Here, Zhou et al. investigated how oncogenic superenhancers (SE), which are found near oncogenes and control cancer gene expression, are regulated. The results demonstrate an essential role for INO80-dependent chromatin remodeling in SE function by showing that INO80 is required for SE-mediated oncogenic transcription and tumor growth in melanoma.
Keywords: INO80, chromatin remodeler, oncogenic transcription, superenhancer
Abstract
Superenhancers (SEs) are large genomic regions with a high density of enhancer marks. In cancer, SEs are found near oncogenes and dictate cancer gene expression. However, how oncogenic SEs are regulated remains poorly understood. Here, we show that INO80, a chromatin remodeling complex, is required for SE-mediated oncogenic transcription and tumor growth in melanoma. The expression of Ino80, the SWI/SNF ATPase, is elevated in melanoma cells and patient melanomas compared with normal melanocytes and benign nevi. Furthermore, Ino80 silencing selectively inhibits melanoma cell proliferation, anchorage-independent growth, tumorigenesis, and tumor maintenance in mouse xenografts. Mechanistically, Ino80 occupies >90% of SEs, and its occupancy is dependent on transcription factors such as MITF and Sox9. Ino80 binding reduces nucleosome occupancy and facilitates Mediator recruitment, thus promoting oncogenic transcription. Consistently, genes co-occupied by Ino80 and Med1 are selectively expressed in melanomas compared with melanocytes. Together, our results reveal an essential role of INO80-dependent chromatin remodeling in SE function and suggest a novel strategy for disrupting SEs in cancer treatment.
Genomic regulatory elements have become increasingly important in understanding the molecular foundations of cancer and developing potential strategies for cancer therapies (Mansour et al. 2014; Weinhold et al. 2014; Melton et al. 2015). Among them, superenhancers (SEs) are genomic regions with an unusually high density of enhancer markers such as H3K27 acetylation (H3K27ac) and the Mediator complex (Heinz et al. 2015; Pott and Lieb 2015). Representing a small fraction of all enhancers, SEs are critically involved in the expression of cell type-specific genes important for development and disease (Chapuy et al. 2013; Hnisz et al. 2013; Loven et al. 2013; Whyte et al. 2013; Chipumuro et al. 2014; Kwiatkowski et al. 2014). In cancer, SEs are enriched at genes with known oncogenic function, and genomic rearrangements often encompass SEs (Affer et al. 2014; Groschel et al. 2014; Northcott et al. 2014; Walker et al. 2014). However, how SEs are regulated and maintained in cancer remains to be elucidated.
Melanoma is among the most common cancers in the United States. Activation of the MAPK pathway, such as by mutations in BRAF or NRAS, is found in the majority of patients. In addition, recent studies have revealed a complex involvement of epigenetic mechanisms in melanoma development and progression. Particularly, factors involved in chromatin regulation, such as the chromatin remodeling complexes, were found to play important roles in controlling the gene expression program in melanoma (van den Hurk et al. 2012; Mehrotra et al. 2014). The ATP-dependent chromatin remodelers are capable of remodeling chromatin structure by sliding, ejecting, or exchanging nucleosomes and are therefore critically engaged in chromatin organization, accessibility, and gene regulation (Ho and Crabtree 2010; Piatti et al. 2011). Although chromatin remodelers are often found to be mutated in cancers, including melanoma (Roberts and Orkin 2004; Mayes et al. 2014; The Cancer Genome Atlas Network 2015), the mechanisms by which they contribute to carcinogenesis and tumor maintenance are not fully understood.
INO80 belongs to the INO80 subfamily of the chromatin remodeling complexes (Morrison and Shen 2009; Watanabe and Peterson 2010). It has been implicated in many crucial cellular functions, including transcription regulation, DNA replication and repair, telomere maintenance, and chromosome segregation. Deletion of Ino80, the SWI/SNF ATPase in the complex, resulted in early embryonic lethality (Min et al. 2013). In addition, Ino80 depletion inhibited cell proliferation and anchorage-independent growth. Interestingly, INO80 haploinsufficiency had no impact on tumor incidence and latency in p53-null mice. Instead, it altered the tumor spectrum and increased the percentage of invasive sarcomas (Min et al. 2013). Despite these findings, the involvement of INO80 in cancer is still much less well characterized compared with the SWI/SNF chromatin remodelers (Masliah-Planchon et al. 2015).
Recently, INO80 has been found to selectively activate pluripotency genes in embryonic stem cells (ESCs) by maintaining an open chromatin structure at promoter-proximal enhancers (Wang et al. 2014). As genes and pathways important for ESC maintenance are often reactivated in cancer (Kim and Orkin 2011), we set out to investigate whether and how INO80 may be involved in tumorigenesis. Here, we show that INO80 indeed plays an essential role in melanoma proliferation and tumorigenesis. It occupies SEs and promotes oncogenic transcription by facilitating nucleosome depletion and Mediator recruitment. Our data define a critical role of INO80-mediated chromatin remodeling in cancer development and the regulation of SEs.
Results
Ino80 is highly expressed in melanoma
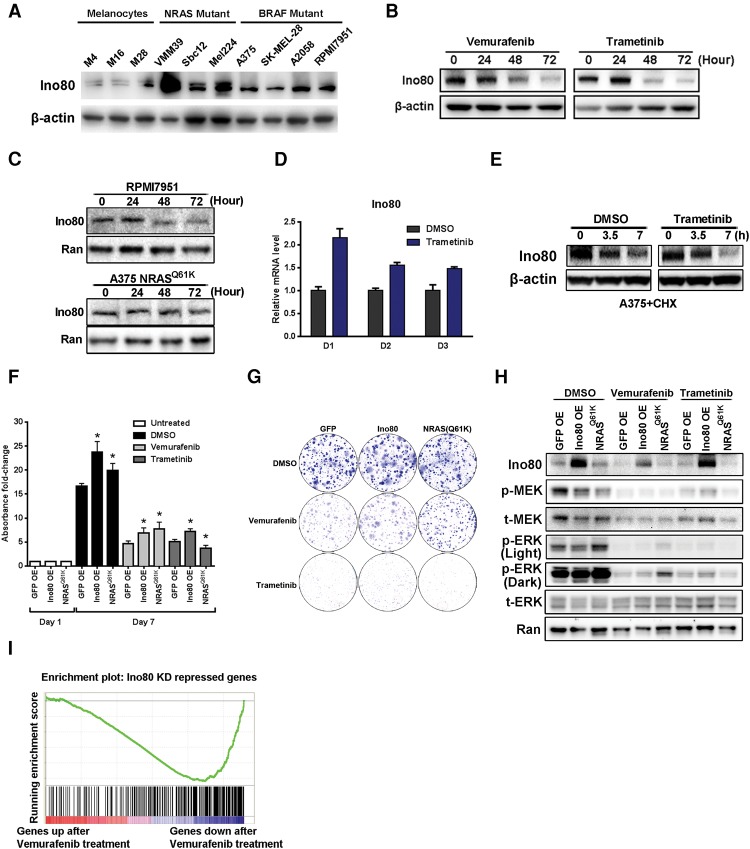
To test the role of INO80 in melanoma and oncogenic SE regulation, we examined the expression of its subunits during melanoma progression. Based on data published by The Cancer Genome Atlas Network (2015), several INO80 subunits show elevated mRNA levels in metastatic melanoma compared with primary melanoma (Supplemental Fig. S1A), and elevated expression was associated with poor prognosis (Supplemental Fig. S1B). In addition, we found that the protein level of Ino80, the core SWI/SNF ATPase of the complex, was also increased in primary melanomas compared with benign nevi in patient samples, as determined by immunohistochemical staining with a validated antibody (Supplemental Fig. S1C; Wang et al. 2014). Finally, we found that Ino80 protein levels were higher in melanoma cell lines harboring BRAF or NRAS mutations, the most frequent oncogenic mutations found in melanoma, compared with primary normal melanocytes (Fig. 1A). Consistent with these results, chromatin immunoprecipitation (ChIP) followed by high through-put sequencing (ChIP-seq) showed prominent peaks of enhancer markers such as H3K27ac, H3K4me1, and Med1 as well as RNA polymerase II (Pol II) near the Ino80 transcription start site (TSS) in melanoma cells (Supplemental Fig. S1D), indicative of active transcription of the Ino80 gene. Together, these observations demonstrated a clear correlation between Ino80 expression and melanoma progression.
