Abstract
Grouping genes as operons is an important genomic feature of prokaryotic organisms. The comprehensive understanding of the operon organizations would be helpful to decipher transcriptional mechanisms, cellular pathways, and the evolutionary landscape of prokaryotic genomes. Although thousands of prokaryotes have been sequenced, genome-wide investigation of the evolutionary dynamics (division and recombination) of operons among these genomes remains unexplored. Here, we systematically analyzed the operon dynamics of Rhodococcus jostii RHA1 (RHA1), an oleaginous bacterium with high potential applications in biofuel, by comparing 340 prokaryotic genomes that were carefully selected from different genera. Interestingly, 99% of RHA1 operons were observed to exhibit evolutionary events of division and recombination among the 340 compared genomes. An operon that encodes all enzymes related to histidine biosynthesis in RHA1 (His-operon) was found to be segmented into smaller gene groups (sub-operons) in diverse genomes. These sub-operons were further reorganized with different functional genes as novel operons that are related to different biochemical processes. Comparatively, the operons involved in the functional categories of lipid transport and metabolism are relatively conserved among the 340 compared genomes. At the pathway level, RHA1 operons found to be significantly conserved were involved in ribosome synthesis, oxidative phosphorylation, and fatty acid synthesis. These analyses provide evolutionary insights of operon organization and the dynamic associations of various biochemical pathways in different prokaryotes.
Keywords: operon, Rhodococcus jostii RHA1, evolutionary dynamics, pathway
Background
In prokaryotic genomes, an operon is a functional unit of multiple neighboring genes under the control of a single promoter and terminator.1,2 Typically, about half of the protein-coding genes are organized into operons, representing one of the main strategies of gene organization, regulation, and transcription in prokaryotes.3–5 The functions and transcriptions of many operons have been studied extensively because of which extensive biological insight has been achieved. For example, the studies of two operons that are related to tryptophan6 and histidine7 syntheses have revealed new and sophisticated mechanisms of transcription control. Additionally, genes grouped in operons are widely found to have similar biological functions, indicating that clustering of genes involved in a biosynthetic route is a common feature of prokaryotic genomes.2,8,9 Furthermore, the information of operon organization, regulation, interactions, and dynamics have been used for identifying functionally linked genes,10–12 annotating gene functions,13,14 explaining the genome expansion/reduction,15,16 and facilitating the synthetic modification of biochemical processes.17–20
Operon organizations are considered to be well maintained even across phylogenetically distant genomes, as the proximity of functionally related genes offers more efficient regulation.1,4 Dynamic events such as division or recombination are also widely observed,6–8,21,22 suggesting that some operons might be a recent invention of evolution and others might result from convergent evolution. For example, a detailed examination of the repABC operon revealed that each member of this operon has its own evolutionary dynamics.23 Evolutionary models of operons such as tryptophan24 have been proposed to study their abundance, distribution of sizes, and evolutionary dynamics over time.25 Thus, a better understanding of the genome-wide operon organization and their dynamics among a large number of genomes will provide essential information not only for understanding experimental designs but also for understanding the evolutionary organization of prokaryotic genomes.
Experimental determination of operons is time consuming, and therefore, several computational methods have been presented to predict genome-wide operons by using a number of genomic/genetic features,26–29 including intergenic distance,30,31 conservation of gene order,32,33 functional relationships,34 and transcriptional data.35 These methods have achieved high accuracies based on the validations of experimentally defined operons,26,29 eg, 90.2% and 93.7% in Bacillus subtilis (B.subtilis) and Escherichia coli genomes, respectively.30 With more genomes sequenced, the applications of these methods have allowed high-quality predicted operons and broad coverage of prokaryotic genomes. So far, the experimentally validated as well as computationally predicted operons of thousands of sequenced prokaryotes have been collected in several operon databases,36–40 providing the opportunity to comprehensively understand the operons of prokaryotic genomes.
Although abundant information on operons is available, there is lack of genome-wide comparison of operons to understand their evolutionary dynamics based on the landscape of prokaryotic genomes. As a part of our demonstration, we analyzed the operon dynamics of Rhodococcus jostii RHA1 (RHA1) among 340 prokaryotic genomes. RHA1 is a soil actinomycete with exceptional abilities to synthesize, store, and degrade large types of lipids.41–44 It has become a model bacterium to understand the pathway of lipid metabolism for biofuel development.43,45–47 In this work, the aims of analyzing RHA1 operons are twofold: one is to provide insights of the evolutionary dynamics of RHA1 operons based on a diverse set of prokaryotic genomes and the second is to discover whether operon evolution contributed to the exceptional ability of lipid metabolism in RHA1 cells. We compared all RHA1 operons and their organization with 340 genomes to understand their dynamic evolution. Subsequently, we categorized the functional conservation of RHA1 operons and found that the operons related to lipid transport and metabolism are significantly conserved.
