Abstract
This manuscript describes the basics of proteomic and metabolic profiling of blood serum and plasma from patients with psychiatric disorders. It will also explain the rationale behind the use of these bodily fluids, due to the need for user-friendly and rapid tests in clinics with simple sampling procedures. It has become evident over the last 15 years or so that psychiatric disorders are whole-body diseases and the bloodstream is a means of molecular transport that therefore provides a conduit for two-way communication with the brain. Here we also describe some of the basic biomarker findings from studies of serum or plasma from patients with psychiatric disorders like schizophrenia, major depression, and bipolar disorder. Finally, we will discuss potential future advancements in this area, which include the development of hand-held devices containing miniature proteomic and metabolic assays which can be used for facilitating diagnoses in a point-of-care setting and yield results in less than 15 minutes from a single drop of blood.
Keywords: biomarker, metabolomics, proteomics, psychiatric disorders, serum
Introduction
The use of blood serum and plasma has increased over more than a decade in molecular profiling studies of psychiatric disorders. The rationale stems from the fact that, although psychiatric disorders appear to be generated in the brain, the effects of these diseases can be seen throughout the whole body. Long-standing scientific and medical research has shown that the brain is integrated in virtually all physiological functions of the body, and this is reflected in the composition of blood proteins and other bioactive molecules. One of the best examples is the well-known fight-or-flight reflex (Laborit, 1976). This begins with a perception of danger, mediated by a region of the brain called the amygdala, which stimulates the release of a hormone called corticotrophin releasing factor (CRF) from the hypothalamus, which, in turn, causes increased secretion of adrenocorticotrophic hormone (ACTH) from the pituitary into the blood. Next, the ACTH circulates throughout the blood and acts on the adrenal glands to release the steroid hormone cortisol. The increased circulating cortisol levels cause an increase in blood pressure as well as a rise in blood glucose concentrations to serve as the fuel for the muscular reactions required in the response. The final stage completes the loop as the elevated cortisol levels exert a negative feedback effect on the hypothalamic-pituitary-adrenal (HPA) axis through down-regulation of glucocorticoid receptors in the brain and pituitary.
But it is not just the effects on the HPA axis which can give an indication of brain function. There are countless physiological circuits of the body that can be monitored by changes in blood molecules, including the response to food intake (Mastorakos and Zapanti, 2004), pregnancy (Smith and Thomson, 1991), the inflammation response (Späth-Schwalbe et al., 1994; Straub et al., 2011), and the presence of metabolic disorders such as insulin resistance and diabetes (Pasquali et al., 2006; Reagan, 2007). In fact, impaired functioning of many hormonal systems of the body, such as the insulin signalling cascade, can contribute to impaired brain functions, including memory and cognitive deficits (Solas et al., 2010). In support of the whole-body concept of psychiatry, recent investigations have shown that molecular signatures for schizophrenia, major depressive disorder, and bipolar disorder can be detected in the circulating blood. Most of these molecules converge on the metabolic and inflammatory pathways, although changes in other molecules representing antioxidant and growth factor pathways have also been found. This is highly useful since blood can be taken from living patients in a minimally invasive way during progressive stages of the disease or following treatment, whereas other tissues, such as those in the brain, are only available for analysis at autopsy.
Clinical Need
There is currently a need for molecular biomarkers in studies of major psychiatric disorders, since these are expected to help improve diagnosis and treatment outcomes. However, research in this field has thus far not lived up to its potential. This is most likely due to the fact that psychiatric diseases are still classified using diagnostic concepts which could be subjective. Furthermore, these disorders are heterogeneous and sometimes overlap in their aetiology and symptoms. The availability of accurate biomarker tests for specific psychiatric disorders could be used to classify at-risk individuals, as well as those with the full-blown illness. Furthermore, the availability of tests that can be used to monitor treatment or even predict treatment response would help to improve patient outcomes. It is important to state that this molecular approach should not be used as a replacement for the current clinical methods, but as an add-on feature. The old ways do work, just not as effectively as expected.
The above movement towards incorporation of molecular tests is in line with the National Institute of Mental Health Research Domain Criteria (RDoC) project, which aims to develop a classification system for mental health disorders that can be linked to the underlying dysfunctional pathways (Cuthbert and Insel, 2013). This is important, since improved understanding of these pathways will lead to more targeted treatments. With this in mind, the RDoC project aims to: (1) integrate multiple scientific disciplines to identify basic behavioral components of psychiatric disorders that can be associated with neurobiological dysfunction; (2) identify features of these components which can be used to define the normal and pathological states; and (3) establish and combine standardized and validated methods to produce molecular, environmental, behavioral, and neurobiological profiles of these disorders for improved classification.
Why Choose Proteomics and Metabolomics To Study Psychiatric Disorders?
Biomarkers are physical characteristics that can be measured and used as an indication of physiological factors such as good health, disease, toxicity or drug response (Biomarkers Definitions Working Group et al., 2001). For practical purposes, it is important that a biomarker can be measured with high accuracy and reproducibility, within a short time frame, and at an affordable cost. Ideally, a biomarker should also reflect in some way the underlying nature of the disorder or condition being measured. In psychiatry, various types of biomarker measurements have emerged, including those derived from brain imaging analyses, psychiatric tests, and electrophysiological responses. This review will focus on proteomic and metabolomic profiles, or “fingerprints,” in blood samples. In other words, we will be describing techniques that use changing patterns of blood molecules as a means of diagnosis. Since changes in physiological states are dynamic in nature, they are likely to introduce alterations in numerous proteins and metabolites that converge on similar pathways. Therefore, it would be most useful to apply proteomic and metabolomic methods, as they can target tens to hundreds of molecules simultaneously, which should result in a more complete fingerprint and therefore a better understanding of the affected pathways. This is like the difference between fishing using specific bait for only one type of fish compared with trawling using a wide net, which can lead to the capture of many different types of fish.