Figure 1.
Ino80 is required for melanoma growth in vitro. (A) Western blot showing the protein level of Ino80 in melanoma cell lines and primary melanocytes. β-Actin was used as the loading control. (B,C) Western blot showing Ino80 expression after drug treatment. (B) A375 cells were treated with 0.5 µM vemurafenib or 1 nM trametinib, and cells were harvested at the indicated time points after treatment. β-Actin was used as the loading control. (C) RPMI-7951- and NRASQ61K-overexpressing A375 cells were treated with 0.5 µM DMSO or vemurafenib, and cells were harvested at the indicated time points. Ran was used as the loading control. (D) Ino80 mRNA expression in A375 cells after treatment with 1 nM trametinib. Relative expression was determined by RT-qPCR, and β-actin was used for normalization. (E) Ino80 protein stability in A375 cells. Cells were treated with 50 µg/mL cycloheximide and harvested at the indicated time points. Ino80 protein was detected by Western blot. (F–H) Ino80 overexpression promotes melanoma cell growth and clonogenicity. A375 cells were transduced with lentivirus expressing GFP, Ino80, or NRASQ61K and subjected to 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (F) and two-dimensional colony formation assay (G) to determine the impact of Ino80 overexpression. NRASQ61K overexpression is known to confer resistance to vemurafenib (but not trametinib) and serves as a positive control. (H) Ino80, ERK, and MEK expression and ERK and MEK phosphorylation were determined by Western blot. (I) Gene set enrichment analysis (GSEA) showing that down-regulated genes after vemurafenib treatment were highly enriched for those that were down-regulated after Ino80 silencing (see the Materials and Methods for more details).
Intriguingly, treatment of BRAF mutant melanoma cell lines with either of two commonly used FDA-approved anti-melanoma drugs—the BRAF inhibitor vemurafenib or the MEK inhibitor trametinib—led to a reduction in Ino80 protein levels (Fig. 1B). In contrast, vemurafenib treatment had no effect on cells that are resistant to the drug (Fig. 1B,C). Thus, Ino80 expression appears to be highly sensitive to the inhibition of upstream driver mutations. To understand how the driver kinases regulate Ino80, we first examined Ino80 mRNA expression after trametinib treatment. Surprisingly, MEK inhibition resulted in slightly elevated levels of Ino80 mRNA (Fig. 1D), suggesting that the MAPK pathway may regulate Ino80 post-translationally. To test the hypothesis, we inhibited protein synthesis with cycloheximide and examined the Ino80 protein level. Indeed, we found that trametinib treatment enhanced the degradation of the Ino80 protein (Fig. 1E). Interestingly, trametinib affected Ino80 stability only in BRAF, but not NRAS, mutant cells (Supplemental Fig. S1E), coinciding with its effect on their growth. Furthermore, Ino80 overexpression enhanced melanoma cell growth and clonogenecity in both the absence and presence of the inhibitors (Fig. 1F,G) without activating the MAPK pathway (Fig. 1H). Finally, gene set enrichment analysis (GSEA) showed that genes down-regulated after vemurafenib treatment were highly enriched for those that were down-regulated after Ino80 silencing (Fig. 1I). Together, these results support the notion that Ino80 may be a critical downstream effector of the driver kinases during melanoma tumorigenesis and development.
Ino80 is required for melanoma growth
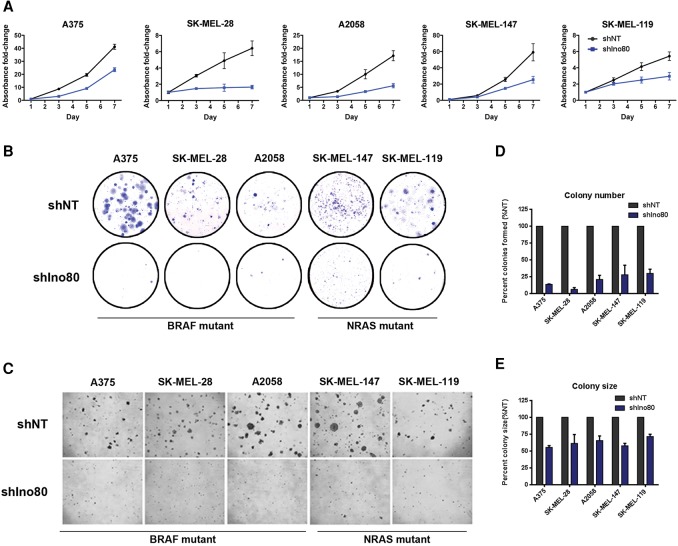
To test whether INO80 is directly required for melanoma development, we silenced Ino80 with lentiviral-based shRNAs in melanoma cell lines (Supplemental Fig. S2A,B). We found that Ino80 silencing resulted in a significant inhibition of cell growth in both BRAF (A375, SK-MEL-28, and A2058) and NRAS (SK-MEL-147 and SK-MEL-119) mutant melanoma cell lines, based on the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Fig. 2A). In contrast, growth of primary melanocytes was not strongly affected (Supplemental Fig. S2C). Furthermore, Ino80 depletion dramatically inhibited both clonogenesis and anchorage-independent growth in all tested melanoma cell lines, as judged by colony formation and soft agar assays (Fig. 2B–E). The residual clones formed from Ino80 shRNA virus transduced cells show no Ino80 silencing (Supplemental Fig. S3A,B), suggesting that they may originate from those that escaped Ino80 silencing. To minimize the possibility of off-target effects, we confirmed the above results by two additional shRNAs in A375 (Supplemental Fig. S3C–F). Together, these results indicate that INO80 is selectively required for the maintenance of melanoma cells but not melanocytes in vitro. To test whether Ino80 can act as an oncogene, we examined the effect of Ino80 overexpression in melanocytes. We found that Ino80 overexpression does not activate the MAPK pathway (Supplemental Fig. S4A). Its overexpression alone, at the level that we were able to achieve, is also not sufficient to drive anchorage-independent growth and thereby malignant transformation in melanocytes (Supplemental Fig. S4B). However, we cannot exclude the possibility that simultaneous overexpression of multiple INO80 subunits may be required.
Figure 2.
Ino80 silencing significantly inhibits melanoma growth in vitro. (A) Growth curves showing the impact of Ino80 silencing on melanoma cell lines. Melanoma cells were transduced with lentiviral-based nontargeting (NT) shRNA (shNT) or Ino80 shRNA (shIno80) and plated on day 1. Cell number (represented by fold change in absorbance) at the indicated time points was determined by MTT assay and normalized to that of day 1. Data were plotted as mean ± SEM from three independent experiments. (B) Representative images of the clonogenesis assay in melanoma cells transduced with NT shRNA or Ino80 shRNA virus. (C) Representative images showing anchorage-independent growth of melanoma cells transduced with NT shRNA or Ino80 shRNA virus. (D,E) Statistical analysis of the anchorage-independent growth assay showing changes in colony number (D) and colony size (E) of cells transduced with Ino80 shRNA relative to NT shRNA control.
