Abstract
Prostate cancer is a leading cause of cancer-related death in Western men. Our understanding of the genetic alterations associated with disease predisposition, development, progression, and therapy response is rapidly improving, at least in part, owing to the development of next-generation sequencing technologies. Large advances have been made in our understanding of the genetics of prostate cancer through the application of whole-exome sequencing, and this review summarises recent advances in this field and discusses how exome sequencing could be used clinically to promote personalised medicine for prostate cancer patients.
Keywords: Prostate Cancer, Whole Exome Sequencing, WES, Personalised Medicine, Androgen, Transcriptome, Metastasis, Cancer progression, DNA damage repair
Introduction
Prostate cancer (PCa) is the most common cancer among men in the UK, with over 40,000 cases diagnosed every year 1. More than 10,000 men die from the disease in the UK per annum, making it the second most common cause of cancer-related death behind only lung cancer. Similarly, in the USA, PCa accounts for just over a quarter of all cancer diagnoses in men, with 220,800 diagnoses and 27,540 deaths from PCa predicted in 2015 2. Statistically significant risk factors associated with PCa include ethnicity, family history of the disease, and age 3, with over 75% of all PCa cases diagnosed in men over 65 years of age 1. Other factors such as cigarette smoking history, lower physical activity, higher body mass index, and height are associated with increased risk of fatal disease 3, 4.
PCa growth is dependent upon the androgen receptor (AR), a ligand-dependent transcription factor. In response to androgen binding (the major AR ligand in prostate is dihydrotestosterone), the AR regulates the expression of target genes/proteins important in PCa growth (e.g. 5– 7). The treatment given to patients with PCa is dependent on the grade and stage of the disease 8. PCa that is contained within the prostate capsule can be removed via surgery to remove the prostate or treated using radiotherapy 9. Given the potential side effects and the fact that low-grade tumours often grow slowly and may not become clinically significant, many patients, especially if they are older, tend to be monitored rather than treated (termed “active surveillance”).
Since androgenic hormones drive prostate tumour growth, therapies that target the androgen signalling pathway are commonly used for tumours that have spread outside the capsule 10. These initial hormonal therapies fall into two categories 11, 12. The first blocks the gonadal production of androgens by pituitary downregulation. This can be achieved using luteinising hormone releasing hormone (LHRH) analogues. They cause an initial spike in androgen levels; however, within 2 weeks, castrate levels of circulating testosterone are achieved due to hyperstimulation of the hypothalamo-pituitary axis. In contrast, anti-androgen therapies (e.g. bicalutamide and enzalutamide) do not reduce androgen levels per se but act directly on the AR; their binding to it results in the AR adopting an inactive conformation with subsequent inhibition of downstream events. The two approaches may be used sequentially with a switch in treatment when the first fails, or simultaneously in complete androgen blockade. However, although such hormone therapy is initially successful in the majority of patients, it invariably eventually fails, with tumours becoming unresponsive to therapy within 1–3 years and tumours progressing to the aggressive stage termed castration-resistant PCa (CRPC). Although in recent years several new therapies have been developed with some licensed for use in CRPC 13, 14, there remain few effective options and the mean survival period for patients with CRPC in 2012 was just 13.5 months 15. There is therefore a great need to identify therapeutic strategies to prevent/treat CRPC but also to develop the means and biomarkers to stratify patients for optimal therapy, and the genetic information from the studies described below is a major step in this process.
Whole-exome sequencing
The human genome consists of approximately 3 × 10 9 base pairs. Only around 1% of these (3 × 10 7 base pairs) is believed to represent coding sequence, but it is estimated that 85% of disease-causing mutations are located in these coding regions of the genome – collectively termed the exome 16, 17. Hence, most studies to date have concentrated on characterizing the exome, initially indirectly, through microarray expression analyses, and now owing to advances in DNA sequencing technologies by whole-exome sequencing (WES). WES, on which this review focuses, has led the way in uncovering mutations in coding regions responsible for many diseases and in practical and economic terms is, for many, more feasible than whole-genome sequencing (WGS), although it will not identify changes in non-coding regions of the genome (see later) 18. Hence, availability of financial resources as well as project-specific requirements (such as the ability to detect splice variants, gene fusions, and non-coding transcripts) are likely to influence decisions on whether to employ WGS, transcriptomic sequencing, WES, or other forms of targeted re-sequencing (such as deep sequencing of targeted gene panels).