Materials and Methods
Selection of 341 bacterial genomes
To properly compare operon structures among genomes, we first need to carefully select the genomes. Currently, more than 5,000 prokaryotic genomes have been completely sequenced and are available in the NCBI database,48 but they are largely imbalanced and biased to pathogenic bacteria such as E. coli and Mycobacterium smegmatis. Here, we selected only the genome with the largest DNA sequence from each genus of domain bacteria and archaea, where the genus was a well-used evolutionary distance for comparative genomic analysis.49,50 Selecting only the largest genome of a genus will be beneficial not only to procure abundant information of genes but also to avoid the redundancy and imbalance of sequenced genomes among genera. In total, 341 genomes (Supplementary Table 1) of different genera were selected and downloaded from the NCBI database (NCBI release of August 2015). We used the RHA1 genome as the reference genome and the other 340 genomes as comparing genomes. All the genes/proteins mentioned in the paper are labeled by NCBI Geninfo Identifier numbers or if available by their official symbol names. Usually, one gene of a bacterial genome codes one protein, so we did not differentiate between gene and protein throughout the paper.
Comparative and phylogenetic analysis of RHA1 operons
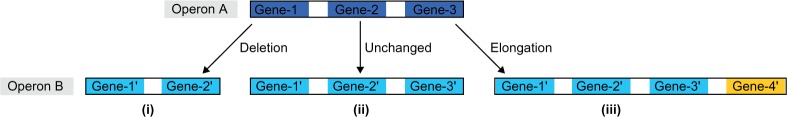
We first aligned each of the 9,145 genes of RHA1 with all the genes of the 340 comparing genomes by using the BLASTP program.51 For an RHA1 gene, we defined its homology gene in another comparing genome as the best-matched gene, which has the smallest e-value. If the e-values of genes of the comparing genome are all greater than 1e-05, then it is considered that no homology has been detected.29 For all of the 341 selected genomes, their possible operons were predicted using the operon prediction program with the default parameters30 and these operons are available at the DOOR database.36,52 The 9,145 RHA1 genes were predicted to belong to 5,556 operons. A single gene is also considered an operon (termed a single-gene operon). Among the 5,556 operons, 55 operons have no homologies found in the 340 comparing genomes, while each of the other 5,501 operons includes at least one gene with homologies found in at least one of these comparing genomes. When comparing an operon A of RHA1 with operon B of another genome, we compared the genes of A and their homologies in B. Three dynamic types of an operon pair A and B were considered: deletion, elongation, and unchanged (Fig. 1). Operon A was defined as unchanged from operon B if all gene homologies of A were all found in operon B and vice versa. If only a subset of gene homologies of A were found in operon B, operon A was called deleted. If the gene homologies of A were all found in operon B and the gene number of A was less than B, B was defined as an elongation of A. As an extreme type of deletion, if no homologies of operon A were found in a comparing genome, A was called absent in this genome. For each of the RHA1 operons, we compared it with all the operons of a comparing genome to detect its dynamic types, and recorded the number of genomes in which this type was observed. We then defined the ratio of deletion (or elongation or unchanged) for an operon as the proportion of the genomes with its deletion (or elongation or unchanged) observed among total genomes with any one of the three dynamic types. To describe the dynamic landscape of 5,556 operons within the 340 genomes, we constructed a 5,556 × 340 matrix (termed as the operon comparative matrix) by setting the state “Elongated” as 2, “Unchanged” as 1, “Deleted” as −1, and “Absent” as 0. A two-way hierarchical clustering method53 was performed on this matrix to analyze the evolutionary similarity of RHA1 operons.
Figure 1.
Schematic view of dynamic changes of operons.
Notes: Illustration of three possible types when comparing operon A and operon B. (i) Deletion: gene-3 was deleted in operon B. (ii) Unchanged: operon A and operon B had similar genes. (iii) Elongation: operon A was elongated to operon B where gene-4′ denotes the newly added gene in operon B.
