Abstract
The nucleotide-binding oligomerization domain-like receptor (Nlrp) 6 maintains gut microbiota homeostasis and regulates antibacterial immunity. We now report a role for Nlrp6 in the control of enteric virus infection. Nlrp6−/− and control mice systemically challenged with encephalomyocarditis virus had similar mortality, however, the gastrointestinal tract of Nlrp6−/− mice exhibited increased viral loads. Nlrp6−/− mice orally infected with encephalomyocarditis virus had increased mortality and viremia compared to controls. Similar results were observed with murine norovirus 1. Nlrp6 bound viral RNA via the RNA helicase Dhx15 and interacted with Mavs to induce type I/III interferons (IFNs) and IFN-stimulated genes (ISGs). These data demonstrate that Nlrp6 functions with Dhx15 as a viral RNA sensor to induce ISGs, and this effect is especially important in the intestinal tract.
Nucleotide oligomerization domain (NOD) like receptors (NLRs) play a central role in the immune response to diverse microorganisms, and react to environmental insults and cellular danger signals (1, 2). Some NLRs contribute to antiviral immunity. NOD2 recognizes ssRNA viruses to induce type I interferons (IFNs) via mitochondrial antiviral-signaling protein (MAVS) (3), and the NLRP3 inflammasome is crucial for the control of diverse viral infections in vivo (4–7). Several NLRs, on the other hand, dampen antiviral immune responses. NLRX1 and NLRC5 negatively regulate type I IFNs and NF-κB signaling via distinct molecular mechanisms (8–12); NLRC3 attenuates Toll-like receptor signaling and the stimulator of interferon genes (STING)-mediated anti-DNA virus immune signaling (13, 14). A role for Nlrp6 in the regulation of antibacterial immune responses has recently been documented (15–18); however, whether Nlrp6 regulates viral infection has not yet been elucidated.
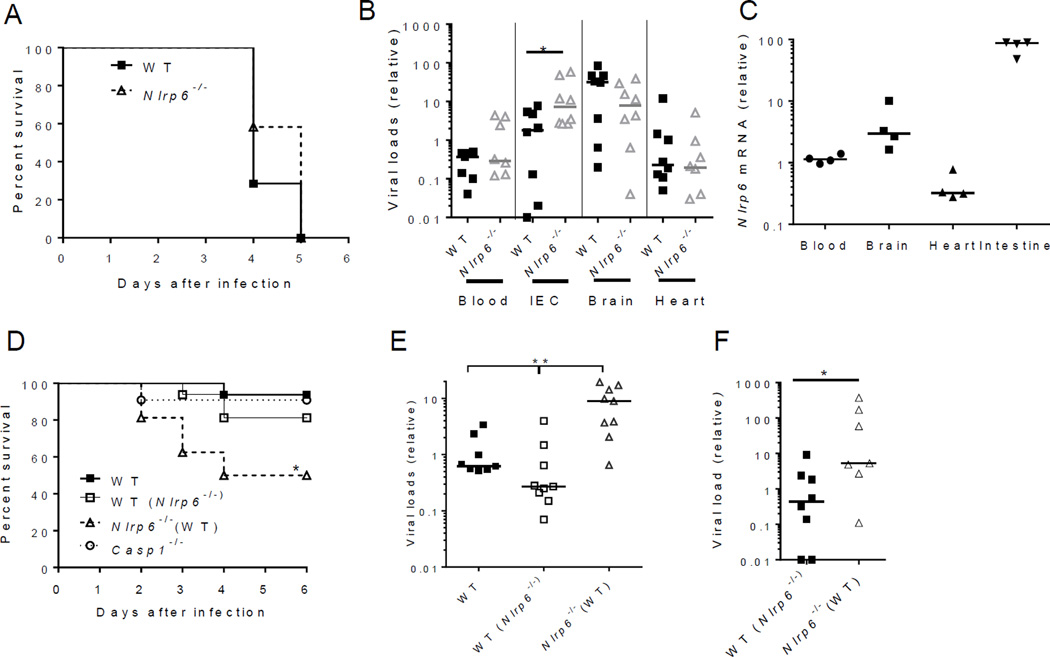
Nlrp6 exhibits a tissue and cell-type specific pattern of expression, with the highest level in intestinal epithelial cells (IECs) (15) (fig. S1 and fig. S2). We therefore determined whether Nlrp6 plays a prominent role in inhibiting enteric virus infection at the intestinal interface. We used a (+) ssRNA virus, encephalomyocarditis virus (EMCV), which is transmitted via the fecal-oral route in nature. We infected both wild-type (WT) and Nlrp6−/− mice with EMCV systemically via intraperitoneal injection, and noted that the survival curve of Nlrp6−/− mice was similar to that of WT animals (Fig. 1A). Viral dissemination was also the same in the blood, brains and hearts of Nlrp6−/− and WT mice. The intestinal viral burden of Nlrp6−/− mice was, however, higher than that of WT animals (Fig. 1B) -- suggesting that Nlrp6 plays an important role in limiting EMCV replication at this location. In support of this, Nlrp6 mRNA expression was much higher in the intestines than other tissues after EMCV infection (Fig. 1C). We therefore reasoned that Nlrp6 prevents systemic infection and mortality when EMCV is delivered orally to its principal site of infection -- the intestine. Indeed, Nlrp6−/− mice were more susceptible to oral infection with EMCV than WT animals (Fig. 1D, see also Fig. 3E).
