Dear Editors
There is compelling evidence for association of rs1344706, an intronic SNP within the zinc-finger protein 804A gene (ZNF804A), with schizophrenia (SZ) and bipolar disorder (BD) (O’Donovan et al., 2008; Williams et al., 2011; Zhang et al., 2012). Recently, some of the potential molecular mechanisms underlying the association between rs1344706 and disease risk were published (Girgenti et al., 2012; Hill and Bray, 2012; Hill et al., 2012; Kim et al., 2012; Okada et al., 2012; Umeda-Yano et al., 2013; Wright et al., 2013). Detection of expression quantitative trait loci (eQTL) is an important step in annotation of genome wide association study (GWAS) results that can provide possible functional links to pathophysiology, and putative use in biomarker studies (Vawter et al., 2011). There has been an over-representation of eQTL in the brain with the results of GWAS studies conducted in SZ (Richards et al., 2012). These recent results and prior attempts to find cis- and trans-regulatory influences on gene expression in SZ (Vawter et al., 2006; Martin et al., 2009) were motivation to further look for cis-regulatory influences on gene expression for the ZNF804A associated SNP.
We provide further independent evidence for the rs1344706 allelic specific expression (ASE) in the dorsolateral prefrontal cortex (DLPFC) of postmortem brains from the Stanley Medical Research Institute (SMRI). The SMRI Array Collection is a collection of postmortem brains from individuals with SZ (n = 35), with BD (n = 34), and psychiatrically normal controls (n = 35). RNA from the DLPFC was available for 99 individuals in the collection. Detailed information about the SMRI Array Collection is available at (http://www.stanleyresearch.org/dnn/Default.aspx?tabid=197). The ASE published method (Guella et al., 2014) was used for all SMRI Array samples that were heterozygous for the rs12476147 exonic SNP, and normalized to the pooled gDNA assay (Guella et al., 2014). This sensitive method is a specific assay for detection of cis-regulatory effects of variants. The ASE assay detected a significant over-expression of the rs12476147 A allele in DLPFC (average 1.12, p-value < 0.0002, 95% confidence interval 1.07–1.17) (Fig. 1) for all 47 subjects pooled. Moreover, cDNA allele ratios were significantly different according to the intronic rs1344706 genotypes (p-value = 0.014), with the SZ risk A allele associated with increased ZNF804A expression (average 1.17, p-value < 0.00002, 95% confidence interval 1.11–1.24) for 27 heterozygous subjects vs. genomic DNA. Further there is a significant difference in cDNA allele ratios between rs1344706 SNP homozygotes (n = 17) and heterozygotes (n = 27) for the expressed A/T allele ratio (p-value = 0.014). This latter result shows that the in phase risk allele for homozygous subjects gives a balanced allelic expression, while heterozygous subjects show an imbalanced expression.
Fig. 1.
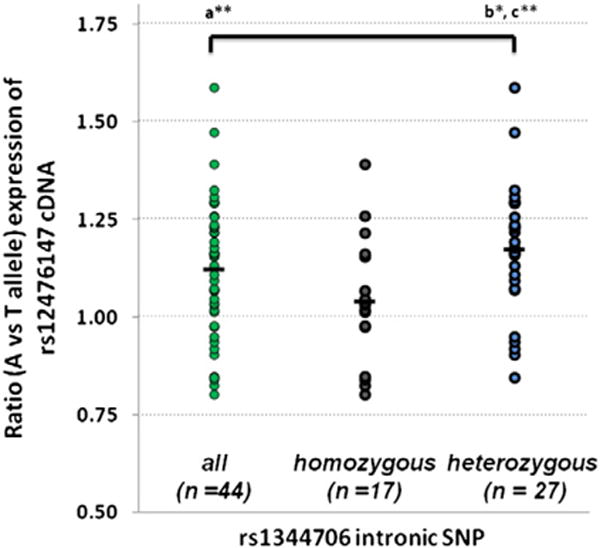
ZNF804A allelic specific expression in DLPFC. Individual data points represent the A/T allele ratio at the expressed rs12476147 SNP. The cDNA ratios for all subjects, and cDNA ratios for the separate groups, rs1344706 homozygotes and rs1344706 heterozygotes, are shown. The mean value for each distribution is also indicated by a horizontal bar. cDNA allele ratios are significantly higher than genomic DNA allele ratios (Guella et al., 2014) (a**, p-value < 0.0002) while there is a significant difference in cDNA allele ratios between rs1344706 SNP homozygotes (n = 17) and heterozygotes (n = 27) for the expressed A/T allele ratio (b*, p-value = 0.014). The heterozygous risk allele showed the highest expression of A/T rs1344706 (c**, p < 0.00002).
