Abstract
Introduction Genomic profiling information is frequently available to oncologists, enabling targeted cancer therapy. Because clinically relevant information is rapidly emerging in the literature and elsewhere, there is a need for informatics technologies to support targeted therapies. To this end, we have developed a system for Automated Identification of Molecular Effects of Drugs, to help biomedical scientists curate this literature to facilitate decision support.
Objectives To create an automated system to identify assertions in the literature concerning drugs targeting genes with therapeutic implications and characterize the challenges inherent in automating this process in rapidly evolving domains.
Methods We used subject-predicate-object triples (semantic predications) and co-occurrence relations generated by applying the SemRep Natural Language Processing system to MEDLINE abstracts and ClinicalTrials.gov descriptions. We applied customized semantic queries to find drugs targeting genes of interest. The results were manually reviewed by a team of experts.
Results Compared to a manually curated set of relationships, recall, precision, and F2 were 0.39, 0.21, and 0.33, respectively, which represents a 3- to 4-fold improvement over a publically available set of predications (SemMedDB) alone. Upon review of ostensibly false positive results, 26% were considered relevant additions to the reference set, and an additional 61% were considered to be relevant for review. Adding co-occurrence data improved results for drugs in early development, but not their better-established counterparts.
Conclusions Precision medicine poses unique challenges for biomedical informatics systems that help domain experts find answers to their research questions. Further research is required to improve the performance of such systems, particularly for drugs in development.
Keywords: precision oncology, targeted therapy, molecular, SemRep, biomedical question answering, pharmacogenomics
INTRODUCTION
Precision oncology, or personalized cancer therapy, involves using the molecular characteristics of a tumor and patient attributes to “personalize” a patient's therapy, with the goal of providing more effective and less toxic cancer treatment.1,2 Therapy can be personalized using different factors, including a patient’s exposure history, preferences, and clinical features. However, genomic profiling is emerging as a popular personalization option that is affordable, increasingly available to cancer patients, and can help select “genomically informed” targeted therapy options. For example, over 300 clinical trials based on targeted therapies (drugs that interfere with specific molecules known to drive cancer growth and survival) are currently ongoing at The University of Texas MD Anderson Cancer Center. To support clinical decisions, domain experts must continuously review the published literature to develop and maintain a knowledge base of cancer-related genes as well as the agents that target these genes or their associated biological pathways. The MD Anderson Cancer Center Personalized Cancer Therapy website (personalizedcancertherapy.org) is one such publically available knowledge base that can serve as a reference for clinicians.3 With both the number of genes and the relevant literature growing rapidly, manual review of the available research is not feasible. Thus, there is a pressing need for informatics technologies to help curators more rapidly retrieve and review relevant biomedical literature in order to identify drugs that target aberrations in cancer-related genes.2 To this end, we have developed a system for the Automated Identification of Molecular Effects of Drugs (AIMED), which leverages semantic information extracted by the SemRep4 and MetaMap5 Natural Language Processing (NLP) systems to impose constraints on searches for evidence of clinically actionable drug-gene relationships.
BACKGROUND AND SIGNIFICANCE
The biomedical literature often contains answers to clinicians’ clinical and research questions.6,7 However, the vast amount of literature accessible by online search tools (eg, MEDLINE/PubMed) as well as the overwhelming number of documents that are often retrieved by searches conducted with those tools, limit clinicians’ ability to find correct answers efficiently, thereby further limiting the extent to which those answers can inform clinical decisions.8,9 Identifying relevant citations in MEDLINE/PubMed can be difficult, and advanced features such as Boolean combinations of Medical Subject Headings (MeSH) terms are seldom used.10,11 Traditionally, document retrieval systems (eg, PubMed) return a list of documents in response to a user’s query. However, this requires manual review of each document. Question answering (QA) systems that return structured knowledge (eg, drug A targets gene B) with links to supporting documents are a desirable alternative to document retrieval systems.12–14
Due to interest in this area, the Text REtrieval Conference (TREC)15 added a QA track in TREC-8 (1999). The TREC Genomics Track (2003–2007)16 focused solely on biomedical content and was one of the largest challenge evaluations in biomedical QA. One task (implemented in 2006 and 2007) was entity-based QA to retrieve passages that specifically answered a question, with links to the original document.17,18 Analysis of the 2006 track showed that concept-based query term normalization and Entrez Gene-based query term expansion were associated with better performance.19 Essie, for example, was the best-performing search engine in TREC Genomics 2003 and one of the best in 2006, demonstrating the utility of Unified Medical Language System (UMLS)-based query term expansion for biomedical information retrieval.20 Overall, to provide accurate answers, most QA systems draw upon curated knowledge sources (such as the UMLS) and leverage the ensuing reasoning capabilities.13 In closed-domain QA systems (such as biomedicine), using domain-specific ontologies and reasoning can improve a system’s performance.21–24 For example, normalizing query terms, expanding synonyms, post-filtering answers, and including an option to specify answer entity types (eg, genes, proteins, diseases, etc.) were associated with higher precision.19,25,26
SemBT is a QA system based on semantic relations extracted from biomedical literature using the SemRep NLP system4 that can answer a wide range of questions, including those regarding gene-drug relationships.27 SemRep depends on both MetaMap5 and knowledge encoded in the UMLS.28 However, the task-domain of precision oncology is different than those that have motivated the development of prior QA systems, because of the importance of emerging knowledge – relevant drug-gene relationships may be recently documented, and existing terminologies may not represent recently developed targeted therapies, rendering ontology-based techniques less effective. Furthermore, knowledge from both the literature (including clinical and cancer biology) and other sources (such as clinical trials or pharmaceutical companies) may be relevant, which presents additional challenges for the technologies employed. For example, pharmaceutical companies do not expose their pipelines as structured data, and extracting that information from web pages complicates the process. In this paper, we describe the design and evaluation of AIMED, a system that aims to identify knowledge that is pertinent to clinical decisions in precision oncology, using information extracted from the literature and other sources with NLP.