In WES, DNA sequences are isolated only from exons, and data analysis compares the patient sequence to that of a reference exome aligning all of the captured exons. The variants found are compared to a control population database containing non-disease-causing variants. After the common variants are filtered out, the data can be compared to the exomes of unaffected individuals or normal tissue to identify disease-associated variants 19. The first exome sequencing study to be reported was performed by Ng et al. 20, in which the exomes of 12 individuals with Freeman–Sheldon syndrome (FSS), a rare dominantly inherited disorder, were sequenced. In agreement with previous reports 21, the study demonstrated that mutations of embryonic myosin heavy chain ( MYH3) were present in patients with the disease. Since this study, the amount of literature published using WES technology, and the range of diseases, has been increasing exponentially. Furthermore, the successful diagnostic rate of rare disease using this technology has now reached 25% 22, with many clinical laboratories offering WES as a cost-effective means of clinical testing and diagnosis 23.
WES is preferably performed on DNA obtained from fresh or frozen patient samples. The issue of obtaining fresh PCa tissue for genome analysis was recently addressed by Menon and colleagues, who used WES to compare fresh samples and formalin-fixed paraffin-embedded (FFPE) material from the same patient 24. The study demonstrated a high degree of overlap in single nucleotide variations in both types of samples, suggesting that FFPE material is a viable option for such studies. This supports previous work by Schweiger et al. and Kerick et al. 25, 26 and is a major consideration for PCa research, since it supports the use of archival FFPE biopsy samples for WES.
The availability of samples from advanced metastatic PCa is historically limited to biopsies taken from the primary tumour; analysis of tumours to, for example, identify mechanisms of therapy resistance has therefore been hampered by lack of material. This situation is improving, with metastatic samples taken post-mortem in systematic approaches such as the expanding “warm autopsy” program developed at the University of Michigan by Rubin, Pienta, and colleagues 27 and recently the move to biopsy metastatic tumours from living patients, as exemplified in the landmark paper from Robinson et al. using both WES and transcriptomic sequencing to characterize genetic lesions in 150 metastatic CRPC patients, both at bone and soft tissue metastatic sites 28. Also, in recent years, there has been a move towards non-invasive sampling (liquid biopsies such as plasma, serum, urine, and semen) to obtain samples for WES analysis and biomarkers in general. For example, Mutaza and colleagues performed WES on circulating cell-free tumour DNA (ccfDNA) obtained from the plasma of patients 29. The study identified a number of mutations associated with drug resistance, e.g. an activating mutation in phosphatidylinositol-4,5-bisphosphate 3-kinase (PIK3CA) found following paclitaxel treatment. The ability to use liquid biopsies in the prostate field will circumvent issues of material availability and is an important development in the field of WES use for personalised medicine.
Identification of gene alterations associated with prostate cancer susceptibility
Since familial PCa was first described in the 1950s 30, there has been a strong search for hereditary mutations linked with the occurrence of the disease. For example, genome-wide association studies (GWAS) and single nucleotide polymorphism (SNP) arrays have identified more than 100 PCa susceptibility loci 31, 32. The majority of these SNPs are located in non-coding regions, and bioinformatic approaches have been used to identify candidate genes affected by these variants 33.
WES has also been used to identify genetic variants in coding regions that correlate with PCa predisposition. The G84E mutation in the homeobox transcription factor HOXB13 is an example of such a SNP strongly associated with early onset familial PCa, confirmed through parallel targeted sequencing of germline DNA from 94 unrelated PCa patients and their families 34– 36. Similarly, both BRCA1 and BRCA2 have been linked to PCa predisposition, although no specific SNP has been identified, rather a variety of genetic alterations (e.g. protein-truncating mutations, in-frame deletions, and missense variants) in the two genes that cause loss of protein function 37, 38. The IMPACT study showed, in fact, that targeting prostate-specific antigen (PSA) screening to men bearing BRCA mutations identified a higher proportion with PCa and that BRCA mutation-positive men are more likely to have an aggressive form of the disease 39.
Rand et al. 40 compared the exomes of 2165 PCa cases and 2034 controls of African ancestry with the aim of identifying protein-coding variations that affect disease risk in this population. Among the significant associations identified were mutations in Secreted Protein Acidic and Rich in Cysteine-Like 1 (SPARCL1) and Protein Tyrosine Phosphatase, Receptor Type, R (PTPRR). SPARCL1 has been shown to have tumour suppressor activity 41, and the alanine to aspartic acid substitution at amino acid 49 is associated with reduced PCa risk (odds ratio [OR] = 0.78, p = 1.8 × 10 -6). In contrast, the substitution identified in PTPRR (Val239Ile), an AR target gene and regulator of the RAS/ERK1/2 pathway 42, was associated with increased risk (OR = 1.62, p = 2.5 × 10 -5). In another WES study of individuals from families with three or more affected individuals, it is notable that several of the changes associated with PCa were in genes linked to DNA damage repair, including three poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) genes 43; this is especially interesting given recent reports of clinical benefit conferred by PARP inhibitors in PCa patients with defects in DNA-repair genes 44.