Fig. 1. Nlrp6 controls EMCV infection of the intestine.
(A) The survival curves of WT and Nlrp6−/− mice infected with EMCV via the intra-peritoneal route (i.p.). N=12/group. Quantitative PCR analyses of (B) EMCV viral loads and (C) Nlrp6 in various tissues 72 hours after infection with EMCV i.p. IEC: isolated intestinal epithelial cells from infected intestines. (D) The survival curves of WT mice, WT mice cohoused with Nlrp6−/− [WT (Nlrp6−/−)], Nlrp6−/− mice cohoused with WT [Nlrp6−/−(WT)] and Casp1−/− mice after oral infection with EMCV. N=10–16/group, *P<0.05 (Log-rank test). Results were pooled from two independent experiments. Quantitative PCR analysis of EMCV loads (E) in the whole blood cells 72 hours after oral infection or (F) intestines 72 hours after i.p. infection. Each symbol in (B), (C), (E) and (F) represents one mouse; small horizontal lines indicate the median of the result. *P < 0.05, **P < 0.01 (nonparametric Mann-Whitney analysis). The data are representative of at least 2–3 independent experiments.
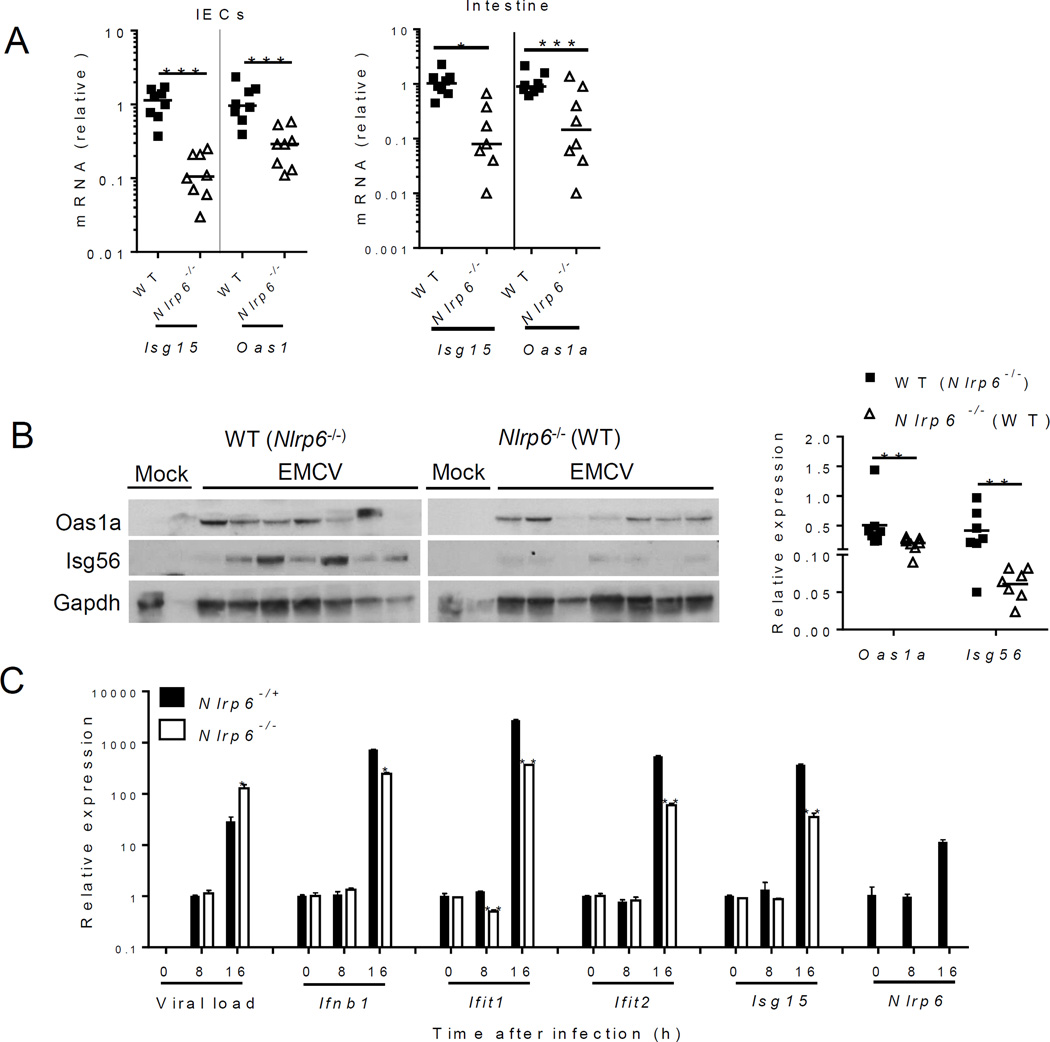
Fig. 3. Nlrp6 regulates type I/III IFN and ISG expression in the intestine.
