Abstract
A disintegrin and metalloproteinases (ADAMs) are a family of cell surface proteases that regulate diverse cellular functions, including cell adhesion, migration, cellular signaling, and proteolysis. Proteolytically active ADAMs are responsible for ectodomain shedding of membrane-associated proteins. ADAMs rapidly modulate key cell signaling pathways in response to changes in the extracellular environment (e.g., inflammation) and play a central role in coordinating intercellular communication within the local microenvironment. ADAM10 and ADAM17 are the most studied members of the ADAM family in the gastrointestinal tract. ADAMs regulate many cellular processes associated with intestinal development, cell fate specification, and the maintenance of intestinal stem cell/progenitor populations. Several signaling pathway molecules that undergo ectodomain shedding by ADAMs [e.g., ligands and receptors from epidermal growth factor receptor (EGFR)/ErbB and tumor necrosis factor α (TNFα) receptor (TNFR) families] help drive and control intestinal inflammation and injury/repair responses. Dysregulation of these processes through aberrant ADAM expression or sustained ADAM activity is linked to chronic inflammation, inflammation-associated cancer, and tumorigenesis.
Keywords: ADAM10, ADAM17, Notch, TNFα, EGFR, intestinal stem cells
INTRODUCTION
A disintegrin and metalloproteinases (ADAMs) are a family of multidomain transmembrane proteins with functions in cell adhesion, migration, cellular signaling, and proteolysis. Proteolytically active ADAMs cleave extracellular domains from type I and type II transmembrane proteins and some glycosylphosphatidylinositol-anchored proteins in a process known as ectodomain shedding. This cleavage event represents an irreversible posttranslational protein modification that defines a protein’s function. ADAM substrates are diverse and include growth factors, cytokines and chemokines, their receptors, cell adhesion molecules, and proteins of the extracellular matrix. For several ADAM substrates, ectodomain shedding is also an initiating and rate-limiting step for sequential cleavage events. This process is called regulated intramembrane proteolysis (RIP) and releases intracellular domains that translocate to the nucleus and regulate gene transcription. The best-studied example of RIP is the sequential processing of Notch receptors, in which ADAM10 is the α-secretase responsible for S2 cleavage and release of the Notch receptor ectodomain. ADAMs are also involved in signaling events associated with IL-6 trans-signaling and exosomes (1–6).
ADAMs can rapidly modulate key cell signaling pathways in response to changes in their extracellular environment. They may act as cellular sensors, providing a mechanism for cells to respond quickly and generate the appropriate responses to different environmental stimuli (e.g., inflammation or cellular stress). ADAM10, ADAM17, and many other ADAMs are ubiquitously expressed in the gastrointestinal tract, which allows different ADAMs to regulate and coordinate cellular communication between different cell types. For example, ADAM-mediated shedding events are thought to be involved in signaling cross talk between intestinal epithelial cells (IECs) and stromal cells in the lamina propria of the gastrointestinal tract. ADAM signaling is fundamental for regulating many cellular processes during intestinal development and homeostasis. Dysregulation of these processes is linked to pathological states, including inflammation and cancer. Analysis of ADAM loss-of-function mouse models has contributed to our initial understanding of the role of ADAMs in these events. This review provides an overview of our current knowledge of proteolytically active ADAMs within the gastrointestinal tract. Given the many potential substrates for ADAMs, only those with direct links to observed phenotypes in gastrointestinal pathophysiology are discussed. Several excellent ADAM reviews provide more detailed descriptions of ADAM biology, activity, and substrate specificity (1–6).