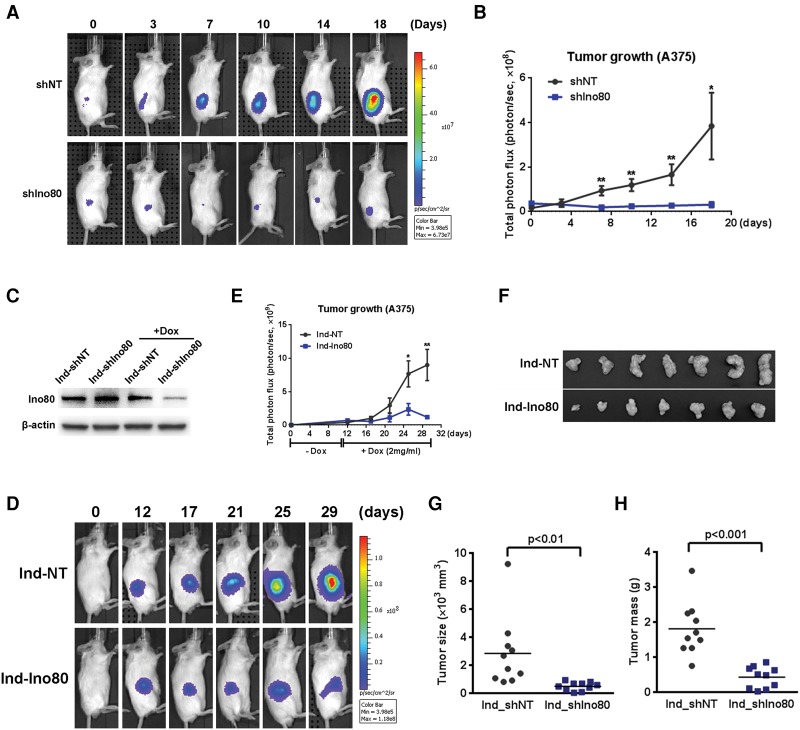
We next used a xenograft mouse model to test whether INO80 is also required for melanoma growth in vivo. We generated A375 cells (BRAF mutant) stably expressing a luciferase reporter gene to monitor tumor growth in live animals by bioluminescence imaging. We transduced the cells with either the nontargeting (NT) or Ino80 shRNA lentivirus and injected the cells subcutaneously into immunocompromised SCID-Beige mice. Strikingly, Ino80 silencing almost completely suppressed tumor growth in vivo, based on bioluminescence imaging (Fig. 3A,B). At 18 d after injection, animals were sacrificed, and the tumors were removed for further analysis. Consistent with the above results, tumors formed by Ino80 silenced cells were much smaller in size compared with those formed from NT shRNA transduced cells (Supplemental Fig. S5A–C) and likely originated from cells that escaped Ino80 silencing, as Ino80 expression was not reduced (Supplemental Fig. S5D). As our results suggested that INO80 may be a critical downstream effector of key driver mutations during melanoma tumorigenesis (Fig. 1B–F), we tested whether INO80 is also important for NRAS mutant melanoma development. Similar to what we observed in BRAF mutant cells, Ino80 silencing by multiple shRNAs suppressed xenograft tumor growth in the NRAS mutant cell line SK-MEL-147 (Supplemental Fig. S6). Together with the observation that Ino80 expression is reduced by BRAF and MEK inhibitors (Fig. 1B), these results strongly suggest that INO80 plays a pivotal role in melanoma growth driven by driver mutations in the MAPK pathway.
Figure 3.
Ino80 silencing significantly inhibits melanoma growth in mouse xenografts. (A) Representative bioluminescence images showing inhibition of tumor formation in mouse xenografts after Ino80 silencing. A375 cells were transduced with either NT shRNA or Ino80 shRNA lentivirus and injected subcutaneously into immunocompromised mice. Tumor cell growth was monitored by bioluminescence imaging at the indicated time points. (B) Statistical analysis of A. Data were plotted as mean ± SEM, where n = 8 in each group. (C) Western blot showing efficient Ino80 silencing by doxycycline (Dox)-inducible shRNA. A375 cells were transduced with NT shRNA or Ino80 shRNA lentivirus, drug-selected, and treated with Dox at 1 µg/mL for 3 d. β-Actin was used as the loading control. (D) Representative bioluminescence images showing the impact of Ino80 silencing on melanoma growth in mouse xenografts. A375 cells were transduced with lentivirus carrying inducible NT shRNA (Ind-shNT) or Ino80 shRNA (Ind-shIno80). Cells were transplanted to SCID-Beige mice subcutaneously, and Dox was administered 12 d after transplantation to induce shRNA expression. Tumor growth was monitored by bioluminescence imaging at the indicated time points. (E) Statistical analysis of tumor bioluminescence. Values were plotted as mean ± SEM, where n = 10 in each group. (F) Representative images showing the size of tumors dissected at the end of the experiment in Figure 2D. (G,H) Analysis of dissected tumor size (G) and mass (H). Data were plotted as mean ± SEM, where n = 10 in each group.
Taking it a step further, we next tested whether Ino80 inhibition can inhibit the growth of established tumors. We transduced A375 melanoma cells with lentivirus expressing doxycycline (Dox)-inducible NT shRNAs or Ino80 shRNAs (Fig. 3C) and transplanted the cells subcutaneously into immune-compromised mice. Tumors were allowed to grow for 12 d to a similar size in both the NT shRNA and Ino80 shRNA groups, after which Dox was administered to the animals to induce the expression of shRNAs. Strikingly, Ino80 silencing strongly inhibited the growth of established tumors, based on bioluminescence imaging (Fig. 3D,E). Consistently, the tumor size and mass in the Ino80 shRNA group were significantly smaller than those in the NT shRNA group at the end of the experiment (Fig. 3F–H). Taken together, the above data strongly suggest that INO80 is required for melanoma growth both in vitro and in vivo.
Ino80 regulates the expression of cancer-related genes
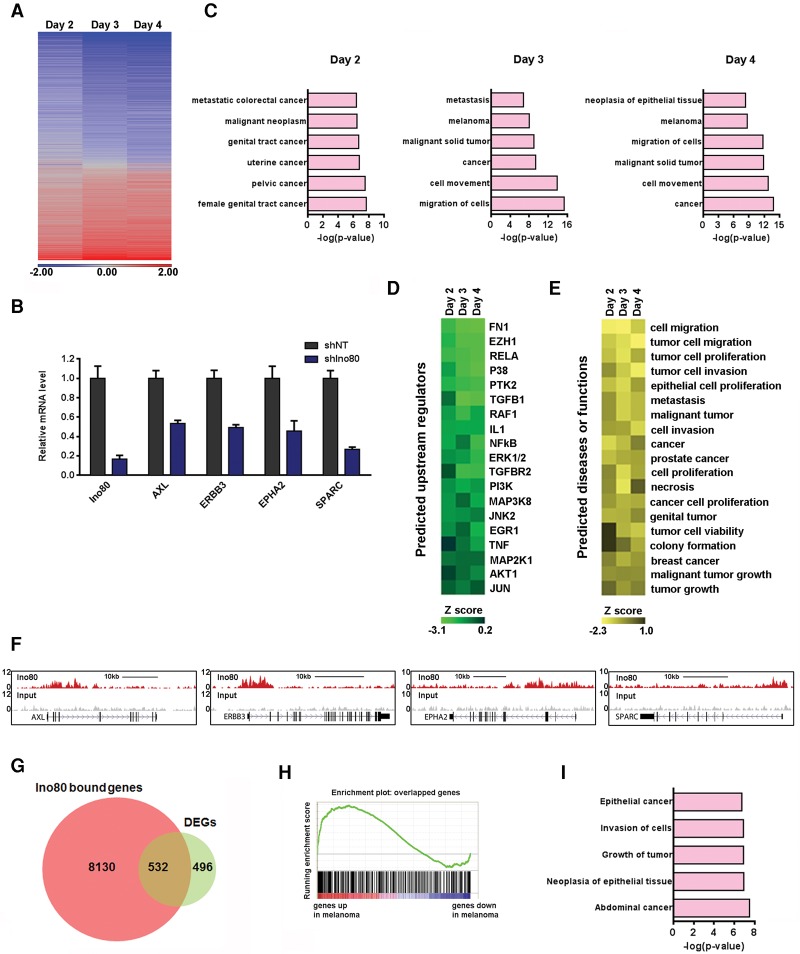
To understand how INO80 regulates melanoma growth at the molecular level, we determined gene expression changes upon Ino80 silencing at multiple time points in A375 by total RNA sequencing (RNA-seq). We found that nearly two-thirds of differentially expressed genes (DEGs) were down-regulated upon Ino80 silencing (Fig. 4A), including many known to play important roles in melanoma such as AXL, ERBB3, EPHA2, and SPARC (Fig. 4B). Consistently, Ingenuity Pathway Analysis (IPA) showed that the down-regulated genes are heavily enriched for those involved in cancer (Fig. 4C). Thus, INO80 is important for oncogene expression in melanoma. Interestingly, Ingenuity Upstream Regulator Analysis suggested that the gene expression changes caused by Ino80 silencing were similar to those caused by down-regulation of signaling pathways highly related to melanoma tumorigenesis, such as ERK1/2, AKT1, and MAP3K8 (Fig. 4D). In addition, the Ingenuity Predicted Biological Consequences showed that the gene expression changes caused by Ino80 silencing were consistent with decreased tumor-related behaviors, including tumor cell proliferation, migration, and invasion (Fig. 4E). Collectively, these analyses suggest that INO80 plays a central role in the expression of genes important for melanoma progression.
