Abstract
An algorithm for intensity-based 3D–2D registration of CT and x-ray projections is evaluated, specifically using single- or dual-projection views to provide 3D localization. The registration framework employs the gradient information similarity metric and covariance matrix adaptation evolution strategy to solve for the patient pose in six degrees of freedom. Registration performance was evaluated in an anthropomorphic phantom and cadaver, using C-arm projection views acquired at angular separation, Δθ, ranging from ~0°–180° at variable C-arm magnification. Registration accuracy was assessed in terms of 2D projection distance error and 3D target registration error (TRE) and compared to that of an electromagnetic (EM) tracker. The results indicate that angular separation as small as Δθ ~10°–20° achieved TRE <2 mm with 95% confidence, comparable or superior to that of the EM tracker. The method allows direct registration of preoperative CT and planning data to intraoperative fluoroscopy, providing 3D localization free from conventional limitations associated with external fiducial markers, stereotactic frames, trackers and manual registration.
Keywords: 3D–2D image registration, target registration error, image-guided surgery
1. Introduction
A surgical guidance system is intended to assist a surgeon in localizing anatomical targets with respect to surgical instruments while helping to avoid injury to adjacent normal tissue. The predominant basis for surgical guidance involves dedicated tracking systems (e.g., optical or electromagnetic (EM) that track the location of predefined markers attached to the patient and surgical instruments. Navigation in the context of 3D images (and surgical planning data therein) is achieved through registration of the coordinate system associated with the tracker with that of the image, most often using manual procedures such as touching predefined fiducials with a pointer (Maurer et al 1997, Youkilis et al 2001, Vahala et al 2001).
Intraoperative fluoroscopy (2D x-ray projections) are fairly common in the operating room—especially in minimally invasive procedures—but the images are most often only qualitatively interpreted, and there is growing interest in the capability to accurately align the 2D data with 3D images and planning (Söderman et al 2005, Hanson et al 2006). Compared to surgical trackers that register a sparse set of features (i.e., fiducial markers), intraoperative images provide rich, up-to-date information that includes accurate depiction of anatomical deformation and resection within the region of interest. However, in the context of surgical guidance, they provide a limited (2D) view of the 3D scene and thereby seem limited in their utility for 3D localization—hence, the use of systems capturing multiple views (e.g., biplane imaging) for interpreting 2D images within a more accurate 3D context. For a human observer, biplane imaging (i.e., projections acquired with angular separation, Δθ ~ 90°) is common, since it presents familiar (e.g., AP and LAT) anatomical views and simplifies the mental correspondence of two projection views with the 3D image and planning context.
Incorporation of preoperative 3D image and planning information into intraoperative 2D images via 3D–2D registration has been extensively investigated, showing utility in increasing the precision and accuracy of interventional radiology, surgery, and radiation therapy (Comeau et al 2000, Fei et al 2003, Wein et al 2005, Micu et al 2006, Birkfellner et al 2007, Hummel et al 2008, Frühwald et al 2009, Markelj et al 2012). Use of multiple views has been suggested, as in tiling images in a composite manner (Penney et al 1998) or using the metric of one image pair per optimizer iteration in an alternating manner (Leventon et al 1997). Previous work in spine surgery—the ‘LevelCheck’ method—computes a 3D–2D registration from a single radiograph, to overlay the locations of target vertebrae as defined in preoperative CT (Otake et al 2012). The method was shown to be robust against the presence of tools, variations in patient positioning, and gross anatomical deformation (Otake et al 2013b). Such methods are designed to assist the surgeon by projecting information from the 3D image onto the 2D intraoperative image, thereby providing a familiar image context that reliably depicts anatomy and the position of interventional devices (Weese et al 1997).
In this study we extend the 3D–2D registration framework underlying the LevelCheck algorithm to provide a basis for 3D surgical guidance similar to that achieved via external tracking systems. We investigate the geometric accuracy and specifically focus on registration using one or two fluoroscopic views acquired at angular separation (Δθ) ranging from ~0° (single perspective) to ~90° (biplane fluoroscopy) and ~180° (opposing views). We furthermore identify the minimum angular separation required to yield accuracy in 3D localization that is equivalent or better to that achieved with a conventional surgical tracker. Distinct contributions of the current work include: (1) extending the utility of intraoperative fluoroscopy from that of qualitative interpretation to that of a reliable roadmap in which 3D image and planning information is accurately resolved; and (2) enabling accurate 3D surgical guidance without trackers—i.e., using the imaging system itself as a tracker (and the patient him/herself as the registration ‘fiducial’). The system potentially absolves the complexities of conventional navigational tools, such as manual setup using fiducials and/or fixation frames, line of sight (in optical tracking), metal artifacts (in EM tracking), additional equipment, and gradual deterioration of the image-to-world registration during the case. Experiments were conducted in anthropomorphic phantom and cadaver to assess the feasibility and accuracy of the proposed 3D–2D guidance method. Studies investigated the accuracy of 3D localization in comparison to that of a commercial EM tracker in various anatomical sites as a function of C-arm magnification, the voxel size in preoperative CT images, the pixel size in intraoperative fluoroscopy images, and—most importantly—the angular separation between projection image pairs used in the 3D–2D registration process.
2. Methods
2.1. 3D–2D registration
2.1.1. Algorithm
The algorithm for 3D–2D registration iteratively solves the transformation of a 3D image (e.g., preoperative or intraoperative CT) such that a 2D projection computed from the 3D image (i.e., a digitally reconstructed radiograph (DRR)) yields maximum similarity to the intraoperative 2D image (e.g., x-ray radiograph acquired via C-arm fluoroscopy). This process amounts to calculation of the six degrees of freedom (DoF) of the patient pose that aligns the preoperative patient image and surgical plan with the actual 2D projection. The basic algorithm was described in detail in (Otake et al (2012) and (2013b)) in application to labeling surgical targets (viz., vertebral levels—ergo, the ‘LevelCheck’ algorithm), and a brief summary is provided below. We extend the process to utilize multiple projections acquired at different C-arm angles, such that a joint optimization provides accurate triangulation for 3D localization (not just 2D overlay).
