Abstract
Basal cells (BC) are the stem/progenitor cells of the human airway epithelium capable of differentiating into secretory and ciliated cells. Notch signaling activation increases BC differentiation into secretory cells, but the role of individual Notch ligands in regulating this process in the human airway epithelium is largely unknown. The objective of this study was to define the role of the Notch ligand JAG1 in regulating human BC differentiation. JAG1 over-expression in BC increased secretory cell differentiation, with no effect on ciliated cell differentiation. Conversely, knockdown of JAG1 decreased expression of secretory cell genes. These data demonstrate JAG1-mediated Notch signaling regulates differentiation of BC into secretory cells.
Keywords: Notch signaling, JAG1, differentiation, human airway epithelium, basal stem/progenitor cells
Introduction
The human airway epithelium, a complex tissue composed of basal, ciliated and secretory cells, provides barrier defense and protection against pathogens and inhaled particulates (1–3). Basal cells (BC) are located throughout the conducting airways and function as the stem/progenitor population of the human airway epithelium capable of differentiating into the other specialized epithelial cell types during turnover and repair (4–11). This is in contrast to the murine airway epithelium whereby BC are restricted to the trachea and absent from more distal regions where other cell types function as stem/progenitors (8,10). The Notch signaling pathway regulates a wide variety of cellular processes, including stem cell renewal and differentiation (12–15). Activation of the Notch signaling cascade is a multi-step process involving binding of one of five plasma membrane bound Notch ligands (DLL1, DLL3, DLL4, JAG1 and JAG2) to one of four Notch receptors (NOTCH1-4) on the plasma membrane of a neighboring cell. Upon ligand binding, the receptor is cleaved at the intracellular transmembrane region, resulting in release of the Notch intracellular domain (NICD) which translocates to the nucleus and, in conjunction with RBPJ, functions as a transcriptional regulator of multiple target genes (16–18). Murine studies have demonstrated that during development and in the adult lung, Notch signaling regulates differentiation of the airway epithelium into the secretory, Clara (also known as club cells), ciliated and neuroendocrine cell types (19–37). In addition, there is increasing evidence that Notch signaling plays a central role in regulating the stem/progenitor capacity of both human and murine BC to differentiate into specific cell types, with specific Notch receptor activity regulating the balance of secretory and ciliated cell differentiation (24,27–30,32,38,39).
The present study was designed to understand the role of individual Notch ligands in regulating human BC differentiation into a mucociliated epithelium. The data demonstrate that JAG1 expression is high in human BC and that modulation of its expression levels during differentiation of BC plays an important role in regulating secretory cell differentiation with no effect on ciliated cell differentiation. These observations have implications for developing novel targets to specifically modulate levels of secretory cells in human airway disorders.
Methods
Sampling the Airway Epithelium and Isolation of Primary Human Airway Basal Cells
Using Institutional Review Board-approved clinical protocols and after obtaining written informed consent, flexible bronchoscopy was performed to collect large airway epithelial cells by brushing the epithelium of healthy nonsmokers (5). Pure populations of basal cells (BC) were obtained and characterized as previously described (5). Both primary and immortalized (BCi-NS1.1 cells) BC were maintained in Bronchial Epithelial Growth Media (BEGM, Lonza, Walkersville, MD) and passaged by seeding at a cell density of 3000 cells/cm2 in BEGM (40).
RNA Sequencing Gene Expression
Nonsmoker primary BC (n=5, at passage 0) and BCi-NS1.1 cells (n=3) were assessed using RNA sequencing on the Illumina HiSeq2500 following TruSeq v2 mRNA library prep. The raw data are publically available at the Gene Expression Omnibus (GEO) site (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE76421. Paired-end reads were processed with STAR (version 2.3.1z13_r470) (41) for alignment to the GRCh37/hg19 human reference genome and RefSeq gene definitions (2014-06-02). Gene expression quantification was performed using Cufflinks (version 2.2) with conversion of aligned reads into fragments per kilobase of exon per million fragments sequenced (FPKM) using the same RefSeq gene definitions to correct for transcript length and coverage depth. The expression level of Notch ligands was assessed with expression characterized as FPKM ≥0.04 in every sample.
Western Analysis
Western analysis was performed usingNuPAGE 4 to 12% Bis-Tris gradient gels (Invitrogen, Carlsbad, CA) (40). The primary antibodies used were JAG1 (1/2000, SC-6011) and GAPDH (1/5000, SC-32233; both from Santa Cruz Biotechnology, Santa Cruz, CA).
TaqMan PCR
The expression of specific genes was assessed using TaqMan quantitative PCR and relative expression levels determined using the dCt method with 18S ribosomal RNA (Cat. No: 4308329, ThermoFisher Scientific, Waltham, MA) as the endogenous control (39). Premade gene assays were obtained from ThermoFisher Scientific: JAG1 (Hs01070032_m1); HES1 (Hs00172878_m1); HES2 (Hs01021800_g1); HES4 (Hs00368353_g1); HES5 (Hs01387463_g1); HES6 (Hs00936587_g1); HEY1 (Hs01114113_m1); HEY2 (Hs00232622_m1); HEYL (Hs00232718_m1); KRT5 (Hs00361185_m1); TP63 (Hs00978343_m1); MUC5AC (Hs01365616_m1); SCGB1A1 (Hs00171092_m1); DNAI1 (Hs00201755_m1); and TEKT1 (Hs00364985_m1).
Air-liquid Interface Culture
To investigate the differentiation of BC, primary (passage 1) and immortalized BC were grown on air-liquid interface (ALI) cultures (40). For histological analysis ALI day 28 trans-well inserts were fixed for paraffin embedding and sectioning (performed by Histoserv, Germantown, MD). For general histology, sections were stained using standard protocols for hematoxylin and eosin (H&E) or Alcian blue (39). For quantification of differentiation at the histological level via Alcian blue or immunofluorescence staining using cell type specific antibodies, a minimum of 10 images equally distributed between both ends of the sectioned membrane were acquired and a minimum of 500 total cells counted for each individual experiment.