Methods
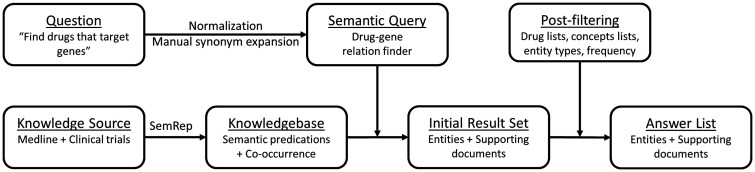
We designed and implemented a semantic QA system (Figure 1) based on a large collection of predications that is publicly available in SemMedDB,29 which is generated by SemRep processing of MEDLINE. Semantic predications in SemMedDB are organized as subject-predicate-object triples, with subjects and objects being UMLS concepts and predicates coming from the UMLS Semantic Network.30
Figure 1:
High-level summary of the QA system built for finding drugs that target genes of interest.
Our semantic queries were developed to find “drugs that target genes,” with oncogenes or their synonyms as the input and a list of drugs potentially targeting those genes as the output. We represented the verb “target” with the predicates INHIBITS, INTERACTS_WITH, COEXISTS_WITH, which were chosen based on our knowledge of the domain and the predication database31 and verified by our preliminary results. Our reference set was the gene-drug knowledge base (henceforth: Gene Sheets) provided and maintained by the Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy (IPCT) Precision Oncology Decision Support team, which included 12 cancer biologists and clinicians (a full team description is available at http://personalizedcancertherapy.org). Each gene sheet contained a list of drugs that are relevant for tumors with alterations in that gene.
We created a modified version of SemRep (henceforth: SemRep_UTH) by updating its data files to the then-latest version of the UMLS (2013AB) (as explained in the SemMedDB_UTH Database Outline32) and altering these data files to ensure that all drugs in the National Cancer Institute (NCI) thesaurus were considered. In all experiments, this version of SemRep was used to normalize gene and drug names from our reference set to UMLS concepts.
To evaluate the utility of existing knowledge resources, we ran a preliminary experiment (Experiment 1), in which, using SemRep_UTH, we created a small database of predications derived from a sample of MEDLINE abstracts and clinical trial descriptions (henceforth: SemMedDB_Local) to run the query (see Supplementary Appendix for details) on information from one Gene Sheet (PIK3CA) and compared the results with those from the official version of SemMedDB.33 PIK3CA was chosen as the starting point for the project because it was a current focus of discussion at IPCT, and a substantial amount of related literature was already available. The results of this preliminary experiment informed the construction of query parameters and constraints in the context of a development set consisting of four Gene Sheets (PIK3CA, NRAS, KRAS, MET), which were chosen because they were among the first Gene Sheets developed by the IPCT and, consequently, were available for development purposes while the remainder of the reference set was constructed. The development set also included the downstream genes in their respective cancer-related pathways and their known synonyms, as specified in each respective Gene Sheet. At the time of the study, there were a total of 21 Gene Sheets, and so the developed system was evaluated using the remaining 17 Gene Sheets. Table 1 summarizes the query parameters and constraints used with the development set as well as the options available for each.
Table 1:
Parameters of the System, as Apply to the Query and the Answers
| Parameter Name | Description | Options |
|---|---|---|
| Semantic relationship | Type of relationship between drug and gene required for retrieval. | Predications, sentence-level co-occurrence, document-level co-occurrence |
| Food and Drug Administration (FDA) filter | Accept drugs that appear on a list of FDA-approved drugs. The list was obtained from the FDA website (FDA.gov) and normalized using SemRep_UTH. | Yes or No |
| Clinical trials (CT) filter | Accept drugs found in the “intervention” field from ClinicalTrials.gov, normalized using SemRep_UTH. | Yes or No |
| Phase filter | Accept drugs either passing the FDA filter (marketed) or the CT filter for trials with a phase of, at most, x (phases 1–3). | Phase 1,1/2, 2, 3, or Marketed |
| National Cancer Institute (NCI) thesaurus filter | Return drugs that appear in the Pharmacological Substance branch of the NCI thesaurus hierarchy. | Yes or No |
| Frequency filter | Minimum number of extracted relationships (predication or co-occurrence) required before the drug is returned. | One to Many (eg, 5) |
| Predication filter | For predications, retrieve only drugs that occur in relationships with the target gene of predicate type x. | INHIBITS, INTERACTS_WITH, COEXISTS_WITH |
| Semantic type filter | Semantic types of drugs to retain. | aapp, antb, clnd, horm, imft, nnon, opco, orch, phsu |
aapp, amino acid, peptide, or protein; antb, antibiotic; clnd, clinical drug; horm, hormone; imft, immunologic factor; nnon, nucleic acid, nucleoside, or nucleotide; opco, organophosphorus compound; orch, organic chemical; phsu, pharmaceutical substance.