CT images are first converted from Hounsfield units (HU) to linear attenuation coefficients (μ, units of mm−1) based on the coefficient of water at an effective energy of the CT acquisition, and the intraoperative x-ray projections (radiography or fluoroscopy) are log-normalized. The similarity between the intraoperative radiograph (pF, fixed image) and a DRR (pM, moving image) is then defined in terms of the gradient information (GI) metric (Pluim et al 2000):
| (1) |
where i, j are pixel indices within the image domain Ω, and the gradient (g) is
| (2) |
The weighting function (w) favors small gradient angles (i.e., alignment of edges):
| (3) |
The use of GI was motivated in part by its robustness against potential mismatches between the images, since the min(*) operator ensures both images present strong gradients. Therefore, GI only accrues information that is common in both the radiograph and the DRR, and gradients present in only one of the images (e.g., a surgical device) do not contribute to GI. Similarly with respect to anatomical deformation, the similarity metric provides a degree of robustness by ignoring inconsistent gradients between deformed tissues and instead relies upon consistent information presented by locally rigid structures. Additional robustness to motion and deformation is offered by an optional GI mask to differentially weight regions of the image (e.g., higher weight in the center than the periphery) and a multi-start method in the optimization process (Otake et al 2013b).
When multiple (N) projections are provided, we sum the respective similarity measures (Russakoff et al 2005, Jomier et al 2006, Turgeon et al 2005, Sadowsky et al 2011, Zollei et al 2001, Lemieux 1994), such that
| (4) |
Taking the sum of GI values is equivalent to a composite approach (Penney et al 1998) in which multiple images are considered one large image, and a single similarity measure is computed. A more in-depth comparison of alternative approaches is the subject of possible future research. The optimization problem was therefore to solve for the six DoF transform maximizing GI:
| (5) |
where the 3D–2D registration ( ) was solved by an iterative search of the translation (tx, ty, tz ) and rotation (rx, ry, rz ) parameters, with (x, y, z) coordinates defined in figure 1. The covariance matrix adaptation evolution strategy (CMA-ES) (Hansen et al 2009) was employed, which generates a population (λ) of random sample points around the current estimate at each iteration, and evaluates the objective function per sample. The population is generated according to a multivariate normal distribution, where the mean and covariance of the distribution are updated per generation such that it is approximately aligned with the gradient direction of the objective. A major advantage of this stochastic approach is its robustness against local minima, with nominal parameter values shown in table 1. The convergence of CMA-ES can be slow and require a large number of function evaluations, but the method is amenable to parallel evaluation as implemented on graphics processing units (GPU) and described in Otake et al (2012) and below. It may also be implemented in a multi-resolution framework, which limits the local search space. Considering the application of interest in this study, in which projections are acquired in a consecutive manner throughout the procedure, each registration can initialize the next to within a local neighborhood of the solution. With the benefit of robust initialization via global search, the search space was limited to within ± 10 {mm,°} (as described below), and we did not employ a multi-resolution scheme.
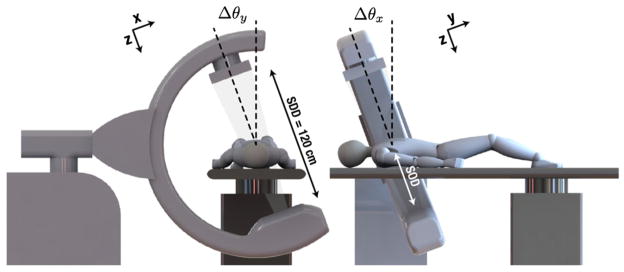
Figure 1.
Illustration of mobile C-arm geometry, coordinate frames and angulations (Δθ about the longitudinal or lateral axis) to achieve projection image pairs.
Table 1.
Summary of 3D–2D registration parameters and nominal values.
| Parameter | Symbol | Nominal value |
|---|---|---|
| Initial registration | – | ~5–10 mm PDE |
| Optimizer population (offspring) size | λ | 50 |
| Optimizer step size (standard deviation) | σ | 5 {mm,°} |
| Optimizer (upper/lower) bounds | – | ± 10 {mm,°} |
| Optimizer stopping criterion (fitness) | – | 0.01 {mm,°} |
| Separation between projection views | Δθ | 0°–180° |
| C-arm geometric magnification | m | 2.0 |
| CT voxel size (slice thickness) | az | 0.6 mm |
| Fluoro pixel size | bu,v | 0.6 mm |
2.1.2. Implementation
The algorithm was implemented utilizing the parallel computation capabilities of GPUs. The basic implementation was based on previous work (Otake et al 2012) and (Otake et al 2013b), where DRRs are generated via forward projection of 3D images using parallelized ray-tracing algorithms. A linear projection operator was used due to its simplicity and amenability to GPU (i.e., efficient use of hardware-accelerated interpolation—texture fetching), using a step size equal to the voxel size. The Siddon projection algorithm (Siddon 1985) in which the analytically exact line integral was computed by accumulating the intersection length between the ray and intersecting voxels, was also implemented for use in experiments where the slice thickness was varied (section 2.2.3) to remove potential bias from step-size selection. Finally, the CMA-ES algorithm allowed computation of each sample of a generation in parallel, where GI per generation was computed in parallel on GPU.
A number of parameters governing the registration process are summarized in table 1. Although the workflow envisioned (i.e., consecutive acquisition of fluoro shots without major changes in the anatomical scene) allows for robust initialization (i.e., the previous solution initializes the next), an initial global registration is still required at the beginning of the process. This global search was solved in previous work (Otake et al 2012) and (Otake et al 2013b), including conditions of strong deformation between the preoperative CT and intraoperative fluoroscopy, and its reported accuracy of ~5–10 mm projection distance error (PDE) was used as the basis for initialization in studies reported below. Initial registrations were thus obtained by randomly perturbing all six DoFs such that they produced at least 5 mm PDE. The optimizer step size and upper/lower bounds were selected accordingly, searching within ± 10 {mm,°} for translation and rotation, respectively, with a standard deviation of 5 {mm,°}.
The optimization was terminated when the change at each coordinate was less than the stopping criterion. To ensure repeatable convergence, the stopping criteria and population size were tested over a range of 0.01–0.1 mm and 10–100, respectively. Both resulted in a reproducible transform (e.g., TRE with a standard deviation of 6 × 10−3 mm), thus demonstrating convergence.
2.2. Experiments
2.2.1. Imaging systems
Fluoroscopic images were acquired using a mobile C-arm incorporating a flat-panel detector and a motorized orbit (Siewerdsen et al 2005). The C-arm geometry was calibrated using a spiral BB phantom (Navab 1996), and the resulting calibration was represented as projection matrices (Galigekere et al 2003) composed of intrinsic and extrinsic parameters describing the 3D position of the x-ray source with respect to the detector. The calibration parameters were also used to quantify the C-arm magnification , where SDD and SOD denote the source-detector and source-object distance, respectively, as marked in figure 1.