Gene alterations associated with prostate cancer development/progression
A number of genetic alterations have been correlated with PCa development and progression. Perhaps unsurprisingly, the recent comprehensive profiling study of metastatic CRPC using WES and transcriptomic sequencing found mutations in genes in the AR signalling pathway in over 71% of cases: the majority were in the AR gene itself 28. Since Taplin et al.’s original report of AR mutation and Visakorpi et al.’s report of AR amplification in advanced PCa, many more have been published 45, 46. These alterations are rare in early stages of the disease and appear in response to selective pressure resulting from the hormone therapies administered, allowing the receptor to continue to drive growth in CRPC 10, 47– 53. There are many reports of missense mutations leading to amino acid substitutions, usually within the ligand-binding region, which broaden ligand specificity to allow activation by, for example, adrenal androgens, glucocorticoids, and even therapeutic antiandrogens 54– 58. More recently, constitutively active AR splice variants have been identified, which circumvent the requirement for ligand 59, 60. To date, at least 20 such variants have been described, all of which lack the ligand-binding domain, are nuclear in the absence of ligand, and have been reported to have constitutive ligand-independent activity, although some studies suggest they still require the presence of the full-length receptor for activity 60– 64. As well as alterations of AR, mutations of other components of the AR signalling pathway have been found to correlate with disease progression. In their landmark study, Grasso et al. compared the exomes of 50 heavily treated metastatic CRPC tumours with 11 high-grade treatment-naïve non-metastatic tumours 65, and a number of genes encoding proteins that interact with, and/or regulate the activity of, the AR were found to be altered. For example, mixed-lineage leukemia protein 2/histone-lysine N-methyltransferase 2D (MLL2/KMT2D), a histone-modifying enzyme that interacts with the AR, was mutated in 8.6% of cancers. Alterations were also found in FOXA1 (3.4% of cases), a pioneer factor that de-compacts DNA allowing genomic access of nuclear receptors, including the AR 66. The majority of FOXA1 mutations and indels identified were in the carboxy-terminal transactivation domain, and functional assays demonstrated that these alterations enhanced tumour growth. In support of other studies, AR mutations and an increase in copy number were also identified in the majority of patients with advanced disease 65. In addition, the recent Robinson et al. paper highlighted that 71.3% of metastatic CRPC tumours carried AR pathway mutations, the majority in the AR itself but others in e.g. AR cofactors (NCoR1/2) and, again, the pioneer factor FOXA1 28.
The gene often cited as most frequently mutated in primary PCa, encoding Speckle-type POZ protein (SPOP), is mutated in over 10% of primary prostate tumours 67. Mutations in this gene appear early in development and impact the ability of the cell to repair DNA damage, leading to genomic instability 68. Other targets of SPOP include the AR and the ERG oncogene, confirming the importance of this gene in driving tumour progression 69.
Phosphatase and Tensin homolog (PTEN) is found mutated at a similar frequency in primary PCa (10%), while PTEN deletion occurs in up to 70% of surgically treated cancers and over 60% of metastatic prostate tumours 70– 73. PTEN mutations or copy loss leads to increased PI3K/Akt signalling, which translates into cell survival and proliferation, e.g. through ligand-independent activation of the AR signaling pathway 74. It has been hypothesised that PTEN deletion creates genomic instability that then facilitates other alterations, e.g. the TMPRSS-ERG fusion commonly found in PCa 75, 76. The TMPRSS-ERG gene originates from fusion between the TMPRSS2 promoter, which is androgen responsive, and the ERG gene 77. ERG is a member of the ETS family of transcription factors, which has roles in numerous processes including cell proliferation, apoptosis, differentiation, angiogenesis, and invasiveness. This gene fusion causes the oncogene ERG to be under the control of the androgen inducible TMPRSS2 promoter, which appears to have a subsequent bearing upon tumour progression 78. Although ERG is the most common fusion partner, other ETS genes (notably ETV1 and ETV5) can be fused to the TMPRS22 promoter in prostate tumours, and also mutations in ETS gene family members have been identified in tumours, prompting speculation that some of these may have tumour suppressive function 65, 79.
Loss of function of tumour suppressors is also a common event in PCa development and progression, and those frequently lost are p53, retinoblastoma (Rb), and NK3 transcription factor related, locus 1 (NKX3.1) 80– 82. Dysregulation of the tumour suppressor p53 is rarer in PCa than other tumour types, at around 5–10% in primary tumours, but increases to around 50% in metastatic CRPC 28, 67, 83. A recent study, using candidate gene exome sequencing, suggested that patients with dominant negative p53 mutations had the worst outcome, and this feature by itself has independent prognostic relevance for patients 84. NKX3.1 is an androgen-regulated gene known to regulate prostate organogenesis during embryogenesis and is expressed throughout adult life, where it regulates ductal function and secretion 85. Complete loss of NKX3.1 expression is evident in 5% of benign prostatic hyperplasias, 20% of high-grade prostatic intraepithelial neoplasias, 34% of hormone-refractory PCa, and 78% of metastases, supporting a role in disease progression 65, 82. In contrast to p53 and Rb, NKX3.1 tumour suppressor activity is restricted to the prostate and its loss of activity is usually due to absent protein expression rather than inactivating mutations 86.