We used SemRep_UTH to process MEDLINE and ClinicalTrials.gov to create a modified version of SemMedDB. We used the “summary” and “full description” sections from 183 260 trials (entire set) downloaded from ClinicalTrials.gov in January 2015 (henceforth: CTDescs) as well as 23 537 576 PubMed abstracts (entire set) downloaded in August 2014 (henceforth: PMAbstracts) as our knowledge sources and processed them using SemRep_UTH to create a version of SemMedDB, which was required for our semantic query. We also extracted all UMLS concepts identified by SemRep_UTH (which uses MetaMap for concept extraction and normalization) regardless of whether or not they resulted in a predication. We used those concepts to capture sentence- and document-level co-occurrences and represented this information as triples (eg, X CO-OCCURS_WITH Y). Thus, the co-occurrence data could be combined with predications. The resulting predication database is known as SemMedDB_UTH. A version of this database that only contains MEDLINE-derived predications (without co-occurrences) is hosted by the National Library of Medicine.32
Because we were interested in finding clinically available drugs (that could be used to treat patients), we only included drugs that were available via clinical trials (CT filter) or were Food and Drug Administration (FDA)-approved (FDA filter). Furthermore, drugs available via clinical trials were associated with the trial phase (ie, phase 1, 1/2, 2, 3, with phase 1/2 involving both phases 1 and 2).34 Using phase information allowed us to limit the data source for query evaluation. To calculate precision and recall, each drug was considered within its phase category only, because different development phases required different parameter settings. For example, to evaluate query performance for phase 3, drugs from other phases were eliminated from the result set, and performance was calculated against the same phase drugs from the reference set. We hypothesized that the optimal strategy to identify drugs in each phase would depend on the number of drugs in this phase as well as the amount of available information, with less published literature and clinical trials for early-phase drugs and more for late-phase or marketed drugs. We also used information from the NCI thesaurus, extracted from UMLS 2013AB, to only retain drugs that were mentioned under the Pharmacologic Substance branch of the NCI thesaurus hierarchy as they appeared in the UMLS.
Parameter Selection
Published information about potentially useful drugs may be scarce, and the annotators wanted a system that would identify any potentially useful drug. Thus, we emphasized recall over precision. We used the F2 measure (a variant of the F-measure that emphasizes recall) as the single measure of choice to determine the best set of parameters within each drug phase category. The F2 measure is calculated as:
The parameters determined in the development phase were also used for the evaluation (Experiment 2). Specifically, the optimal data source for marketed and phase 3 drugs was the semantic predications alone. We included results of the semantic types pharmaceutical substance (phsu) and organic chemical (orch), retaining results for which at least five predication instances were found. For phases 2 and 1/2, we also included sentence-level co-occurrence, and for phase 1, we used both predications and document-level co-occurrence (with co-occurrence based on the identification of concepts by MetaMap).
Evaluation Set
We used a set of 17 genes (ABL1, AKT1, ALK, BRAF, CDK4, CDK6, EGFR, ERBB2, FGFR1, FGFR2, FLT3, KDR, KIT, PDGFRA, RET, ROS1, SMO) as our evaluation set and processed them using the optimal parameters from the development set, including the query parameters and results filtering (Experiment 3).
RESULTS
In the preliminary experiment, precision and recall were 0.06 and 0.29, respectively, with the standard version of SemMedDB, and 0.09 and 1.0, respectively, with the modified version SemMedDB_Local, demonstrating significant improvement in recall and emphasizing the need for customized knowledge resources in this domain. Table 2 shows the query results for the different experiments. The parameters were optimized for the best F2 for each drug phase in development, and the same settings were used for the evaluation. The results of the evaluation were compared with those from the standard version of SemMedDB. We found 3- to 4-fold improvements in recall, precision, F1, and F2 (0.12, 0.05, 0.07, 0.09, respectively, with SemMedDB and 0.39, 0.21, 0.27, 0.33, respectively, with SemMedDB_UTH). Some actual examples of the drugs returned by the system are: Sirolimus (true positive), Lovastatin (false positive), AZD9291 (false negative – reason: no concept unique identifier [CUI] found in the UMLS), c-Met Inhibitor LY2801653 (ostensibly false positive, later found to be a true positive during the manual evaluation). A breakdown of the recall errors is discussed in the next section.