Analyzing the functional conservation of operons
We used the Clusters of Orthologous Groups (COG) to classify the genes/operons of RHA1 into 17 functional categories.54 We defined an operon belonging to a functional category if most of the genes of this operon belong to this category. If each of the genes has a different COG, the operon is classified into the category “S: Function Unknown”. We then classified the 5,556 operons into 17 COG categories (operon groups). For each of the 5,556 operons, we calculated the number of genomes that the operon was kept Unchanged. Clearly, the greater the Unchanged number is for an operon, the more conserved it is among the 340 genomes. We then tested for each operon group (termed as X) if it is significantly conserved with all the other 16 operon groups (termed as Y). Mathematically, suppose there are m and n operons in X and Y, we can achieve two vectors (x1,x2, …,xm) and (y1,y2, …,ym), where xi, i = 1,2, …,m and yi, i = 1,2, …,n are the number of genomes that operons were kept unchanged. Since the operon numbers the operon numbers of X and Y are usually different, we performed the two samples Kolmogorov–Smirnov test (K–S test) between them to test if they are significantly different or not. For each category, this statistical procedure can be considered a test based on sampling with replacement. We also classified the 17 categories into four super functional groups: information storage and processing, cellular processes and signaling, metabolism, and the poorly characterized group all according to the COG database.54 Similar to the statistical procedure used above, we tested whether the operons of a super functional group are more significantly conserved than the collection of operons from the other three groups. Gene phylogenetic analysis was performed using MEGA4.0.55 Functional enrichment analyses of gene sets were performed by utilizing the DAVID database.56
Analyzing the operon conservation of pathways
All of the pathways from the 341 genomes were downloaded from the KEGG database (released in August 2015).57,58 There are a total of 109 pathways from RHA1, which include 4,148 operons. Similar to the operon analysis of COG functional categories mentioned above, we tested the conservation of the 109 pathways by using K–S test individually.
Results
Comparative analysis of RHA1 operons with 340 prokaryotic genomes
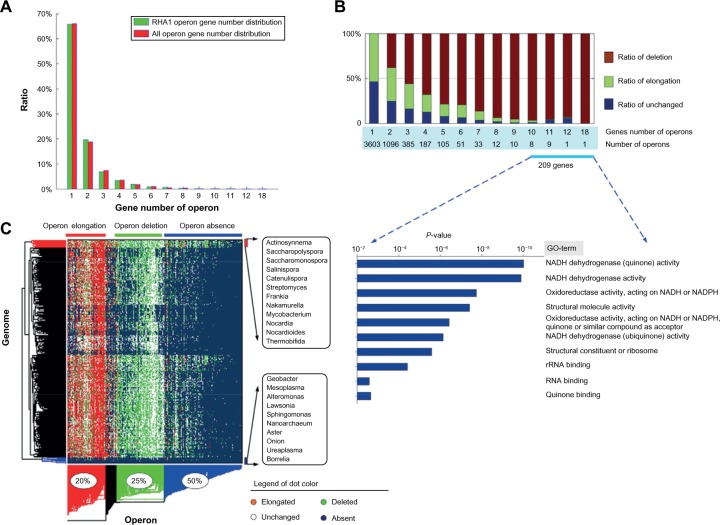
To investigate the evolutionary dynamics of operon structures, a comparative and phylogenetic analysis of RHA1 operons was performed on all operons of the 340 comparing prokaryotic genomes. In the RHA1 genome, the 9,145 genes were organized as 5,556 operons, and the gene number distribution of these operons is similar to the distribution of the total 701,360 operons from the 340 comparing genomes (Fig. 2A). The ratio of operons with at least two genes among the total 341 genomes is ~35%, which is consistent with previous operon analysis.2,59 For an RHA1 operon, three possible dynamic types are considered if it is observed to be partially deleted, elongated, or unchanged in another comparing genome (“Materials and methods” section). In total, 99% (5,501) of the 5,556 operons were observed with deletion or elongation types among at least one of the comparing 340 genomes. The 3,603 single-gene operons were frequently observed to be elongated by combining with different genes, achieving the highest ratio of elongation as 52.19% (Fig. 2B). For larger operons, the ratio of elongation decreased, whereas the ratio of deletion increased. Surprisingly, we found that 19 larger operons (Supplementary Table 2), each with at least 10 genes, remained unchanged in a relatively high ratio of genomes, indicating that these operons were highly conserved in more genomes. We then performed functional enrichment analysis on the 209 genes of these 19 operons and found that they were mainly involved in 10 categories including NADH activities, rRNA/RNA binding, and structural constituents of ribosomes (lower figure, Fig. 2B). Five of these 10 categories are significantly related to NADH activity (P < 1e-06, Fisher’s exact test), suggesting that the molecular functions of NADH activities are evolutionarily conserved among prokaryotic organisms.