Proteomic and Metabolomic Technologies
Most researchers now consider the study of proteins (proteomics) and metabolites (metabolomics) in a large-scale manner to be among the most informative reflections of biological functions, considering that these molecules actually carry out or respond to most processes of the body. Furthermore, the majority of drugs in use today are designed to turn on or turn off proteins, such as receptors or enzymes, or induce metabolic changes, which can be seen as turnover of various proteins and small molecules. The term “proteome” was coined by Marc Wilkins in 1994 while he was a PhD student working on the concept of the expressed proteins of a genome (Wilkins et al., 1996). But what does proteome actually mean? The proteome is the set of expressed proteins by a cell, tissue, or organism, in a given moment, under a given condition. As a reminder, each cell in an organism contains the same genomes, but only some of these are expressed as proteins, depending on, for example, the type of cell, the time of day, the presence of disease, or responses to drugs or toxins. The word “metabolome” was first used in a 1998 publication by Tweeddale, Notley-McRobb, and Ferenci as the total metabolite pool in a cell, tissue, or organism (Tweeddale et al., 1998). Proteomics and metabolomics are, respectively, the methods that study the proteome and the metabolome.
The rationale for looking at the blood is that most biomarker studies of psychiatric disorders are stymied by the difficulties and the sheer inappropriateness of accessing and studying brain samples from living patients. However, the identification of correlating biomarkers in the periphery is the approach being taken by most researchers today, since these can reflect brain function and because blood-based tests are more amenable to clinical studies. Several psychiatric disorder studies aiming to identify molecular biomarker candidates have now been carried out using multi-protein arrays, mass spectrometry, or nuclear magnetic resonance (NMR) profiling platforms. These techniques enable the simultaneous detection of hundreds of proteins or small molecules, some of which can be altered in the disease state and therefore used as potential biomarker fingerprints.
Proteomic Techniques
Multiplex Immunoassay
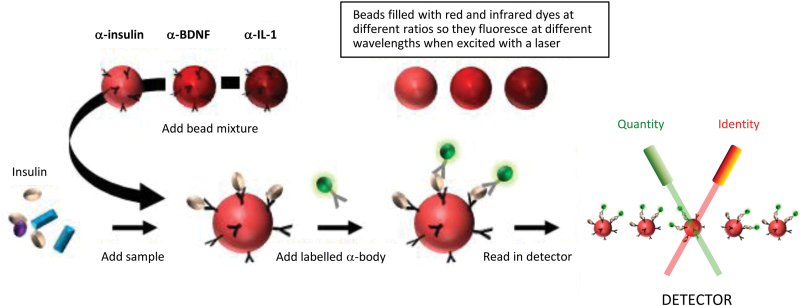
Blood serum and plasma contain many important bioactive or regulatory molecules, such as hormones, growth factors, and cytokines. However, most of these are present only at low concentrations and therefore require detection by high-sensitivity platforms. One of the best methods to achieve this is multiplex immunoassay (Fulton et al., 1997; Figure 1). These assays are constructed and carried out using serum or plasma samples as follows: (1) micro-beads are loaded with different ratios of red and infrared dyes to give unique fluorescent signatures; (2) a capture antibody is attached to the fluorescent bead surface such that each specific antibody is attached to a bead with a specific signature; (3) the sample is added and a target molecule (in this case, Figure 1 shows insulin as the target) binds to antibody-bead conjugate; (4) a fluorescently-labelled detection antibody is added which binds to the target molecule in a sandwich format; and (5) the beads are streamed though a reader and analyzed by two lasers for identification and quantification of the analyte present. For the last step, the lasers can identify which analytes are present by measuring the unique fluorescent signature of each bead and can determine the quantity of analytes present by measuring the amount of the fluorescent tag associated with each bead (proportional to the analyte concentration). In the example shown, it would be possible to determine the concentrations of insulin, brain-derived neurotrophic factor, and interleukin 1 simultaneously, as each antibody-bead conjugate would have a distinct signature. Thus far this method has been used in many disease areas, including the study of multiple psychiatric disorders, such as schizophrenia, major depressive disorder, and bipolar disorder (Chan et al., 2014).
Figure 1.
Multiplex immunoassay. Samples are added to dye-coded microsphere-antibody conjugates that target specific proteins. After incubation with a second antibody containing a fluorescent label, the mixtures are streamed through a reader, which uses lasers for identification of the antibody-microsphere conjugates and quantitation of the bound analytes.