Table 2:
Query Results for the Different Experiments
| Scope | DB | Drug Phase | FDA/CT, NCI | Source | Freq. | Predicates | Drug ST | Doc. | Drug | Recall | Prec. | F1 | F2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experiment 1: Preliminary 1 Gene Sheet, 7 reference drugs | SemMedDB_Local | Mixed | Yes | Pred. | >2 | INHIBITS, INTERACTS_WITH | phsu, antb | 559 | 74 | 1.0 | 0.09 | 0.17 | 0.33 |
| SemMedDB | Mixed | Yes | Pred. | >2 | INHIBITS, INTERACTS_WITH | phsu, antb | 540 | 35 | 0.29 | 0.06 | 0.1 | 0.16 | |
| Experiment 2: Parameter Selection 4 Gene Sheets, 115 reference drugs | SemMedDB_UTH | Marketed | Yes | Pred. | >4 | INHIBITS, INTERACTS_WITH, COEXISTS_WITH | phsu, orch | 624 | 50 | 0.86 | 0.12 | 0.21 | 0.39 |
| 3 | Yes | Pred. | – | INHIBITS, INTERACTS_WITH, COEXISTS_WITH | – | 242 | 42 | 0.79 | 0.26 | 0.39 | 0.56 | ||
| 2 | Yes | CoOccSen | – | – | – | 1466 | 125 | 0.69 | 0.18 | 0.29 | 0.44 | ||
| 1/2 | Yes | CoOccSen | – | – | – | 993 | 25 | 0.45 | 0.20 | 0.28 | 0.36 | ||
| 1 | Yes | CoOccDoc | – | – | – | 544 | 99 | 0.39 | 0.20 | 0.26 | 0.33 | ||
| All Phases: | 3869 | 341 | 0.56 | 0.19 | 0.28 | 0.4 | |||||||
| Experiment 3: Evaluation 17 Gene Sheets, 276 reference drugs | SemMedDB_UTH | Marketed | Yes | Pred. | >4 | INHIBITS, INTERACTS_WITH, COEXISTS_WITH | phsu, orch | 2251 | 80 | 0.69 | 0.3 | 0.42 | 0.55 |
| 3 | Yes | Pred. | – | INHIBITS, INTERACTS_WITH, COEXISTS_WITH | – | 299 | 61 | 0.35 | 0.3 | 0.32 | 0.34 | ||
| 2 | Yes | CoOccSen | – | – | – | 4723 | 205 | 0.5 | 0.17 | 0.25 | 0.36 | ||
| 1/2 | Yes | CoOccSen | – | – | – | 3875 | 40 | 0.29 | 0.18 | 0.22 | 0.26 | ||
| 1 | Yes | CoOccDoc | – | – | – | 1609 | 129 | 0.25 | 0.19 | 0.22 | 0.24 | ||
| All Phases: | 12757 | 515 | 0.39 | 0.21 | 0.27 | 0.33 | |||||||
| SemMedDB | Marketed | No | Pred. | – | INHIBITS, INTERACTS_WITH, COEXISTS_WITH | phsu, orch | 1730 | 661 | 0.46 | 0.02 | 0.04 | 0.09 | |
| 3 | No | Pred. | – | INHIBITS, INTERACTS_WITH, COEXISTS_WITH | phsu, orch | 1730 | 661 | 0.17 | 0.01 | 0.02 | 0.04 | ||
| 2 | No | Pred. | – | INHIBITS, INTERACTS_WITH, COEXISTS_WITH | phsu, orch | 1730 | 661 | 0.1 | 0.01 | 0.02 | 0.04 | ||
| 1/2 | No | Pred. | – | INHIBITS, INTERACTS_WITH, COEXISTS_WITH | phsu, orch | 1730 | 661 | 0.04 | 0.002 | 0.004 | 0.01 | ||
| 1 | No | Pred. | – | INHIBITS, INTERACTS_WITH, COEXISTS_WITH | phsu, orch | 1730 | 661 | 0.01 | 0.002 | 0.003 | 0.01 | ||
| All Phases: | 1730 | 661 | 0.12 | 0.05 | 0.07 | 0.09 | |||||||
FDA, Food and Drug Administration; NCI, National Cancer Institute. DB, Database used to run the queries; FDA/CT, NCI, Filters used to refine the results; Freq., Frequency filter; Drug ST, Drug semantic type (antb, ; orch, organic chemical; phsu, pharmaceutical substance); Doc., Number of documents returned; Drug, Number of concepts returned by the query; Prec., Precision; F1, Harmonic mean; F2, A variant of F1, emphasizing recall.
Error Analysis for the Evaluation Set
Of all the false negative results, 19% were not found in the original knowledge sources (PMAbstracts, CTDescs). SemRep did not identify a CUI for 24% of the false negative results, suggesting that they did not appear in the UMLS data files used to extract concepts. Drug filters (FDA/CT, NCI) were responsible for 30% of the false negatives. Those drugs were either absent from the source vocabularies, or their manually designated phases were different from those specified in the filter (eg, a drug that was in phase 1 trials at the time that the reference set was created was in phase 2 trials at the time the evaluation was performed). Because all the queries were phase-based, the phase specified for the drug in the reference set had to match the one specified in the FDA/CT filter, or the drug would either be found but not matched against the reference set (wrongly marked as false positive instead of true positive) or eliminated altogether (false negative); 23% of the missing drugs would have been found if we had used a less-restrictive approach (ie, sentence-level co-occurrence instead of predications, document-level rather than sentence-level co-occurrence). Finally, 4% of the marketed drugs were excluded by either the frequency or semantic type filter.
Manual Evaluation
To test the hypothesis that some ostensibly false positive results were actually relevant, three domain experts from the IPCT scientific team each reviewed 50 of the retrieved drugs. For each drug, experts were provided with: the normalized concept name, the targeted gene, a random selection of up to 10 source excerpts that were one or more sentences long, and a link to the source document for each excerpt. To facilitate the manual evaluation, drug and gene names were highlighted. For the document-level co-occurrence results, all the sentences from the original document that contained the terms in question were provided. Drugs were picked randomly and were equally distributed across the five phase categories (ie, 1, 1/2, 2, 3, and marketed). Each evaluator was provided with 40 unique drugs and 10 drugs that were also reviewed by two other evaluators (five each, see Table 4), to assess interobserver agreement. Thus, a total of 135 drugs were evaluated. Each evaluator assigned a score of 1, 2, or 3 to each source excerpt (Table 3).