Fluoroscopic images were acquired in a continuous orbit (Δθy in figure 1) spanning 178° yielding 200 projections at roughly equal increments of Δθy = 0.9°. Each projection image was 7682 pixels with 0.388 mm2 isotropic pixel size. Projections acquired in this manner served two purposes: (1) any two projections (separated by different Δθ) could be used to assess the angular dependence of the geometric accuracy; and (2) each scan could be used to reconstruct a cone-beam CT (CBCT) image of the target anatomy. CBCT images were reconstructed using the Feldkamp algorithm (Feldkamp et al 1984), at 2563 voxels with 0.6 mm3 isotropic voxel size to provide reliable definition of true target locations.
2.2.2. Measurement of registration accuracy
The first set of experiments investigated the accuracy of registration with different projective view pairs (varying both in angulation and angular separation) in comparison to conventional tracking methods. A human torso cadaver (81 year old male) was used, and experiments spanned three anatomical sites—thoracic (R1), abdominal (R2) and pelvic (R3) regions as shown in figure 2, with projections acquired at nominal C-arm magnification (m = 2). A total of 27 plastic fiducial divots were affixed to the surface of the cadaver and were used to register a surgical tracker. The preoperative CT scan was performed immediately prior to target implantation and fluoroscopy with the cadaver taped to a carbon fiber board. The scan was acquired with a routine body protocol—120 kVp tube voltage, 103 mAs (with automatic exposure control), and reconstructed with voxel size (0.71 × 0.71 × 0.6) mm3 using the B30f kernel (Somatom Definition Flash, Siemens Healthcare).
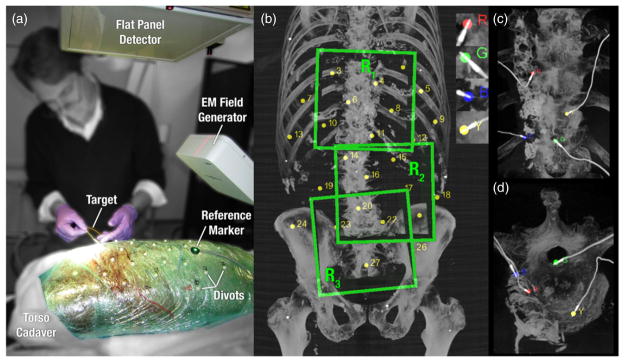
Figure 2.
Illustration of: (a) experimental setup and materials in the cadaver study; (b) three anatomical regions spanning the thorax, abdomen and pelvis, along with surface fiducials used for tracker registration; and (c) MIP renderings (in the R1 thoracic region about the spine) showing the four implanted EM coils (labeled R, G, B, Y) used as targets.
An EM tracker (Aurora, NDI, Waterloo ON) was employed as a basis of comparison to conventional surgical guidance. Targets consisted of four markers manufactured using five DoF EM sensor coils (figures 2(c) and (d)) implanted in the cadaver to provide location information both via the EM tracker and in CBCT for analysis of registration accuracy. The targets were implanted in various regions of the cadaver using a cannula and taking care to minimize deformation during implantation. The markers were not present in preoperative image so as not to bias the registration. The compact marker and thin electrical wires did not significantly perturb the surrounding anatomy and cast little or no metal artifacts in CBCT. In comparison to optical trackers, where rigid, needle-like tools would be used with markers placed outside of the body, tracking the targets directly at the tooltip was hypothesized to circumvent errors due to tool deflection, tooltip calibration, and operator variability.
Registration of the EM tracker and CBCT used two six DoF EM sensors embedded in a pointer tool and a reference marker attached to the cadaver surface to account for possible specimen motion by left-multiplication of the recorded poses with the inverse of reference pose. Recordings of the 27 surface fiducials and each of the four EM target locations were repeated ten times, each capturing ~100 samples, in each of the three regions in the cadaver. The median of the resulting distributions was taken as an outlier-rejecting estimate of the tracked locations, and each was transformed by the inverse of the reference marker to account for potential specimen motion.
The targets were localized in CBCT using an automated, reproducible method for definition of ground truth in evaluating the accuracy of both the tracker and the 3D–2D registration algorithm. An initial manual segmentation about each marker defined the region of interest, large enough to contain the full extent of an EM coil. Thresholding was applied to separate the high-intensity coil from the underlying tissue, followed by connected component labeling to reduce noise by eliminating patches with small areas. The midline of the roughly cylindrical marker was extracted by a thinning operation, and the distal tool tip was identified. Finally, half the length of the coil (2 mm) was traversed backwards from the tip along the shaft to localize the target center as defined by manufacturer specifications. This method for localizing targets depended on just a single tunable parameter—the threshold value—and we validated that the location of the target center measured was invariant to <0.1 mm for a broad range of intensity threshold (~1800–2200 HU).
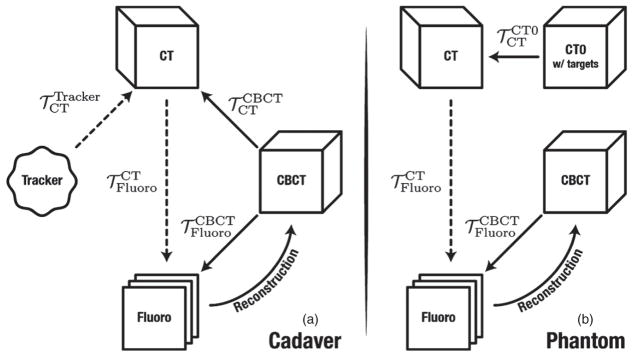
The coordinate transforms involved in the experiment are depicted in figure 3, where the 3D–2D registration solution is denoted . For the cadaver studies, the tracker was registered to the preoperative CT as in common clinical practice, using the surface fiducials recorded by the EM pointer tool and segmented in CT. To serve as ground truth, the location of the four EM targets implanted in each anatomical region were mapped from CBCT to CT by a 3D–3D rigid registration ( ) using adaptive stochastic gradient descent optimization (Klein et al 2008) of normalized correlation coefficient. The CBCT and CT images were histogram-equalized to reduce susceptibility of the registration to voxel value intensity differences. The CBCT image was reconstructed from the acquired fluoroscopy images, hence sharing the same coordinate frame ( ). The projective geometry from CBCT to fluoroscopy is given by the projection matrix ℘Fluoro obtained from the C-arm calibration.
Figure 3.
Flowcharts depicting the coordinate transforms for (a) cadaver and (b) phantom studies. Dotted lines signify the final registrations used for both fluoroscopy and tracker guidance.