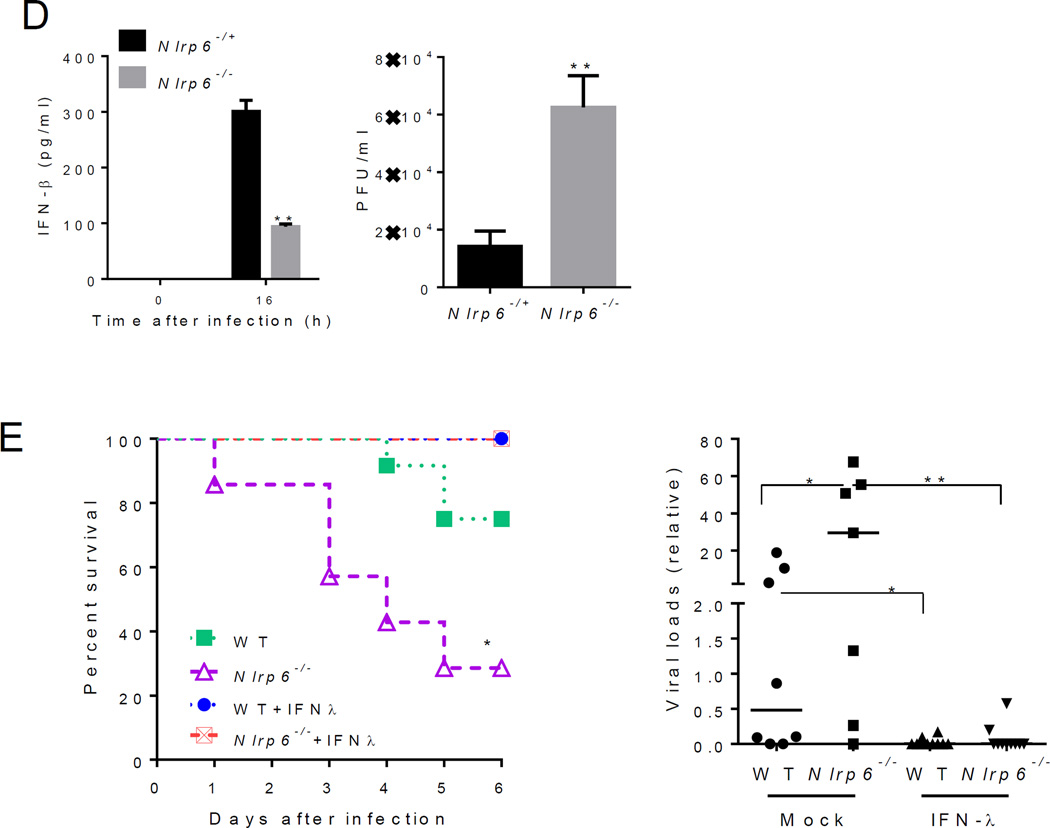
In (A) to (C), mouse tissues were analyzed on day 3 after intraperitoneal (i.p.) infection with EMCV. (A) Quantitative PCR analyses of selected ISG mRNA expression in IECs and whole intestine. (B) Immunoblotting analyses of ISG protein abundance in whole intestine of co-housed mice. Right panel indicates relative ISG abundance normalized to a house keeping protein, Gapdh. (C) Quantitative PCR analyses of cellular EMCV loads and immune gene expression in MEFs after EMCV infection (MOI=0.1). (D) ELISA of IFN-β concentrations in the culture medium of MEFs after EMCV infection and quantification of infectious viral particles in the culture medium 16 hours after EMCV infection. (E) Left panel: the survival curves of WT and Nlrp6−/− mice treated with 0.9% saline (mock) or 25 µg of recombinant mouse IFN-λ2 4h before oral infection with EMCV. Right panel: quantitative PCR analysis of EMCV loads in the whole blood cells 72 hours after infection. N=7–10/group, *P<0.05 (Log-rank test). In (A), and (E) the data are normalized with mouse beta actin and are presented as fold change over the mean of the results of WT (Mock-WT in E) mice. Each band/dot represents an animal. The horizontal lines in the figures indicate the median of the results. *P < 0.05, **P < 0.01 and ***P<0.001 (nonparametric Mann-Whitney analysis). In (C) and (D), bars: mean + S.E.M, n=3. *, P<0.05; **, P<0.01 (unpaired students’ t-test). ). The data are representative of /pooled from at least two independent experiments.