Immunofluorescence
Immunofluorescence staining was performed on paraffin embedded cross-sections or direct top-staining of the ALI membrane (39). The following primary antibodies were used: JAG1 (5 µg/ml; SC-6011; Santa Cruz Biotechnology); β-tubulin IV (ciliated cell; 5 µg/ml; MU178-UC; Biogenex, Fremont, CA); KRT5 (2 µg/ml; PA1-37974; ThermoFisher Scientific); and SCGB1A1 (5 µg/ml; RD181022220; BioVendor LLC, Candler, NC). Isotype matched IgG (Jackson Immunoresearch Laboratories, West Grove, PA) was the negative control. To visualize the antibody binding, Alexa Fluor® 555 Donkey Anti-Goat IgG (A-21432; Invitrogen) and Alexa Fluor® 546 Goat Anti-Rabbit IgG (A-11035; Invitrogen) labeled secondary antibodies were used. The cells were counterstained with DAPI to identify cell nuclei.
Generation of Lentiviruses
For over-expression of JAG1, the full-length cDNA sequence was PCR amplified using specific primers and cloned into the multiple cloning site of pCDH-MSCV-MCS-EF1α-GFP (CD711B-1, System Biosciences, Mountain View, CA) via EcoRI and BamHI restriction sites. Knockdown of JAG1 expression was performed using the GIPz lentiviral plasmid expressing GFP and either control scrambled or JAG1 specific shRNAmir (shRNA1 RHS4430-99294390 and shRNA2 RHS4430-101073041, GE Dharmacon, Lafayette, CO). For expression of Cherry-Picker, the pLVX-CherryPicker Control plasmid was used (632580, Clontech Laboratories, Mountain View, CA). Recombinant replication deficient lentiviruses pseudotyped with the VSVg envelope were generated in 293A cells and titered as previously described (39).
BCi-NS1.1 cells were infected with recombinant lentiviruses at an equal multiplicity of infection (MOI=5) overnight in BEGM supplemented with 2 µg/ml of polybrene. The following day the infectious media was removed and the cells expanded in BEGM to generate sufficient numbers before culturing on ALI as described above.
Isolation of CherryPicker-Positive Cells
For isolation of CherryPicker expressing cells, the CherryPicker Assay Kit was used (632570, Clontech Laboratories). ALI day 28 co-cultures were trypsinized for 6 min to remove the cells from the trans-well insert and generate single cell suspensions. Following neutralization, the cells were centrifuged in a 15 ml tube (23°C, 8 min, 250 × g) and washed once with PBS. The cells (5 × 105) were then incubated with 10 µl of CherryPicker antibody (0.5 mg/ml) in a 1.5 ml tube for 30 min, 23°C with gentle inversion to mix the cells every 10 min. The cells were then washed twice with PBS (23°C, 8 min, 250 × g following each wash) and resuspended in 1000µl of Wash Buffer (1X) provided in the kit. Next, pre-washed magnetic beads (20 µl per 5 × 105 cells) were added to each cell suspension followed by incubation for 30 min, 23°C with rotation. Once completed, the tubes were placed on the magnetic stand until the cells were pulled to the wall of the tube to allow removal of the supernatant. The cells were then resuspended with 1000 µl of Wash Buffer (1X) and incubated for 5 min, 23°C with rotation followed by application to the magnetic stand to repeat the wash process. The cells were washed a total of five times and then processed to isolate RNA and generate cyto-preparations to confirm purity.
Statistical Analysis
Statistical comparisons were calculated using an unpaired, 2-tailed Student’s t test with unequal variance. A p value <0.05 was considered significant.
Results
Expression of Notch Signaling Pathway Ligands in Human Airway Basal Cells
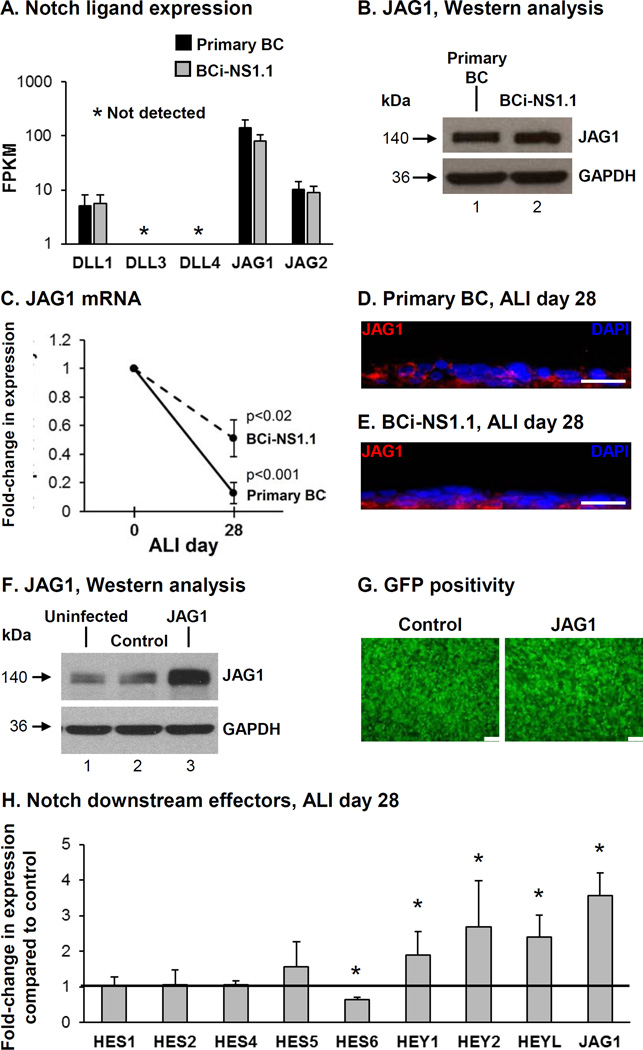
Expression of each Notch ligand (DLL1, DLL3, DLL4, JAG1 and JAG2) was analyzed by RNA sequencing in primary BC and hTERT immortalized BC (BCi-NS1.1 cells) (40). Both primary BC and the BCi-NS1.1 cells expressed the Notch ligands DLL1, JAG1 and JAG2 at comparable levels (Figure 1A). The ligands DLL3 and DLL4 were not expressed. In both primary BC and the BCi-NS1.1 cells, JAG1 was the highest expressed ligand. Western analysis demonstrated equal levels of JAG1 protein in primary BC and BCi-NS1.1 cells (Figure 1B). Based on the observation that JAG1 was the highest expressed Notch ligand in BC we focused our study on investigating its role in regulating BC differentiation. Analysis of JAG1 expression levels during differentiation of BC on air-liquid interface (ALI) culture demonstrated