Two-Dimensional Gel Electrophoresis
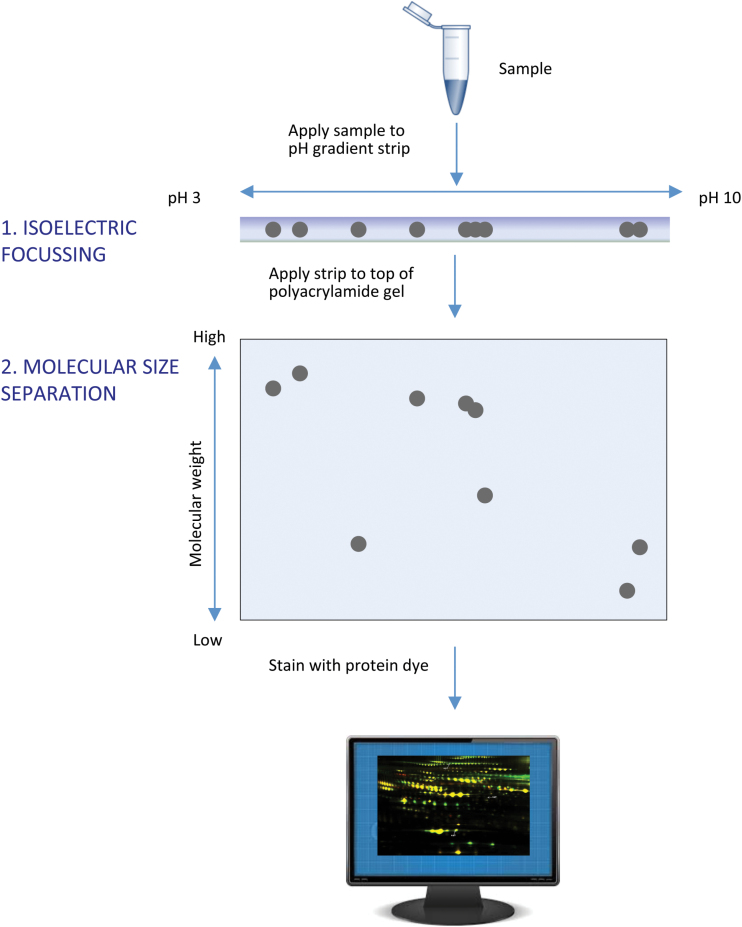
Patrick O’Farrell first presented the two-dimensional gel electrophoresis (2DE) method in 1975, and this technique singlehandedly revolutionized the discovery of biomarkers (O’Farrell, 1975). Just before completion of the human genome project in 2001, 2DE remained the method of choice for comparative proteome analyses. The method works as follows: (1) proteins in the samples are applied to a strip gel and separated according to their isoelectric point (the point at which no net charge on the protein occurs) using isoelectrofocusing; (2) they are separated according to their apparent molecular weight by application of the strip onto a sodium dodecylsulphate polyacrylamide gel and electrophoresed again; and (3) the protein spots in the gels can be visualized with any number of stains (such as Coomassie Blue or Sypro Ruby) and then quantitated using an imaging software (Figure 2). The resulting effect can be visually striking and often resembles a constellation or a small galaxy of stars.
Figure 2.
Two-dimensional gel electrophoresis. Proteins are extracted from a tissue or other sample and then separated by electrophoresis in two dimensions. The first dimension is isoelectric focusing, during which proteins are separated according to their isoelectric points (the state of zero net charge). The second dimension is detergent-based gel electrophoresis, during which the proteins are separated according to apparent molecular weight. The resulting protein spots can be subjected to image analysis and quantitated.
2DE techniques allow the study of intact proteins, but there are some problems with analyses of blood serum or plasma samples. This occurs mainly due to the fact that blood contains a massive concentration range of proteins spanning more than 10 orders of magnitude (Anderson and Anderson, 2002). This means that very abundant proteins such as albumin would appear as large blobs on the gels and eclipse less abundant proteins such as the cytokines. For this reason, other methods began to be used increasingly for proteome analysis.
Mass Spectrometry
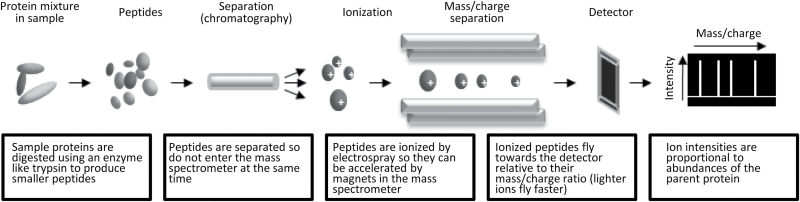
Just before the end of the human genome project, the shotgun mass spectrometry revolution began as a more sensitive and higher throughput approach for proteomic biomarker identification (Link et al 1999). These methods are called “shotgun” as the protein mixtures can be cleaved with enzymes to generate smaller protein fragments called peptides (resembling shotgun pellets), which are the actual analytes in this approach. This is done since most intact proteins are too large and complex in their structure to be analyzed directly in a mass spectrometer. In the next phase, the peptides are separated according to their physiochemical properties by liquid chromatography so that they do not enter the mass spectrometer at the same time. As the peptides leave the column and enter the mass spectrometer on-line, they are ionized by electrospray, which is basically the application of an electric charge to evaporate all the fluids, leaving the peptides in a changed plasma state. This allows the peptide ions to be accelerated by magnets in the mass spectrometer and the peptides speed towards a detector, essentially according to their precise mass (heavy peptide ions move slower than light ones). Quantitation can be achieved since the amount of a given peptide hitting the detector per unit of time is proportional to the quantity of that peptide and, therefore, the corresponding parent protein (Figure 3). Simultaneously, the sequence of the peptide can be determined by streaming in a gas like nitrogen, which bombards the peptides, causing them to break into smaller pieces (not shown in the figure). Determining the masses of these pieces can then allow researchers to derive the amino acid sequences that make up the peptides, which could then be used to search a protein database to obtain the identification of the parent protein. This method is also compatible with metabolomic analysis. In this case, there is no need for enzymatic cleavage of the molecules, as these are already of a manageable size. In this approach, the sample can be infused directly into the mass spectrometer, the quantity determined as above, and the identity deciphered through molecular weight comparisons with known standards.
Figure 3.
Main stages of mass spectrometry profiling. This method can screen hundreds to thousands of proteins or small molecules in one run, depending on whether or not a pre-fractionation step is applied. In the case of proteomics, the method requires enzymatic digestion of the proteins to produce smaller manageable peptides. This step is omitted in metabolomics approaches since the analytes in question are already of a manageable size.