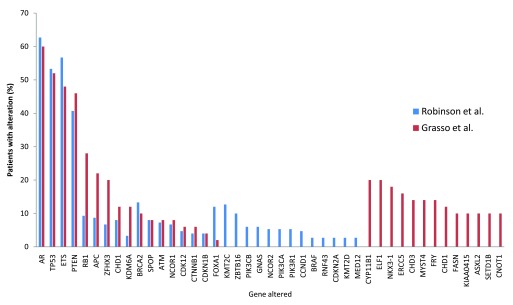
When comparing WES studies, some common alterations are evident. Taking the top 30 hits each from two recent comparable exome studies of advanced metastatic PCa, 17 genes were found to be altered in both studies 28, 65 ( Figure 1). For example, alterations in AR, TP53, ETS fusion, PTEN, and RB1 were common to both studies. However, a number of other alterations were found in only one study, some with a relatively high incidence rate (e.g. Grasso et al. found CYP11B1 to be altered in 20% of patients 65). The discrepancies between studies are likely to be resolved as the number of tumours sequenced increases but are also likely to represent the significant heterogeneity associated with PCa. Inevitably, key driver mutations will be found in multiple studies. Further comparison with other larger sequencing studies such as the 100,000 human genome project 87 will aid in the identification of the key/driver mutations and variants important in PCa initiation or progression, as these will be enriched in PCa compared to other diseases and the general population.
Figure 1. Common genetic alterations associated with advanced metastatic prostate cancer identified in two major whole-exome sequencing studies.
The top 30 genetic alterations found in each of the studies by Robinson et al. and Grasso et al. 28, 65 were compared.
The use of whole-exome sequencing for personalised medicine
There is an urgent need to stratify patients according to which therapeutic is likely to be most effective, to increase drug efficacy, and to reduce over-treatment and unnecessary side effects. WES holds great promise in this regard and has already been demonstrated to be a useful tool in terms of determining the cause of resistance to therapeutics or indeed why certain unconventional treatments may benefit patients with a particular cancer for which they would not normally be given. An example of this was an unexpected finding in a study conducted by Beltran and colleagues 88, which used WES to analyse the exomes of 97 patients with a range of metastatic cancers. One of the tumours analysed was from a PCa patient found to have an exceptional response to cisplatin treatment. Exome sequencing identified that the DNA repair protein FANCA had reduced expression and activity as a result of somatic hemizygous deletion and a partial loss of function as a result of a germline missense mutation in the second allele; subsequent assays demonstrated that loss of FANCA function was associated with platinum hypersensitivity, thus providing a rationale for the patient’s clinical response to an unconventional treatment. The widespread application of prospective WES in personalised medicine was also demonstrated in this study, since the authors were able to identify therapeutics (approved or in development), for 94% of the patients, expected to be effective given the exome profile generated 88. Robinson et al. reported a similar rate of “actionable” mutations in their study of metastatic CRPC, i.e. mutations on the basis of which informed treatment advice could be offered, including BRCA or ATM mutations that indicate use of PARP inhibitors 28. To date, each exome or transcriptome sequencing study of the prostate identifies a number of mutations unique to that study, while also a notable number of common mutations and/or affected genes. This provides both evidence for the key pathways and driver mutations in PCa and potential information leading to the effective application of a wide range of therapeutics.
Concluding remarks
The launch of the International Cancer Genome Consortium 89 ( https://icgc.org) in 2008 paved the way for genome studies on over 50 cancer types and through the use of sequencing approaches, including WES, has significantly improved our understanding of the genomic, transcriptomic, and epigenomic changes associated with different tumour types. Repositories such as this are providing a valuable resource for researchers in the field. In comparison to complete genome sequencing, WES provides information only about alterations in the coding sequence; however, it is cost effective and the reduced data analysis associated with WES means that it is likely to continue to be a valuable tool for PCa research. It also holds great promise in the clinic, having the potential to assist and inform personalised medicine for men with the disease. Undoubtedly in the future, as transcriptomic approaches become more widely used, they will lead to similar advances relating to the non-coding portion of the genome; microRNAs, long non-coding RNAs, other non-coding RNAs, and epigenetic changes are also likely to yield markers for tumour classification as well as actionable mutations. Despite the seemingly unlimited potential of WES, the frequent lack of knowledge of the functional consequences of such gene alterations is an issue that requires addressing in subsequent functional studies. These will be invaluable in characterising the phenotypic consequences of these gene alterations and are likely to yield findings that can be exploited for therapeutic gain.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Nigel Mongan, The University of Nottingham, Nottingham, UK
Iain McEwan, University of Aberdeen, Aberdeen, UK
Funding Statement
The authors are grateful for support from Prostate Cancer UK (S12-026) and Cancer Research UK (C42671/A12990) during the writing of this review.