2.2.3. CT slice thickness and detector binning
Due to variations in CT scanner technology and clinical scan protocols used at various institutions, we sought to characterize the dependence of the 3D–2D registration algorithm on the voxel size (specifically, the slice thicknesses, denoted az) in the preoperative CT image. Most scanners in current practice yield axial (x, y) voxel size ~0.5 mm; however, slice thickness commonly varies from ~0.5–2 mm, depending on the scanner and scan protocol. In addition, considering the behavior of the GI similarity metric, we investigated the dependence of registration accuracy on detector pixel size (i.e., denoted bu,v ), recognizing that coarser voxels and pixels likely tradeoff accuracy for reconstruction speed.
The study used the same cadaver data as described above, repeating the analysis with voxel and pixel size binned across the range 0.6–4.8 mm and 0.3–2.4 mm, respectively. The preoperative CT was binned with linear interpolation along the longitudinal direction to simulate thicker slices, whereas the intraoperative fluoro images were binned with bicubic interpolation to minimize artifacts. Although a linear forward projection was found adequate in previous studies (where voxel and pixel size were reasonably fine), the more accurate Siddon projection algorithm was used in this experiment to avoid potential bias associated with the coarser voxels.
2.2.4. C-arm magnification
A final experiment investigated the effect of C-arm magnification on registration accuracy. We hypothesized competing effects in the influence of magnification: (i) degradation at lower magnification (i.e., extended SOD) due to reduced triangulation between projection image pairs of a given Δθ; (ii) degradation at higher magnification (i.e., reduced SOD) due to smaller field of view (FoV) and reduced image content and GI with which to work; and (iii) an optimum at middle magnification. Various magnification (m = 1.6–2.8) was achieved as illustrated in figure 4 by shifting the C-arm isocenter with respect to the patient (and table support).
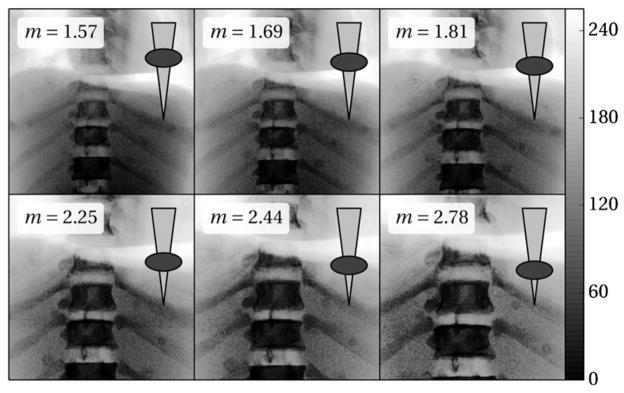
Figure 4.
PA radiographs of the chest phantom (histogram-equalized for purpose of display) at varying C-arm magnification. Note the reduction in FoV as the magnification is varied from m = 1.6 to 2.8. The superimposed ellipse (body) and triangle (x-ray beam) illustrate the change in table height giving the corresponding magnification.
The study employed an anthropomorphic chest phantom upon which 17 plastic fiducial divots were randomly affixed on the anterior and posterior surface. The rigid surface allowed us to extend the previous two studies (sections 2.2.2 and 2.2.3) to a wide range of magnification, with the CBCT FoV containing a subset of the fiducials for truth definition. Two preoperative CT scans of the phantom were acquired with the same parameters as for the cadaver—before and after placement of the fiducials. The skin fiducials in this experiment served as segmentable targets, and the CT without the targets was used in registration to avoid potential bias due to gradients from the fiducial themselves. Meanwhile, the CT with fiducials (referred to as CT0 in figure 3(b)) was used only for purposes of analysis. The target locations were manually segmented in CBCT ten times independently to minimize intra-operator variability. The coordinate transforms are depicted in figure 3, with only minor differences from the cadaver experiment: the EM tracker was not used, and targets defined in CT0 were mapped to CT employing the same rigid registration method used to solve for .
2.3. Characterization of registration accuracy
A common metric of 3D–2D registration accuracy is the PDE defined by the distance between the ‘true’ and estimated target positions on the 2D projection image (van de Kraats et al 2005). Referring to figure 3(b), the PDE of 3D–2D registration may be expressed as:
| (6a) |
and the PDE of tracker registration error as:
| (6b) |
where 〉*〈 is the mean, and ||*||2 is the L2 norm operator. The transforms and were fixed as described above and assumed to contribute negligible contribution to the measured error. The primary motivation of computing the PDE was to ensure that the initial registration was sufficiently far from the solution, as we assumed initialization within 5–10 mm PDE by the global search strategy with multi-resolution framework (as in Otake et al (2012)) and Otake et al (2013b)). The main disadvantage of this metric with respect to 3D localization is that it does not describe errors along the ‘depth’ direction of the x-ray beam (i.e., normal to the detector). For small angular separation between projection views, this is expected to be the direction with the highest error due to limited depth resolution.
We therefore characterized the 3D registration accuracy in terms of the target registration error (TRE) as used predominantly in image-guidance literature (Fitzpatrick et al 1998). For the two cases considered above, the TRE may be expressed as:
| (7a) |
| (7b) |
whereas the TRE for the phantom (magnification) experiments directly use the targets defined in CT:
| (8) |
While PDE is the distance error in the fluoroscopic image plane (i.e., at the detector), TRE is the 3D distance error in the domain of the CBCT image (alternatively, the patient).
In the context of 3D surgical guidance, a primary objective of the experiment was to determine the minimum angular separation between projection views yielding 3D registration accuracy (i.e., TRE) at least as good as the EM tracker. We therefore measured the minimum Δθ for which the TRE was <2 mm with 95% confidence intervals as upper bounds.
3. Results
3.1. Effect of angular separation (Δθ) between projection views
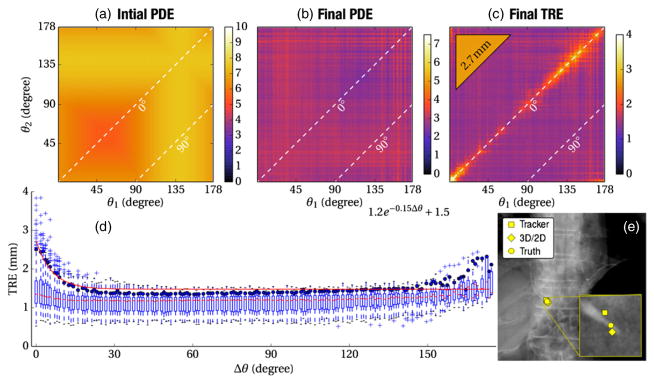
The accuracy of target localization achieved by 3D–2D registration is shown in figure 5 in terms of PDE and TRE. Data were aggregated over each anatomical site (thorax, abdomen, and pelvis), all of which demonstrated approximately the same magnitude and trends. The initial system pose is shown in figure 5(a), obtained by perturbing a coarse, manual fiducial-based registration over all six DoFs to yield an average PDE of ~5–10 mm, with variations among different views attributable to projection of the 3D error. This corresponds to the scenario in which a robust, multi-start registration is assumed to have already run (e.g., at the beginning of the case), and the task at hand is to provide 3D–2D guidance in successive fluoro shots between which perturbation of the system pose is within ~10 mm. The axes in figures 5(a)–(c) represent the C-arm angle between any two projections, such that the diagonal (dotted) line represents 0° separation (3D–2D registration based on a single projection), and the second dotted line represents registration using projections separated by Δθ = 90° (biplane fluoroscopy). Symmetry about the 0° line is due to invariance of the order of projections used.