Prior studies examined the potential effect of rs1344706 on ASE among SNPs in high LD with rs1344706 (rs12476147 and rs4667001) in the postmortem cortical brain of non-mentally ill controls (Williams et al., 2011; Hill and Bray, 2012). These studies did not find a direct effect of rs1344706 genotype on ZNF804A allelic expression, which may lead one to erroneously conclude that the observed allelic expression imbalance was not directly attributable to rs1344706, at least in the adult brain. However, in agreement with our results, Riley et al. (2010) showed significant association of the rs1344706 risk A-allele with higher total ZNF804A expression in the adult DLPFC of SMRI controls (Riley et al., 2010), whereas the opposite effect has recently been described in SMRI SZ patients (Schultz et al., 2013) where ZNF804A gene expression was found to be significantly lower with an increasing number of risk A-alleles in the prefrontal cortex of patients. Thus, the ZNF804A expression profile appears to be different between SZ patients and healthy controls. One explanation might be differences in epistatic interactions resulting from different genetic backgrounds between patients and healthy controls. Genetic pleiotropism has been previously reported in human complex diseases, including psychiatric diseases (Consortium, 2013) which might also explain some differences between studies of different subjects. Further, some studies do not report actual allele specific expression.
Buonocore et al. (2010) indicated no substantial allele imbalance observed for rs12476147 in the DLPFC of adult postmortem brain when considering only eight subjects (Buonocore et al., 2010). When Buonocore et al. (2010) considered all of the individual brain regions in a pooled analysis, they did find over-expression of A/T ratio in their study, and this effect was strongest in the hippocampus, nucleus accumbens, temporal cortex, parietal cortex, amygdala, hippocampus, and caudate regions. Thus, their results could be compatible with our present results, except they did not find evidence of A/T over-expression in the DLPFC, while with larger sample sizes, and using a sensitive TaqMan assay to detect allelic expression imbalance in our two studies, we are able to replicate the ZNF804A allelic expression data from our first study (Guella et al., 2014).
We also cannot presently exclude that there will be trans-regulatory effects that could either modify the cis-regulatory ZNF804A variant, or act independently of the rs1344706 polymorphism, such as a miRNA that can bind at ZNF804A or an effector of ZNF804A transcription, and alter the observed allelic specific expression pattern in adult DLPFC during development. Furthermore, it is noteworthy that, in contrast with our data, rs1344706 SZ risk allele was significantly associated with a reduced ZNF804A allelic expression in the second-trimester fetal brain tissue (Hill and Bray, 2012), a discrepancy which might be explained by the reversal of fetal expression trajectories (Colantuoni et al., 2011). The findings of Hill and Bray (2012) suggested that rs1344706 risk-allele contributes to decreased ZNF804A expression in the second-trimester fetal postmortem brains (Hill and Bray, 2012); an effect which might also be explained by a strong acting trans-regulatory effector. It is possible that the evidence provided in our study of adult DLPFC could suggest alternative modes of rs1344706 activity in the adult brain compared to these fetal brain results. The actions of cis- and trans-regulatory effects have been suggested before for complex traits, and it has been thought, perhaps due to limited evidence, that cis-regulatory variants are stronger in effect size than trans-regulators, while easier to consider from a computational standpoint. For example, the number of possible trans-regulatory effects across the genome acting at a single locus is enormous, and the corrections needed are substantial, so most researchers have avoided these complications, and settled upon a ‘limited’ analysis of cis-regulatory effects. For the field of neuropsychiatry, analysis of both cis-acting and trans-acting regulators is recommended to fully understand genetic signals acting across the life span. At this point, the effects of trans-regulators may need consideration, and might have been entirely missed, to have a more complete understanding and integration of results of expression of ZNF804A during development and across brain regions.