a significant decrease in expression at ALI day 28 compared to day 0 for both primary BC (0.1-fold, p<0.001) and BCi-NS1.1 cells (0.5-fold, p<0.02; Figure 1C). Immunofluorescence staining of JAG1 protein at ALI day 28 for both primary and immortalized BC demonstrated strong staining of JAG1 in BC and reduced staining in the luminal differentiated cell populations (Figure 1D, E). Together, these data suggest that expression of JAG1 is enriched in BC and there is down-regulation of JAG1 expression in differentiated cells. Based on the knowledge that BCi-NS1.1 cells retain the stem/progenitor phenotype of primary BC and have comparable JAG1 expression levels and kinetics, we used BCi-NS1.1 cells as a model to further study the role of JAG1 in regulating Notch dependent differentiation of BC into a mucociliated epithelium.
Figure 1.
Expression of the Notch ligand JAG1 in human airway basal cells (BC). A. RNAseq analysis of Notch ligand (DLL1, DLL3, DLL4, JAG1 and JAG2) expression in primary nonsmoker airway BC and immortalized BC (BCi-NS1.1 cells). Data shown represents the average FPKM expression from n=5 independent primary BC samples (black bars) and n=3 independent passages of BCi-NS1.1 cells (grey bars). Error bars indicate standard deviation. B. Western analysis for JAG1. Lane 1 – Primary BC; and lane 2 – BCi-NS1.1 cells. GAPDH was used as a loading control. C. mRNA expression of JAG1 during BC differentiation on air-liquid interface (ALI) culture. TaqMan PCR analysis to assess expression of JAG1 at ALI day 0 and day 28 for primary BC (solid line) and BCi-NS1.1 cells (dashed line). Lines indicate the mean fold-change of mRNA expression compared to day 0 ALI cells from n=5 independent experiments, each performed in triplicate. Error bars indicate standard deviation. D-E. Immunofluorescence assessment of JAG1 (red) and DAPI (nuclei, blue) at ALI Day 28. D. Primary BC and E. BCi-NS1.1 cells. Scale bar 20 µm. F-H. Over-expression of JAG1 increases Notch signaling activity. BCi- NS1.1 cells were infected with lentivirus expressing GFP (control) or GFP and JAG1 (JAG1) and subsequently cultured on ALI culture for 28 days to assess the role of JAG1. F. Western analysis for JAG1. Lane 1 – Uninfected cells; lane 2 – Control cells and lane 3 – JAG1 cells. GAPDH was used as a loading control. G. Efficiency of lentivirus infection (GFP positivity) at ALI day 0. Scale bar 100 µm. H. Over-expression of JAG1 modulates Notch activation and expression of Notch downstream effectors. TaqMan PCR analysis to assess mRNA expression for JAG1 and Notch pathway downstream effectors (HES1, HES2, HES4, HES5, HES6, HEY1, HEY2, and HEYL) at ALI day 28. Bars indicate the mean fold-change of mRNA expression compared to control cells from n=4 independent experiments, each performed in triplicate. Error bars indicate standard deviation. Asterisks (*) indicate p<0.05.
Modulation of JAG1 Expression Levels during Basal Cell Differentiation
To investigate the effect of modulating JAG1 expression levels on BC differentiation, BCi-NS1.1 cells were infected with either lentivirus expressing GFP (control) or lentivirus expressing GFP and JAG1 (JAG1) for subsequent culture on ALI. To verify over-expression of JAG1, Western analysis was performed on cell lysates of uninfected BCi-NS1.1 cells or cells infected with control or JAG1 lentivirus (Figure 1F). Equal infectivity of each virus was confirmed by GFP positivity of the cultures (Figure 1G). At ALI day 28, the cells were harvested to confirm that over-expression of JAG1 mRNA was maintained in cells infected with JAG1 lentivirus vs control (3.6-fold, p<0.005; Figure 1H). Based on the knowledge that activation of Notch signaling results in multiple downstream responses in human airway BC (38,39), we analyzed the changes in expression of multiple Notch downstream effectors in differentiating BC following over-expression of JAG1. The results demonstrated no significant changes in expression of HES1 (1.1-fold), HES2 (1.1-fold), HES4 (1.1-fold) and HES5 (1.6-fold) in cells over-expressing JAG1 vs control (all p>0.2, Figure 1H). However, there were significant changes in expression of HES6 (0.6-fold), HEY1 (1.9-fold), HEY2 (2.7-fold) and HEYL (2.4-fold) demonstrating increased Notch signaling activity in response to JAG1 over-expression (all p<0.05; Figure 1H).