Metabolomic Techniques
1H-NMR Spectroscopy
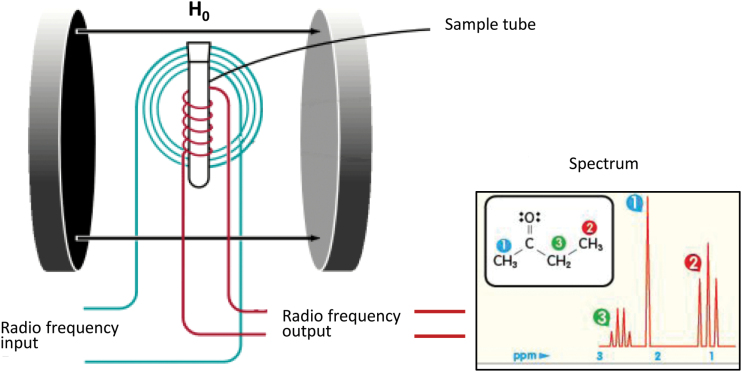
Although mass spectrometry can also be employed, 1H-NMR is chiefly used for metabolomic and small molecule analyses and does not require separation or pre-fractionation of the molecules in some cases. A key advantage of this approach is the analytical reproducibility and sheer simplicity of the sample preparation step. However, this method is less sensitive compared to mass spectrometry–based approaches (Griffin, 2003; Beckonert et al., 2007). The technique gives information about the structural properties of molecules and is therefore suited for identification purposes. 1H (proton) NMR works by tracking the behavior of protons when they are introduced into a very strong magnetic field. In this scenario, the nuclei of the protons on molecules line up with the magnetic field, similar to the way that a compass needle aligns with the magnetic field of the Earth. However, the NMR magnetic field is around four orders of magnitude stronger than that enveloping the Earth. The NMR procedure begins with the application of radio-frequency pulses to the sample. This stimulates the nuclei to rotate away from their equilibrium position so that they can rotate around the axis of the magnetic field. The frequency of the rotation is related to the chemical and physical environment of the atom within the molecule. This means that by using different combinations of radio pulses it is possible to determine how each atom interacts with other atoms in the same molecule, yielding the structure and therefore the identity of the molecule (Figure 4). NMR can also be used to monitor relative changes in the levels of key analytes such as amino acids, neurotransmitters, and neurotransmitter precursors or metabolites, making it useful for biomarker studies of psychiatric disorders.
Figure 4.
Principles of the proton nuclear magnetic resonance method for determining molecular structure.
Molecular Fingerprints in Psychiatric Disorders
More than 100 studies have now been performed that have looked at proteomic and metabolomic biomarker changes in the serum or plasma of patients with psychiatric disorders (Sethi and Brietzke, 2015; Chan et al., 2014; Nascimento and Martins-de-Souza, 2015). It should be noted that the most informative and useful biomarker test suggested by clinicians would be one that could distinguish one psychiatric disorder from another. This would appear to be difficult at first glance since virtually all of these disorders display changes in circulating proteins and small molecules involved in similar molecular pathways, such as hormone signalling (adiponectin, adrenocorticotrophic hormone, cortisol, insulin, leptin, prolactin, testosterone), energy metabolism (cholesterol, glucose, lactate), growth factor (brain-derived neurotrophic factor, epidermal growth factor, nerve growth factor), inflammation (cytokines, antibodies, acute phase response proteins), and oxidative/reduction pathways (glutathione, malondialdehyde, methylglyoxal, dehydroepiandrosterone; Lo et al., 2009; Bicikova et al., 2011; Yang et al., 2013; Chan et al., 2014; Huang et al., 2014; Huang and Lin, 2015; Table 1). Although some of these biomarkers have been reported to be altered in one or two of these disorders (suggesting that these may be unique for that particular disorder), the main problem lies in the fact that these have not been compared across the disorders in the same study. Of course, this would require a massive integration of efforts across multiple clinical locatoins to acquire a sufficient number of samples to add enough statistical power to the investigation. Most importantly, it will also require adoption of the same standard operating procedures for all of these locations regarding stages of the procedure such as patient recruitment and assessment, sample collection and storage, as well as standardization of the proteomic and metabolic platforms and data analysis methods used.
Table 1.
Pathways and Associated Molecule or Molecular Class Found to be Altered in Psychiatric Disorders
| Pathway | Molecule | Platform |
|---|---|---|
| Hormone signalling | Adiponectin | MIA |
| Adrenocorticotropic hormone | MIA | |
| Cortisol | MIA | |
| Insulin | MIA | |
| Leptin | MIA | |
| Prolactin | MIA | |
| Testosterone | MIA, LC-MS/MS | |
| Energy metabolism | Cholesterol | MIA, LC-MS/MS |
| Glucose | NMR | |
| Lactate | NMR | |
| Apolipoprotein AI | MIA, 2DGE | |
| Apolipoprotein AIV | MIA, 2DGE | |
| Growth factor | Brain-derived neurotrophic factor | MIA |
| Epidermal growth factor | MIA | |
| Nerve growth factor | MIA | |
| Inflammation | Cytokines | MIA |
| Antibodies | MIA, 2DGE, LC-MS/MS | |
| Acute phase response proteins | MIA, 2DGE, LC-MS/MS | |
| Transferrin | 2DGE | |
| Fibrinogen | 2DGE | |
| Oxidative/reduction | Glutathione | NMR |
| Methylglyoxal | NMR | |
| Dehydroepiandrosterone | NMR | |
| Glutathione peroxidase | MIA, 2DGE, LC-MS/MS | |
| Superoxide dismutase | MIA, 2DGE, LC-MS/MS | |
| Protein synthesis | Protein disulfide isomerase | 2DGE |
| T-complex protein subunit beta | 2DGE |
Molecular class was used for the inflammation-related molecules given the high number of molecules affected in this class. 2DGE, two-dimensional gel electrophoresis; LC-MS/MS, liquid chromatography tandem mass spectrometry; MIA, multiplex immunoassay; NMR = 1H-nuclear magnetic resonance spectroscopy.