Figure 5.
Effect of angular separation on 3D–2D registration accuracy. (a) The initial PDE prior to registration. (b) The PDE following 3D–2D registration. (c) The TRE following 3D–2D registration, (d) TRE plotted versus the angular separation (Δθ) between projection views. (e) Example PA fluoroscopy image of the cadaver overlaid by the target position as assessed by (circle) truth, (diamond) 3D–2D registration and (square) the EM tracker.
Figure 5(b) shows the PDE following 3D–2D registration, converging in all cases to a solution with <3 mm PDE irrespective of the projection views used. Slight variation among particular views is due to anatomical structure presenting more or less GI from different perspectives. These results are consistent with (Otake et al 2012) and show the ability to overlay image or planning data in fluoroscopy within 3 mm (at the detector plane; ~1.5 mm at C-arm isocenter). In terms of PDE, this level of registration accuracy is irrespective of angular separation and can be accomplished in any single projection view (i.e., the diagonal with Δθ = 0°).
The same data are analyzed in terms of TRE in figure 5(c), illustrating one of the central results of this experiment: TRE is highest (~3–4 mm) in a small angular range about Δθ = 0° (i.e., reduced capability for 3D localization in a single shot due to limited depth resolution) and falls rapidly to a level of TRE ~1.5 mm over a large range of angular separations. Registration using biplane projections (Δθ = 90°) yields a low TRE, with broad expanse in Δθ over which the TRE is just as low. The narrow band about Δθ = 0° for which TRE is elevated is not reflected in the PDE measurements (figures 5(a) and (b)), since PDE does not capture errors in-depth resolution. For comparison, the mean TRE for the EM tracker is shown in the triangular inset (upper left corner) on the same colorscale: TRE = 2.7 mm consistent with other work (Wood et al 2005). Overall, 3D–2D registration yields higher accuracy than the tracker given at least a small angular separation between projection views. In fact, the TRE achieved by 3D–2D registration from a single projection (Δθ = 0°) was about the same as the tracker.
Figure 5(d) aggregates the TRE from figure 5(c) as a function of Δθ. At each point, a boxplot is shown marking the median (horizontal line), first and third quartile (box), and range (whiskers) in TRE, with outliers marked by a + symbol. An exponential fit to the 95% confidence interval is also shown, computed using nonlinear least squares. The results suggest that a TRE <2 mm can be achieved with 95% confidence for angular separation Δθ of at least ~10°, with no further improvement in TRE (~1.6 mm) beyond Δθ ~20°. Slight trends and variations (all less than experimental error) over the range Δθ ~20°–90°and beyond are likely due to varying strength of anatomical gradients from different perspectives, varying levels of quantum noise, and stochasticity of CMA-ES. Interestingly, opposing views (Δθ → 180°) yield similarly low TRE, exploiting magnification effects in extended anatomy.
Finally, figure 5(e) illustrates a representative fluoroscopic view and registration of a single target in the region of the thoracic vertebrae. The zoomed inset shows the tip of the target, with true location marked by a circle. The location indicated by 3D–2D registration (Δθ = 45°) is within ~1.5 mm of truth, while that of the tracker is within ~3 mm, representative of the median values in the plots of figure 5.
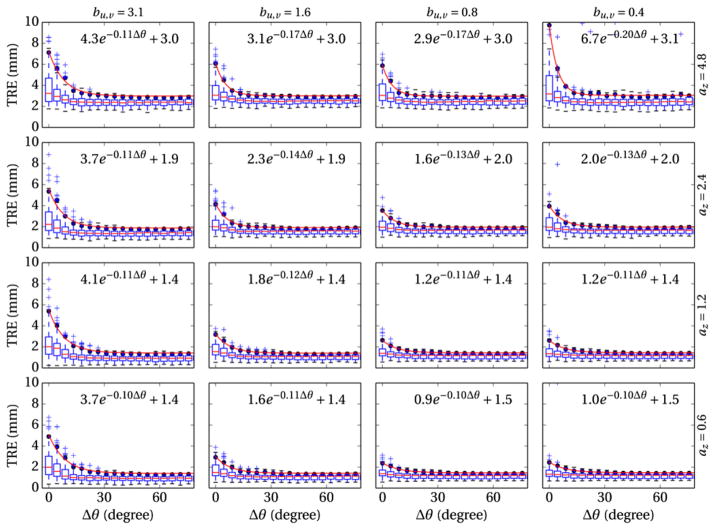
3.2. Effect of CT slice thickness and detector binning
The effect of CT slice thickness (az) and detector pixel size (bu,v) on 3D–2D registration accuracy is summarized in figure 6. Overall, larger voxel and pixel size (upper left of figure 6) degrade TRE and increase the minimum angular separation (Δθmin) between projection views. Specifically, larger pixel size extends the exponential falloff in TRE versus Δθ, thus increasing Δθmin, whereas larger voxel size increases the offset in minimum achievable TRE. An increase in the number of outliers (instability in the registration result) is also observed when the voxel and pixel size are mismatched—e.g., small pixel sizes and large voxel sizes in the upper right of figure 6. This effect appears to be a result of unmatched gradients in the projection image (cf, DRR), which create extraneous local minima around the actual solution. This suggests an optimum (or at least good practice) in which voxel and pixel size are reasonably matched; for example, in cases of thick CT slices, TRE improves when pixels are binned to yield a comparable aperture size.
Figure 6.
Effect of CT slice thickness (rows, az = 0.6–4.8 mm) and pixel binning (columns, bu,v = 0.3–2.4 mm) on the TRE of 3D–2D registration.