PROTEOLYTICALLY ACTIVE ADAMs
ADAMs: Domain Structure and Biosynthesis
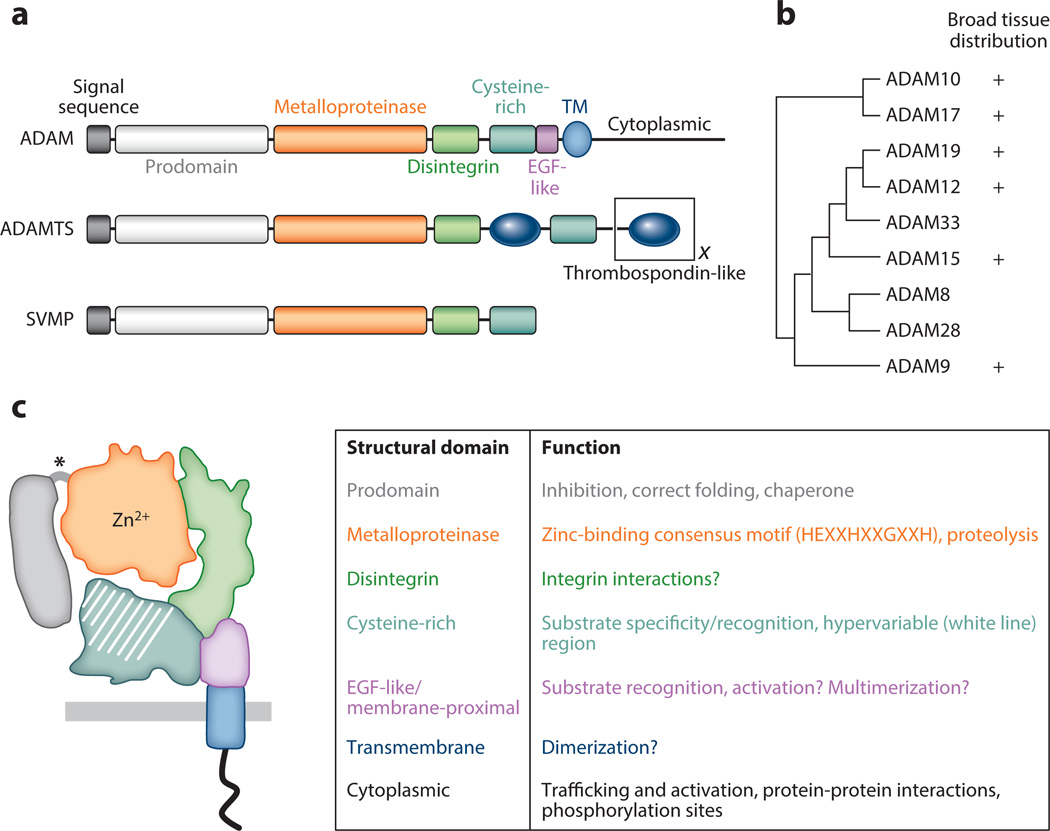
ADAMs belong to the adamalysin subfamily of metzincin metalloproteinases, which includes the snake venom metalloproteinases and ADAMs containing thrombospondin motifs (ADAMTSs) (1, 2, 7). The ADAM family of type I transmembrane proteins is defined by a distinct modular structure, distinguishing them from other adamalysins (Figure 1a). In humans, 22 functional ADAMs have been identified, but only 12 of these family members have proteolytic activity (ADAM8, 9, 10, 12, 15, 17, 19, 20, 21, 28, 30, and 33). The functional roles of mammalian proteolytically active ADAMs are reflected, in part, by their relative amino acid sequence homology (Figure 1b) and by their tissue distribution. In the gastrointestinal tract, the ubiquitously expressed ADAMs, especially ADAM10 and 17, are the most biologically relevant; however, the expression of other broadly distributed ADAMs can be upregulated or misexpressed in disease states including inflammation and cancer.
Figure 1.