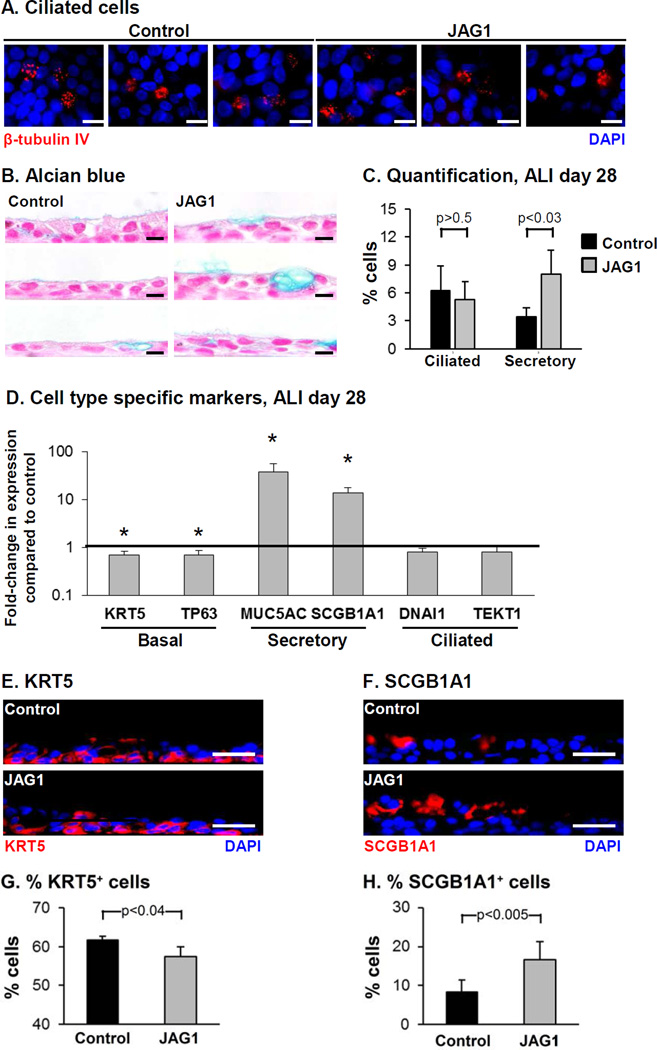
The effects of JAG1 over-expression on the differentiation capacity of BC into ciliated cells and secretory cells was assessed at the histological level by immunofluorescence staining of ciliated cells and Alcian blue staining for mucus producing secretory cells (Figure 2A, B). Both control and JAG1 over-expressing cells differentiated into a mucociliated epithelium, however over-expression of JAG1 resulted in a significant increase in the number of secretory cells (3.4% control vs 8.0% JAG1, p<0.03) with no significant effect on the number of ciliated cells (6.3% control vs 5.3% JAG1, p>0.5, Figure 2C). To further characterize the effects of JAG1 overexpression on BC differentiation, the expression of cell type-specific markers was analyzed (Figure 2D). Relative to control cells, JAG1 over-expression resulted in a significant decrease in expression of the BC markers KRT5 and TP63 (both 0.7-fold; p<0.05). Immunofluorescence staining of KRT5 demonstrated a significant decrease in the number of KRT5 positive cells in response to JAG1 over-expression (61.7% control vs 57.4% JAG1, p<0.04) consistent with the mRNA data (Figure 2D, E, G). Similarly, consistent with the Alcian blue staining, JAG1 overexpression resulted in a significant increase in expression of the mucus producing secretory cell marker MUC5AC (37.4-fold, p<0.05). In addition, significant increased expression of the nonmucus producing secretory cell marker SCGB1A1 was also observed in JAG1 over-expressing cells (13.7-fold, p<0.05) which was further validated at the histological level with increased numbers of SCGB1A1 positive cells (8.2% control vs 16.6% JAG1, p<0.005; Figure 2D, F, H). No significant change in expression of the ciliated cell markers DNAI1 and TEKT1 (both 0.8- fold, both p>0.1) was observed, further confirming the histological data (Figure 2A, C). Together, these data demonstrate that JAG1-dependent Notch activation specifically promotes differentiation of BC into secretory cells.
Figure 2.
Over-expression of JAG1 modulates Notch signaling activity and increases secretory cell differentiation. BCi-NS1.1 cells were infected with lentivirus expressing GFP (control) or GFP and JAG1 (JAG1) and subsequently cultured on air-liquid interface (ALI) culture for 28 days to assess the role of JAG1 on BC differentiation. A. Immunofluorescence staining of β- tubulin IV-positive ciliated cells. β-tubulin IV (ciliated cell, red) and DAPI (nuclei, blue). B. Alcian blue staining. C. Quantification of ciliated cells and Alcian blue positive secretory cells. The data are the mean for n=4 independent experiments; error bars indicate standard deviation. A, B. Scale bar 10 µm. D. Over-expression of JAG1 increases expression of secretory cell genes. TaqMan PCR analysis to assess mRNA expression of basal cell markers (KRT5 and TP63); secretory cell markers (MUC5AC and SCGB1A1) and ciliated cell markers (DNAI1 and TEKT1) at ALI day 28. Bars indicate the mean fold-change of mRNA expression compared to control cells from n=4 independent experiments, each performed in triplicate. Error bars indicate standard deviation. Asterisks (*) indicate p<0.05. E-H. Sections of ALI day 28 membranes for both control and JAG1 cells were stained with cell type-specific markers to quantify differentiation. E. Immunofluorescence staining of KRT5 positive cells. KRT5 (basal cell, red) and DAPI (nuclei, blue). F. Immunofluorescence staining of SCGB1A1 positive cells. SCGB1A1 (secretory cell, red) and DAPI (nuclei, blue). G. Quantification of KRT5 positive cells. H. Quantification of SCGB1A1 positive cells. The data for G and H are the mean for n=4 independent experiments; error bars indicate standard deviation. E, F. Scale bar 20 µm.