[version 1; referees: 2 approved]
References
- 1. CRUK: Cancer mortality for common cancers.2014. [cited 2014 6/11/14]. Reference Source [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A: Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 3. Giovannucci E, Liu Y, Platz EA, et al. : Risk factors for prostate cancer incidence and progression in the health professionals follow-up study. Int J Cancer. 2007;121(7):1571–8. 10.1002/ijc.22788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Möller E, Wilson KM, Batista JL, et al. : Body size across the life course and prostate cancer in the Health Professionals Follow-up Study. Int J Cancer. 2016;138(4):853–65. 10.1002/ijc.29842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brooke GN, Gamble SC, Hough MA, et al. : Antiandrogens act as selective androgen receptor modulators at the proteome level in prostate cancer cells. Mol Cell Proteomics. 2015;14(5):1201–16. 10.1074/mcp.M113.036764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brooke GN, Culley RL, Dart DA, et al. : FUS/TLS is a novel mediator of androgen-dependent cell-cycle progression and prostate cancer growth. Cancer Res. 2011;71(3):914–24. 10.1158/0008-5472.CAN-10-0874 [DOI] [PubMed] [Google Scholar]
- 7. Gamble SC, Chotai D, Odontiadis M, et al. : Prohibitin, a protein downregulated by androgens, represses androgen receptor activity. Oncogene. 2007;26(12):1757–68. 10.1038/sj.onc.1209967 [DOI] [PubMed] [Google Scholar]
- 8. Trewartha D, Carter K: Advances in prostate cancer treatment. Nat Rev Drug Discov. 2013;12:823–4. 10.1038/nrd4068 [DOI] [PubMed] [Google Scholar]
- 9. CRUK: About hormone therapy for prostate cancer.2014. Reference Source [Google Scholar]
- 10. Brooke GN, Bevan CL: The role of androgen receptor mutations in prostate cancer progression. Curr Genomics. 2009;10(1):18–25. 10.2174/138920209787581307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goktas S, Crawford ED: Optimal hormonal therapy for advanced prostatic carcinoma. Semin Oncol. 1999;26(2):162–73. [PubMed] [Google Scholar]
- 12. Heidenreich A, Bastian PJ, Bellmunt J, et al. : EAU guidelines on prostate cancer. Part II: Treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur Urol. 2014;65(2):467–79. 10.1016/j.eururo.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 13. Crawford ED, Higano CS, Shore ND, et al. : Treating Patients with Metastatic Castration Resistant Prostate Cancer: A Comprehensive Review of Available Therapies. J Urol. 2015;194(6):1537–47. 10.1016/j.juro.2015.06.106 [DOI] [PubMed] [Google Scholar]
- 14. Ferraldeschi R, Welti J, Luo J, et al. : Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects. Oncogene. 2015;34(14):1745–57. 10.1038/onc.2014.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hirst CJ, Cabrera C, Kirby M: Epidemiology of castration resistant prostate cancer: a longitudinal analysis using a UK primary care database. Cancer Epidemiol. 2012;36(6):e349–53. 10.1016/j.canep.2012.07.012 [DOI] [PubMed] [Google Scholar]
- 16. Botstein D, Risch N: Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33Suppl:228–37. 10.1038/ng1090 [DOI] [PubMed] [Google Scholar]
- 17. Majewski J, Schwartzentruber J, Lalonde E, et al. : What can exome sequencing do for you? J Med Genet. 2011;48(9):580–9. 10.1136/jmedgenet-2011-100223 [DOI] [PubMed] [Google Scholar]
- 18. Rabbani B, Tekin M, Mahdieh N: The promise of whole-exome sequencing in medical genetics. J Hum Genet. 2014;59(1):5–15. 10.1038/jhg.2013.114 [DOI] [PubMed] [Google Scholar]
- 19. Prows CA, Tran G, Blosser B: Whole exome or genome sequencing: nurses need to prepare families for the possibilities. J Adv Nurs. 2014;70(12):2736–45. 10.1111/jan.12516 [DOI] [PubMed] [Google Scholar]
- 20. Ng SB, Turner EH, Robertson PD, et al. : Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–6. 10.1038/nature08250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Toydemir RM, Rutherford A, Whitby FG, et al. : Mutations in embryonic myosin heavy chain ( MYH3) cause Freeman-Sheldon syndrome and Sheldon-Hall syndrome. Nat Genet. 2006;38(5):561–5. 10.1038/ng1775 [DOI] [PubMed] [Google Scholar]
- 22. Seaby EG, Pengelly RJ, Ennis S: Exome sequencing explained: a practical guide to its clinical application. Brief Funct Genomics. 2015; pii: elv054. 10.1093/bfgp/elv054 [DOI] [PubMed] [Google Scholar]
- 23. Williams ES, Hegde M: Implementing genomic medicine in pathology. Adv Anat Pathol. 2013;20(4):238–44. 10.1097/PAP.0b013e3182977199 [DOI] [PubMed] [Google Scholar]