Table 4:
The Distribution of Drugs Among Reviewers
| Drug Count (Drug Number) | Reviewer(s) | Agreement | Details |
|---|---|---|---|
| 40 (1−40) | 1 | ||
| 40 (41−80) | 2 | ||
| 40 (81−120) | 3 | ||
| 5 (121−125) | 1 and 2 | 5/5 (100%) | Both evaluators gave a score of 2 to all five drugs. |
| 5 (126−130) | 1 and 3 | 3/5 (60%) | Both evaluators gave three of the drugs a score of 2.Evaluator 1 gave one drug a score of 2, and evaluator 2 gave the same drug a score of 3. Evaluator 1 gave another drug a score of 3, and evaluator 2 gave this drug a score of 2. |
| 5 (131−135) | 2 and 3 | 5/5 (100%) | Both evaluators gave a score of 3 to three of the drugs and a score of 2 to the other two drugs. |
Table 3:
The Scoring System That Evaluator Used to Score the Drug Lists
| Score | Description |
|---|---|
| 3 | Evidence exists to add to reference set (Gene Sheets). |
| Criteria: | |
| Either: | |
| |
| 2 | Gene name or its alias is mentioned with the drug or its synonym, but evidence is not sufficient to add to reference set. |
| Categories | |
| High relevance: | |
| |
| Low relevance: | |
| |
| No relevance: | |
| |
| 1 | No mention of the drug and/or gene or its alias. |
Of the 135 drugs that were reviewed, 35 (26%) received a score of 3, 82 (61%) received a score of 2, and 18 (13%) received a score of 1. Interobserver agreement was 100% (reviewers 1 and 2), 100% (reviewers 2 and 3), and 60% (reviewers 1 and 3). The drugs used to assess interobserver agreement were different for each reviewer pair. Table 4 shows a summary of the distribution of drugs among the reviewers.
Most of the manually reviewed results were given a score of 2, which meant that they were relevant for review, but the level of evidence did not merit inclusion in the reference set (Gene Sheets). This group of drugs was retrospectively divided into three subcategories (high relevance – useful to communicate to clinicians but not recommended as therapy; low relevance; and no relevance), based on curator feedback. Approximately 26% of the ostensibly false positive results were in fact true positives. If this finding were consistent across the entire evaluation set, the re-estimated precision and recall would be 0.29 and 0.55, respectively (vs the current values of 0.21 and 0.39, respectively). However, we cannot exclude the possibility that there are other relevant drugs that were neither retrieved by the system, nor recognized as such by our team of curators. In this case, the system’s recall may be overestimated.
DISCUSSION
We developed and evaluated the AIMED system, which is intended to help curators create and maintain drug-gene association knowledge bases for precision oncology. At first glance, the recall, precision, and F2 achieved by AIMED are relatively modest. However, manual review of the ostensibly false positive results showed that 26% were actually true positives and an additional 61% were appropriate for review, but there was insufficient evidence to include these in the gold standard.
Precision oncology is rapidly evolving, and scientists at cancer centers spend a significant amount of time and effort maintaining knowledge bases that directly affect clinical decision-making processes.2 The need for precision oncology decision support knowledge bases has been recognized by other researchers. My Cancer Genome is an example of a knowledge base that provides precision-oncology-related resources.35 Unlike our work, however, My Cancer Genome relies on manual curation without the aid of informatics. Similarly, the Drug-Gene Interaction database (DGIdb) is a database of potentially druggable genes aggregated from multiple other resources, including My Cancer Genome and other manually curated databases.36 In contrast, we created an automated system. Though its results were not sufficiently accurate for use in direct clinical decision support, AIMED can be used to support curation efforts in this domain.
We found that recall was higher for marketed drugs (0.69) than those in early development phases (eg, phases 1, 1/2, and 2), and we were able to show that, of the ostensibly false positive answers generated by AIMED, the majority (87%) were considered to be relevant for review by IPCT curators, and, of that majority, 43% (26% of the total) were subsequently determined to be candidates for addition to the Gene Sheets, supporting the hypothesis that a QA system might benefit expert annotators. Gene Sheets constitute a knowledge base maintained by the IPCT for clinical decision support. Consequently, Gene Sheets will be disseminated as part of the decision support that IPCT provides.
Moreover, what would constitute optimal system performance is not well defined. The system retrieved information that expert curators considered to be clinically relevant. This includes drugs that were included on the manually constructed Gene Sheets (as evident in the precision, recall, and F-measures) as well as a substantial number of additional drugs (over 25% of the ostensibly false positive results) and drugs that could be relevant to guidelines for other reasons (eg, components of combination therapies and drugs that are known to be ineffective in the context of specific aberrations). On the one hand, this strongly supports the utility of our system as an aid for curators for the purposes of guideline development and maintenance. Further, traditional recall/precision evaluations may not fully reflect a system’s utility. To some extent, this is due to the nature of the field, because it is constantly evolving (exemplified by the progression of drugs through the development phases during the course of this work) and no gold standard is likely to be complete, or remain complete for long. As the system tries to extend its coverage by improving recall, lower precision, as an inevitable consequence, may limit its usefulness.
Typically, medical QA systems that follow an evidence-based medicine approach try to provide answers supported by extensive evidence. Semantic predications from SemRep have been used, for example, to identify therapies for certain diseases or drugs that inhibit genes.27,37 Also, ontology-based semantic knowledge modeling allows for reasoning across different domains and the incorporation of metadata, such as provenance or trust data, into core biomedical knowledge.38–41 In our case, gene names and their synonyms as well as all the drug names from the FDA/CT filters and Gene Sheets were normalized to UMLS CUIs (and/or Entrez Gene IDs, for genes). This allowed us to unambiguously filter answers using FDA/CT and NCI filters and also apply the semantic type filter (in some cases). Such techniques have been shown to be associated with higher precision,19,25,26 which is consistent with our results.