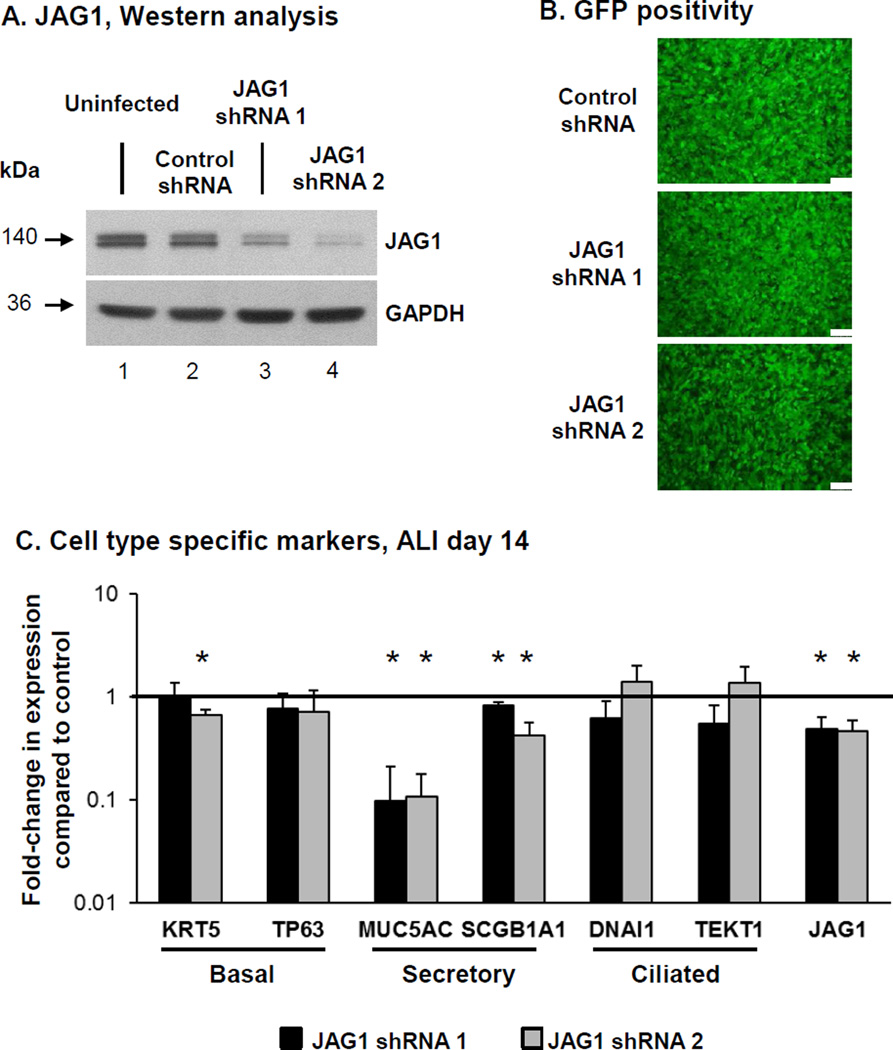
To further confirm the role of JAG1 mediated Notch signaling on BC differentiation, we assessed the effect of JAG1 knockdown. BCi-NS1.1 cells were infected with either lentivirus expressing GFP and scrambled shRNA (control shRNA) or lentivirus expressing GFP and two different JAG1 specific shRNA (JAG1 shRNA 1 and 2), and the modified cells then cultured on ALI. Knockdown of JAG1 at the protein levels for both JAG1 specific shRNA was confirmed by Western analysis (Figure 3A) and equal infectivity of each virus was confirmed by GFP positivity of the cultures (Figure 3B). Due to loss of shRNA-mediated JAG1 silencing following long term ALI culture (data not shown), the cells were harvested at ALI day 14 to assess the impact of JAG1 knockdown on BC differentiation. TaqMan analysis confirmed knockdown of JAG1 at the mRNA level was maintained at ALI day14 in cells infected with both JAG1 specific shRNA vs control shRNA lentivirus (both 0.5-fold, p<0.05; Figure 3C). To further characterize the effects of JAG1 knockdown on BC differentiation, the expression of cell type-specific markers was analyzed (Figure 3C). Relative to control shRNA cells, JAG1 knockdown with both shRNA resulted in a significant decrease in expression of the secretory cell markers MUC5AC (0.1-fold, p<0.006 for both shRNA) and SCGB1A1 (0.8-fold shRNA 1 and 0.4-fold shRNA 2; both p<0.05). Apart from a significant decrease in expression of the BC marker KRT5 (0.7-fold, p<0.03) in JAG1 shRNA 2 expressing cells relative to control, no significant (all p>0.1) difference was observed in expression of the BC marker TP63 (0.8-fold shRNA 1 and 0.7-fold shRNA 2) and ciliated cell markers DNAI1 (0.6-fold shRNA 1 and 1.4-fold shRNA 2) and TEKT1 (0.6-fold shRNA 1 and 1.4-fold shRNA 2, Figure 3C). Overall, these data demonstrate that JAG1-dependent Notch signaling specifically regulates differentiation of BC into secretory cells with no significant effect on differentiation into ciliated cells.
Figure 3.
Knockdown of JAG1 expression decreases expression of secretory cell genes. BCi- NS1.1 cells were infected with lentivirus expressing control scrambled shRNAmir and GFP (control shRNA) or two different JAG1 specific shRNAmir and GFP (JAG1 shRNA 1 and 2) and subsequently cultured on air-liquid interface (ALI) culture for 14 days to assess the role of JAG1 on BC differentiation. A. Western analysis for JAG1. Lane 1 – Uninfected cells; lane 2 – Control shRNA cells; lane 3 – JAG1 shRNA 1 cells and lane 4 – JAG1 shRNA 2 cells. GAPDH was used as a loading control. B. Efficiency of lentivirus infection (GFP positivity) at ALI day 0. Scale bar 100 µm. C. Knockdown of JAG1 expression decreases expression of secretory cell genes. TaqMan PCR analysis to assess mRNA expression of basal cell markers (KRT5 and TP63); secretory cell markers (MUC5AC and SCGB1A1), ciliated cell markers (DNAI1 and TEKT1) and JAG1 at ALI day 14. Bars indicate the mean fold-change of mRNA expression compared to control shRNA cells from n=3 independent experiments, each performed in triplicate. Error bars indicate standard deviation. Asterisks (*) indicate p<0.05.