Figure 4.
INO80 regulates genes important for melanoma progression. (A) Heat map showing gene expression changes upon Ino80 knockdown in A375 cells at the indicated time points. DEGs were defined as those with a fold change of >1.5 and a false discovery rate (FDR) of <0.05, and the total number of DEGs on days 2, 3, and 4 were 1028, 1956, and 2116, respectively. (B) RT-qPCR showing down-regulation of genes important in melanoma after Ino80 silencing. Gene expression was normalized to β-actin, and values are plotted as mean ± SEM from three independent experiments. (C) IPA showing the enrichment of cancer-related genes in the DEGs upon Ino80 silencing at day 2, 3, or 4. (D) Heat map showing the upstream regulators responsible for the DEGs upon Ino80 knockdown as predicted by IPA. (E) Heat map showing the biological consequences or associated diseases downstream from the DEGs upon Ino80 knockdown as predicted by IPA. (F) Genome browser tracks showing Ino80 occupancy near selected melanoma genes AXL, ERBB3, EPHA2, and SPARC in A375 cells. (G) Venn diagram showing the overlap between DEGs after Ino80 knockdown on day 2 and Ino80-bound genes. (H) GSEA showing that the 532 INO80 targets were enriched for genes that are highly expressed in melanoma versus melanocytes. (I) IPA of the 532 Ino80 target genes. Selected top categories are shown.
To identify genes directly regulated by INO80, we carried out ChIP-seq against Ino80 in A375 melanoma cells. We identified 33,687 Ino80 peaks with high confidence (false discovery rate [FDR] of <10−6), which can be assigned to the 8662 nearest genes. Representative genome tracks showed that Ino80 occupies genomic regions associated with genes critical for melanoma development, including AXL, ERBB3, EPHA2, and SPARC (Fig. 4F). Globally, Ino80-occupied genes were significantly enriched in those that are highly expressed in melanoma (Supplemental Fig. S7A). Of the 1028 DEGs immediately down-regulated after Ino80 silencing, 532 show Ino80 occupancy in the vicinity and are likely direct targets of INO80 (Fig. 4G). Again, these INO80 targets were significantly enriched for genes that are highly expressed in melanoma compared with normal melanocytes (Fig. 4H) and were also enriched for genes involved in cancer development (Fig. 4I). Therefore, these data support the notion that INO80 may directly regulate oncogenic gene expression in melanoma cells.
Ino80 regulates oncogenic SEs
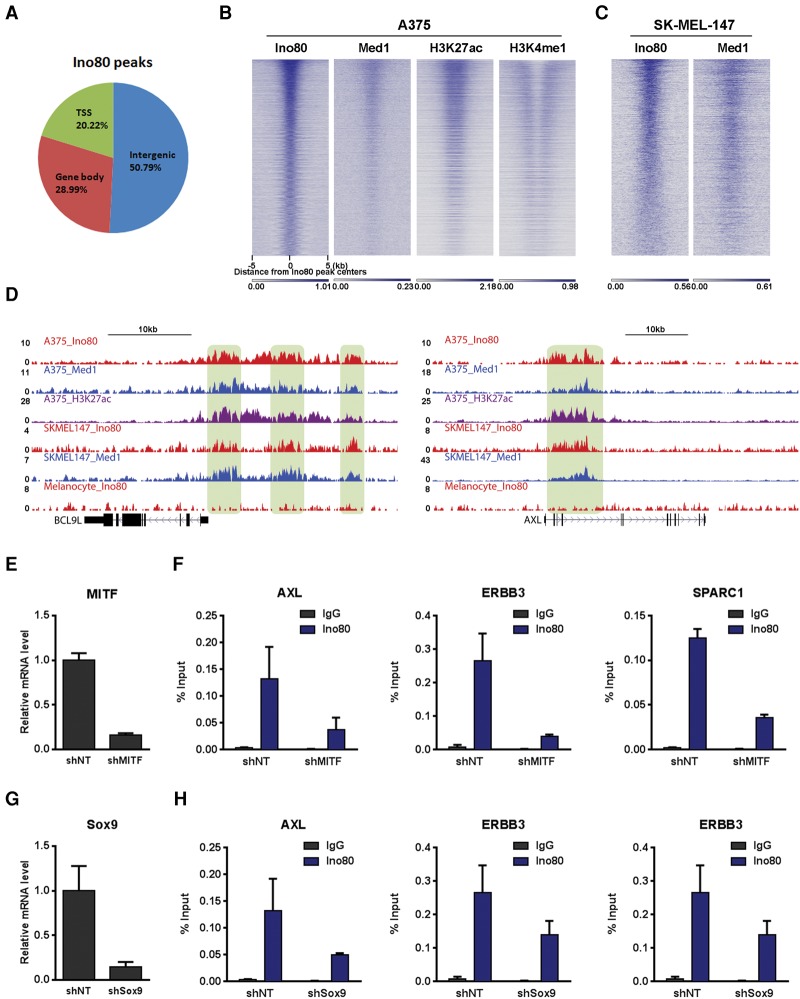
To further understand how INO80 regulates genes involved in melanoma tumorigenesis, we examined the genomic localization of Ino80 peaks and compared Ino80 localization with transcriptional regulatory sequences such as promoters and enhancers. We found that Ino80 mainly localized near intergenic regions (Fig. 5A). Furthermore, Ino80 occupancy significantly overlapped and positively correlated with enhancer and SE markers H3K4me1, H3K27ac, and Med1 in both BRAF and NRAS mutant cells (Fig. 5B,C). These results are consistent with the idea that INO80 occupies enhancers in melanoma cells. Importantly, Ino80, H3K27ac, and Med1 colocalized at enhancer regions associated with oncogenic genes such as BCL91 and AXL. Furthermore, this colocalization was observed in only melanoma cells but not primary melanocytes (Fig. 5D), suggesting that dynamic Ino80 occupancy at enhancers may contribute to the differential expression of oncogenes in melanoma. As chromatin remodelers can often be recruited by transcription factors (Iwafuchi-Doi and Zaret 2014; Laurette et al. 2015), we examined whether Ino80 occupancy is dependent on transcription factors involved in melanoma development, such as MITF and Sox9. Indeed, we found that Ino80 genomic occupancy was impaired upon MITF or Sox9 silencing (Fig. 5E–H), suggesting that the transcription factors may facilitate the recruitment of INO80 to regions near melanoma-related genes.
Figure 5.