The performance of a knowledge-based system depends on the accuracy and breadth of the source knowledge.27,42,43 This is also consistent with our findings, as we showed that the default predications from SemMedDB were only modestly useful for finding emerging medications. Their utility was greatly enhanced by updating SemRep’s source vocabulary and adding predications from other knowledge sources (ie, clinical trials) or co-occurrence data. Further, we enriched the underlying ontology by modifying the data files that SemRep was using to include suppressed drug names from the NCI thesaurus. Although that technique helped with some drug categories, for drugs from lower development phases, we had to further relax the constraints by including co-occurrence data. Nonetheless, this was done in a controlled fashion, because all the elements of co-occurrence were normalized concepts that were generated by the same ontology-driven system. Biomedical QA can benefit from combining different knowledge-based and statistical methods.44 For example, CQA-1.0 (Clinical Question Answering 1.0) combined UMLS-based concept recognition with supervised machine learning techniques in its knowledge extractors,45 and MiPACQ (the Multi-source Integrated Platform for Answering Clinical Questions) combined semantic annotations with machine-learning-based re-ranking.46
In general, QA systems involve three distinct processes: question processing, document processing, and answer processing.47 In this experiment, we attempted question processing by expanding the query terms through the inclusion of gene synonyms that were part of the information contained in a Gene Sheet. However, we did not attempt to process natural language questions. We also did not attempt to implement constraints related to the source of the answer provided by the system, such as limiting the results to specific contexts or domains (eg, certain cancer types). Moreover, answer processing was limited to filtering the results, whereas it has been shown that more sophisticated statistical methods, such as relevance ranking, can improve average precision.45 One third of the false negative results were negative because they were eliminated by the drug filters, either because they were not found or because their phase did not match its phase in ClinicalTrials.gov. This further emphasizes that maintaining knowledge bases is an ongoing process that can benefit from automated systems.
It must be noted that our intended users were annotators, rather than clinicians. Although systems such as UpToDate48 provide direct support for clinical decision making, they are not automatically generated. For example, UpToDate is a continuously updated textbook whose entries are authored and maintained by humans. Due primarily to the limitations of the current state of NLP, automated processing of the biomedical literature into structured knowledge provided directly to clinicians without human supervision is not yet advisable.
Future directions for this work include exploring methods that could improve precision, such as more accurate post-filtering, ranking49 and clustering50 of results as well as methods that could improve recall, such as incorporating logic and reasoning;24 incorporating distributional statistics to estimate semantic relatedness;51 using additional ontologies or using the most recent version of existing ones; and including additional knowledge sources, such as drug company websites, or genetic pathway information. It must be noted that although SemRep was not perfectly accurate (Kilicoglu et al. reported a precision of 0.75 and a recall of 0.64 in a recent evaluation52), we were able to show that the information it retrieved was, nonetheless, very valuable. However, because increasing the breadth of our queries would inevitably increase the size of the result set, and, hence, the burden on curators, the development of an interface that permits users to adjust query constraints in accordance with their preferences regarding workload and completeness is an immediate priority. To put AIMED into routine use, the underlying vocabularies and knowledge sources need to be updated regularly (eg, every 6 months for the UMLS, every week for RxNorm). Further, user-facing applications must be created to let the annotators rank results by relevance, customize reports, adjust parameters, and collaboratively process results.
CONCLUSION
Precision oncology can benefit from QA systems that help manage clinically relevant knowledge. More research is required to determine the factors that affect the performance of knowledge-based QA systems in constantly evolving biomedical domains such as precision oncology as well as the extent to which the incorporation of open-domain QA methods can improve their performance. Research in this area will help create models and more efficient solutions.
CONTRIBUTORS
S.F., T.C., F.M.B., H.X., A.M.J., and E.V.B. conceived the methods. S.F., J.Z., A.A., and T.C. implemented and tested the software used to collect data and perform the analyses. A.M.J., V.H., A.M.B., B.C.L., N.S.S., and Y.K. performed the manual review and helped with the cancer biology aspect of the project. S.F., T.C., and E.V.B. drafted the original version of manuscript. All authors read and agreed with the analysis and the manuscript.
FUNDING
This study was supported in part by National Institute of General Medicine Services (NIGMS) grant R01 GM103859, National Cancer Institute (NCI) U01 CA180964, Sheikh Bin Zayed Al Nahyan Foundation, Cancer Prevention Research Institute of Texas (CPRIT) Precision Oncology Decision Support Core RP150535, National Center for Advanced Translational Sciences (NCATS) grant UL1 TR000371 (Center for Clinical and Translational Sciences), the Bosarge Foundation, and the MD Anderson Cancer Center Support grant (NCI P30 CA016672).
COMPETING INTERESTS
None.