Figure 2.
Comparing RHA1 operons with those of 340 prokaryotic genomes.
Notes: (A) The gene number distribution of RHA1 operons and the operon union of 340 comparing genomes. (B) The statistical analysis of three dynamic types of the 5,501 RHA1 operons. The ratio of deletion, elongation, and unchanged was calculated as an average of the corresponding ratios for all operons with the same gene numbers. Functional enrichment analysis of 209 genes of the 19 larger operons (Supplementary Table 2) was performed using the DAVID database.56 (C) Clustered results of RHA1 operons and 340 comparing genomes. For an operon, its dynamic events in genomes are presented as different colored dots. The dominant dynamic events of operons were manually marked as Elongation, Deletion, and Absence. The operons were mainly clustered into three groups as elongation (red), deletion (green), and absence (blue), and their proportions of operons are noted in elliptical circles, respectively. The genomes of 12 genera with close evolutionary distances are clustered into one group. The bottom lists 10 genomes where most of the RHA1 operons are absent.
To further understand the bias of the three dynamic types, we constructed a 5,556 × 340 matrix to record the dynamic types of each operon within the 340 genomes. We analyzed the matrix using a two-way clustering method and manually annotated the operon clusters with their dominant dynamic events (elongation, deletion, or absence) among the 340 genomes. We found that all 5,556 operons were clustered into four groups. Approximately 20% of operons tend to elongate, 25% of operons tend to be deleted, and 5% of operons are mixed, either having deletions or elongations. This sums up more than 50% of the RHA1 operons, whose dynamic events may contribute to obtaining novel biological functions or regulatory modules in the different genomes. We also checked the genomes within different clusters, and found that they are relatively congruent with taxonomic classification from the NCBI database (as an example, see the 12 genera in Fig. 2C).
Evolutionarily dynamics of the His-operon: a case study
We selected an RHA1 operon for detailed analysis of its dynamic events among compared genomes. This operon is composed of 11 genes, including eight enzymes related to histidine biosynthesis, one suhB protein, and two hypothetical proteins (termed as “His-operon”). After investigating and clustering the His-operon with their homologies, we found that the two hypothetical genes (with Geninfo Identifier number of 111018037 and 111018041) were almost absent in 340 comparing genomes, suggesting that they could be newly obtained from the RHA1 genome (Fig. 3A). The His-operon turned out to be divided into several sub-operons in other genomes, even in two strains with close evolutionary distance to RHA1, M. smegmatis MC2 155 (Mycobacterium) and Nocardia farcinica IFM 10152 (Nocardia). In the Mycobacterium genome, the His-operon was divided into two sub-operons: one keeps the main body of the His-operon and the other includes one separated gene 111018040 (hisC1). In the Nocardia genome, the His-operon was divided into three operons, where the main body of the His-operon in Mycobacterium was further separated into two smaller operons (Fig. 3A). Interestingly, different recombination events of hisC1 with other genes are observed in Mycobacterium and Nocardia (Fig. 3B). In Mycobacterium, hisC1 has a homologous gene 118470683 that was recombined with four genes annotated as MaoC, CoA transferase, amidohydrolase 3, and DNA-binding protein in an operon. Meanwhile, the homologies of these four genes in RHA1 are all separated from each other along the chromosome (indicated in red color, Figure 3B). In Nocardia, hisC1 has a homologous gene 54026425 that was recombined with three genes annotated as two ccrB proteins and one phosphoglucomutase in an operon (indicated in green color, Fig. 3B). The homologies of these three genes in RHA1 are also grouped into an operon. Comparatively, the four genes in Mycobacterium have no sequence similarity with the three genes in Nocardia. These evidences suggest that the recombination events of hisC1 in Mycobacterium and Nocardia could be independent after the divergence of the two strains and could be involved in different cellular functions.
Figure 3.
Evolutionary dynamics of His-operon.
Notes: (A) Heatmap of the homologies of the 11 His-operon genes among the 340 selected genomes. The genomes clustered by using the e-values of the 11 His-operon genes that were obtained from the BLASTP program.51 These 11 genes were enriched in 25 genera (red cluster) and almost absent in 47 genera (blue cluster). Operon structures of the His-operon are shown for 25 genomes. Homologous genes are depicted in identical colors. (B) Divided operon structures of His-operon in M. smegmatis MC2 155 and N. farcinica IFM 10152. The e-values between homologous genes are noted adjacent to the gene boxes.