Ino80 occupies genomic regions near melanoma-related genes. (A) Ino80 peak distribution in the genome. (B,C) Heat maps showing Ino80, Med1, H3K27ac, and H3K4me1 occupancy in the BRAF mutant A375 cells (B) or NRAS mutant SK-MEL-147 cells (C). Genes were sorted based on the Ino80 signal around the Ino80 peak center. (D) Genome browser showing Ino80, Med1, and H3K27ac occupancy near representative SE genes in A375 and SK-MEL-147 melanoma cells and in primary melanocytes. (E–H) Dependency of Ino80 occupancy on MITF and Sox9. MITF or Sox9 was silenced by lentiviral shRNAs in A375 cells (E,G), and the Ino80 occupancy at selected genes was determined by ChIP-qPCR (F,H).
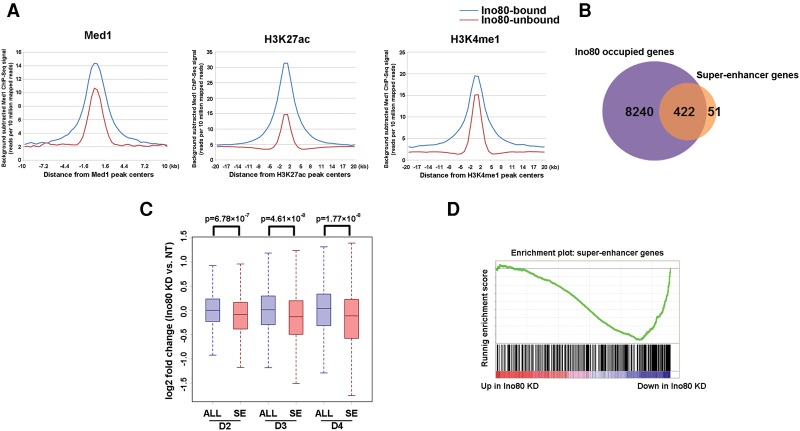
Intriguingly, Ino80 occupancy strongly correlated with that of the SE markers Med1 and H3K27ac (Fig. 5B,C). In addition, Ino80-bound regions displayed higher signals for Med1 and H3K27ac compared with Ino80-unbound regions (Fig. 6A; Supplemental Fig. S7B). Thus, we set out to test the hypothesis that INO80 may regulate oncogenic SEs. We used the Med1 ChIP-seq to define SEs in melanoma cells. This definition resulted in a total of 1084 SEs associated with 473 genes. These SE-associated genes include many well-characterized genes important for melanoma development such as SOX10 and AXL (Supplemental Fig. S7C) and are highly enriched for genes involved in cancer proliferation and invasion (Supplemental Fig. S7D). Notably, ∼90% (422 out of 473) SE-associated genes were occupied by Ino80 (Fig. 6B). Moreover, SE-associated genes, but not regular enhancer (RE)-associated-genes, were down-regulated upon Ino80 silencing (Fig. 6C; Supplemental Fig. S7E). Consistently, GSEA showed that SE genes were significantly enriched in Ino80 knockdown-repressed genes (Fig. 6D). Collectively, these results support the model that INO80 directly occupies SEs to promote the expression of oncogenic genes in melanoma.
Figure 6.
Ino80 occupies oncogenic SE genes and promotes their expression. (A) Average ChIP-seq read density of Med1, H3K27ac, and H3K4me1 at Ino80-bound and Ino80-unbound genes. (B) Venn diagram showing the overlap between Ino80-bound and SE genes. (C) Box plots showing gene expression changes for all genes (ALL) versus SE genes upon Ino80 silencing in A375 cells at the indicated time points. The P-values were calculated by Student's t-test. (D) GSEA showing that SE genes were preferentially enriched for those that are down-regulated after Ino80 silencing.
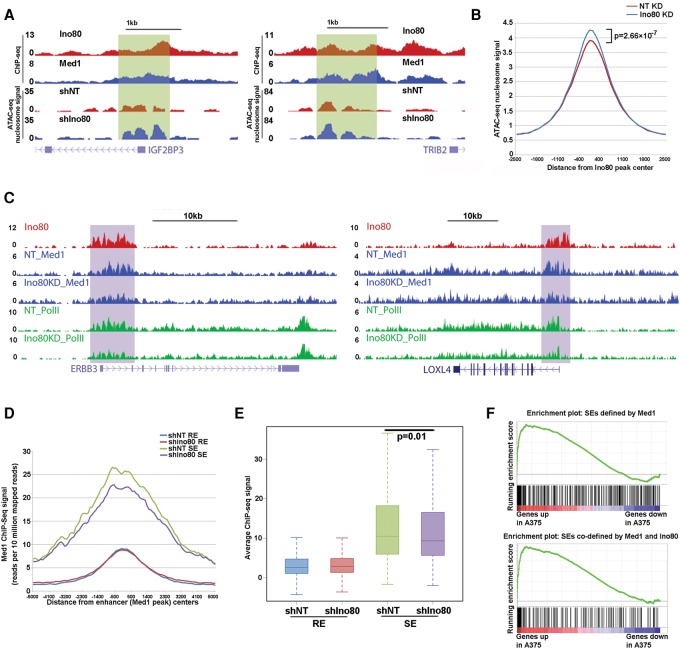
As INO80 is a chromatin remodeler and can regulate nucleosome occupancy and turnover, we hypothesized that INO80 may regulate oncogenic SEs via the action of chromatin remodeling. To test this, we determined nucleosome occupancy in melanoma cells before and after Ino80 depletion using the recently developed assay for transposase-accessible chromatin (ATAC) with high-throughput sequencing (ATAC-seq) (Buenrostro et al. 2015). We found that Ino80 silencing led to significant increases in nucleosome occupancy at Ino80-bound regions (Fig. 7A,B), suggesting that INO80 may normally promote nucleosome depletion. Interestingly, Ino80-dependent nucleosome depletion occurs at both SE- and RE-associated genes (Supplemental Fig. S7F,G). To test how INO80-mediated chromatin remodeling affects SE function, we examined the impact of Ino80 depletion on the occupancy of Med1, a component of the Mediator complex and a commonly used SE marker. Indeed, Med1 occupancy at SEs was significantly compromised upon Ino80 silencing (Fig. 7C). In contrast, Med1 occupancy at REs was not affected (Fig. 7D,E). This result is consistent with the observation that Ino80 silencing resulted in the down-regulation of SE-associated, but not RE-associated, genes (Fig. 6C; Supplemental Fig. S7E). Because SEs are bound by an unusually high density of transcription factors and enhancer markers, it is possible that they are more sensitive to changes in the chromatin state. Together, these results support the model that INO80 promotes oncogene transcription by regulating nucleosome occupancy and the assembly of oncogenic SEs in melanoma cells (Supplemental Fig. S8).
Figure 7.
INO80 regulates nucleosome occupancy and Mediator recruitment at oncogenic SEs. (A) Genome tracks showing nucleosome occupancy at SEs near melanoma-related genes in NT versus Ino80 knockdown A375 cells. (B) Average ATAC-seq nucleosome-associated read density at Ino80-bound regions in NT versus Ino80 knockdown A375 cells. The P-values were calculated by Student's t-test. (C) Genome browser tracks showing Med1 and Pol II occupancy at SEs near selected melanoma-related genes in NT or Ino80 knockdown A375 cells. (D) Average ChIP-seq read density of Med1 around the Med1 peak center on SEs versus REs in NT or Ino80 knockdown A375 cells. (E) Box plot of Med1 ChIP-seq read density on SEs versus REs in NT or Ino80 knockdown A375 cells. (F) GSEA of SE-associated genes in genes that are highly expressed in melanoma compared with melanocytes. SEs are defined by Med1 occupancy alone or Med1 and Ino80 co-occupancy.