Proteolytically active A disintegrin and metalloproteinases (ADAMs): structure and function. (a) ADAMs are members of the adamalysin subfamily of metzincin metalloproteinases, which also includes ADAMs containing thrombospondin motifs (ADAMTSs) and snake venom metalloproteinases (SVMPs). Different ADAMTSs contain variable numbers of thrombospondin-like motifs (represented by the black X) and other functional domains at the C terminus (not shown). Most ADAMs are synthesized as transmembrane (TM) proteins, whereas all ADAMTSs and SVMPs are secreted proteins. Another important difference between the proteins is that all SVMPs and ADAMTSs are predicted to be catalytically active, but only ~60% of ADAMs have intact metalloproteinase domains capable of proteolytic activity. (b) Phylogenetic tree built from aligned full-length amino acid sequences (left; tree adapted from References 2 and 7) and the tissue distribution of all human proteolytically active ADAMs (right). (c) ADAM domain structure and function. (Left) Color representation of structural domains: prodomain, metalloproteinase domain, disintegrin domain, cysteine-rich domain, EGF-like/proximal membrane domain, transmembrane domain, and cytoplasmic domain. Putative furin-like cleavage sites between the prodomain and metalloproteinase domain are shown by an asterisk. (Right) Summary of domain functions. Many ADAMs display broad tissue distribution, but detailed analysis of their cellular expression in different tissues is lacking.
The ADAM domain structure includes a prodomain, a metalloproteinase domain, a disintegrin domain, a cysteine-rich domain, an EGF-like (or membrane-proximal) domain, a transmembrane domain, and a cytoplasmic domain. Each domain has distinct functions, briefly summarized in Figure 1c. The biosynthesis, trafficking, and posttranslational modifications of ADAMs through the secretory pathway are highly regulated events that can impact the production and activity of mature proteolytically active ADAMs on multiple levels. In particular, the metalloproteinase domain of catalytically active ADAMs contains a conserved active site sequence within a globular structure, which, together with the disintegrin and cysteine-rich domains, forms a C-shaped structure. The hypervariable region of the cysteine-rich domain is juxtaposed and in close contact with the catalytic site of the metalloproteinase domain (1, 2). Recent studies have demonstrated that the hypervariable region is important for substrate recognition and the regulation of catalytic activity. The extensive molecular surface of this region may be involved in controlling other protein-protein interactions important for ADAM activity. Investigations into the regulation of ADAM activity are ongoing.
As type I transmembrane proteins, ADAMs contain an N-terminal signal sequence that directs newly synthesized protein into the endoplasmic reticulum (ER);maturation in the Golgi apparatus follows. Recently, researchers discovered that ER resident proteins iRhom1 and iRhom2 regulate the trafficking of immature ADAM17 [also termed TNFα-converting enzyme (TACE)] from the ER into the Golgi apparatus (8–11). iRhom loss of function inhibits the maturation of ADAM17, blocking its proteolytic activity. This iRhom-dependent mechanism of regulated ER trafficking appears specific to ADAM17. In the gastrointestinal tract, ADAM17 activity is probably regulated by different iRhom isoforms in a cell context–dependent manner (12). Interestingly, ADAM10 contains other ER-retention and basolateral sorting signals within its cytoplasmic domain, and the protein was recently shown to interact with specific TspanC8 proteins that regulate its trafficking and maturation (13–15).
Proteolytically Active ADAMs: Mechanisms of Action
For cell surface molecules susceptible to ADAM-mediated processing, the extracellular domain of the molecule is cleaved at a site in close proximity to the outer face of the transmembrane domain. The cleaved ectodomain is released into the extracellular environment. The membrane-anchored remnant contains a short residual extracellular stalk attached to the transmembrane and cytoplasmic domains, which is retained in the plasma membrane. Most ADAM substrates are signaling molecules that act as growth factors, cytokines/chemokines, receptors, and cell adhesion molecules. Ectodomain shedding not only alters the cell surface fate of a candidate substrate but also, more importantly, directly modulates cell function and signaling potential through juxtacrine, autocrine, and paracrine pathways (Figure 2a,b). Ectodomain shedding can also deplete signaling molecules, such as receptors, from the plasma membrane and thereby reduce functional cell surface receptor signaling. Soluble receptor ectodomains released from the surface of cells can also act as decoy receptors (Figure 2a,b). By contrast, ectodomain cleavage of the IL-6 receptor (IL-6R) produces soluble IL-6R (sIL-6R), which, in complex with IL-6, has agonistic properties. This mode of IL-6/sIL-6R signaling has been termed IL-6 trans-signaling and mediates proinflammatory responses (Figure 2c) (16). Another important function of ADAM-mediated ectodomain shedding is the regulation of cell adhesion molecules involved in homotypic (e.g., E-cadherin) and heterotypic (e.g., VCAM1) cell-cell interactions (Figure 2d).