In conclusion, with a powerful ASE assay we are able to replicate our previous findings of the rs1344706 risk allele being associated with allelic imbalance of ZNF804A (Guella et al., 2014). However, the existing evidence for ZNF804A allelic gene expression in the adult and fetal human brain is still conflicting, and other SNPs that are in strong LD in this region, or trans-regulatory variants could theoretically give rise to the pattern of over-expression observed. The present data when combined with our prior results of DLPFC includes 91 heterozygous subjects, and both studies show similar findings. Replication of these results in other brain regions and age groups in a large sample size with sensitive methodology is required to determine whether this cis-acting variant confers risk through the cis-regulatory variant shown in our adult data. The significant allelic expression imbalance of ZNF804A in the DLPFC (Guella et al., 2014) provides functional legitimacy for rs1344706 as the cis-regulatory variant directly responsible for this allelic expression imbalance.
Acknowledgments
RNA from postmortem brain tissue was donated by The Stanley Medical Research Institute Brain Collection.
Role of funding source
The authors have no competing funding sources to disclose for this research.
Footnotes
Contributors
IG designed and carried out the laboratory experiments, analyzed the data, interpreted the results and wrote the paper. MPV participated in the conception of the study, interpretation of the results, in writing the manuscript, and supervised the entire study.
Conflict of interest
The authors declare no conflict of interest.
All authors read and approved the final manuscript.
References
- Buonocore F, Hill MJ, Campbell CD, Oladimeji PB, Jeffries AR, Troakes C, Hortobagyi T, Williams BP, Cooper JD, Bray NJ. Effects of cis-regulatory variation differ across regions of the adult human brain. Hum Mol Genet. 2010;19(22):4490–4496. doi: 10.1093/hmg/ddq380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, C.-D.G.o.t.P.G. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgenti MJ, LoTurco JJ, Maher BJ. ZNF804a regulates expression of the schizophrenia-associated genes PRSS16, COMT, PDE4B, and DRD2. PLoS One. 2012;7(2):e32404. doi: 10.1371/journal.pone.0032404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guella I, Sequeira A, Rollins B, Morgan L, Myers RM, Watson SJ, Akil H, Bunney WE, DeLisi LE, Byerley W, Vawter MP. Evidence of allelic imbalance in the schizophrenia susceptibility gene ZNF804A in human dorsolateral prefrontal cortex. Schizophr Res. 2014;152(1):111–116. doi: 10.1016/j.schres.2013.11.021. http://dx.doi.org/10.1016/j.schres.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ, Bray NJ. Evidence that schizophrenia risk variation in the ZNF804A gene exerts its effects during fetal brain development. Am J Psychiatry. 2012;169(12):1301–1308. doi: 10.1176/appi.ajp.2012.11121845. [DOI] [PubMed] [Google Scholar]
- Hill MJ, Jeffries AR, Dobson RJ, Price J, Bray NJ. Knockdown of the psychosis susceptibility gene ZNF804A alters expression of genes involved in cell adhesion. Hum Mol Genet. 2012;21(5):1018–1024. doi: 10.1093/hmg/ddr532. [DOI] [PubMed] [Google Scholar]