Alterations in microbiota and inflammasome activation are two potential processes that may influence the ability of Nlrp6−/− mice to control intestinal EMCV infection. The intestinal microbial ecology of Nlrp6−/− mice is different from that of WT mice (15), which could impact antiviral immunity. We therefore cohoused mice for 4 weeks before EMCV infection, which we previously showed was sufficient to equilibrate the microbiota between WT and Nlrp6−/− mice. WT and Nlrp6−/− mice had similar levels of TM7 and Prevotellacae bacteria (15) after co-housing (fig. S3A), indicating stabilization of the microbiota. Nlrp6−/− mice, however, died of EMCV infection more rapidly than WT and co-housed WT animals (Fig. 1D); and viremia was ~10-fold higher in Nlrp6−/− than WT animals (Fig. 1E). When inoculated systemically via intraperitoneal injection, EMCV loads in the intestines of co-housed Nlrp6−/− mice were also over 10-fold higher than those of co-housed WT animals (Fig. 1F). Similar survival results were noted for Nlrp6−/− and Nlrp6+/+ littermates (fig. S3B). These data demonstrate that the increased viral susceptibility of Nlrp6−/− mice is not a result of altered intestinal microbial ecology. To extend our finding further, we examined another enteric virus, murine norovirus 1(MNV-1), a (+) ssRNA virus. MNV-1 was rapidly cleared by the innate immune system in WT mice (19); but persisted much longer in Nlrp6−/−(fig. S3, C to E). Nlrp6 initiates inflammasome signaling via caspase-1. We therefore determined whether Nlrp6 requires caspase-1 to control EMCV at the intestinal epithelia. In agreement with a previous report (20), following EMCV challenge, the survival of Casp1−/− and WT mice was similar (Fig. 1D). These data suggest that intestinal Nlrp6 controls EMCV infection by an alternative mechanism.
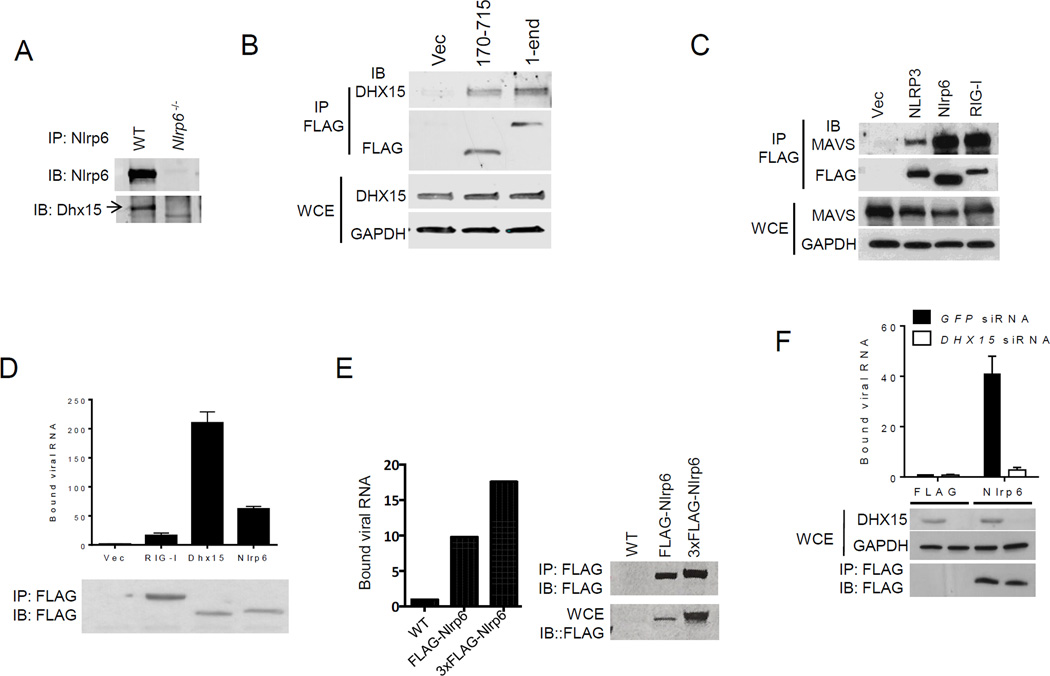
To understand how Nlrp6 contributes to antiviral innate immune responses, we used an Nlrp6 antibody to immunoprecipitate Nlrp6 binding partners from mouse primary IECs, and a FLAG-Nlrp6 overexpression system in HEK293T cells. We identified DEAH (Asp-Glu-Ala-His) box helicase 15 (Dhx15) by mass spectrometry (fig. S4), and confirmed it using a specific antibody to Dhx15 (Fig. 2A and fig. S5A). GST-Dhx15 expressed in E. coli pulled down FLAG-Nlrp6 expressed using a mammalian in vitro translation system (fig. S5B), suggesting a direct interaction. Nlrp6 is comprised of 3 functional domains, an N-terminal pyrin domain (PYD), a NACHT domain and C-terminal leucine rich repeat domain (LRR). Each individual domain failed to bind Dhx15 when compared to full-length Nlrp6 (fig. S5C). A fragment encompassing the NATCH and NACHT-associated domain (NAD) interacted with Dhx15 (Fig. 2B). NLRP3, a close relative of Nlrp6, did not interact with Dhx15, demonstrating specificity (fig. S5C).