- 24. Menon R, Deng M, Rüenauver K, et al. : Somatic copy number alterations by whole-exome sequencing implicates YWHAZ and PTK2 in castration-resistant prostate cancer. J Pathol. 2013;231(4):505–16. 10.1002/path.4274 [DOI] [PubMed] [Google Scholar]
- 25. Schweiger MR, Kerick M, Timmermann B, et al. : Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (FFPE) tumor tissues for copy-number- and mutation-analysis. PLoS One. 2009;4(5):e5548. 10.1371/journal.pone.0005548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kerick M, Isau M, Timmermann B, et al. : Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med Genomics. 2011;4:68. 10.1186/1755-8794-4-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rubin MA, Putzi M, Mucci N, et al. : Rapid ("warm") autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6(3):1038–45. [PubMed] [Google Scholar]
- 28. Robinson D, van Allen EM, Wu Y, et al. : Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–28. 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murtaza M, Dawson S, Tsui DW, et al. : Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–12. 10.1038/nature12065 [DOI] [PubMed] [Google Scholar]
- 30. Gianferrari L, Arrigoni G, Cresseri A, et al. : [Genetic and clinico-statistical research on neoplasms of the prostate]. Acta Genet Med Gemellol (Roma). 1956;5(2):224–33. [PubMed] [Google Scholar]
- 31. Goh CL, Schumacher FR, Easton D, et al. : Genetic variants associated with predisposition to prostate cancer and potential clinical implications. J Intern Med. 2012;271(4):353–65. 10.1111/j.1365-2796.2012.02511.x [DOI] [PubMed] [Google Scholar]
- 32. Eeles RA, Olama AA, Benlloch S, et al. : Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45(4):385–91, 391e1–2. 10.1038/ng.2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thibodeau SN, French AJ, McDonnell SK, et al. : Identification of candidate genes for prostate cancer-risk SNPs utilizing a normal prostate tissue eQTL data set. Nat Commun. 2015;6: 8653. 10.1038/ncomms9653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ewing CM, Ray AM, Lange EM, et al. : Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366(2):141–9. 10.1056/NEJMoa1110000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Breyer JP, Avritt TG, McReynolds KM, et al. : Confirmation of the HOXB13 G84E germline mutation in familial prostate cancer. Cancer Epidemiol Biomarkers Prev. 2012;21(8):1348–53. 10.1158/1055-9965.EPI-12-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akbari MR, Trachtenberg J, Lee J, et al. : Association between germline HOXB13 G84E mutation and risk of prostate cancer. J Natl Cancer Inst. 2012;104(16):1260–2. 10.1093/jnci/djs288 [DOI] [PubMed] [Google Scholar]
- 37. Kote-Jarai Z, Leongamornlert D, Saunders E, et al. : BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer. 2011;105(8):1230–4. 10.1038/bjc.2011.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maia S, Cardoso M, Paulo P, et al. : The role of germline mutations in the BRCA1/2 and mismatch repair genes in men ascertained for early-onset and/or familial prostate cancer. Fam Cancer. 2016;15(1):111–21. 10.1007/s10689-015-9832-x [DOI] [PubMed] [Google Scholar]
- 39. Bancroft EK, Page EC, Castro E, et al. : Targeted prostate cancer screening in BRCA1 and BRCA2 mutation carriers: results from the initial screening round of the IMPACT study. Eur Urol. 2014;66(3):489–99. 10.1016/j.eururo.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rand KA, Rohland N, Tandon A, et al. : Whole-exome sequencing of over 4100 men of African ancestry and prostate cancer risk. Hum Mol Genet. 2016;25(2):371–81. 10.1093/hmg/ddv462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xiang Y, Qiu Q, Jiang M, et al. : SPARCL1 suppresses metastasis in prostate cancer. Mol Oncol. 2013;7(6):1019–30. 10.1016/j.molonc.2013.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Munkley J, Lafferty NP, Kalna G, et al. : Androgen-regulation of the protein tyrosine phosphatase PTPRR activates ERK1/2 signalling in prostate cancer cells. BMC Cancer. 2015;15:9. 10.1186/s12885-015-1012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnson AM, Zuhlke KA, Plotts C, et al. : Mutational landscape of candidate genes in familial prostate cancer. Prostate. 2014;74(14):1371–8. 10.1002/pros.22849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mateo J, Carreira S, Sandhu S, et al. : DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373(18):1697–708. 10.1056/NEJMoa1506859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taplin ME, Bubley GJ, Shuster TD, et al. : Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med. 1995;332(21):1393–8. 10.1056/NEJM199505253322101 [DOI] [PubMed] [Google Scholar]