Functional conservation of RHA1 operons
Although dynamic types of operons are widely observed among the 340 genomes, we also noticed that a number of operons tend to remain unchanged (see the columns dominated with white spots in Fig. 2C). We investigated on the functions of these conserved operons, which are of great interest, since these well-maintained operons could contribute to important cellular processes and thus be essential for the evolution of prokaryotic organisms. We categorized all the genes in the 5,556 operons into COG categories, and then associated each operon to the COG category that the majority of its genes belonged to. We then checked the operons of four super COG functional categories (“Materials and methods” section) and found that the metabolism group is the most conserved. In detail, the operons of the metabolism group remained unchanged in an average of 99.5 of 430 genomes, which is significantly larger than the average number (47.5) for the operons of the other three groups (P = 8.12e-189, K-S test). The metabolism super group includes eight basic categories (Table 1). All of them were tested to be significantly conserved (P < 1e-03, K–S test), where the category of lipid transport and metabolism is the most conserved (P = 1.87e-39).
Table 1.
Statistical analysis of eight metabolism categories.
| COG | FUNCTIONAL DESCRIPTION | P VALUE |
|---|---|---|
| I | Lipid transport and metabolism | 1.87E-39 |
| C | Energy production and conversion | 1.87E-33 |
| E | Amino acid transport and metabolism | 2.87E-25 |
| G | Carbohydrate transport and metabolism | 5.14E-19 |
| Q | Secondary metabolites biosynthesis, transport and catabolism | 1.15E-17 |
| H | Coenzyme transport and metabolism | 2.04E-13 |
| P | Inorganic ion transport and metabolism | 1.42E-10 |
| F | Nucleotide transport and metabolism | 4.42E-04 |
Functional investigation of the conserved operons of RHA1 pathways
We also tested the functional conservation of operons based on biological pathways. From the KEGG pathway database,58 we downloaded 109 pathways for the RHA1 genome, which included 4,148 operons. All these pathways can be divided into three super groups: metabolism (92 pathways, 3,792 operons), genetic information processing (13 pathways, 218 operons), and environmental information processing (4 pathways, 138 operons). By analyzing the operons of these 109 pathways, we found that five pathways are significantly conserved, including ribosome (1.85e-06), oxidative phosphorylation (3.08e-06), fatty acid biosynthesis (5.51e-05), polyketide sugar unit biosynthesis (2.15e-04), and peptidoglycan biosynthesis (8.39e-04) (Table 2). The conservation of the ribosome pathway and metabolism pathways further confirmed our previous analysis based on COG functional categories and is consistent with earlier evolutionary studies of prokaryotic operons.2,59
Table 2.
Statistical analysis of five RHA1 pathways.
| KEGG | FUNCTION | P VALUE |
|---|---|---|
| 03010 | Ribosome | 1.85E-06 |
| 00190 | Oxidative phosphorylation | 3.08E-06 |
| 00061 | Fatty acid biosynthesis | 5.51E-05 |
| 00523 | Polyketide sugar unit biosynthesis | 2.15E-04 |
| 00550 | Peptidoglycan biosynthesis | 8.39E-04 |
Discussion
The evolution of operons has been well studied in microorganisms such as E. coli; however, there is lack of genome-wide comparison of operon organization among a large number of prokaryotic genomes. Here, we have systematically categorized the conservation of RHA1 operons based on their dynamic types among the 340 compared genomes. The deletion and elongation of RHA1 operons are widely observed among diverse genomes, indicating that the organization of genes belonging to the same biological pathway followed different routes in different prokaryotes. Furthermore, the clustering analysis of the total 341 genomes based on the dynamic types of RHA1 operons largely matches with the taxonomic results from the NCBI database, suggesting that the majority of operons are inherited vertically.
Although a large amount of research and data are available regarding the structure, distribution, and functions of operons, the formation and dynamics of operons are still unclear. Our results confirmed that recombination events (such as deletion and elongation) are widely observed for most operons, supporting a highly dynamic view of operon formation and evolution. Divergent evolutionary events, including horizontal gene transfer,23 point mutations, and homologous recombination,2 have been hypothesized to be major force to drive operon formation and dynamics. Thus, it is interesting to further investigate the different rates of how these evolutionary events are involved in operon recombination among prokaryotic genomes. Our results also suggest that there could be a high false-positive ratio of identifying functionally linked genes or annotating gene functions13,14 using the information of operon organization, since genes performing different functions can form an operon and the operon structures are dynamically changing. As this is the case, we may need to integrate more different/independent information (such as co-evolution of genes,60 transcriptome)11 and employ better mathematical models to improve the precision of predictions.