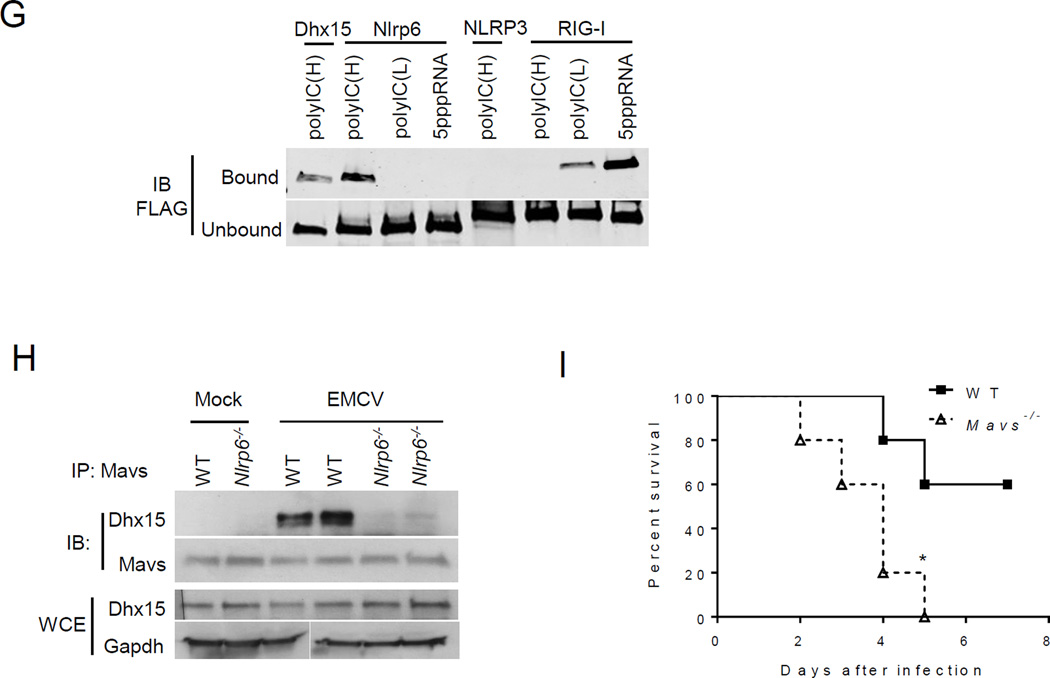
Fig. 2. Nlrp6 binds viral RNA via Dhx15.
(A) Co- immunoprecipitation (IP) of Nlrp6 with Dhx15 from WT and Nlrp6−/− mouse intestinal epithelial cells using an anti-Nlrp6 antibody. IB: immunoblotting. (B) Co-IP of FLAG-Nlrp6 NACHT+NAD (amino residues 170–715) and the full-length (1–end) with endogenous DHX15 from HEK293T cells overexpressing FLAG-tagged proteins using an anti-FLAG antibody. (C) Co-IP of FLAG-tagged proteins with endogenous MAVS from HEK293T cells as in (B). WCE, whole cell extract. RIG-I, retinoic acid inducible gene 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (D) Quantitative PCR analyses of viral RNA bound by FLAG-tagged proteins from EMCV-infected and FLAG fusion protein-expressing HEK293T cells. The data are presented as fold increase over vector (FLAG). (E) Binding of endogenous Nlrp6 to viral RNA. Left chart, quantitative PCR analyses of viral RNA bound by endogenous FLAG-Nlrp6 in IECs. Right panel: immunoblots of FLAG-Nlrp6 in WCE and IP. 3xFLAG-Nlrp6 denotes 3 FLAG motifs tagged to Nlrp6. (F) Quantitative PCR analyses of EMCV RNA bound by FLAG-Nlrp6 from GFP or DHX15 siRNA-treated HEK293T cells. The lower panel: immunoblots of WCE and IP. (G) Immunoblots showing FLAG-tagged proteins (purified from HEK293T) bound by biotin-labeled RNA analogues. PolyIC(H): high molecular weight (1.5–8kb), polyIC(L): low molecular weight (0.2–1kb). (H) Co-IP of Mavs with Dhx15 from IECs of WT and Nlrp6−/− mice infected with EMCV using a rabbit anti-Mavs antibody. (I) The survival curves of WT and Mavs−/− mice after oral infection with EMCV. N=5/group; *, p<0.05 (Log-rank test). The data are representative of at least two independent experiments.