- Kim AH, Parker EK, Williamson V, McMichael GO, Fanous AH, Vladimirov VI. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophr Res. 2012;141(1):60–64. doi: 10.1016/j.schres.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MV, Rollins B, Sequeira PA, Mesen A, Byerley W, Stein R, Moon EA, Akil H, Jones EG, Watson SJ, Barchas J, DeLisi LE, Myers RM, Schatzberg A, Bunney WE, Vawter MP. Exon expression in lymphoblastoid cell lines from subjects with schizophrenia before and after glucose deprivation. BMC Med Genomics. 2009;2:62. doi: 10.1186/1755-8794-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40(9):1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Okada T, Hashimoto R, Yamamori H, Umeda-Yano S, Yasuda Y, Ohi K, Fukumoto M, Ikemoto K, Kunii Y, Tomita H, Ito A, Takeda M. Expression analysis of a novel mRNA variant of the schizophrenia risk gene ZNF804A. Schizophr Res. 2012;141(2–3):277–278. doi: 10.1016/j.schres.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF, Sanders AR, Purcell S, Visscher PM, Craddock N, Owen MJ, Holmans P, O’Donovan MC. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012;17(2):193–201. doi: 10.1038/mp.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO, Fanous AH, Vladimirov V, O’Neill FA, Walsh D, Kendler KS. Replication of association between schizophrenia and ZNF804A in the Irish Case–Control Study of Schizophrenia sample. Mol Psychiatry. 2010;15(1):29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CC, Nenadic I, Riley B, Vladimirov VI, Wagner G, Koch K, Schachtzabel C, Muhleisen TW, Basmanav B, Nothen MM, Deufel T, Kiehntopf M, Rietschel M, Reichenbach JR, Cichon S, Schlosser RG, Sauer H. ZNF804A and cortical structure in schizophrenia: in vivo and postmortem studies. Schizophr Bull. 2013 doi: 10.1093/schbul/sbt123. http://dx.doi.org/10.1093/schbul/sbt123 (First published online: September 27, 2013) [DOI] [PMC free article] [PubMed]
- Umeda-Yano S, Hashimoto R, Yamamori H, Okada T, Yasuda Y, Ohi K, Fukumoto M, Ito A, Takeda M. The regulation of gene expression involved in TGF-beta signaling by ZNF804A, a risk gene for schizophrenia. Schizophr Res. 2013;146(1–3):273–278. doi: 10.1016/j.schres.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Atz ME, Rollins BL, Cooper-Casey KM, Shao L, Byerley WF. Genome scans and gene expression microarrays converge to identify gene regulatory loci relevant in schizophrenia. Hum Genet. 2006;119(5):558–570. doi: 10.1007/s00439-006-0172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawter MP, Mamdani F, Macciardi F. An integrative functional genomics approach for discovering biomarkers in schizophrenia. Brief Funct Genomics. 2011;10(6):387–399. doi: 10.1093/bfgp/elr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L, Georgieva L, Williams NM, Morris DW, Quinn EM, Giegling I, Ikeda M, Wood J, Lencz T, Hultman C, Lichtenstein P, Thiselton D, Maher BS, Malhotra AK, Riley B, Kendler KS, Gill M, Sullivan P, Sklar P, Purcell S, Nimgaonkar VL, Kirov G, Holmans P, Corvin A, Rujescu D, Craddock N, Owen MJ, O’Donovan MC. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry. 2011;16(4):429–441. doi: 10.1038/mp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C, Turner JA, Calhoun VD, Perrone-Bizzozero N. Potential Impact of miR-137 and its targets in schizophrenia. Front Genet. 2013;4:58. doi: 10.3389/fgene.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Yan JD, Valenzuela RK, Lu SM, Du XY, Zhong B, Ren J, Zhao SH, Gao CG, Wang L, Guo TW, Ma J. Further evidence for the association of genetic variants of ZNF804A with schizophrenia and a meta-analysis for genome-wide significance variant rs1344706. Schizophr Res. 2012;141(1):40–47. doi: 10.1016/j.schres.2012.07.013. [DOI] [PubMed] [Google Scholar]