Molecular Tests to Improve Treatment Response
Previous studies have shown that basic measurements of physical characteristics, like waist circumference and body mass index, or testing the levels of fat-based molecules, such as triglycerides and high density lipoproteins, can be used with some accuracy to predict whether or not a patient with schizophrenia will develop metabolic side effects in response to antipsychotic treatment (Jin et al., 2010). Therefore, a test which incorporates measurements such as these would be useful for identifying those patients who would be most likely to benefit from add-on treatments with antidiabetic drugs, as these would help to minimize the metabolic side effects such as insulin resistance or weight gain. A proteomic study showed that schizophrenia patients with higher serum levels of the pituitary hormone prolactin have a better outcome after long-term treatment with antipsychotics (Shrivastava et al., 2012). Another molecular profiling study found that the circulating levels of prolactin, the metabolism-related fatty acid binding protein, and the inflammation pathway molecules ferritin, C-reactive protein, myoglobin, complement factor H, and interleukin-16 could be used to predict improvement in positive symptoms of schizophrenia (psychosis, delusions, hallucinations) following antipsychotic treatment (Schwarz et al., 2012). The same study also found that measurement of the levels of insulin and the inflammation marker matrix metalloproteinase 2 were useful for prediction of improvements in negative symptoms (depression, flattened effect, listlessness). The same study also found that if a patient had low levels of the hormones insulin and leptin in combination with high levels of the inflammatory protein transforming growth factor–beta, they were more likely to have a relapse more rapidly than those patients who did not have this same molecular pattern. Finally, a recent study showed that pre-treatment levels of molecules involved in fatty acid metabolism could be used to predict response to treatment with the antipsychotic olanzapine (Tomasik et al., 2015). It should be stressed that although these studies show considerable promise, further investigations are required to confirm the findings and then translate measurement of these analytes into a user-friendly format for testing in a clinical setting.
Future Prospects
In the near future, it is conceivable that the increasing use of circulating proteomic and metabolomic biomarkers will lead to more extensive biological signatures in people that reflect the physiological changes occurring in health and disease. It is clear from the literature that some blood-based biomarker candidates have now been replicated across several studies of psychiatric disorders and converge mostly on the pathways of hormonal metabolism and inflammation. Also, recent studies have begun to emerge that identify biomarker candidates to predict psychiatric disease development in at-risk individuals. Some of these tests have shown sensitivities of approximately 90%. This includes tests for schizophrenia (Chan et al., 2014; Perkins et al., 2014; Table 2), major depressive disorder (Gottschalk et al., 2015; Table 3), and bipolar disorder (Haenisch et al., 2015; Table 4). The exciting point of all of these studies is that they demonstrate the presence of a disease signature in at-risk individuals months to years before full clinical manifestation. However, one drawback of all of these investigations is that each was carried out using retrospective sample collections. It will be important to validate the findings using new sample collections in prospective investigations.
Table 2.
Identification of Serum Biomarkers for Prediction of Schizophrenia Before Disease Onset
| Molecular pathway | Analyte | Publication |
|---|---|---|
| Inflammation | Alpha-2 macroglobulin | Chan et al., 2015 |
| Beta-2 microglobulin | Chan et al., 2015 | |
| Carcinoembryonic antigen | Chan et al., 2015 | |
| Chemokine (C-C) motif ligand 8 | Perkins et al., 2014 | |
| Haptoglobin | Chan et al., 2015 | |
| Immunoglobulin-A | Chan et al., 2015 | |
| Immunoglobulin-E | Perkins et al., 2014 | |
| Interleukin-1 receptor antagonist | Chan et al., 2015 | |
| Interleukin-beta | Perkins et al., 2014 | |
| Interleukin-10 | Chan et al., 2015 | |
| Interleukin-7 | Perkins et al., 2014 | |
| Interleukin-8 | Perkins et al., 2014; Chan et al., 2015 | |
| Interleukin-13 | Chan et al., 2015 | |
| Macrophage migration inhibitory factor | Chan et al., 2015 | |
| Matrix metalloproteinase 7 | Perkins et al., 2014 | |
| Receptor for advanced glycosylation end products | Chan et al., 2015 | |
| Serum glutamic oxaloacetic transaminase | Chan et al., 2015 | |
| Tenascin C | Chan et al., 2015 | |
| Uromodulin | Perkins et al., 2014 | |
| Von Willebrand factor | Chan et al., 2015 | |
| Hormone signalling | Cortisol | Perkins et al., 2014 |
| Follicle-stimulating hormone | Chan et al., 2015 | |
| Growth hormone | Perkins et al., 2014 | |
| Leptin | Chan et al., 2015 | |
| Pancreatic polypeptide | Chan et al., 2015 | |
| Resistin | Chan et al., 2015 | |
| Testosterone (total) | Chan et al., 2015 | |
| Thyroid stimulating hormone | Perkins et al., 2014; Chan et al., 2015 | |
| Growth factor signalling | AXL receptor tyrosine kinase | Chan et al., 2015 |
| Insulin-like growth factor-binding protein 2 | Chan et al., 2015 | |
| Stem Cell Factor | Perkins et al. 2014; Chan et al., 2015 | |
| Clotting cascade | Angiotensin-converting enzyme | Chan et al., 2015 |
| Factor VII | Perkins et al., 2014; Chan et al., 2015 | |
| Lipid transport | Apolipoprotein-D | Perkins et al., 2014 |
| Malonaldehyde-modified low density lipoprotein | Perkins et al., 2014 |
The table shows the molecular pathways and specific molecules covered by these tests as well as the associated publications.