Dhx15 is a putative pre-mRNA-splicing factor and ATP-dependent RNA helicase, and modulates antiviral immune responses via MAVS, an adaptor protein for RIG-I like receptors (RLRs) (21, 22). We reasoned that the Nlrp6-Dhx15 complex might use MAVS to trigger type I interferon (IFN) responses. Indeed FLAG-Nlrp6 bound endogenous MAVS, as did Nlrp3 (23) and RIG-I (24–27) (Fig. 2C). The negative controls FLAG-NLRC5 (11) or Nlrp10 did not co-precipitate with MAVS (fig. S5D), confirming the specificity of the Nlrp6-MAVS interaction. Since Dhx15 is a putative RNA helicase and viral RNA sensor (22), we then determined whether Nlrp6-Dhx15 forms a viral RNA sensing complex. Both Nlrp6 and Dhx15 showed high affinity for viral RNA (Fig. 2D and fig. S6A). The Nlrp6 NACHT domain was sufficient for RNA binding, but weaker than full-length Nlrp6 (fig. S6B). To exclude non-specific binding due to overexpression, we examined endogenous Nlrp6 binding to viral RNA in WT and FLAG-Nlrp6 knock-in mice (fig. S2). Both Nlrp6 and FLAG-Nlrp6 was co-immunoprecipitated with EMCV RNA from infected IECs (Fig. 2E, fig. S6C). Since the RNA binding capacity of Dhx15 was much greater than that of Nlrp6, we reasoned that Nlrp6-RNA binding was dependent on Dhx15. Indeed, the amount of Nlrp6-bound viral RNA was reduced significantly in Dhx15 siRNA-treated cells (Fig. 2F). In contrast, Dhx15-RNA binding was not altered in Nlrp6−/− cells (fig. S6D). Like Dhx15 (22), Nlrp6 bound only RNA but not DNA viruses (fig. S6E). To assess the nature of viral RNA bound by Nlrp6, we tested several synthetic RNA analogues. Nlrp6 preferably bound the long dsRNA analogue- polyinosinic:polycytidylic acid (poly I:C) (Fig. 2G). To provide in vivo evidence for a functional interaction between MAVS, Dhx15 and Nlrp6, we examined MAVS-Dhx15 interactions in WT and Nlrp6−/− IECs. Consistent with a previous report (22), MAVS binding to Dhx15 was enhanced by EMCV infection in WT IECs, but the interaction was weaker in Nlrp6−/− (Fig. 2H). Mavs−/− mice were also much more susceptible to EMCV administered orally when compared with WT mice (Fig. 2I). These data suggest that the Dhx15-Nlrp6-MAVS axis plays an important role in restricting EMCV infection of the intestine.
To validate a role for Nlrp6 in Dhx15-Mavs-mediated antiviral immunity, we examined the expression of type I/III IFN-induced genes (ISGs). The mRNA and protein expression of a number of ISGs was reduced in Nlrp6−/− IECs compared to WT (fig. S7A, Fig. 3A and B). Although both type I and III IFNs can elicit antiviral responses, type III IFNs are particularly critical for controling viral infection in IECs (28–30). IFN-λ (also known as IL-28a) protein and mRNA, and Ifnb mRNA were reduced in Nlrp6-/- intestines after EMCV infection (fig. S7B). ISG mRNA amounts were, however, similar in other WT and Nlrp6−/− tissues (fig. S8).
To assess whether the Nlrp6-caspase-1 inflammasome regulates antiviral immunity in the intestine, we compared ISG expression in Nlrp6−/− with Casp1−/− and WT mice. The viral loads and ISG expression were similar in the intestines of Casp1−/− and WT mice (fig. S9), demonstrating an inflammasome-independent antiviral mechanism for Nlrp6. In support of the in vivo findings, EMCV loads in Nlrp6−/− embryonic fibroblasts (MEFs) were 6-fold higher than those in Nlrp6+/− cells at 16 h after infection; while antiviral gene expression was significantly lower (Fig. 3C and D, fig. S10A and B). We also observed a decrease in poly (I:C)-induced Ifnb1 expression in Nlrp6−/− compared to Nlrp6-/+ MEFs (fig. S10C). In agreement with the results from Nlrp6−/− cells, overexpression of Nlrp6 enhanced Ifnb1 and Il6 expression modestly (fig. S11). All these data demonstrate a pivotal role for Nlrp6 in inducing type I/III IFNs and ISGs. Type III IFNs are particularly critical for control of viral infection of IECs (28–30). Indeed, exogenous IFN-λ fully protected WT and Nlrp6−/− mice against lethal EMCV infection and reduced viremia significantly (Fig. 3E). We next determined whether the antiviral function of Nlrp6 is specific for RNA viruses. Neither herpes simplex virus-1 (HSV-1) titers nor Ifnb1 expression in Nlrp6−/− was different from those in Nlrp6-/+ cells (fig. S12A). IFN-α, polyd(A:T) or lipopolysaccharide-induced ISGs or cytokine expression in Nlrp6−/− was also similar to that in Nlrp6-/+ MEFs (fig. S12, B to D).