RHA1 is known as a “lipid factory” for its high ability of synthesis and storage of diverse lipids.41,61 Our results provide potential evidence to explain its exceptional ability of lipid processes. First, we found that genes involved in the highly conserved operons mainly participate in eight COG functional categories of metabolism. Specially, several larger and conserved operons are functionally enriched in NADH dehydrogenase activity and the ribosome complex. Second, the His-operon is well maintained as a whole-pathway operon, while its members are separated and recombined with different genes as new operons in diverse organisms. In general, we found that most of the operons related to metabolism tend to keep more gene members since they are often observed to be deleted in the 340 compared genomes. Based on the hypothesis that the genes in an operon are usually regulated as a unit, operons that embraced more functionally related members could provide high efficiency in biochemical processes.1,20 Therefore, the completeness of the RHA1 operons could be contributing to its high ability of lipid processes. RHA1 has been considered to have a high potential in biofuel development.41,47 To define its main pathways of lipid metabolism, such as triacylglycerol synthesis, a large number of transcriptomic analysis and biochemical experiments have been performed.44,61 Our comparative evaluations of the dynamic organizations of RHA1 operons could help to understand the pathways of lipid synthesis by mining combined operons among different genomes, and thus to improve the development of biofuel.
Supplementary Material
Supplementary Table 1. Detailed information of 341 genomes (xls file).
Supplementary Table 2. Gene list and annotation of 19 operons (xls file).
Acknowledgments
The authors gratefully acknowledge Prof. Pingsheng Liu for his insightful suggestions on the functional analysis.
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1816 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported in partial by grants from the National Natural Science Foundation of China (Grant No. 61273228 and 61572358) and the Natural Science Foundation of Tianjin Science and Technology Committee (Grant No. 15JCYBJC46600 and 16JCYBJC23600). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: YC, SZ. Analyzed the data: YC, DG. Wrote the first draft of the manuscript: YC, DG. Made critical revisions and approved the final version: YC, DG, KE, SZ. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Jacob F, Perrin D, Sanchez C, Monod J. Operon: a group of genes with the expression coordinated by an operator. C R Hebd Seances Acad Sci. 1960;250:1727–9. [PubMed] [Google Scholar]
- 2.Fondi M, Emiliani G, Fani R. Origin and evolution of operons and metabolic pathways. Res Microbiol. 2009;160(7):502–12. doi: 10.1016/j.resmic.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: genomic analyses and predictions. Proc Natl Acad Sci U S A. 2000;97(12):6652–7. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou D, Yang R. Global analysis of gene transcription regulation in prokaryotes. Cell Mol Life Sci. 2006;63(19–20):2260–90. doi: 10.1007/s00018-006-6184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hijum SA, Medema MH, Kuipers OP. Mechanisms and evolution of control logic in prokaryotic transcriptional regulation. Microbiol Mol Biol Rev. 2009;73(3):481–509. doi: 10.1128/MMBR.00037-08. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie G, Keyhani NO, Bonner CA, Jensen RA. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol Mol Biol Rev. 2003;67(3):303–42. doi: 10.1128/MMBR.67.3.303-342.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alifano P, Fani R, Lio P, et al. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev. 1996;60(1):44–69. doi: 10.1128/mr.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Daruvar A, Collado-Vides J, Valencia A. Analysis of the cellular functions of Escherichia coli operons and their conservation in Bacillus subtilis. J Mol Evol. 2002;55(2):211–21. doi: 10.1007/s00239-002-2317-1. [DOI] [PubMed] [Google Scholar]