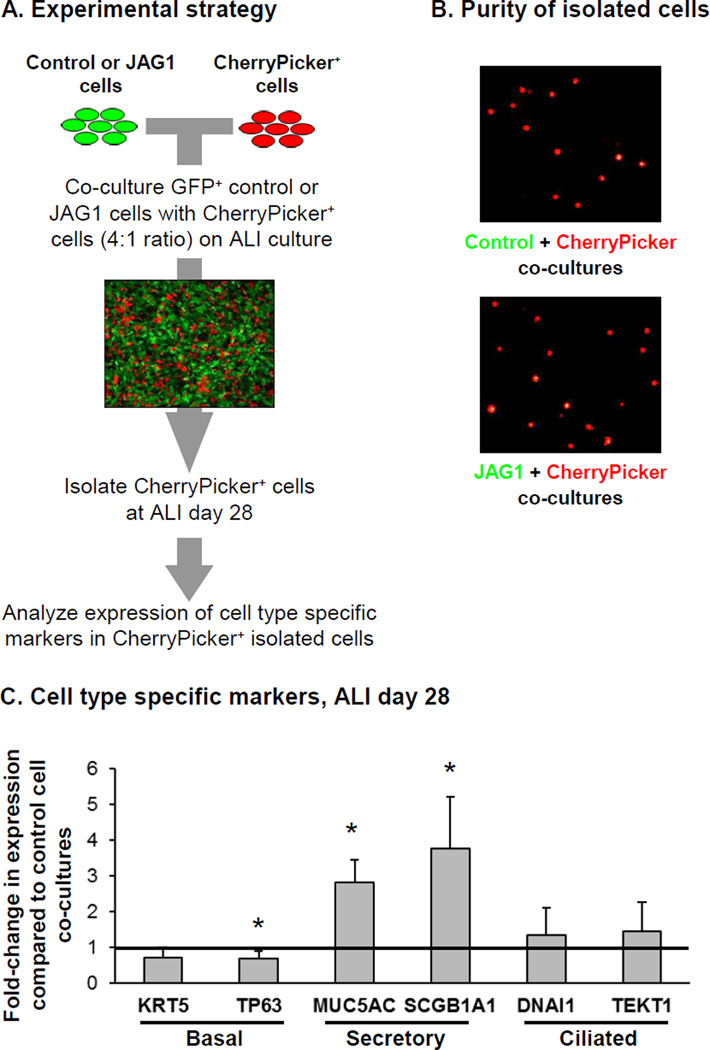
JAG1-Mediated Intercellular Signaling
The Notch signaling pathway is known to act through direct cell to cell interactions, and thus we assessed whether over-expression of JAG1 in BC could activate Notch signaling in neighboring cells and induce secretory cell differentiation. To answer this question BCi-NS1.1 cells were infected with a lentivirus expressing the chimeric membrane-anchored CherryPicker fluorescent protein which could then can be captured on magnetic beads via a specific antibody. CherryPicker+ BCi-NS1.1 cells were subsequently co-cultured on ALI with either GFP+ control or JAG1 over-expressing BCi-NS1.1 cells at a ratio of 4:1 (GFP:mCherry) for 28 days (Figure 4A). At ALI day 28, the cells were harvested and the CherryPicker+ cells isolated by magnetic bead separation from each co-culture with purity of the isolated CherryPicker+ cells confirmed by fluorescent microscopy which demonstrated the presence of only mCherry+ cells and absence of GFP+ cells (Figure 4B). Following isolation, the differentiation of the mCherry+ BC in response to co-culture with control or JAG1 over-expressing cells was assessed by analyzing expression of cell type specific markers. Relative to co-culture with control cells, CherryPicker+ cells co-cultured with JAG1 over-expressing cells had a significant decrease in expression of the BC marker TP63 (0.7-fold, p<0.03) and an increase in expression of the secretory cell markers MUC5AC and SCGB1A1 (2.8-fold and 3.8-fold, respectively; both p<0.01). No significant changes in expression of the BC marker KRT5 (0.7-fold, p>0.08) and ciliated cell markers DNAI1 and TEKT1 (1.3-fold, p>0.4 and 1.4-fold, p>0.3, respectively) were observed (Figure 4C). These data demonstrate that over-expression of JAG1 in BC can activate Notch signaling in neighboring BC and promote differentiation into secretory cells.
Figure 4.
JAG1-mediated Notch signaling promotes secretory cell differentiation on neighboring cells. BCi-NS1.1 cells were infected with lentivirus expressing GFP (control) or GFP and JAG1 (JAG1) and subsequently co-cultured on air-liquid interface (ALI) culture for 28 days with BCi- NS1.1 infected with lentivirus expressing CherryPicker at a ratio of 4:1. Following 28 days of ALI culture, the cells were harvested and CherryPicker positive cells were isolated by magnetic bead separation for subsequent analysis to quantify expression of differentiation related genes. A. Schematic representation of experimental strategy. B. Immunofluorescence analysis of magnetic bead separated cells to demonstrate purity of isolated cells from ALI day 28 co-cultures. Control and JAG1 cells (green) and CherryPicker cells (red). C. TaqMan PCR analysis to assess mRNA expression of basal cell markers (KRT5 and TP63); secretory cell markers (MUC5AC and SCGB1A1) and ciliated cell markers (DNAI1 and TEKT1) at ALI day 28. Bars indicate the mean fold-change of mRNA expression compared to CherryPicker positive cells isolated from Control cell co-cultures from n=4 independent experiments. Error bars indicate standard deviation. Asterisks (*) indicate p<0.05.