Table 3.
Identification of Serum Biomarkers for Prediction of Major Depression Before Disease Onset.
The table shows the molecular pathways and specific molecules covered by these tests as well as the associated publications
| Molecular pathway | Analyte | Publication |
|---|---|---|
| Inflammation | Carcinoembryonic antigen | Haenisch et al., 2015 |
| CD5 | Haenisch et al., 2015 | |
| CD40 | Haenisch et al., 2015 | |
| Cystatin C | Haenisch et al., 2015 | |
| ENRAGE | Haenisch et al., 2015 | |
| Growth-regulated alpha protein | Haenisch et al., 2015 | |
| Interleukin-1 receptor antagonist | Haenisch et al., 2015 | |
| Interleukin-10 | Haenisch et al., 2015 | |
| Macrophage inflammatory protein-1 beta | Haenisch et al., 2015 | |
| Matrix metalloproteinase-3 | Haenisch et al., 2015 | |
| Matrix metalloproteinase-7 | Haenisch et al., 2015 | |
| Matrix metalloproteinase-9, total | Haenisch et al., 2015 | |
| Receptor for advanced glycosylation end products | Haenisch et al., 2015 | |
| Serum amyloid P-component | Haenisch et al., 2015 | |
| Tumor necrosis factor receptor-Like 2 | ||
| Growth factor signalling | Hepatocyte growth factor | Haenisch et al., 2015 |
| Lipid transport | Apolipoprotein A1 | Haenisch et al., 2015 |
| Apolipoprotein A2 | Haenisch et al., 2015 | |
| Lipoprotein (a) | Haenisch et al., 2015 |
Table 4.
Identification of Serum Biomarkers for Prediction of Bipolar Disorder Before Disease Onset
| Molecular pathway | Analte | Publication |
|---|---|---|
| Inflammation | Collagen IV | Gottschalk et al., 2015 |
| Vascular cell adhesion molecule 1 | Gottschalk et al., 2015 | |
| Vitronectin | Gottschalk et al., 2015 | |
| Growth factor signalling | Insulin-like growth factor binding protein 3 | Gottschalk et al., 2015 |
| Receptor tyrosine kinase AXL | Gottschalk et al., 2015 |
The table shows the molecular pathways and specific molecules covered by these tests as well as the associated publications.
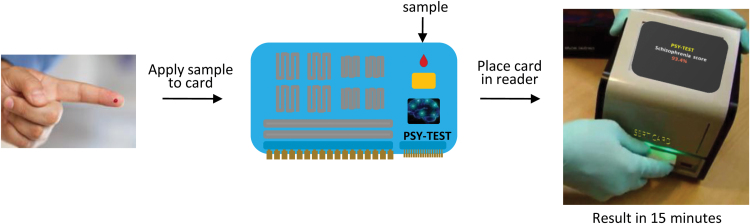
Another point to consider is that all of the existing prototype tests for diagnosis, disease prediction, and drug treatment response monitoring have been devised using current proteomic and metabolomic platforms. These are typically comprised of multiple large components that require considerable technical expertise and knowledge for correct operation, and most do not return results rapidly enough to be of use in point-of-care scenarios. In line with this, a market has emerged for the development of novel and more effective diagnostic and treatment approaches. Multiplex biomarker tests have now been developed that use the multiplex immunoassay system on diagnostic devices, which are approximately the size of a credit card. These feature a high degree of modularity, allowing various assay types and formats. These tests have many advantages over existing approaches, including that they are user friendly, require no expertise, are rapid, and have a low cost. Such a test has been developed by the scientists at the Fraunhofer Institute in Germany and normally involves application of blood drops into a small chamber in a card, which is inserted into a book-sized reader to produce the diagnosis “score” in less than 15 minutes (Schumacher et al., 2012; Figure 5). The idea is that these devices, which feature analyses of both proteomic and small molecule biomarkers, could be used directly in a doctor’s office, with the result returned within the timeframe of the visitation period. Thus, a major benefit will be the rapid turnover, which will help to cut down on waiting times for lab test results, which often take days to weeks using the larger platforms. Furthermore, these devices could contain a USB port for connection to computers and other devices for transmission of data. This opens up a new realm of possibilities.
Figure 5.
Credit card-sized diagnostic tests of proteomic/metabolomic biomarkers for diseases which can be achieved in less than 15 minutes using a single drop of blood. This approach has been pioneered by the scientists from the Fraunhofer Institute.
Point-of-care-type tests that are already in use include those used to measure blood gases and ions, as well as biomarkers of cardiac problems, diabetes, pregnancy, specific infectious diseases, or the presence of alcohol or drugs of abuse. These devices usually consist of a test strip, lateral flow, slides, flow-through tubes, and cassettes, and they often have built-in meters and displays for easy visualization of the test results. Another requirement of point-of-care devices which can measure proteomic and metabolomic biomarkers is mobile communication and internet access so the data can be organized, analysed, and formatted for simple and manageable presentation to the end users. This would allow testing using real-time multiplexed sensors of proteomic and metabolomic biomarkers, combined with artificial intelligence and mobile communication systems for the analysis and display of the results.