- 9.Fani R, Fondi M. Origin and evolution of metabolic pathways. Phys Life Rev. 2009;6(1):23–52. doi: 10.1016/j.plrev.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Moreno-Hagelsieb G. The power of operon rearrangements for predicting functional associations. Comput Struct Biotechnol J. 2015;13:402–6. doi: 10.1016/j.csbj.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korbel JO, Jensen LJ, von Mering C, Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat Biotechnol. 2004;22(7):911–7. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Yang L, Ding Y, et al. Tracing evolutionary footprints to identify novel gene functional linkages. PLoS One. 2013;8(6):e66817. doi: 10.1371/journal.pone.0066817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Halgamuge SK, Kells CI, Tang SL. Gene function prediction based on genomic context clustering and discriminative learning: an application to bacteriophages. BMC Bioinformatics. 2007;8(Suppl 4):S6. doi: 10.1186/1471-2105-8-S4-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Mao F, Li G, Xu Y. Genome-wide discovery of missing genes in biological pathways of prokaryotes. BMC Bioinformatics. 2011;12(Suppl 1):S1. doi: 10.1186/1471-2105-12-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Touchon M, Rocha EP. Coevolution of the organization and structure of prokaryotic genomes. Cold Spring Harb Perspect Biol. 2016;8(1):a018168. doi: 10.1101/cshperspect.a018168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunez PA, Romero H, Farber MD, Rocha EP. Natural selection for operons depends on genome size. Genome Biol Evol. 2013;5(11):2242–54. doi: 10.1093/gbe/evt174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24(8):1027–32. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Rubio JJ, Padmanabhan S, Lazaro JM, Salas M, Murillo FJ, Elias-Arnanz M. Operator design and mechanism for CarA repressor-mediated down-regulation of the photoinducible carB operon in Myxococcus xanthus. J Biol Chem. 2004;279(28):28945–53. doi: 10.1074/jbc.M403459200. [DOI] [PubMed] [Google Scholar]
- 19.Winkler ME, Ramos-Montanez S. Biosynthesis of histidine. EcoSal Plus. 2009;3(2):1–34. doi: 10.1128/ecosalplus.3.6.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smanski MJ, Bhatia S, Zhao D, et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat Biotechnol. 2014;32(12):1241–9. doi: 10.1038/nbt.3063. [DOI] [PubMed] [Google Scholar]
- 21.Nemergut DR, Knelman JE, Ferrenberg S, et al. Decreases in average bacterial community rRNA operon copy number during succession. ISME J. 2015;10(5):1147–56. doi: 10.1038/ismej.2015.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ream DC, Bankapur AR, Friedberg I. An event-driven approach for studying gene block evolution in bacteria. Bioinformatics. 2015;31(13):2075–83. doi: 10.1093/bioinformatics/btv128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castillo-Ramirez S, Vazquez-Castellanos JF, Gonzalez V, Cevallos MA. Horizontal gene transfer and diverse functional constrains within a common replication-partitioning system in Alphaproteobacteria: the repABC operon. BMC Genomics. 2009;10:536. doi: 10.1186/1471-2164-10-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santillan M, Mackey MC. Dynamic regulation of the tryptophan operon: a modeling study and comparison with experimental data. Proc Natl Acad Sci U S A. 2001;98(4):1364–9. doi: 10.1073/pnas.98.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cutter AD, Agrawal AF. The evolutionary dynamics of operon distributions in eukaryote genomes. Genetics. 2010;185(2):685–93. doi: 10.1534/genetics.110.115766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brouwer RW, Kuipers OP, van Hijum SA. The relative value of operon predictions. Brief Bioinform. 2008;9(5):367–75. doi: 10.1093/bib/bbn019. [DOI] [PubMed] [Google Scholar]
- 27.Baxevanis AD. An overview of gene identification: approaches, strategies, and considerations. Curr Protoc Bioinformatics. 2004 doi: 10.1002/0471250953.bi0401s6. Chapter 4:Unit41. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Su Z, Dam P, Palenik B, Xu Y, Jiang T. Operon prediction by comparative genomics: an application to the Synechococcus sp. WH8102 genome. Nucleic Acids Res. 2004;32(7):2147–57. doi: 10.1093/nar/gkh510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ermolaeva MD, White O, Salzberg SL. Prediction of operons in microbial genomes. Nucleic Acids Res. 2001;29(5):1216–21. doi: 10.1093/nar/29.5.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dam P, Olman V, Harris K, Su Z, Xu Y. Operon prediction using both genome-specific and general genomic information. Nucleic Acids Res. 2007;35(1):288–98. doi: 10.1093/nar/gkl1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price MN, Huang KH, Alm EJ, Arkin AP. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 2005;33(3):880–92. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogozin IB, Makarova KS, Murvai J, et al. Connected gene neighborhoods in prokaryotic genomes. Nucleic Acids Res. 2002;30(10):2212–23. doi: 10.1093/nar/30.10.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a finger-print of proteins that physically interact. Trends Biochem Sci. 1998;23(9):324–8. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 34.Taboada B, Verde C, Merino E. High accuracy operon prediction method based on STRING database scores. Nucleic Acids Res. 2010;38(12):e130. doi: 10.1093/nar/gkq254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortino V, Smolander OP, Auvinen P, Tagliaferri R, Greco D. Transcriptome dynamics-based operon prediction in prokaryotes. BMC Bioinformatics. 