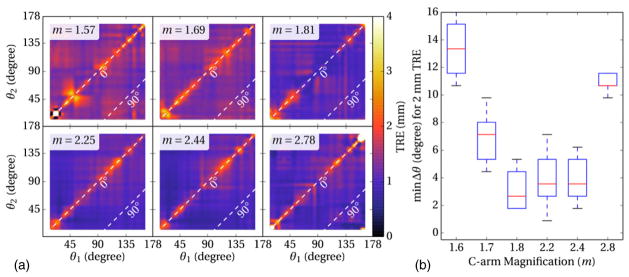
3.3. Effect of C-arm magnification
Finally, figure 7 shows the TRE measured as a function of angular separation and C-arm magnification (m = 1.6–2.8). Each subplot of figure 7(a) shows the same effect as in figure 5(c)—a narrow band about Δθ = 0° for which TRE is elevated, and a broad expanse over which TRE is relatively constant. The mean TRE was slightly lower overall in comparison to the cadaver studies, attributable to the rigidity of the phantom. Aggregating the results as in figure 5(d) and plotting the minimum angular separation (Δθmin) for which TRE <2 mm ( ± 0.25 mm) yields the plot in figure 7(b), suggesting a slight increase in TRE (alternatively, an increase in Δθmin) at lower magnification due to reduced triangulation. Registration performance appears to be optimized at m ~2.2, followed by an increase in TRE (alternatively, in Δθmin) at higher magnification due to reduced FoV (reduced GI). Overall, the nominal C-arm magnification of m ~2.0 (e.g., the value for rotational angiography or CBCT) is near optimal for 3D–2D registration.
Figure 7.
Effects of C-arm magnification on 3D–2D registration accuracy. (a) TRE as a function of projection view angles. (b) Minimum angular separation required to achieve TRE better than 2 mm measured as a function of C-arm magnification.
4. Discussion
The central result of this work is the fairly small angular separation (Δθ~ 10°) in projection views required to achieve 3D localization accuracy (TRE <2 mm) comparable or superior to that of conventional surgical tracking systems. The 3D–2D registration method yielded such accuracy across a very broad range in angular separation, with views at Δθ ~ 15° providing equivalent accuracy to Δθ~ 90° (biplane). Interestingly, even 3D–2D registration from a single projection (Δθ = 0°) performed approximately as well as the EM tracker (TRE ~2.5–3 mm), exploiting magnification effects in extended anatomy to yield 3D localization from just a single view. The result invites analogy to depth perception in natural vision with a fairly small optical baseline (Foley 1980), where in this case, the registration algorithm takes the place of biological neural processing of depth cues and stereovision.
While PDE is a prevalent metric for 3D–2D registration accuracy, TRE was shown to better characterize 3D localization, particularly in the range of small angular separation in which localization suffers from limited depth resolution. Cadaver experiments demonstrated that Δθ ~10° angular separation was adequate to obtain TRE comparable or superior to that of commercial surgical trackers with 95% confidence. Nominal registration parameters were identified and drawn from previous work (Otake et al 2012), and other parameters that may vary across surgical procedures were investigated, including CT slice thickness, detector pixel size, and C-arm magnification.
The work is not without limitations. Experiments were limited to a single cadaver and chest phantom. Although the data involved multiple scans, projection views, and anatomical sites, more data would be desirable for further validation. Current experiments focused on body and/or spine surgery (chest, abdomen, and pelvis sites), and future work will examine extension to intracranial surgery (e.g., ventricular shunt and deep-brain stimulation device placement) and extremities surgery (e.g., femoral nailing and joint implants). Current implementation requires an encoded or geometrically calibrated C-arm, and extending this approach to less reproducible imaging platforms may be possible using uniformly distributed anatomical landmarks as in Jain and Fichtinger (2006)) or even mobile radiography with nine DoFs between the source and detector (Otake et al 2013a). The current methodology does not provide ‘tool tracking’ (cf, a pointer with fiducials as with conventional surgical trackers), and work underway includes automatic detection of devices (Burschka et al 2005, Niethammer et al 2006, Herman et al 2009) to render the tip and trajectory of surgical tools in a manner similar to conventional navigation. The experiments assumed a robust initialization by virtue of registration as described in by Otake et al (2013b), which demonstrated accuracy ~5 mm or better even in the presence of large deformations associated with patient positioning (e.g., prone versus supine patient setup with varying levels of spine curvature). That work includes the same similarity metric and optimization framework as used here, with the addition of an optimal GI mask (to weight certain regions of the image more than others) and a multi-start strategy in the global optimization. The current work employed neither the GI mask nor the multi-start method and specifically addressed a guidance scenario in which registration is updated in successive fluoroscopic views for which deformation between shots is less than ~10 mm. Future work will consider not only gross anatomical deformation—to which conventional tracking is similarly susceptible—but also respiratory motion directly. For example, a preoperative 4D CT image (as common in image-guided radiotherapy) can be used in extension of the same framework to a 4D–2D registration that additionally solves for the respiratory phase. Future work will also consider 3D–2D registration on intraoperative 3D images (e.g., CBCT acquired using the same C-arm as for fluoroscopy), which will help to minimize the effect of deformations arising from discrepancies between the 3D and 2D images, recognizing the challenges of object truncation, artifacts, and CBCT HU inaccuracy in the DRR calculation.
The work potentially extends the utility of x-ray fluoroscopy from that of qualitative depiction to one of quantitative guidance. By incorporation of the same prior information as in conventional navigation (viz., a 3D CT image and planning data), but without the need for trackers, fiducial markers, and stereotactic frames, accurate 3D localization is possible from projections acquired at a small (~10°) angular separation. The result suggests the potential of 3D guidance based on 3D–2D registration with or without conventional trackers. In such a scenario, the imager is the tracker, and the patient is the fiducial.
The workflow by which 3D–2D guidance might be achieved is somewhat different from that of conventional navigation. Specifically, the method does not operate in real-time (~1–5 s registration time on the current GPU implementation), and it involves the delivery of radiation dose. With respect to the first point, the step-by-step presentation of guidance information with each fluoro shot is a reasonable match to the surgeon’s natural workflow, and the real-time (~30 fps) nature of conventional tracking systems is not essential in practice; ‘snapshot guidance’ may suffice. With respect to the second point, the radiation dose in image-guided procedures must be minimized. The method described herein is intended to work within the context of fluoroscopically guided procedures, leveraging images that are already acquired for visualization of surgical progress to provide 3D guidance. In scenarios where a coarse level of localization accuracy is sufficient (e.g., TRE ~3 mm, comparable to that of the EM tracker), the results suggest the capability to perform 3D guidance in a single projection (Δθ = 0°), implying no increase in radiation dose beyond that already employed for fluoroscopic visualization. In scenarios where a higher degree of accuracy is required (e.g., TRE ~1.6 mm), a second projection view is required (Δθ ~10° or more), implying a factor of 2 increase in total dose if the second view is acquired at dose equal to the first. Work underway investigates registration accuracy from data in which the second view is at significantly reduced dose, hypothesizing that the algorithm is sufficiently robust to quantum noise, and the increase in total dose would be incremental. Also, the guidance information provided in each fluoro shot may actually reduce the surgeon’s need for repetitive fluoro shots (i.e., reduce total fluoro time), since s/he would rely less on qualitative image interpretation by virtue of quantitative localization. Finally, there is, of course, the scenario in which such 3D–2D guidance is deployed in concert with conventional tracking, integrating fluoroscopy with navigation in a manner that leverages each to maintain accuracy throughout the case and overcome the shortfalls of the other (e.g., line of sight versus radiation dose).