Finally, we asked whether INO80 occupancy can be used to improve the current definition of SEs. Instead of using all Med1 peaks, we redefined SEs using only Med1 peaks that were co-occupied by Ino80. We found that this new definition produced a list of SEs that were better associated with genes selectively expressed in melanomas compared with melanocytes (Fig. 7F). This result suggested that INO80 may be an integral component of SEs. Collectively, our results reveal a critical role of the INO80 chromatin remodeling complex in the determination of cell type-specific gene expression in cancer.
Discussion
SEs play critical roles in driving the expression of cell identity genes (Hnisz et al. 2013; Loven et al. 2013; Whyte et al. 2013; Chipumuro et al. 2014). Therefore, uncovering key factors that dictate the dynamic regulation of SEs will provide effective ways to manipulate cell fates during development and disease. In this study, we identified INO80 as a novel and important component of SEs. Using INO80 and Med1 occupancy, we defined the SEs in melanoma cells. Compared with SEs defined based on Med1 occupancy alone, those co-occupied by INO80 and Med1 are better enriched for genes specifically expressed in melanoma and may provide new insights into melanoma tumorigenesis. More importantly, we showed that INO80-mediated chromatin remodeling is required for Med1 occupancy and the full activity of SEs in melanoma. Consistent with our findings, SEs have been proposed to be sensitive to perturbation of chromatin-based mechanisms (Pott and Lieb 2015). In addition, nucleosome positioning and chromatin remodeling are known to influence the activity of REs (Hu et al. 2011; Laurette et al. 2015), and nucleosome occupancy changes over key regulatory regions during cell fate transition (West et al. 2014). Thus, our data reveal a critical role of chromatin remodeling in the assembly and function of SEs and identify INO80 as a potential target for the regulation of cell type-specific gene expression in disease and development. It is worth noting that Ino80 depletion was previously reported to inhibit proliferation and anchorage-independent growth of oncogene transformed mouse embryonic fibroblasts. However, INO80 haploinsufficiency had no impact on tumor incidence and latency in p53 deletion mice (Min et al. 2013). It is possible that there may still be a sufficient amount of Ino80 in the heterozygote mice to allow tumorigenesis, and different cancers or different cell types may show different dependency on the expression level of Ino80.
In addition to the driver mutations (Hodis et al. 2012; The Cancer Genome Atlas Network 2015), recent studies have begun to reveal the roles of epigenetic mechanisms in the regulation of oncogene expression, tumorigenesis, and tumor maintenance (Lee et al. 2014). For example, mutations in the SWI/SNF chromatin remodeling complex were found to be prevalent in many cancers, and the complex itself was suggested to function as a tumor suppressor (Gonzalez-Perez et al. 2013; Shain and Pollack 2013). In both melanocytes and melanoma, the SWI/SNF complex interacts with master transcription factors and promotes cell differentiation and survival (Mehrotra et al. 2014). In comparison, we showed that INO80 silencing reduces oncogenic transcription and selectively inhibits the growth of both BRAF and NRAS mutant melanoma cells but not primary melanocytes. Thus, our study provides a novel and potentially promising strategy to inhibit cancer progression by targeting the INO80 chromatin remodeling complex, which may complement and overcome challenges in existing targeted therapies (Marzuka et al. 2015).
Finally, we found that INO80 selectively occupies oncogenic SEs in melanoma cells but not primary melanocytes, suggesting that chromatin remodeling may act as a critical step during the establishment of oncogenic SEs during tumorigenesis. In addition, our data showed that driver kinase inhibition reduced Ino80 expression. At the same time, Ino80 silencing impairs BRAF and NRAS mutant cell growth and tumorigenesis. Thus, the driver kinases may hijack INO80 as a means to establish oncogenic SEs and promote tumor formation. Consistent with this notion, a recent report showed that SEs provide a platform for signaling pathways to regulate genes that control cell identity during tumorigenesis (Hnisz et al. 2015). We propose that INO80 may play an important role in bridging upstream signaling pathways and the transcriptional activation of oncogenes.
Materials and methods
Cell lines
Human melanoma cells (A375, SK-MEL-28, A2058, SK-MEL-147, and SK-MEL-119) were purchased from the Tissue Culture Facility at the University of North Carolina at Chapel Hill. All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% penicillin/streptomycin (Invitrogen). Human melanocytes were purchased from Life Technologies and maintained in Medium 254 with the addition of human melanocyte growth supplement (HMGS) (Life Technologies).
Antibodies
The following antibodies were used in this study: Ino80 (Proteintech, 24819-1-AP) and β-actin (Sigma, A2228) for Western blotting; Ino80 (Proteintech, 18810-1-AP) for immunohistochemistry staining; and Ino80 (Proteintech, 18810-1-AP), H3K4me1 (Active Motif, 39297), H3K27ac (Abcam, ab4729), Med1 (Bethyl Laboratories, A300-793A), and RNA Pol II (Santa Cruz Biotechnology, SC-899X) for ChIP.
Human samples
Approval for this study was obtained from the ethics committee of Wuhan University. All study samples were obtained from patients between 2009 and 2014 who were treated at Renmin Hospital of Wuhan University (Wuhan, China), with written informed consent from the patients. Two pathologists (J.P. Yuan and Y.B. Huang) reconfirmed the histopathological features of these samples.
Immunohistochemistry staining
Paraffin-embedded human melanoma or nevus samples were immunostained using standard protocols. Sections were incubated with anti-Ino80 antibody (1:100; Proteintech, 18810-1-AP), horseradish peroxidase-conjugated goat anti-rabbit IgG, and 3,3-diaminobenzidine successively. Afterward, sections were counterstained with hematoxylin. Representative images from six melanoma samples and three nevus samples are shown.
Western blotting
Cells were lysed in cell lysis buffer (Thermo Scientific) containing 1× protease inhibitor tablet (Roche). The lysates were subsequently sonicated and quantified by BCA assay (Life Technologies). Forty micrograms of proteins was loaded and separated by SDS-PAGE on 4%–12% Bis-Tris gel (Life Technologies) and transferred to nitrocellulose membranes. The membranes were blocked by 5% milk in TBS-T and then incubated with primary antibodies overnight at 4°C. Afterward, membranes were incubated with the appropriate secondary antibodies and developed using the ECL detection kit (GE Healthcare Bio-Sciences).
RNA isolation, reverse transcription, and RT-qPCR
Total RNA was isolated from cells using the GeneJet RNA purification kit (Thermo Scientific), and 0.5 µg of total RNA was reverse-transcribed to generate cDNA using the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. qPCR was performed using the SsoFast EvaGreen supermix (Bio-Rad) on the Bio-Rad CFX-384 or CFX-96 real-time PCR detection system. Actin was used for normalization. Primers used in the study are listed in Supplemental Table S1.
Cell proliferation assay
Standard MTT proliferation assays were performed. Cells were plated at the appropriate densities (1 × 103 to 2 × 103 cells per well) in 96-well plates. The next day (day 1), 5 mg/mL MTT/PBS was added to each well to one-tenth the original culture volume and incubated for 4 h at 37°C. Medium was carefully removed, and 150 µL of 0.04 N HCl in absolute isopropanol was added to each well to solubilize the resulting formazan and shaken for 15 min at room temperature. Data were collected with a Synergy 2 multimode plate reader (BioTek). The procedures were repeated on days 3, 5, and 7. Data were normalized to the average value of day 1, and growth curves were generated from the average of all experiments.
Clonogenic assay
Cells were plated in triplicate at the appropriate cell densities for each cell line (100–500 cells per well) in six-well plates and cultured for 2–4 wk for colony formation. Resulting colonies were washed once with PBS, fixed, and stained with 0.05% (w/v) crystal violet in 50% methanol and 10% acetic acid for 30 min at room temperature. Colonies were carefully rinsed with distilled water until background staining of the wells was minimal. Plates were air-dried and scanned. Staining intensities were quantified with ImageJ (version 1.45s).