Discussion
Basal cells (BC) function as stem/progenitor cells of the human airway epithelium that differentiate into secretory and ciliated cells to replenish the airway epithelium during normal turnover and repair (4–11). Based on the knowledge the Notch signaling pathway is expressed in the human airway epithelium and Notch signaling activity plays a central role in regulating BC differentiation into secretory and ciliated cells (24,27–30,32,38,39,42), the present study was designed to understand the role of individual Notch ligands in regulating BC differentiation into a mucociliated epithelium. The data demonstrate that expression of JAG1 is highly enriched in BC and that its expression decreases in the differentiated cell populations of the mucociliated epithelium. Furthermore, modulation of its expression levels in BC plays an important role in regulating secretory cell differentiation and BC expressed JAG1 can signal to neighboring cells to regulate secretory cell differentiation.
These are the first studies identifying a role for JAG1 in regulating human airway BC differentiation. Recent studies in murine BC have demonstrated that JAG ligands play an important role in regulating differentiation of the airway epithelium (24,27). For example, Mori et al (24) demonstrated JAG1 and 2 ligands regulate differentiation of BC by generating undifferentiated airway progenitors in a NOTCH3-dependent manner that become terminally differentiated into secretory or ciliated cells in response to a secondary Notch activation signal. In addition, Pardo- Saganta et al (27) has shown that BC serve as a niche for their daughter cells by providing a continuous supply of the JAG2 ligand to maintain the secretory cell progenitor pool. Taken together, our data suggests a model whereby JAG1 expression in human airway BC provides a ligand stimulus to activate Notch signaling in other BC to promote secretory cell differentiation, with high levels of JAG1 expression increasing secretory cell differentiation and low levels of JAG1 decreasing secretory cell differentiation. Based on our prior studies demonstrating that constitutive activation of NOTCH1 and NOTCH3 signaling increases secretory cell differentiation of primary basal cells (39) we hypothesize that either one or both of these receptors are engaged by JAG1 on the neighboring cell to increase secretory cell differentiation.
Interestingly, our data demonstrated that modulation of JAG1 expression levels in BC had no impact on ciliated cell differentiation, suggesting that JAG1 specifically regulates secretory cell differentiation of the human airway epithelium. In conjunction with a study demonstrating miRNA-mediated repression of DLL1-NOTCH1 signaling is required for differentiation of human airway epithelial cells into ciliated cells (43), we hypothesize that Notch-dependent differentiation of the human airway into secretory and ciliated cells is regulated by a distinct set of ligands. Treatment of adult mice with specific therapeutic antibodies that block JAG1-mediated Notch signaling decreased the number of SCGB1A1+ secretory cells in the airway epithelium under homeostatic conditions and reversed goblet cell metaplasia in ovalbumin challenged mice (44). Therefore, in conjunction with our data, the blockade of JAG1-mediated Notch signaling may be a viable therapeutic strategy to specifically modulate levels of secretory cells in human airway disorders.
Acknowledgments
We thank N. Mohamed for help in preparing this manuscript. These studies were supported, in part, by R01HL107882; R01HL107882-S; P20 HL113443, and R01HL1189541. KG was supported, in part by a NYSTEM Fellowship. Research reported in this publication was supported by NIH and the Family Smoking Prevention and Tobacco Control Act. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Food and Drug Administration.
Role of sponsor / funder: The funders of this study had no role in the design and conduct of the study; not in the collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Footnotes
Conflict of interest: The authors declare no potential conflict of interest.
References
- 1.Crystal RG, Randell SH, Engelhardt JF, Voynow J, Sunday ME. Airway epithelial cells: current concepts and challenges. Proc. Am Thorac. Soc. 2008;5(7):772–777. doi: 10.1513/pats.200805-041HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knight DA, Holgate ST. The airway epithelium: structural and functional properties in health and disease. Respirology. 2003;8(4):432–446. doi: 10.1046/j.1440-1843.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 3.Tam A, Wadsworth S, Dorscheid D, Man SF, Sin DD. The airway epithelium: more than just a structural barrier. Ther. Adv. Respir Dis. 2011;5(4):255–273. doi: 10.1177/1753465810396539. [DOI] [PubMed] [Google Scholar]
- 4.Evans MJ, Van Winkle LS, Fanucchi MV, Plopper CG. Cellular and molecular characteristics of basal cells in airway epithelium. Exp. Lung Res. 2001;27(5):401–415. doi: 10.1080/019021401300317125. [DOI] [PubMed] [Google Scholar]
- 5.Hackett NR, Shaykhiev R, Walters MS, et al. The human airway epithelial basal cell transcriptome. PLoS. One. 2011;6(5):e18378. doi: 10.1371/journal.pone.0018378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackett TL, Shaheen F, Johnson A, et al. Characterization of side population cells from human airway epithelium. Stem Cells. 2008;26(10):2576–2585. doi: 10.1634/stemcells.2008-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajj R, Baranek T, Le NR, Lesimple P, Puchelle E, Coraux C. Basal cells of the human adult airway surface epithelium retain transit-amplifying cell properties. Stem Cells. 2007;25(1):139–148. doi: 10.1634/stemcells.2006-0288. [DOI] [PubMed] [Google Scholar]
- 8.Hogan BL, Barkauskas CE, Chapman HA, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15(2):123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rock JR, Onaitis MW, Rawlins EL, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad Sci. U.S.A. 2009;106(31):12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 2010;3(9–10):545–556. doi: 10.1242/dmm.006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staudt MR, Buro-Auriemma LJ, Walters MS, et al. Airway Basal stem/progenitor cells have diminished capacity to regenerate airway epithelium in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2014;190(8):955–958. doi: 10.1164/rccm.201406-1167LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: turning over new leaves. Cell. 2007;128(3):445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hori K, Sen A, rtavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126(Pt 10):2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr. Top. Dev. Biol. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 16.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138(17):3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 17.Borggrefe T, Liefke R. Fine-tuning of the intracellular canonical Notch signaling pathway. Cell Cycle. 2012;11(2):264–276. doi: 10.4161/cc.11.2.18995. [DOI] [PubMed] [Google Scholar]