The embodiments by which 3D–2D guidance might be achieved remain to be fully explored. One involves a ‘rocking’ C-arm in which fluoro shots are acquired with the C-arm angulating automatically across Δθ—for example, the first at 0° (PA) for visualization and fusion, and the second purely for 3D registration. A second embodiment suggests a novel configuration of two x-ray sources on the same C-arm, offset in either the angular or orbital direction by Δθ to provide dual-projection views without C-arm motion. A third involves any number of multi-source, multi-detector, fixed or mobile systems—e.g., a pair of overhead x-ray tubes directed at a detector beneath the operating table. The utility of such embodiments is the subject of possible future work, depending first on the assessment of natural workflow, radiation dose, and key applications as mentioned above.
Acknowledgments
This research was supported by academic-industry partnership with Siemens Healthcare (XP Division Erlangen Germany). The authors thank Mr Ronn Wade (University of Maryland Anatomy Board) for assistance with cadaver specimens, Dr Elliot McVeigh (Department of Biomedical Engineering, Johns Hopkins University), Dr Jonathan Lewin and Ms Lori Pipitone (Department of Radiology, Johns Hopkins University) for support and research infrastructure, and finally Dr Ziya L Gokaslan and Dr Jean-Paul Wolinsky (Department of Neurosurgery, Johns Hopkins Medical Institute) for valuable discussions relating to clinical requirements.
References
- Birkfellner W, Figl M, Kettenbach J, Hummel J, Homolka P, Schernthaner R, Nau T, Bergmann H. Rigid 2D/3D slice-to-volume registration and its application on fluoroscopic CT images. Med Phys. 2007;34:246. doi: 10.1118/1.2401661. [DOI] [PubMed] [Google Scholar]
- Burschka D, et al. Navigating inner space: 3-D assistance for minimally invasive surgery. Robot Auton Syst. 2005;52:5–26. [Google Scholar]
- Comeau RM, Sadikot AF, Fenster A, Peters TM. Intraoperative ultrasound for guidance and tissue shift correction in image-guided neurosurgery. Med Phys. 2000;27:787. doi: 10.1118/1.598942. [DOI] [PubMed] [Google Scholar]
- Fei B, Duerk JL, Boll DT, Lewin JS, Wilson DL. Slice-to-volume registration and its potential application to interventional MRI-guided radio-frequency thermal ablation of prostate cancer. IEEE Trans Med Imaging. 2003;22:515–25. doi: 10.1109/TMI.2003.809078. [DOI] [PubMed] [Google Scholar]
- Feldkamp LA, Davis LC, Kress JW. Practical cone-beam algorithm. J Opt Soc Am A. 1984;1:612–9. [Google Scholar]
- Fitzpatrick JM, West JB, Maurer CR. Predicting error in rigid-body point-based registration. IEEE Trans Med Imaging. 1998;17:694–702. doi: 10.1109/42.736021. [DOI] [PubMed] [Google Scholar]
- Foley JM. Binocular distance perception. Psychol Rev. 1980;87:411–34. [PubMed] [Google Scholar]
- Frühwald L, Kettenbach J, Figl M, Hummel J, Bergmann H, Birkfellner W. A comparative study on manual and automatic slice-to-volume registration of CT images. Eur Radiol. 2009;19:2647–53. doi: 10.1007/s00330-009-1452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galigekere RR, Wiesent K, Holdsworth DW. Cone-beam reprojection using projection-matrices. IEEE Trans Med Imaging. 2003;22:1202–14. doi: 10.1109/TMI.2003.817787. [DOI] [PubMed] [Google Scholar]
- Hansen N, Niederberger ASP, Guzzella L, Koumoutsakos P. A method for handling uncertainty in evolutionary optimization with an application to feedback control of combustion. IEEE Trans Evol Comput. 2009;13:180–97. [Google Scholar]
- Hanson GR, Suggs JF, Freiberg AA, Durbhakula S, Li G. Investigation of in vivo 6DOF total knee arthoplasty kinematics using a dual orthogonal fluoroscopic system. J Orthop Res. 2006;24:974–81. doi: 10.1002/jor.20141. [DOI] [PubMed] [Google Scholar]
- Herman A, Dekel A, Botser IB, Steinberg EL. Computer-assisted surgery for dynamic hip screw, using surgix, a novel intraoperative guiding system. Int J Med Robot. 2009;5:45–50. doi: 10.1002/rcs.231. [DOI] [PubMed] [Google Scholar]
- Hummel J, Figl M, Bax M, Bergmann H, Birkfellner W. 2D/3D registration of endoscopic ultrasound to CT volume data. Phys Med Biol. 2008;53:4303–16. doi: 10.1088/0031-9155/53/16/006. [DOI] [PubMed] [Google Scholar]
- Jain A, Fichtinger G. C-arm tracking and reconstruction without an external tracker. Med Image Comput Comput Assist Interv. 2006;4190:494–502. doi: 10.1007/11866565_61. [DOI] [PubMed] [Google Scholar]
- Jomier J, Bullitt E, Van Horn M, Pathak C, Aylward SR. 3D/2D model-to-image registration applied to TIPS surgery. Med Image Comput Comput Assist Interv. 2006;4191:662–9. doi: 10.1007/11866763_81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Pluim JPW, Staring M, Viergever MA. Adaptive stochastic gradient descent optimisation for image registration. Int J Comput Vis. 2008;81:227–39. [Google Scholar]
- Lemieux L. A patient-to-computed-tomography image registration method based on digitally reconstructed radiographs. Med Phys. 1994;21:1749. doi: 10.1118/1.597276. [DOI] [PubMed] [Google Scholar]
- Leventon M, Wells WM, III, Grimson WEL. Multiple view 2D-3D mutual information registration. Image Underst Workshop. 1997;1:625–9. [Google Scholar]
- Markelj P, Tomaževič D, Likar B, Pernuš F. A review of 3D/2D registration methods for image-guided interventions. Med Image Anal. 2012;16:642–61. doi: 10.1016/j.media.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Maurer CR, Fitzpatrick JM, Wang MY, Galloway RL, Maciunas RJ, Allen GS. Registration of head volume images using implantable fiducial markers. IEEE Trans Med Imaging. 1997;16:447–62. doi: 10.1109/42.611354. [DOI] [PubMed] [Google Scholar]