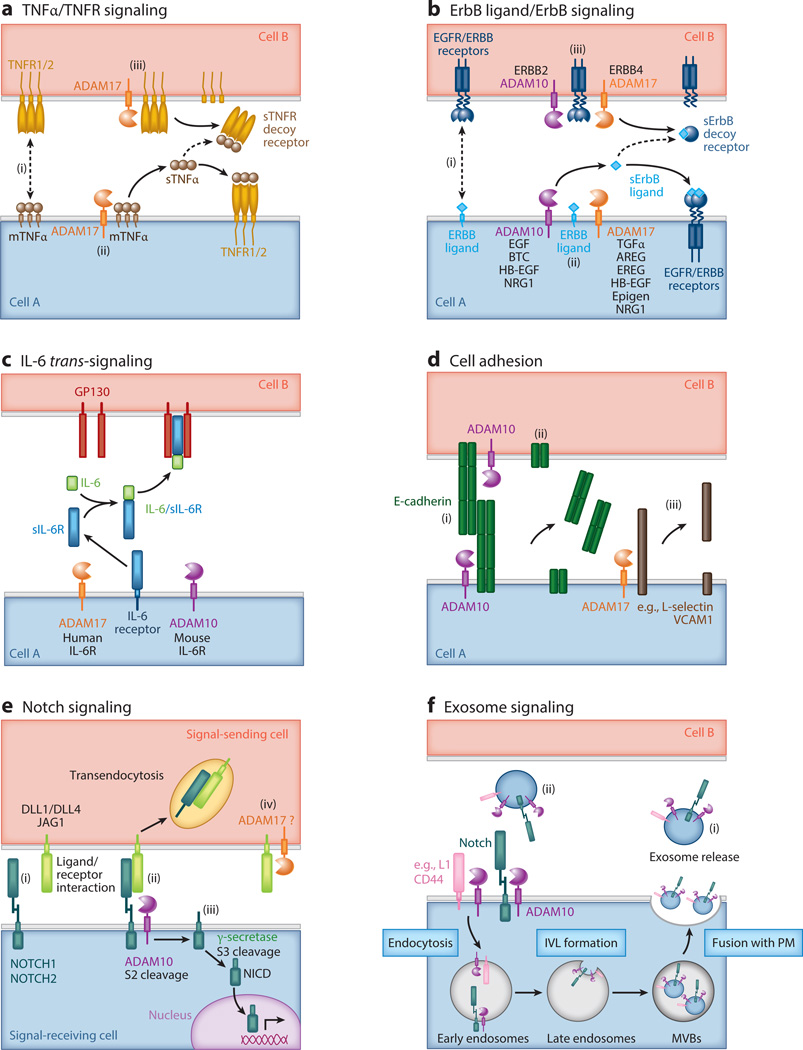
Figure 2.