REFERENCES
- 1.Garraway LA, Verweij J, Ballman KV, et al. Precision oncology: An overview. J Clin Oncol. 2013;31:1803–1805. [DOI] [PubMed] [Google Scholar]
- 2.Meric-Bernstam F, Farhangfar C, Mendelsohn J, et al. Building a personalized medicine infrastructure at a major cancer center. J Clin. 2013;31:1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson A, Zeng J, Bailey AM, et al. The right drugs at the right time for the right patient: the MD Anderson precision oncology decision support platform. Drug Discov Today 2015;20:1433–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36:462–477. [DOI] [PubMed] [Google Scholar]
- 5.Aronson AR. Effective Mapping of Biomedical Text to the UMLS Metathesaurus: The MetaMap Program. http://skr.nlm.nih.gov/papers/references/metamap_01AMIA.pdf. Accessed July 16, 2015. [PMC free article] [PubMed] [Google Scholar]
- 6.Westbrook JI, Coiera EW, Gosling AS. Do online information retrieval systems help experienced clinicians answer clinical questions? J Am Med Inform Assoc. 2005;12:315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westbrook JI, Gosling AS, Coiera EW. Do clinicians use online evidence to support patient care? A study of 55,000 clinicians. J Am Med Inform Assoc. 2004;11:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hersh WR, Crabtree MK, Hickam DH, et al. Factors associated with successful answering of clinical questions using an information retrieval system. Bull Med Libr Assoc. 2000;88:323. [PMC free article] [PubMed] [Google Scholar]
- 9.Hersh WR, Crabtree MK, Hickam DH, et al. Factors associated with success in searching MEDLINE and applying evidence to answer clinical questions. JAMIA. 2002;9:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haynes RB, McKibbon KA, Walker CJ, et al. Online access to MEDLINE in clinical settings. A study of use and usefulness. Ann Intern Med. 1990;112:78–84. [DOI] [PubMed] [Google Scholar]
- 11.Herskovic JR, Tanaka LY, Hersh W, et al. A day in the life of PubMed: analysis of a typical day’s query log. J Am Med Inform Assoc. 2007;14:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hersh WR, SpringerLink (Online service). Information Retrieval: a Health and Biomedical Perspective. 3rd ed. New York, NY: Springer; 2009. [Google Scholar]
- 13.Athenikos SJ, Han H. Biomedical question answering: a survey. Comput Methods Programs Biomed. 2010;99:1–24. [DOI] [PubMed] [Google Scholar]
- 14.Voorhees EM. Question Answering in TREC. In: Proceedings of the Tenth International Conference on Information and Knowledge Management. New York, NY, USA: ACM; 2001: 535–537. [Google Scholar]
- 15. National Institute of Standards and Technology (NIST). Text REtrieval Conference (TREC). http://trec.nist.gov. Accessed August 7, 2015.
- 16. Oregon Health & Science University (OHSU). TREC Genomics Track. http://skynet.ohsu.edu/trec-gen. Accessed August 7, 2015.
- 17.Hersh W, Voorhees E. TREC genomics special issue overview. http://skynet.ohsu.edu/∼hersh/ir-09-trecgen.pdf. Accessed August 2, 2015. [Google Scholar]
- 18.Hersh W, Cohen AM, Roberts P, et al. TREC 2006 Genomics Track Overview. Published Online First: 2006. http://www.citeulike.org/group/1014/article/972764. Accessed August 7, 2015. [Google Scholar]
- 19.Rekapalli HK, Cohen AM, Hersh WR. A comparative analysis of retrieval features used in the TREC 2006 Genomics Track passage retrieval task. In: AMIA… Annual Symposium Proceedings/AMIA Symposium. AMIA Symposium. 2006. 620–4. http://europepmc.org/abstract/med/18693910. Accessed August 5, 2015. [PMC free article] [PubMed] [Google Scholar]
- 20.Ide NC, Loane RF, Demner-Fushman D. Essie: a concept-based search engine for structured biomedical text. J Am Med Inform Assoc. 2007;14:253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H, Sable C. Being Erlang Shen: identifying answerable questions. http://cluster.cis.drexel.edu:8080/sofia/resources/QA.Data/PDF/M_2005_IJCAI_Yu_and_Sable_Being_Erlang_Shen–Identifying_Answerable_Questions-0520589825/M_2005_IJCAI_Yu_and_Sable_Being_Erlang_Shen–Identifying_Answerable_Questions.pdf. Accessed August 15, 2015. [Google Scholar]
- 22.Rinaldi F, Dowdall J, Schneider G. Answering questions in the genomics domain. Citeseer 2004. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.498.3835&rep=rep1&type=pdf. Accessed August 15, 2015. [Google Scholar]
- 23.Zweigenbaum P. Question answering in biomedicine. In: Proceedings Workshop on Natural Language Processing for Question Answering, EACL. Citeseer 2003. 1–4. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.14.9942&rep=rep1&type=pdf#page=10. Accessed August 3, 2015. [Google Scholar]
- 24.Zweigenbaum P. Knowledge and reasoning for medical question-answering. In: Proceedings of the 2009 Workshop on Knowledge and Reasoning for Answering Questions. Association for Computational Linguistics; 2009:1–2. http://dl.acm.org/citation.cfm?id=1697289. Accessed August 15, 2015. [Google Scholar]
- 25.Moldovan D, Ca MP, Harabagiu S, et al. Performance issues and error analysis in an open-domain question answering system. ACM Trans Inf Syst. 2003;21:133–154. [Google Scholar]
- 26.Hersh W, Cohen AM, Ruslen L, et al. TREC 2007 Genomics Track Overview. Published Online First: 2007. http://trec.nist.gov/pubs/trec16/t16_proceedings.html. Accessed August 7, 2015. [Google Scholar]
- 27.Hristovski D, Dinevski D, Kastrin A, et al. Biomedical question answering using semantic relations. BMC Bioinformatics. 2015;16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilicoglu H, Shin D, Fiszman M, et al. SemMedDB: a PubMed-scale repository of biomedical semantic predications. Bioinformatics. 2012;28:3158–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. At M. The UMLS Semantic Network. Proc Annu Symp Comput Appl Sic Med Care Symp Comput Appl Med Care. 1989;503–507.