- Micu R, Jakobs TF, Urschler M, Navab N. A new registration/visualization paradigm for CT-fluoroscopy guided RF liver ablation. Med Image Comput Comput Assist Interv. 2006;4190:882–90. doi: 10.1007/11866565_108. [DOI] [PubMed] [Google Scholar]
- Navab N. Dynamic geometrical calibration for 3D cerebral angiography. Proc SPIE. 1996;2708:361–70. [Google Scholar]
- Niethammer M, Tannenbaum A, Angenent S. Dynamic active contours for visual tracking. IEEE Trans Autom Control. 2006;51:562–79. doi: 10.1109/TAC.2006.872837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otake Y, Schafer S, Stayman JW, Zbijewski W, Kleinszig G, Graumann R, Khanna AJ, Siewerdsen JH. Automatic localization of vertebral levels in x-ray fluoroscopy using 3D–2D registration: a tool to reduce wrong-site surgery. Phys Med Biol. 2012;57:5485–508. doi: 10.1088/0031-9155/57/17/5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otake Y, Wang AS, Stayman JW, Kleinszig G, Vogt S, Khanna AJ, Wolinsky J-PP, Gokaslan ZL, Siewerdsen JH. Verification of surgical product and detection of retained foreign bodies using 3D–2D registration in intraoperative mobile radiographs. CARS: 27th Int. Congress and Exhibit. Computer Assisted Radiology and Surgery; Heidelberg, Germany. 2013a. pp. 185–6. [Google Scholar]
- Otake Y, Wang AS, Stayman JW, Uneri A, Kleinszig G, Vogt S, Khanna AJ, Gokaslan ZL, Siewerdsen JH, Groves M. Robust 3D–2D image registration: application to spine interventions and vertebral labeling in the presence of anatomical deformation. Phys Med Biol. 2013b;58:8535–53. doi: 10.1088/0031-9155/58/23/8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney GP, Weese J, Little JA, Desmedt P, Hill DL, Hawkes DJ. A comparison of similarity measures for use in 2-D-3-D medical image registration. IEEE Trans Med Imaging. 1998;17:586–95. doi: 10.1109/42.730403. [DOI] [PubMed] [Google Scholar]
- Pluim JP, Maintz JB, Viergever MA. Image registration by maximization of combined mutual information and gradient information. IEEE Trans Med Imaging. 2000;19:809–14. doi: 10.1109/42.876307. [DOI] [PubMed] [Google Scholar]
- Russakoff DB, Rohlfing T, Adler JR, Maurer CR. Intensity-based 2D–3D spine image registration incorporating a single fiducial marker. Acad Radiol. 2005;12:37–50. doi: 10.1016/j.acra.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Sadowsky O, Lee J, Sutter EG, Wall SJ, Prince JL, Taylor RH. Hybrid cone-beam tomographic reconstruction: incorporation of prior anatomical models to compensate for missing data. IEEE Trans Med Imaging. 2011;30:69–83. doi: 10.1109/TMI.2010.2060491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddon RL. Fast calculation of the exact radiological path for a three-dimensional CT array. Med Phys. 1985;12:252–5. doi: 10.1118/1.595715. [DOI] [PubMed] [Google Scholar]
- Siewerdsen JH, Moseley DJ, Burch S, Bisland SK, Bogaards A, Wilson BC, Jaffray DA. Volume CT with a flat-panel detector on a mobile, isocentric C-arm: pre-clinical investigation in guidance of minimally invasive surgery. Med Phys. 2005;32:241–54. doi: 10.1118/1.1836331. [DOI] [PubMed] [Google Scholar]
- Söderman M, Babic D, Homan R, Andersson T. 3D roadmap in neuroangiography: technique and clinical interest. Neuroradiology. 2005;47:735–40. doi: 10.1007/s00234-005-1417-1. [DOI] [PubMed] [Google Scholar]
- Turgeon G-A, Lehmann G, Guiraudon G, Drangova M, Holdsworth D, Peters T. 2D–3D registration of coronary angiograms for cardiac procedure planning and guidance. Med Phys. 2005;32:3737. doi: 10.1118/1.2123350. [DOI] [PubMed] [Google Scholar]
- Vahala E, Ylihautala M, Tuominen J, Schiffbauer H, Katisko J, Yrjänä S, Vaara T, Ehnholm G, Koivukangas J. Registration in interventional procedures with optical navigator. J Magn Reson Imaging. 2001;13:93–8. doi: 10.1002/1522-2586(200101)13:1<93::aid-jmri1014>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Van de Kraats EB, Penney GP, Tomazevic D, van Walsum T, Niessen WJ. Standardized evaluation methodology for 2-D–3-D registration. IEEE Trans Med Imaging. 2005;24:1177–89. doi: 10.1109/TMI.2005.853240. [DOI] [PubMed] [Google Scholar]
- Weese J, Penney GP, Desmedt P, Buzug TM, Hill DLG, Hawkes DJ. Voxel-based 2-D/3-D registration of fluoroscopy images and CT scans for image-guided surgery. IEEE Trans Inf Technol Biomed. 1997;1:284–93. doi: 10.1109/4233.681173. [DOI] [PubMed] [Google Scholar]
- Wein W, Roeper B, Navab N. 2D/3D registration based on volume gradients. Proc SPIE. 2005;5747:144–50. [Google Scholar]
- Wood BJ, et al. Navigation with electromagnetic tracking for interventional radiology procedures: a feasibility study. J Vasc Interv Radiol. 2005;16:493–505. doi: 10.1097/01.RVI.0000148827.62296.B4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youkilis AS, Quint DJ, McGillicuddy JE, Papadopoulos SM. Stereotactic navigation for placement of pedicle screws in the thoracic spine. Neurosurgery. 2001;48:771–8. doi: 10.1097/00006123-200104000-00015. [DOI] [PubMed] [Google Scholar]
- Zollei L, Grimson E, Norbash A, Wells W. 2D–3D rigid registration of X-ray fluoroscopy and CT images using mutual information and sparsely sampled histogram estimators. CVPR’2001: Proc IEEE Computer Society Conf on Computer Vision and Pattern Recognition. 2001;2:II-696–II-703. [Google Scholar]