Diverse extracellular signaling pathways are regulated by A disintegrin and metalloproteinase (ADAM)-mediated shedding events. (a) TNFα/TNFR signaling pathway. ADAM17 is the principal protease responsible for the cleavage of the TNFαligand and both TNFR1 and TNFR2 receptors, all of which are type II transmembrane proteins. (i ) Membrane-anchored TNFα (mTNFα) precursors engage in juxtacrine signaling with cell surface TNFRs, particularly high-affinity TNFR2 receptors. (ii ) ADAM17 cleaves mTNFαto release soluble TNFαligand (sTNFα) that can bind to cell surface TNFRs in an autocrine or paracrine manner. (iii ) ADAM17 can also cleave TNFRs, reducing the level of functional TNFR signaling at the cell surface. Soluble TNFRs (sTNFRs) can act as decoy receptors by sequestering sTNFα. (b) ErbB ligand/ErbB receptor signaling pathway. Multiple ADAMs, particularly ADAM10 and ADAM17, are responsible for cleaving different ErbB ligands and ErbB receptors, which are all type I transmembrane proteins. Both ADAM10 and ADAM17 cleave several ErbB ligands, such as HB-EGF and neuregulin (NRG1), under different experimental conditions. Mouse genetic loss-of-function studies have implicated other ADAMs (e.g., ADAM9 and ADAM19) in the processing of ErbB ligands (not shown). (i ) Membrane-anchored ErbB ligand precursors, such as HB-EGF, engage in juxtacrine signaling with cell surface ErbB receptors. (ii ) ADAM10 and ADAM17 cleave different membrane-anchored ErbB ligands, releasing soluble ligands that function in an autocrine or paracrine manner. (iii ) ADAM10 and ADAM17 can cleave ERBB2 and ERBB4 receptors, respectively, reducing functional ErbB receptor signaling at the cell surface. Soluble ErbB receptors can act as decoy receptors, and soluble ERBB2 (sERBB2) can reduce the therapeutic efficacy of neutralizing antibodies against ERBB2 receptors. (c) IL-6 trans-signaling. The IL-6 receptor (IL-6R) can be cleaved by ADAM10 and ADAM17, but ADAM specificity is species dependent. In classical IL-6 signaling, IL-6 binds to cell surface IL-6R, which recruits two molecules of GP130 into a functional ligand/receptor signaling complex. However, in IL-6 trans-signaling, IL-6R is cleaved by either ADAM10 or 17 to release soluble IL-6R (sIL-6R) that can bind to soluble IL-6 ligand. The IL-6/sIL-6R complex has high affinity for cells expressing GP130 receptor and activates them. Classic IL-6 signaling mediates the activation of anti-inflammatory and regenerative pathways, whereas IL-6 trans-signaling is primarily observed in inflammatory and stress conditions. (d ) Cell adhesion. ADAM10 and ADAM17 cleave different cell adhesion molecules that alter cell-cell interactions. (i ) Homotypic E-cadherin protein interactions are involved in maintaining adherens junction formation between epithelial cells. ADAM10 cleaves E-cadherin, resulting in (ii ) decreased cell-cell interaction and altered epithelial cell migration. (iii ) L-selectin and VCAM1 are examples of cell adhesion molecules involved in leukocyte rolling and adhesion to endothelial cells (not shown). (e) Canonical Notch signaling. ADAM10 initiates the processing and activation of the Notch receptors. (i ) Notch ligand expressed on the signal-sending cell engages the Notch receptor on the signal-receiving cell. (ii ) Normally, the negative regulatory region (NRR) within the Notch receptor masks the α-secretase (S2) cleavage site close to the transmembrane domain. Notch ligand binding to its receptor is proposed to confer a conformational change in the NRR domain, allowing ADAM10 to access the Notch S2 cleavage site. ADAM10 is responsible for cleavage of the NOTCH1, -2, and -3 receptors. (iii ) The Notch remnant is subject to intramembrane proteolysis, in which the γ-secretase complex cleaves within the intramembrane domain at the S3 cleavage site to release the Notch intracellular domain (NICD) into the cytoplasm. After translocation into the nucleus, the NICD associates with other transcriptional cofactors and activates expression of Notch-responsive genes such as Hes1. (iv) Under certain experimental conditions, Notch ligands can be subject to extracellular cleavage by ADAM proteases, such as ADAM17. Notch ligand processing may limit active ligand availability, or it may be involved in ligand sequential processing or bidirectional signaling by the ligand intracellular domain. Alternatively, soluble Notch ligand may bind to and activate Notch receptors via a noncanonical pathway. ( f ) Exosome signaling. ADAM10 and substrates such as L1, CD44, and Notch are enriched in exosomes, providing a mechanism for short- and long-range cellular communication. ADAM10 and its substrates on the cell surface are trafficked through the endosomal compartment and then enriched in intraluminal vesicles (ILVs) produced within multivesicular bodies (MVBs). Upon MVB fusion with the plasma membrane (PM), IVLs are released as exosomes into the extracellular environment. (i ) L1 and CD44 can be shed from ADAM10-expressing exosomes into the extracellular space (not shown). (ii ) Exosomes can also interact with cells at distant cellular sites. In addition, ADAMs may be expressed on ectosomes generated by outward budding of the PM (not shown).