- 46. Visakorpi T, Hyytinen E, Koivisto P, et al. : In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9(4):401–6. 10.1038/ng0495-401 [DOI] [PubMed] [Google Scholar]
- 47. Taplin ME, Rajeshkumar B, Halabi S, et al. : Androgen receptor mutations in androgen-independent prostate cancer: Cancer and Leukemia Group B Study 9663. J Clin Oncol. 2003;21(14):2673–8. 10.1200/JCO.2003.11.102 [DOI] [PubMed] [Google Scholar]
- 48. Lamb DJ, Puxeddu E, Malik N, et al. : Molecular analysis of the androgen receptor in ten prostate cancer specimens obtained before and after androgen ablation. J Androl. 2003;24(2):215–25. 10.1002/j.1939-4640.2003.tb02665.x [DOI] [PubMed] [Google Scholar]
- 49. Evans BA, Harper ME, Daniells CE, et al. : Low incidence of androgen receptor gene mutations in human prostatic tumors using single strand conformation polymorphism analysis. Prostate. 1996;28(3):162–71. [DOI] [PubMed] [Google Scholar]
- 50. Marcelli M, Ittmann M, Mariani S, et al. : Androgen receptor mutations in prostate cancer. Cancer Res. 2000;60(4):944–9. [PubMed] [Google Scholar]
- 51. Antonarakis ES, Lu C, Wang H, et al. : AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–38. 10.1056/NEJMoa1315815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Romanel A, Gasi Tandefelt D, Conteduca V, et al. : Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7(312):312re10. 10.1126/scitranslmed.aac9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ware KE, Garcia-Blanco MA, Armstrong AJ, et al. : Biologic and clinical significance of androgen receptor variants in castration resistant prostate cancer. Endocr Relat Cancer. 2014;21(4):T87–T103. 10.1530/ERC-13-0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Green SM, Mostaghel EA, Nelson PS: Androgen action and metabolism in prostate cancer. Mol Cell Endocrinol. 2012;360(1–2):3–13. 10.1016/j.mce.2011.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hay CW, McEwan IJ: The impact of point mutations in the human androgen receptor: classification of mutations on the basis of transcriptional activity. PLoS One. 2012;7(3):e32514. 10.1371/journal.pone.0032514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Veldscholte J, Berrevoets CA, Ris-Stalpers C, et al. : The androgen receptor in LNCaP cells contains a mutation in the ligand binding domain which affects steroid binding characteristics and response to antiandrogens. J Steroid Biochem Mol Biol. 1992;41(3–8):665–9. [DOI] [PubMed] [Google Scholar]
- 57. Brooke GN, Parker MG, Bevan CL: Mechanisms of androgen receptor activation in advanced prostate cancer: differential co-activator recruitment and gene expression. Oncogene. 2008;27(21):2941–50. 10.1038/sj.onc.1210955 [DOI] [PubMed] [Google Scholar]
- 58. Joseph JD, Lu N, Qian J, et al. : A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov. 2013;3(9):1020–9. 10.1158/2159-8290.CD-13-0226 [DOI] [PubMed] [Google Scholar]
- 59. Chan SC, Li Y, Dehm SM: Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem. 2012;287(23):19736–49. 10.1074/jbc.M112.352930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hu R, Dunn TA, Wei S, et al. : Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69(1):16–22. 10.1158/0008-5472.CAN-08-2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hu R, Isaacs WB, Luo J: A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate. 2011;71(15):1656–67. 10.1002/pros.21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dehm SM, Schmidt LJ, Heemers HV, et al. : Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68(13):5469–77. 10.1158/0008-5472.CAN-08-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li Y, Chan SC, Brand LJ, et al. : Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73(2):483–9. 10.1158/0008-5472.CAN-12-3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Watson PA, Chen YF, Balbas MD, et al. : Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107(39):16759–65. 10.1073/pnas.1012443107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grasso CS, Wu YM, Robinson DR, et al. : The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487(7406):239–43. 10.1038/nature11125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang YA, Yu J: Current perspectives on FOXA1 regulation of androgen receptor signaling and prostate cancer. Genes Dis. 2015;2(2):144–51. 10.1016/j.gendis.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Barbieri CE, Baca SC, Lawrence MS, et al. : Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–9. 10.1038/ng.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Boysen G, Barbieri CE, Prandi D, et al. : SPOP mutation leads to genomic instability in prostate cancer. eLife. 2015;4: pii: e09207. 10.7554/eLife.09207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. An J, Ren S, Murphy SJ, et al. : Truncated ERG Oncoproteins from TMPRSS2-ERG Fusions Are Resistant to SPOP-Mediated Proteasome Degradation. Mol Cell. 2015;59(6):904–16. 10.1016/j.molcel.2015.07.025 [DOI] [PubMed] [Google Scholar]