A movement towards mobile phone applications is also underway in molecular diagnostics. Mobile phones have now become virtually ubiquitous, with a number somewhere around the 6 billion mark for actual subscriptions (most of the world’s population; http://mobithinking.com/mobile-marketing-tools/latest-mobile-stats/a#subscribers). The smartphone combines normal voice and text messaging services with information technology, supporting applications, and sensors, as well as internet access and connectivity with other devices, allowing for new medical applications (Klasnja and Pratt, 2012). In fact, smartphone-based interventions have already shown initial promise in improving individual outcomes in a variety of health conditions, diseases, and habit control, and thus have high potential in the future treatment and management of psychiatric disorders. For example, they could be used to improve the attendance of patients at their medical appointments, provide more rapid diagnosis and treatment, and improve communication, along with having a positive impact on behaviors such as smoking cessation and increased medication compliance, as well as clinical enhancements such as lowering of stress levels. Most of these involve automated text or voicemail reminders of encouragement on preventative behaviors or self-management.
Potentially the most exciting recent developments have included the construction of multiplex immunoassay tests on a hand-held colorimetric micro-plate reader, which uses 3D-printed opto-mechanics for illumination of miniature reaction wells on a light-emitting diode optical fibre array (Berg et al., 2015). In this approach, the resulting images can even be transmitted to central servers for diagnostic results output within 1 minute. Thus far, this mobile platform has been tested in a clinical microbiology laboratory using mumps, measles, and herpes simplex I and II virus immunoglobulin tests, and these had accuracies greater than 98%. We should expect that similar tests for other diseases such as psychiatric disorders will be available in the near future based on analysis of validated proteomic and metabolomic biomarkers. However, this is still some distance in the future, as many stages such as validation testing are required to ensure that only the most robust biomarker candidates are incorporated into these formats. In this way, the diagnosis and treatment of psychiatric disorders can move forward into the 21st century in pace with that of other medical areas such as cardiovascular disease, diabetes, and cancer. Most importantly, this will lead to more personalized medicine approaches (Evers, 2009), which will enable treatment of the right patients with the right drug at the right time for better outcomes.
Statement of Interest
The authors do not have any conflicts of interest.
Acknowledgments
The Laboratory of Neuroproteomics, UNICAMP is funded by FAPESP (São Paulo Research Foundation) grant number 13/08711-3.
References
- Anderson NL, Anderson NG. (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 1:845–867. [DOI] [PubMed] [Google Scholar]
- Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK. (2007) Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc 2: 2692–2703. [DOI] [PubMed] [Google Scholar]
- Berg B, Cortazar B, Tseng D, Ozkan H, Feng S, Wei Q, Chan RY, Burbano J, Farooqui Q, Lewinski M, Di Carlo D, Garner OB, Ozcan A. (2015) Cellphone-based hand-held micro-plate reader for point-of-care testing of enzyme-linked immunosorbent assays. ACS Nano 9:7857–7866. [DOI] [PubMed] [Google Scholar]
- Bicikova M, Hampl R, Hill M, Ripova D, Mohr P, Putz Z. (2011) Neuro- and immunomodulatory steroids and other biochemical markers in drug-naive schizophrenia patients and the effect of treatment with atypical antipsychotics. Neuro Endocrinol Lett 32:141–147. [PubMed] [Google Scholar]
- Biomarkers Definitions Working Group , Atkinson AJ, Colburn WA, DeGruttola VG, DeMets DL, Downing GJ, Hoth DF, Oates JA, Peck CC, Schooley RT, Spilker BA, Woodcock J, Zeger SL. (2001) Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 69:89–95. [DOI] [PubMed] [Google Scholar]
- Chan MK, Gottschalk MG, Haenisch F, Tomasik J, Ruland T, Rahmoune H, Guest PC, Bahn S. (2014) Applications of blood-based protein biomarker strategies in the study of psychiatric disorders. Prog Neurobiol 122:45–72. [DOI] [PubMed] [Google Scholar]
- Chan MK, Krebs MO, Cox D, Guest PC, Yolken RH, Rahmoune H, Rothermundt M, Steiner J, Leweke FM, van Beveren NJ, Niebuhr DW, Weber NS, Cowan DN, Suarez-Pinilla P, Crespo-Facorro B, Mam-Lam-Fook C, Bourgin J, Wenstrup RJ, Kaldate RR, Cooper JD, Bahn S. (2015) Development of a blood-based molecular biomarker test for identification of schizophrenia before disease onset. Transl Psychiatry 5:e601. doi: 10.1038/tp.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert BN, Insel TR. (2013) Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med 11:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers K. (2009) Personalized medicine in psychiatry: ethical challenges and opportunities. Dialogues Clin Neurosci 11:427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR., Jr (1997) Advanced multiplexed analysis with the FlowMetrix system. Clin Chem 43:1749–1756. [PubMed] [Google Scholar]
- Gottschalk MG, Cooper JD, Chan MK, Bot M, Penninx BW, Bahn S. (2015) Discovery of serum biomarkers predicting development of a subsequent depressive episode in social anxiety disorder. Brain Behav Immun 48:123–131. [DOI] [PubMed] [Google Scholar]
- Griffin JL. (2003) Metabonomics: NMR spectroscopy and pattern recognition analysis of body fluids and tissues for characterisation of xenobiotic toxicity and disease diagnosis. Curr Opin Chem Biol 7:648–654. [DOI] [PubMed] [Google Scholar]