In addition to direct ectodomain-shedding events, ADAMs play an important role in cell surface ligand–induced cleavage events. The best-documented example of this is ADAM10-mediated Notch signaling, in which Notch ligand, expressed on the sending cell, binds to a Notch receptor on the receiving cell (Figure 2e). Another variation on ligand-induced shedding has been described in cell-based assays for Ephrin-Eph signaling. Upon Ephrin ligand binding, a conformational change releases steric hindrance between ADAM10 and the Eph receptor. This generates a new exosite interaction between the Eph/Ephrin complex and permits ADAM10-mediated ephrin cleavage in trans (17, 18). Similar to ADAM10-mediated Notch processing, this is a ligand-mediated conformational switch that provides precise control of ADAM10 sheddase activity. ADAM10-mediated Notch signaling is also the prototypic example for RIP (Figure 2e). For most RIP substrates, ADAMs are the α-secretases responsible for ectodomain processing and generation of small membrane-bound remnants. These remnants are susceptible to intramembrane proteolysis by a γ-secretase complex or other signal peptide peptidase–like proteases. The released intracellular domain may have signaling activity, but, for many ADAM substrates, the function of further sequential processing is to promote the removal of membrane-anchored remnants from the cell surface via endocytosis and lysosomal degradation (1, 2).
ADAM-mediated shedding events have also been reported in exosomes, providing a unique mechanism for short- and long-range intercellular communication (Figure 2f). Exosome trafficking can alter ADAM substrate specificity, as reported for ADAM10-mediated constitutive and stimulus-induced L1 and CD44 shedding (19). In addition, a recent analysis of exosomes produced by TIMPless fibroblasts [fibroblasts lacking all four endogenous tissue inhibitor of metalloproteinase (TIMP) genes] demonstrated that ADAM10-enriched exosomes promote motility in cancer cells (20). Together, these studies suggest that ADAM-mediated shedding events from exosomes may be an important intercellular signaling pathway.
ADAMs: Substrate Specificity and Constitutive Versus Regulated Processing
Numerous type I and II transmembrane proteins are susceptible to proteolytic cleavage by ADAM proteases. Because of the shallow cleavage pocket in the ADAM metalloproteinase domain, no strong sequence homology has been observed in the substrate cleavage site; this makes it difficult to predict ADAM substrate specificity. As outlined in Figure 2, a substrate can be cleaved by one specific ADAM or several different ADAMs. In fact, ADAM specificity depends on several parameters, including species differences and whether the substrate undergoes constitutive or stimulated shedding. ADAM substrate specificity may differ between normal developmental and physiological conditions and those observed in different disease states. Loss-of-function studies have revealed ADAM redundancy, which suggests a potential hierarchy in both ADAM usage and substrate selection. A recent analysis of B cell–specific Adam10-deficient mice demonstrated that defects in secondary lymphoid organ development result from a compensatory increase in Adam17 expression and TNFα signaling (21). Thus, further analysis of ADAM specificity, redundancy, and compensation is required to fully understand the biological functions of different ADAMs in vivo.