Soft agar assay
Cells were resuspended in 0.4% agar in complete medium at 1 × 104 cells per well in six-well plates and layered on top of 0.6% agar as described previously (Shutes et al. 2004). After 14 d, colonies were stained with 2 mg/mL MTT dissolved in PBS for 1 h at 37°C. Plates were scanned, and colonies were quantified with ImageJ.
Mouse xenograft experiments
All animal experiments were conducted following protocols approved by the Institutional Animal Care and Use Committee of the National Institute of Environmental Health Sciences. A375 cells or SK-MEL-147 cells (2 × 106 cells) carrying a luciferase reporter plasmid (Promega) with either lentiviral-based NT shRNA or Ino80 shRNA were injected subcutaneously into the flanks of 8- to 10-wk-old SCID-Beige mice (Charles River Laboratories). For experiments with preformed tumors, A375 cells transduced with either inducible NT shRNA or inducible Ino80 shRNA lentivirus were injected subcutaneously into the flanks of SCID-Beige mice, and cells were allowed to grow for 12 d to form tumors. After that, 2 mg/mL Dox was administered orally in the drinking water to induce the expression of corresponding shRNAs. For bioluminescence imaging experiments, 15 mg/mL D-luciferin (Caliper LifeSciences) was injected intraperitoneally based on 10 µL per gram of body weight. At 10–15 min after injection, luciferase signal was detected by a Xenogen IVIS imaging system (STTARR). Mice were sacrificed at the indicated time points, and tumor mass and size were measured. Tumor size was calculated based on a formula described previously (Tomayko and Reynolds 1989).
RNA-seq
A375 cells were transduced with NT (NT shRNA) or Ino80 (Ino80 shRNA) lentiviral-based shRNAs. Total RNA was extracted from cells using the GeneJet RNA purification kit (Thermo Scientific). One microgram of RNA was used for sequencing library generation using the TruSeq RNA library preparation kit version 2 (Illumina) according to the manufacturer's instructions. All libraries were sequenced on the NextSeq sequencer (Illumina) using the 75-nucleotide (nt) paired-end sequencing protocol.
ChIP-seq
Ino80, H3K4me1, H3K27ac, Med1, and RNA Pol II ChIP were performed as described previously (Whyte et al. 2013). Briefly, A375 or SK-MEL-147 cells were fixed using 1% formaldehyde for 10 min, and 0.125 M glycine was added to stop the fixation. Cells were harvested, and DNA was fragmented to 300–500 base pairs (bp) by sonication with a microtip attached to a Misonix 3000 sonicator. Immunoprecipitation was performed with antibodies conjugated to Dynabeads protein G beads (Life Technology). ChIP DNA was eluted, reverse-cross-linked, extracted by phenol/chloroform, and precipitated.
For the Med1 and Pol II ChIP in Figure 4, A375 cells were transduced with NT shRNA or Ino80 shRNA viruses. After 48 h, cells were harvested for ChIP against Med1 or Pol II, based on the procedure described above. For ChIP-seq, 1 ng of ChIP DNA or input DNA was used to generate sequencing libraries using the Nextera XT DNA sample preparation kit (Illumina). Libraries were sequenced on the NextSeq sequencer (Illumina) using the 75-nt paired-end sequencing protocol. Two biological replicates were performed for each ChIP-seq sample, and reads were combined for analysis.
ATAC-seq
ATAC-seq (Buenrostro et al. 2015) was performed with a modified protocol. Briefly, A375 cells were transduced with lentiviral shRNAs. At 48 h after transduction, 80,000 cells were harvested and washed twice with cold PBS. Cells were lysed with 25 µL of CSK buffer (10 mM PIPES at pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.1 % Triton X-100) for 5 min on ice. After lysis, cell nuclei were spun down, and 25 µL of transposase solution (12.5 µL of 2× TD buffer, 5 µL of Tn5 transposase, 7.5 µL of nuclease free water) was added. Transposition reactions were incubated for 30 min at 37°C. After incubation, DNA was purified with the MinElute PCR purification kit (Qiagen) and subsequently amplified by PCR. The PCR reaction was set up as follows: 10 µL of eluted DNA, 2.5 µL of 25 µM index primer 1, 2.5 µL of 25 µM index primer 2, 25 µL of New England Biolabs 2× PCR master mix, and 5 µL of nuclease-free water. PCR was run with a 12-cycle program (5 min at 72°C, 30 sec at 98°C, 10 sec at 98°C, 30 sec at 63°C, and 1 min at 72°C; repeat steps 2–5 for another 11 cycles). The quality of amplified DNA was examined on a Bioanalyzer, and the DNA was subsequently sequenced on a NextSeq sequencer by 75-nt paired-end sequencing.
RNA-seq data analysis
Raw reads were filtered to include only those with a median Phred quality score of ≥20 and trimmed with Galore! version 0.2.8 to remove adapter sequences. The average mate inner distance and variance of the fragment lengths were estimated by first aligning a subset of the reads to the hg19 known gene transcriptome using Bowtie version 0.12.8 with the following parameters: -v 2 -m 1 -X 1000. Next, the CollectInsertSizeMetrics tool from Picard tools suite version 1.86 was used to obtain the mean and variance estimates for the fragment lengths, and the average mate inner distance was calculated based on the mean fragment length and the read length. The gene model was defined using RefGene annotations downloaded from the University of California at Santa Cruz (UCSC) browser on February 5, 2013. The filtered and trimmed reads were aligned to the hg19 genome assembly using TopHat2 version 2.0.4 with the mate inner distance, mate standard deviation, and gene model parameters defined from the previous steps and the following additional parameter: -g 10. The expression was quantified using CuffLinks version 2.0.2 with the gene model to obtain FPKM (fragments per kilobase per million mapped fragments) measurements for each gene. Only genes with an FPKM value of ≥0.1 in all samples were kept for downstream analysis.
Public RNA-seq data for seven melanoma cell lines from Gene Expression Omnibus (GEO; GSE46817) and public RNA-seq data for one melanocyte cell line from GEO (GSM1138580) were processed and aligned, and FPKM values were calculated in the same way as the other RNA-seq data. Genes were filtered to include only those with an FPKM value of ≥1 in all samples, and quantile normalization was performed. The log2 fold change of the average of the normalized melanoma FPKM values over the melanocyte FPKM value was calculated for each gene.
GSEA between vemurafenib treatment and Ino80 silencing
Gene expression data for vemurafenib-treated A375 cells were obtained from GEO (GSE42872). For genes with multiple probes, a single expression measurement was calculated by taking the mean across all probes. P-values were calculated for each gene using a two-sample Student's t-test, and FDR values were corrected for multiple comparisons using the Benjamini-Hochberg method. Vemurafenib target genes were defined as those that were down-regulated upon vemurafenib treatment with a fold change of >1.5 and an FDR of <0.01. Gene expression changes after Ino80 silencing were determined as described above.
ChIP-seq analysis
Cutadapt version 1.2.1 was used to trim any reads containing an adapter sequence. The trimmed reads were filtered to include only those with a median Phred quality score of ≥20. The trimmed and filtered reads were aligned to the hg19 genome assembly using Bowtie version 0.12.8 with the following parameters: -v 2 -m 1 --best --strata -I 15 -X 1000. We removed duplicate read pairs where both mates in a pair aligned to the same genomic positions. Coverage tracks were generated from the full-length aligned fragments using the genomecov tool from BEDTools suite version 2.16.2. The depths in the coverage tracks were normalized by multiplying by 10 million and dividing by the total number of aligned read pairs. Peaks were called and assigned to genes using SICER version 1.1 and the following parameters: redundancy_threshold = 100, window_size = 200, gap_size = 800, species = hg19, effective_genome_fraction = 0.77, and FDR = 0.00001. Bound genes were defined by finding the TSS closest to each peak center using RefGene annotations downloaded from the UCSC browser on February 5, 2013, and only those whose peak center was within 20 kb of the TSS were included. Heat maps were generated by calculating the average normalized depth in 50-nt bins that span 5 kb upstream of and downstream from the peak centers. Metagenes were generated by averaging each 50-nt bin across all peaks. Box plots were generated using the average depth within the entire window 5 kb upstream of and downstream from each peak center. This average depth for each peak was used to calculate a P-value between two groups using a two-sample Student's t-test.