- Haenisch F, Cooper JD, Reif A, Kittel-Schneider S, Steiner J, Leweke FM, Rothermundt M, van Beveren NJ, Crespo-Facorro B, Niebuhr DW, Cowan DN, Weber NS, Yolken RH, Penninx BW, Bahn S. (2015) Towards a blood-based diagnostic panel for bipolar disorder. Brain Behav Immun. Advance online publication. Retrieved 13 Oct 2015. doi: 10.1016/j.bbi.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Huang T, Lin CC. (2015) Advances in biomarkers of major depressive disorder. Adv Clin Chem 68:177–204. [DOI] [PubMed] [Google Scholar]
- Huang TL, Sung ML, Chen TY. (2014) 2D-DIGE proteome analysis on the platelet proteins of patients with major depression. Proteome Sci 12:1. doi: 10.1186/1477-5956-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H Meyer J Mudaliar S Henry R Khandrika S Glorioso DK, Kraemer H, Jeste D (2010) Use of clinical markers to identify metabolic syndrome in antipsychotic-treated patients. J Clin Psychiatry 71:1273–1278. [DOI] [PubMed] [Google Scholar]
- Laborit H. (1976) On the mechanism of activation of the hypothalamo--pituitary--adrenal reaction to changes in the environment (the ‘alarm reaction’). Resuscitation 5:19–30. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd (1999) Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol 17:676–682. [DOI] [PubMed] [Google Scholar]
- Lo LH, Huang TL, Shiea J. (2009) Acid hydrolysis followed by matrix-assisted laser desorption/ionization mass spectrometry for the rapid diagnosis of serum protein biomarkers in patients with major depression. Rapid Commun Mass Spectrom 23:589–598. [DOI] [PubMed] [Google Scholar]
- Klasnja P, Pratt W. (2012) Healthcare in the pocket: mapping the space of mobile-phone health interventions. J Biomed Inform 45:184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastorakos G, Zapanti E. (2004) The hypothalamic-pituitary-adrenal axis in the neuroendocrine regulation of food intake and obesity: the role of corticotropin releasing hormone. Nutr Neurosci 7:271–280. [DOI] [PubMed] [Google Scholar]
- Nascimento JM, Martins-de-Souza D. (2015) The proteome of schizophrenia. NPJ Schizophrenia 1: 14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell PH. (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Pasquali R, Vicennati V, Cacciari M, Pagotto U. (2006) The hypothalamic-pituitary-adrenal axis activity in obesity and the metabolic syndrome. Ann NY Acad Sci 1083:111–128. [DOI] [PubMed] [Google Scholar]
- Perkins DO Jeffries CD Addington J Bearden CE Cadenhead KS Cannon TD, Cornblatt BA, Mathalon DH, McGlashan TH, Seidman LJ, Tsuang MT, Walker EF, Woods SW, Heinssen R (2014) Towards a psychosis risk blood diagnostic for persons experiencing high-risk symptoms: preliminary results from the NAPLS project. Schizophr Bull 41:419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan LP. (2007) Insulin signaling effects on memory and mood. Curr Opin Pharmacol 7:633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E, Guest PC, Steiner J, Bogerts B, Bahn S. (2012) Identification of blood-based molecular signatures for prediction of response and relapse in schizophrenia patients. Transl Psychiatry 2:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher S, Nestler J, Otto T, Wegener M, Ehrentreich-Förster E, Michel D, Wunderlich K, Palzer S, Sohn K, Weber A, Burgard M, Grzesiak A, Teichert A, Brandenburg A, Koger B, Albers J, Nebling E, Bier FF. (2012) Highly-integrated lab-on-chip system for point-of-care multiparameter analysis. Lab Chip 12:464–473. [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Johnston M, Bureau Y, Shah N. (2012) Baseline serum prolactin in drug-naive, first-episode schizophrenia and outcome at five years: is it a predictive factor? Innov Clin Neurosci 9:17–21. [PMC free article] [PubMed] [Google Scholar]
- Sethi S, Brietzke E. (2015) Omics-based biomarkers: application of metabolomics in neuropsychiatric disorders. Int J Neuropsychop. Advance online publication. Retrieved 9 Oct 2015. pii: pyv096. doi: 10.1093/ijnp/pyv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Thomson M. (1991) Neuroendocrinology of the hypothalamo-pituitary-adrenal axis in pregnancy and the puerperium. Baillieres Clin Endocrinol Metab 5:167–186. [DOI] [PubMed] [Google Scholar]
- Solas M, Aisa B, Mugueta MC, Del Río J, Tordera RM, Ramírez MJ. (2010) Interactions between age, stress and insulin on cognition: implications for Alzheimer’s disease. Neuropsychopharmacology 35:1664–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Späth-Schwalbe E, Born J, Schrezenmeier H, Bornstein SR, Stromeyer P, Drechsler S, Fehm HL, Porzsolt F. (1994) Interleukin-6 stimulates the hypothalamus-pituitary-adrenocortical axis in man. J Clin Endocrinol Metab 79:1212–1214. [DOI] [PubMed] [Google Scholar]
- Straub RH, Buttgereit F, Cutolo M. (2011) Alterations of the hypothalamic-pituitaryadrenal axis in systemic immune diseases - a role for misguided energy regulation. Clin Exp Rheumatol 29:S23-31 [PubMed] [Google Scholar]
- Tomasik J, Schwarz E, Lago SG, Rothermundt M, Leweke FM, van Beveren NJ, Guest PC, Rahmoune H, Steiner J, Bahn S. (2015) Pretreatment levels of the fatty acid handling proteins H-FABP and CD36 predict response to olanzapine in recent-onset schizophrenia patients. Brain Behav Immun. Advance online publication. Retrieved 2 Nov 2015. doi: 10.1016/j.bbi.2015.10.019. [DOI] [PubMed] [Google Scholar]
- Tweeddale H, Notley-McRobb L, Ferenci T. (1998) Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J Bacteriol 180:5109–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF, Williams KL. (1996) Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev 13:19–50. [DOI] [PubMed] [Google Scholar]
- Yang J, Chen T, Sun L, Zhao Z, Qi X, Zhou K, Cao Y, Wang X, Qiu Y, Su M, Zhao A, Wang P, Yang P, Wu J, Feng G, He L, Jia W, Wan C. (2013) Potential metabolite markers of schizophrenia. Mol Psychiatry 18:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]