerg Med. 2010;17(10):1104–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. ArcGIS [website]. 2015. http://www.arcgis.com/features/. Accessed July 7, 2015.
- 50.Beck AF, Simmons JM, Huang B, et al. Geomedicine: area-based socioeconomic measures for assessing risk of hospital reutilization among children admitted for asthma. Am J Public Health. 2012;102(12):2308–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhai H, Iyer S, Ni Y, et al. Mining a large-scale EHR with machine learning methods to predict all-cause 30-day unplanned readmissions. In Proceedings of the 2nd ASE International Conference on Big Data Science and Computing. 2014. [Google Scholar]
- 52.Kerber R. Chimerge - discretization of numeric attributes. In Aaai-92 Proceedings: Tenth National Conference on Artificial Intelligence. 1992;1992:123–128. [Google Scholar]
- 53.Maslove DM, Podchiyska T, Lowe HJ. Discretization of continuous features in clinical datasets. J Am Med Inform Assoc. 2013;20(3):544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bishop CM. Pattern Recognition and Machine Learning. Springer Science + Business Media, LLC: Singapore. 2006. [Google Scholar]
- 55.Shawe-Taylor J, Christianini N. Kernel Methods for Pattern Analysis. Cambridge University Press: Cambridge. 2004. [Google Scholar]
- 56.Breiman L. Random forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 57.Altman DG, Bland JM. Diagnostic tests. 1: Sensitivity and specificity. BMJ. 1994;308(6943):1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altman DG, Bland JM. Diagnostic tests 2: Predictive values. BMJ. 1994;309(6947):102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rice JA. Mathematical Statistics and Data Analysis, 3rd edn. Duxbury Advanced: California. 2006. [Google Scholar]
- 60.Deleger L, Lingren T, Ni Y, et al. Preparing an annotated gold standard corpus to share with extramural investigators for de-identification research. J Biomed Inform. 2014;50:173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shalev-Shwartz S. Online learning and online convex optimization. Foundations Trends Mach Learn. 2012;4(2):107–194. [Google Scholar]