ATAC-seq analysis
Cutadapt version 1.2.1 was used to trim any reads containing an adapter sequence. The trimmed reads were filtered to include only those with a median Phred quality score of ≥20. The trimmed and filtered reads were aligned to the hg19 genome assembly using Bowtie version 0.12.8 with the following parameters: -v 2 -m 1 --best --strata -I 15 -X 1000. We removed duplicate read pairs where both mates in a pair aligned to the same genomic position. To adjust the fragments to represent the center of the transposon-binding event, all reads aligning to the plus strand were offset by +4 nt, and those aligning to the minus strand were offset by −5 nt. Read pairs were split into two groups based on fragment length: Those <100 nt were defined as open chromatin, and those between 180 and 247 nt were defined as mononucleosome. Coverage tracks were generated for both groups from the full-length aligned fragments using the genomecov tool from BEDTools suite version 2.16.2. The depth in the coverage tracks were normalized by multiplying by 10 million and dividing by the total number of aligned read pairs in each group. Further normalization of the nucleosome group was carried out by subtracting the normalized open chromatin signal from it. Metagenes and P-values were calculated in the same way as for the ChIP-seq data.
SE definition
The Med1 ChIP-seq data were used to identify SEs based on the method described previously (Whyte et al. 2013). Briefly, a rank was assigned to each of the Med1 peaks based on their SICER FDR values. Next, a plot with rank on the X-axis and −log10(FDR value) on the Y-axis was created, normalizing the plot such that each axis ranged between 0 and 1. Finally, the point in the curve where the slope transitioned to >1 was identified. All of the peaks that fell beyond this transition point were defined as SEs. SE-associated genes were defined by finding the TSSs closest to each SE center, only including those whose center was within 20 kb of the TSS.
Accession numbers
The GEO accession number for the data reported in this study is GSE 82334.
Supplementary Material
Acknowledgments
We thank the Epigenomics core facility at National Institute of Environmental Health Sciences for providing the Illumina sequencing services. This study was supported in part by the Intramural Research Program of the National Institutes of Health, the National Institute of Environmental Health Sciences (Z01ES102745 to G.H.) and National Institutes of Health grants CA042978 and CA161494 (to A.D.C.). B.Z., L.W., A.D.C., and G.H. designed the experiments. B.Z. and L.W. carried out the experiments in melanoma cell lines. L.W. and S.Z. carried out animal experiments. F.H., Y.Z., and C.X. carried out experiments in patient samples. L.W. and P.L. carried out genomic experiments. B.B., D.F., L.H., and L.D. carried out bioinformatics analysis. B.Z., L.W., A.D.C., and G.H. interpreted the data and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.277178.115.
References
- Affer M, Chesi M, Chen WD, Keats JJ, Demchenko YN, Tamizhmani K, Garbitt VM, Riggs DL, Brents LA, Roschke AV, et al. 2014. Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 28: 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21 29 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2015. Genomic classification of cutaneous melanoma. Cell 161: 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. 2013. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24: 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, Abraham BJ, Sharma B, Yeung C, Altabef A, et al. 2014. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159: 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Jene-Sanz A, Lopez-Bigas N. 2013. The mutational landscape of chromatin regulatory factors across 4,623 tumor samples. Genome Biol 14: r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, van der Velden VH, Havermans M, Avellino R, van Lom K, et al. 2014. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell 157: 369–381. [DOI] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. 2015. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16: 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. 2015. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell 58: 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature 463: 474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. 2012. A landscape of driver mutations in melanoma. Cell 150: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q, Gattinoni L, Restifo NP, Huang S, Zhao K. 2011. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res 21: 1650–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. 2014. Pioneer transcription factors in cell reprogramming. Genes Dev 28: 2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Orkin SH. 2011. Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Med 3: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, et al. 2014. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511: 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurette P, Strub T, Koludrovic D, Keime C, Le Gras S, Seberg H, Van Otterloo E, Imrichova H, Siddaway R, Aerts S, et al. 2015. Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. eLife 10.7554/eLife.06857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Murphy GF, Lian CG. 2014. Melanoma epigenetics: novel mechanisms, markers, and medicines. Lab Invest 94: 822–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, et al. 2014. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346: 1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzuka A, Huang L, Theodosakis N, Bosenberg M. 2015. Melanoma treatments: advances and mechanisms. J Cell Physiol 230: 2626–2633. [DOI] [PubMed] [Google Scholar]
- Masliah-Planchon J, Bieche I, Guinebretiere JM, Bourdeaut F, Delattre O. 2015. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol 10: 145–171. [DOI] [PubMed] [Google Scholar]
- Mayes K, Qiu Z, Alhazmi A, Landry JW. 2014. ATP-dependent chromatin remodeling complexes as novel targets for cancer therapy. Adv Cancer Res 121: 183–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra A, Mehta G, Aras S, Trivedi A, de la Serna IL. 2014. SWI/SNF chromatin remodeling enzymes in melanocyte differentiation and melanoma. Crit Rev Eukaryot Gene Expr 24: 151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Reuter JA, Spacek DV, Snyder M. 2015. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet 47: 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JN, Tian Y, Xiao Y, Wu L, Li L, Chang S. 2013. The mINO80 chromatin remodeling complex is required for efficient telomere replication and maintenance of genome stability. Cell Res 23: 1396–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, Shen X. 2009. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol 10: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Lee C, Zichner T, Stutz AM, Erkek S, Kawauchi D, Shih DJ, Hovestadt V, Zapatka M, Sturm D, et al. 2014. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 511: 428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti P, Zeilner A, Lusser A. 2011. ATP-dependent chromatin remodeling factors and their roles in affecting nucleosome fiber composition. Int J Mol Sci 12: 6544–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott S, Lieb JD. 2015. What are super-enhancers? Nat Genet 47: 8–12. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. 2004. The SWI/SNF complex—chromatin and cancer. Nat Rev Cancer 4: 133–142. [DOI] [PubMed] [Google Scholar]
- Shain AH, Pollack JR. 2013. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8: e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutes A, Berzat AC, Cox AD, Der CJ. 2004. Atypical mechanism of regulation of the Wrch-1 Rho family small GTPase. Curr Biol 14: 2052–2056. [DOI] [PubMed] [Google Scholar]
- Tomayko MM, Reynolds CP. 1989. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol 24: 148–154. [DOI] [PubMed] [Google Scholar]
- van den Hurk K, Niessen HE, Veeck J, van den Oord JJ, van Steensel MA, Zur Hausen A, van Engeland M, Winnepenninckx VJ. 2012. Genetics and epigenetics of cutaneous malignant melanoma: a concert out of tune. Biochim Biophys Acta 1826: 89–102. [DOI] [PubMed] [Google Scholar]
- Walker BA, Wardell CP, Brioli A, Boyle E, Kaiser MF, Begum DB, Dahir NB, Johnson DC, Ross FM, Davies FE, et al. 2014. Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J 4: e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X, Miao YL, Zhou B, Han L, Fargo DC, et al. 2014. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14: 575–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Peterson CL. 2010. The INO80 family of chromatin-remodeling enzymes: regulators of histone variant dynamics. Cold Spring Harb Symp Quant Biol 75: 35–42. [DOI] [PubMed] [Google Scholar]
- Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. 2014. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet 46: 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Cook A, Alver BH, Stadtfeld M, Deaton AM, Hochedlinger K, Park PJ, Tolstorukov MY, Kingston RE. 2014. Nucleosomal occupancy changes locally over key regulatory regions during cell differentiation and reprogramming. Nat Commun 5: 4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.