- 70. Feilotter HE, Nagai MA, Boag AH, et al. : Analysis of PTEN and the 10q23 region in primary prostate carcinomas. Oncogene. 1998;16(13):1743–8. 10.1038/sj.onc.1200205 [DOI] [PubMed] [Google Scholar]
- 71. Whang YE, Wu X, Suzuki H, et al. : Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci U S A. 1998;95(9):5246–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang SI, Parsons R, Ittmann M: Homozygous deletion of the PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin Cancer Res. 1998;4(3):811–5. [PubMed] [Google Scholar]
- 73. Chen Z, Trotman LC, Shaffer D, et al. : Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–30. 10.1038/nature03918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Feldman BJ, Feldman D: The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1(1):34–45. 10.1038/35094009 [DOI] [PubMed] [Google Scholar]
- 75. Bismar TA, Yoshimoto M, Vollmer RT, et al. : PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107(3):477–85. 10.1111/j.1464-410X.2010.09470.x [DOI] [PubMed] [Google Scholar]
- 76. Phin S, Moore MW, Cotter PD: Genomic Rearrangements of PTEN in Prostate Cancer. Front Oncol. 2013;3:240. 10.3389/fonc.2013.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tomlins SA, Rhodes DR, Perner S, et al. : Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. 10.1126/science.1117679 [DOI] [PubMed] [Google Scholar]
- 78. Barry Delongchamps N: Prostate cancer: review in 2014. Diagn Interv Imaging. 2014;95(7–8):739–42. 10.1016/j.diii.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 79. Demichelis F, Setlur SR, Beroukhim R, et al. : Distinct genomic aberrations associated with ERG rearranged prostate cancer. Genes Chromosomes Cancer. 2009;48(4):366–80. 10.1002/gcc.20647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bookstein R, Shew JY, Chen PL, et al. : Suppression of tumorigenicity of human prostate carcinoma cells by replacing a mutated RB gene. Science. 1990;247(4943):712–5. 10.1126/science.2300823 [DOI] [PubMed] [Google Scholar]
- 81. Qian J, Hirasawa K, Bostwick DG, et al. : Loss of p53 and c-myc overrepresentation in stage T 2-3N 1-3M 0 prostate cancer are potential markers for cancer progression. Mod Pathol. 2002;15(1):35–44. 10.1038/modpathol.3880487 [DOI] [PubMed] [Google Scholar]
- 82. Bowen C, Bubendorf L, Voeller HJ, et al. : Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 2000;60(21):6111–5. [PubMed] [Google Scholar]
- 83. Bettendorf O, Schmidt H, Staebler A, et al. : Chromosomal imbalances, loss of heterozygosity, and immunohistochemical expression of TP53, RB1, and PTEN in intraductal cancer, intraepithelial neoplasia, and invasive adenocarcinoma of the prostate. Genes Chromosomes Cancer. 2008;47(7):565–72. 10.1002/gcc.20560 [DOI] [PubMed] [Google Scholar]
- 84. Kluth M, Harasimowicz S, Burkhardt L, et al. : Clinical significance of different types of p53 gene alteration in surgically treated prostate cancer. Int J Cancer. 2014;135(6):1369–80. 10.1002/ijc.28784 [DOI] [PubMed] [Google Scholar]
- 85. Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, et al. : Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13(8):966–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kim MJ, Cardiff RD, Desai N, et al. : Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci U S A. 2002;99(5):2884–9. 10.1073/pnas.042688999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Siva N: UK gears up to decode 100,000 genomes from NHS patients. Lancet. 2015;385(9965):103–4. 10.1016/S0140-6736(14)62453-3 [DOI] [PubMed] [Google Scholar]
- 88. Beltran H, Eng K, Mosquera JM, et al. : Whole-Exome Sequencing of Metastatic Cancer and Biomarkers of Treatment Response. JAMA Oncol. 2015;1(4):466–74. 10.1001/jamaoncol.2015.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. International Cancer Genome Consortium. 2016 Reference Source [Google Scholar]