2014;15:145. doi: 10.1186/1471-2105-15-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao F, Dam P, Chou J, Olman V, Xu Y. DOOR: a database for prokaryotic operons. Nucleic Acids Res. 2009;37(Database issue):D459–63. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chetal K, Janga SC. OperomeDB: a database of condition-specific transcription units in prokaryotic genomes. Biomed Res Int. 2015;2015:318217. doi: 10.1155/2015/318217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okuda S, Yoshizawa AC. ODB: a database for operon organizations, 2011 update. Nucleic Acids Res. 2011;39(Database issue):D552–5. doi: 10.1093/nar/gkq1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pertea M, Ayanbule K, Smedinghoff M, Salzberg SL. OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 2009;37(Database issue):D479–82. doi: 10.1093/nar/gkn784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taboada B, Ciria R, Martinez-Guerrero CE, Merino E. ProOpDB: prokaryotic operon database. Nucleic Acids Res. 2012;40(Database issue):D627–31. doi: 10.1093/nar/gkr1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLeod MP, Warren RL, Hsiao WW, et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci U S A. 2006;103(42):15582–7. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren R, Hsiao WW, Kudo H, et al. Functional characterization of a catabolic plasmid from polychlorinated- biphenyl-degrading Rhodococcus sp. strain RHA1. J Bacteriol. 2004;186(22):7783–95. doi: 10.1128/JB.186.22.7783-7795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davila Costa JS, Herrero OM, Alvarez HM, Leichert L. Label-free and redox proteomic analyses of the triacylglycerol-accumulating Rhodococcus jostii RHA1. Microbiology. 2015;161(pt 3):593–610. doi: 10.1099/mic.0.000028. [DOI] [PubMed] [Google Scholar]
- 44.Ding Y, Yang L, Zhang S, et al. Identification of the major functional proteins of prokaryotic lipid droplets. J Lipid Res. 2012;53(3):399–411. doi: 10.1194/jlr.M021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villalba MS, Alvarez HM. Identification of a novel ATP-binding cassette transporter involved in long-chain fatty acid import and its role in triacylglycerol accumulation in Rhodococcus jostii RHA1. Microbiology. 2014;160(pt 7):1523–32. doi: 10.1099/mic.0.078477-0. [DOI] [PubMed] [Google Scholar]
- 46.Montersino S, van Berkel WJ. Functional annotation and characterization of 3-hydroxybenzoate 6-hydroxylase from Rhodococcus jostii RHA1. Biochem Biophys Acta. 2012;1824(3):433–42. doi: 10.1016/j.bbapap.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Zhang C, Shen X, et al. Microorganism lipid droplets and biofuel development. BMB Rep. 2013;46(12):575–81. doi: 10.5483/BMBRep.2013.46.12.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatusova T, Ciufo S, Federhen S, et al. Update on RefSeq microbial genomes resources. Nucleic Acids Res. 2015;43(Database issue):D599–605. doi: 10.1093/nar/gku1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu G, Zhao H, Li C, et al. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet. 2015;11(11):e1005614. doi: 10.1371/journal.pgen.1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lugli GA, Milani C, Turroni F, et al. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol. 2014;80(20):6383–94. doi: 10.1128/AEM.02004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mao X, Ma Q, Zhou C, et al. DOOR 2.0: presenting operons and their functions through dynamic and integrated views. Nucleic Acids Res. 2014;42(Database issue):D654–9. doi: 10.1093/nar/gkt1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28(1):33–6. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 56.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 57.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Memon D, Singh AK, Pakrasi HB, Wangikar PP. A global analysis of adaptive evolution of operons in cyanobacteria. Antonie Van Leeuwenhoek. 2013;103(2):331–46. doi: 10.1007/s10482-012-9813-0. [DOI] [PubMed] [Google Scholar]
- 60.Katara P, Grover A, Sharma V. Phylogenetic footprinting: a boost for microbial regulatory genomics. Protoplasma. 2012;249(4):901–7. doi: 10.1007/s00709-011-0351-9. [DOI] [PubMed] [Google Scholar]
- 61.Hernandez MA, Mohn WW, Martinez E, Rost E, Alvarez AF, Alvarez HM. Biosynthesis of storage compounds by Rhodococcus jostii RHA1 and global identification of genes involved in their metabolism. BMC Genomics. 2008;9:600. doi: 10.1186/1471-2164-9-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Detailed information of 341 genomes (xls file).
Supplementary Table 2. Gene list and annotation of 19 operons (xls file).