- 18.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194(3):237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 19.Boucherat O, Chakir J, Jeannotte L. The loss of Hoxa5 function promotes Notch-dependent goblet cell metaplasia in lung airways. Biol Open. 2012;1(7):677–691. doi: 10.1242/bio.20121701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guha A, Vasconcelos M, Cai Y, et al. Neuroepithelial body microenvironment is a niche for a distinct subset of Clara-like precursors in the developing airways. Proc. Natl. Acad Sci. U.S.A. 2012;109(31):12592–12597. doi: 10.1073/pnas.1204710109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guseh JS, Bores SA, Stanger BZ, et al. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development. 2009;136(10):1751–1759. doi: 10.1242/dev.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito T, Udaka N, Yazawa T, et al. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. 2000;127(18):3913–3921. doi: 10.1242/dev.127.18.3913. [DOI] [PubMed] [Google Scholar]
- 23.Li A, Chan B, Felix JC, et al. Tissue-dependent consequences of Apc inactivation on proliferation and differentiation of ciliated cell progenitors via Wnt and notch signaling. PLoS. One. 2013;8(4):e62215. doi: 10.1371/journal.pone.0062215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori M, Mahoney JE, Stupnikov MR, et al. Notch3-Jagged signaling controls the pool of undifferentiated airway progenitors. Development. 2015;142(2):258–267. doi: 10.1242/dev.116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morimoto M, Liu Z, Cheng HT, Winters N, Bader D, Kopan R. Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. J Cell Sci. 2010;123(Pt 2):213–224. doi: 10.1242/jcs.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morimoto M, Nishinakamura R, Saga Y, Kopan R. Different assemblies of Notch receptors coordinate the distribution of the major bronchial Clara, ciliated and neuroendocrine cells. Development. 2012;139(23):4365–4373. doi: 10.1242/dev.083840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardo-Saganta A, Tata PR, Law BM, et al. Parent stem cells can serve as niches for their daughter cells. Nature. 2015;523(7562):597–601. doi: 10.1038/nature14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pardo-Saganta A, Law BM, Tata PR, et al. Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell. 2015;16(2):184–197. doi: 10.1016/j.stem.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul MK, Bisht B, Darmawan DO, et al. Dynamic changes in intracellular ROS levels regulate airway basal stem cell homeostasis through Nrf2-dependent Notch signaling. Cell Stem Cell. 2014;15(2):199–214. doi: 10.1016/j.stem.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rock JR, Gao X, Xue Y, Randell SH, Kong YY, Hogan BL. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8(6):639–648. doi: 10.1016/j.stem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan L, Aster JC, Sklar J, Sunday ME. Notch-1 regulates pulmonary neuroendocrine cell differentiation in cell lines and in transgenic mice. Am J Physiol Lung Cell Mol. Physiol. 2007;292(2):L500–L509. doi: 10.1152/ajplung.00052.2006. [DOI] [PubMed] [Google Scholar]
- 32.Tata PR, Pardo-Saganta A, Prabhu M, et al. Airway-specific inducible transgene expression using aerosolized doxycycline. Am J Respir Cell Mol. Biol. 2013;49(6):1048–1056. doi: 10.1165/rcmb.2012-0412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development. 2009;136(13):2297–2307. doi: 10.1242/dev.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsao PN, Wei SC, Wu MF, et al. Notch signaling prevents mucous metaplasia in mouse conducting airways during postnatal development. Development. 2011;138(16):3533–3543. doi: 10.1242/dev.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing Y, Li A, Borok Z, Li C, Minoo P. NOTCH1 is required for regeneration of Clara cells during repair of airway injury. Stem Cells. 2012;30(5):946–955. doi: 10.1002/stem.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu K, Moghal N, Egan SE. Notch signaling in lung development and disease. Adv. Exp Med Biol. 2012;727:89–98. doi: 10.1007/978-1-4614-0899-4_7. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Loch AJ, Radtke F, Egan SE, Xu K. Jagged1 is the major regulator of Notch-dependent cell fate in proximal airways. Dev. Dyn. 2013;242(6):678–686. doi: 10.1002/dvdy.23965. [DOI] [PubMed] [Google Scholar]
- 38.Danahay H, Pessotti AD, Coote J, et al. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep. 2015;10(2):239–252. doi: 10.1016/j.celrep.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 39.Gomi K, Arbelaez V, Crystal RG, Walters MS. Activation of NOTCH1 or NOTCH3 signaling skews human airway basal cell differentiation toward a secretory pathway. PLoS. One. 2015;10(2):e0116507. doi: 10.1371/journal.pone.0116507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walters MS, Gomi K, Ashbridge B, et al. Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respir Res. 2013;14:135. doi: 10.1186/1465-9921-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNAseq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tilley AE, Harvey BG, Heguy A, et al. Down-regulation of the notch pathway in human airway epithelium in association with smoking and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2009;179(6):457–466. doi: 10.1164/rccm.200705-795OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcet B, Chevalier B, Luxardi G, et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat Cell Biol. 2011;13(6):693–699. doi: 10.1038/ncb2241. [DOI] [PubMed] [Google Scholar]
- 44.Lafkas D, Shelton A, Chiu C, et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature. 2015;528(7580):127–131. doi: 10.1038/nature15715. [DOI] [PubMed] [Google Scholar]