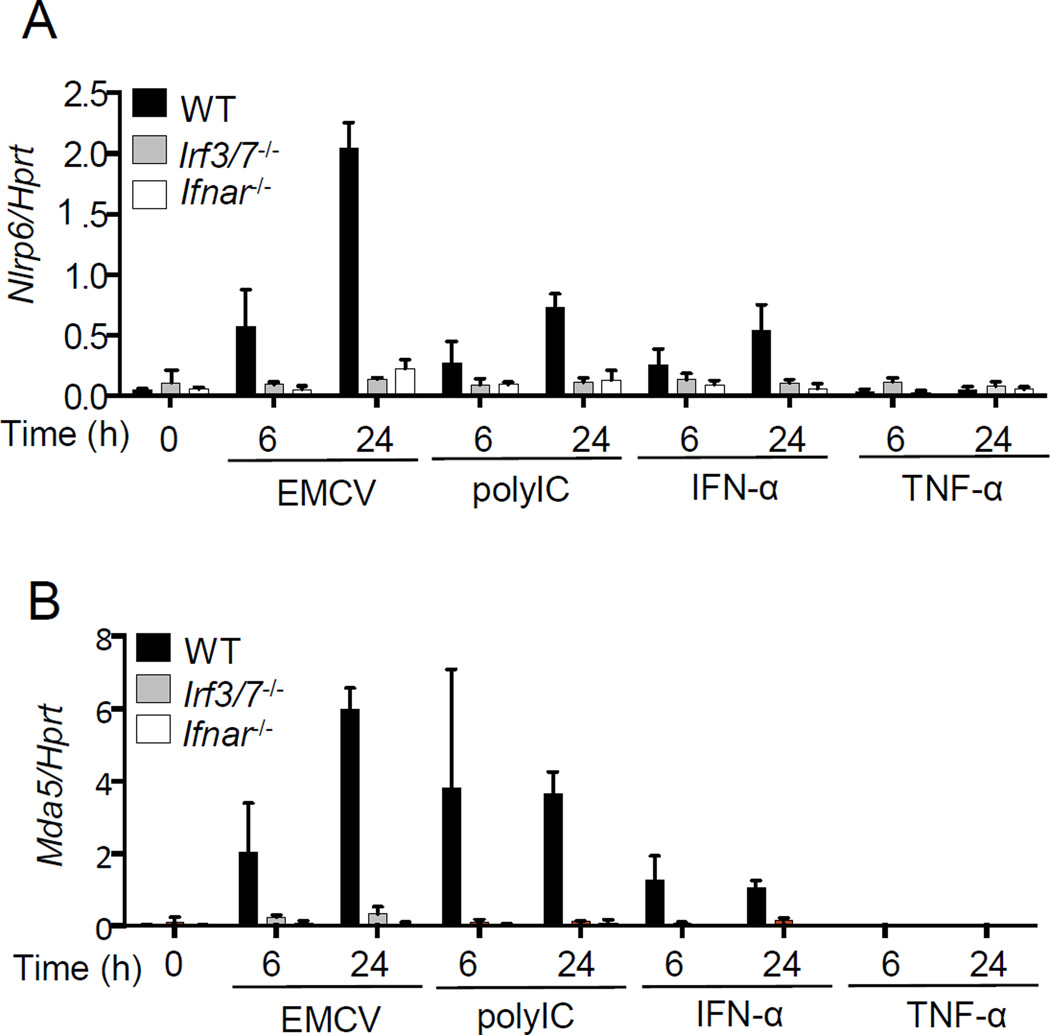
As viral infections, and the ligands that can induce robust type I IFN expression, also up-regulated Nlrp6 expression (Fig. 3C, fig. S10C, fig. S12C and fig. S13), we reasoned that Nlrp6 per se might be an ISG. Indeed, induction of Nlrp6 mRNA expression by EMCV or poly(I:C) treatment was almost abolished in Irf3/7−/− or Ifnar1−/− MEFs. Consistent with this, recombinant IFN-α, but not TNF-α was able to induce Nlrp6 expression vigorously, suggesting that IRF/IFN signaling but not NF-κB signaling controls Nlrp6 expression (Fig. 4, fig. S14A). Nlrp6 mRNA expression was also induced by recombinant IFN-λ2 (fig. S14B). These results indicate that Nlrp6 expression is regulated by type I/III IFNs via IRF3/7.
Fig. 4. Nlrp6 is an ISG.
Quantitative PCR analyses of the transcripts of (A) Nlrp6, (B) Mda5 in WT, Irf3/7−/− and Ifnar−/− MEFs treated with EMCV, polyI:C, recombinant IFN-α or TNF-α. The data are expressed as percentage of a house keeping gene Hprt. Bars: mean + S.D. The data are representative of at least two independent experiments.