- 31. Lister Hill National Center for Biomedical Communications, Cognitive Science Branch. SemMedDB Info. http://skr3.nlm.nih.gov/SemMedDB/dbinfo.html. Accessed April 28, 2015.
- 32. Lister Hill National Center for Biomedical Communications, Cognitive Science Branch. SemMedDB_UTH Database Outline. http://skr3.nlm.nih.gov/SemMedDB/index_uth.html. Accessed August 11, 2015.
- 33. Lister Hill National Center for Biomedical Communications, Cognitive Science Branch. SemMedDB Download. http://skr3.nlm.nih.gov/SemMedDB/download/download.html. Accessed April 28, 2015.
- 34. United States Food and Drug Administration (FDA). The FDA’s Drug Review Process: Ensuring Drugs Are Safe and Effective. http://www.fda.gov/drugs/resourcesforyou/consumers/ucm143534.htm. Accessed September 14, 2015.
- 35. Vanderbilt-Ingram Cancer Center. My Cancer Genome, Genetically Informed Cancer Medicine. http://www.mycancergenome.org. Accessed September 12, 2015.
- 36.Griffith M, Griffith OL, Coffman AC, et al. DGIdb - Mining the druggable genome. Nat Methods. 2013;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42:760–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cruchet S, Gaudinat A, Rindflesch T, et al. What about trust in the Question Answering world. In: AMIA 2009 Annual Symposium. 2009. http://www.hon.ch/Conf/Docs/submittedpaper_QA_HON_NLM.pdf. Accessed August 2, 2015. [Google Scholar]
- 39.Bodenreider O. Provenance information in biomedical knowledge repositories - a use case. In: SWPM. Citeseer 2009. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.154.1164&rep=rep1&type=pdf. Accessed April 28, 2015. [Google Scholar]
- 40.Carroll JJ, Bizer C, Hayes P, et al. Named graphs, provenance and trust. In: Proceedings of the 14th International Conference on World Wide Web. ACM 2005: 613-622. http://dl.acm.org/citation.cfm?id=1060835. Accessed April 28, 2015. [Google Scholar]
- 41.Nguyen V, Bodenreider O, Sheth A. Don’t like RDF reification?: making statements about statements using singleton property. In: Proceedings of the 23rd International Conference on World Wide Web. International World Wide Web Conferences Steering Committee 2014; 759-770. http://dl.acm.org/citation.cfm?id=2567973. Accessed April 28, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez V, Motta E, Uren V, et al. State of the art on semantic question answering. Citeseer 2007. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.107.1922&rep=rep1&type=pdf. Accessed August 9, 2015. [Google Scholar]
- 43.Basili R, Hansen DH, Paggio P, et al. Ontological Resources and Question Answering. In: Workshop on Pragmatics of Question Answering, held in conjunction with NAACL 2004. 1–8. http://forskningsbasen.deff.dk/Share.external?sp=S3f703020-0197-11de-b05e-000ea68e967b&sp=Sku. Accessed August 9, 2015. [Google Scholar]
- 44.Sneiderman CA, Demner-Fushman D, FiszmanM, et al. Knowledge-based methods to help clinicians find answers in MEDLINE. J Am Med Inform Assoc. 2007;14:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demner-Fushman D, Lin J. Answering clinical questions with knowledge-based and statistical techniques. Comput Linguist. 2007;33. http://www.lhncbc.nlm.nih.gov/files/archive/pub2007004.pdf. Accessed August 2, 2015. [Google Scholar]
- 46.Cairns BL, Nielsen RD, Masanz JJ, et al. The MiPACQ Clinical Question Answering System. AMIA Annu Symp Proc. 2011;2011:171–180. [PMC free article] [PubMed] [Google Scholar]
- 47.Hirschman L, Gaizauskas R. Natural language question answering: the view from here. Nat Lang Eng. 2001;7:275–300. [Google Scholar]
- 48. UpToDate. http://www.uptodate.com. Accessed September 14, 2015.
- 49.Salton G, McGill MJ. Introduction to modern information retrieval. Published Online First: 1986. http://www.citeulike.org/group/1808/article/821224. Accessed January 29, 2016. [Google Scholar]
- 50.Hearst MA, Pedersen JO. Reexamining the cluster hypothesis: scatter/gather on retrieval results. In: Proceedings of the 19th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval. ACM; 1996:76–84. http://dl.acm.org/citation.cfm?id=243216. Accessed January 29, 2016. [Google Scholar]
- 51.Pedersen T, Pakhomov SV, Patwardhan S, et al. Measures of semantic similarity and relatedness in the biomedical domain. J Biomed Inform. 2007;40:288–299. [DOI] [PubMed] [Google Scholar]
- 52.Kilicoglu H, Fiszman M, Rosemblat G, et al. Arguments of nominals in semantic interpretation of biomedical text. In: Proceedings of the 2010 Workshop on Biomedical Natural Language Processing. Association for Computational Linguistics; 2010: 46–54. http://dl.acm.org/citation.cfm?id=1869967 Accessed September 14, 2015. [Google Scholar]