The above-mentioned data demonstrate that Dhx15-Nlrp6 senses long dsRNA in the cytoplasm (Fig. 2G), a well-established feature for MDA5. We then determined whether Nlrp6-mediated signaling is also dependent on MDA5. siRNA knockdown of Nlrp6 reduced Ifnb1 and Isg15 mRNA expression following poly I:C stimulation in Mda5−/− MEFs (fig. S15A), suggesting an MDA5-independent antiviral role for Nlrp6. Similar results were noted with Rig-I−/− MEFs (fig. S15B). Nlrp6-RNA binding was unchanged in Mda5−/− or Rig-I−/− MEFs compared to WT (fig. S15C); and there was no interaction between Nlrp6 and MDA5 or RIG-I (fig. S15D). We next examined the relative anti-viral role for MDA5 in the intestine in comparison to Nlrp6. The viral loads in both Nrp6−/− and Mda5−/− IECs were similar, but much higher than those in WT mice (Fig. S15E). These results, in conjunction with the Nlrp6, Dhx15 and MDA5 expression data (fig. S1), suggest that Dhx15-Nlrp6 constitutes the first line of anti-EMCV defense in the intestinal epithelia, while MDA5 is dominant in myeloid cells.
In summary, our results demonstrate that Nlrp6 controls enteric virus infection in the intestine by interacting with a RNA sensor, Dhx15, to trigger MAVS-dependent antiviral responses. This inflammasome-independent response provides a mechanism for Nlrp6 to elicit pleotropic effects in the host, and demonstrates its importance against diverse classes of microbes.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants N01-HHSN272201100019C, AI099625 and AI103807. E.F. and R.A.F. are Investigators of the Howard Hughes Medical Institute. S.Z. was supported by a fellowship from Howard Hughes Medical Institute-The Helen Hay Whitney Foundation.
Footnotes
The data presented in this manuscript are tabulated in the main paper and in the supplementary materials.
References and Notes
- 1.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 2.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 3.Sabbah A, et al. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 2009;206:79. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen IC. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanneganti TD. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 7.Thomas PG. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore CB. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 9.Allen IC, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei Y, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933–946. doi: 10.1016/j.immuni.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui J, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–496. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong Y, et al. Enhanced TLR-induced NF-kappaB signaling and type I interferon responses in NLRC5 deficient mice. Cell research. 2012;22:822–835. doi: 10.1038/cr.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider M, et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol. 2012;13:823–831. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, et al. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity. 2014;40:329–341. doi: 10.1016/j.immuni.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kersse K, Bertrand MJ, Lamkanfi M, Vandenabeele P. NOD-like receptors and the innate immune system: Coping with danger, damage and death. Cytokine Growth Factor Rev. 2011;22:257–276. doi: 10.1016/j.cytogfr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Wlodarska M, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anand PK, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488:389–393. doi: 10.1038/nature11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karst SM, Wobus CE, Lay M, Davidson J, Virgin HWt. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 20.Rajan JV, Rodriguez D, Miao EA, Aderem A. The NLRP3 inflammasome detects encephalomyocarditis virus and vesicular stomatitis virus infection. J Virol. 2011;85:4167–4172. doi: 10.1128/JVI.01687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosallanejad K, et al. The DEAH-Box RNA Helicase DHX15 Activates NF-kappaB and MAPK Signaling Downstream of MAVS During Antiviral Responses. Science signaling. 2014;7:ra40. doi: 10.1126/scisignal.2004841. [DOI] [PubMed] [Google Scholar]
- 22.Lu H, et al. DHX15 Senses Double-Stranded RNA in Myeloid Dendritic Cells. J Immunol. 2014 doi: 10.4049/jimmunol.1303322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawa T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 25.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 26.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Molecular cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Pott J, et al. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A. 2011;108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broquet AH, Hirata Y, McAllister CS, Kagnoff MF. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol. 2011;186:1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 30.Nice TJ, et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wobus CE, et al. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS biology. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, et al. Caspase-12 controls West Nile virus infection via the viral RNA receptor RIG-I. Nat Immunol. 2010;11:912–919. doi: 10.1038/ni.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P, et al. UBXN1 interferes with Rig-I-like receptor-mediated antiviral immune response by targeting MAVS. Cell reports. 2013;3:1057–1070. doi: 10.1016/j.celrep.2013.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P, et al. Matrix metalloproteinase 9 facilitates West Nile virus entry into the brain. J Virol. 2008;82:8978–8985. doi: 10.1128/JVI.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang P, et al. IL-22 signaling contributes to West Nile encephalitis pathogenesis. PLoS One. 2012;7:e44153. doi: 10.1371/journal.pone.0044153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taube S, et al. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J Virol. 2009;83:4092–4101. doi: 10.1128/JVI.02245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.