ADAM proteolytic activity can be regulated at the level of transcription, via alternative splicing, and by posttranslational modification. In general, upregulation of Adam expression is associated with increased ADAM activity. However, the most rapid and efficient way to modulate ADAM proteolytic activity is at the protein level (1, 2, 4, 5). ADAM activity can be regulated by various posttranslational modifications; examples include prodomain cleavage, changes in disulfide bond formation of the ADAM extracellular domain associated with protein disulfide isomerase interactions and altered redox environment, and phosphorylation of the cytoplasmic domain. Autocatalysis and ADAM shedding by other proteases, the regulation of ADAM dimerization/multimerization, interactions with endogenous TIMPs, protein-protein interactions associated with ADAM intracellular trafficking (e.g., tetraspanins and iRhoms), and substrate recognition/presentation all affect ADAM activity (Figure 3a) (1, 2, 4, 5).
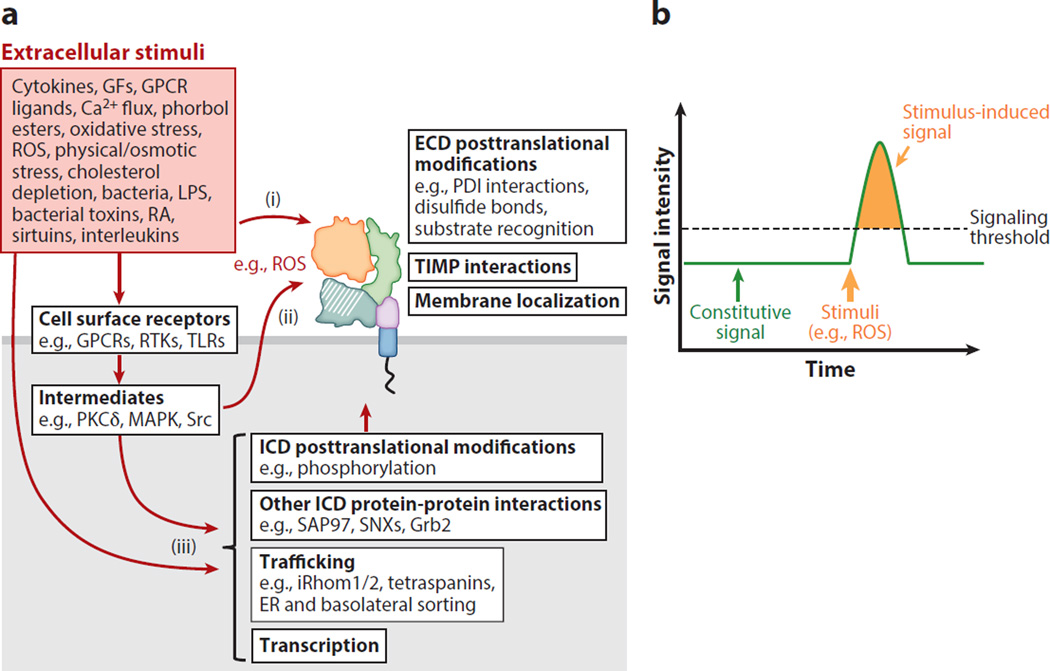
Figure 3.
ADAM proteolytic activity can be regulated at multiple levels. (a) Constitutive and stimulus-induced ADAM-mediated substrate shedding can be regulated at multiple levels. Although constitutive shedding may require functional interactions with the ADAM cytoplasmic domain, it is generally accepted that rapid, stimulus-induced ADAM shedding events result from posttranslational modifications of the ADAM extracellular domain and act independently of the ADAM cytoplasmic domain. (i ) Extracellular stimuli such as ROS can directly interact with the ADAM extracellular domain to regulate proteolytic activity and substrate recognition (22). (ii ) Cell surface receptor activation via signaling intermediates may generate signals (e.g., ROS) that are released into the extracellular environment and act directly on the ADAM extracellular domain. (iii ) Alternatively, cell surface receptor signaling or other extracellular signals can regulate ADAM transcription, trafficking, and protein-protein interactions, in addition to posttranslational modifications of the ADAM intracellular domain. Many of these same signaling pathways are also likely to act on substrates directly, regulating their presentation to and recognition by ADAMs. (b) Schematic of signaling activity generated by a substrate shed under constitutive and stimulus-induced shedding conditions. The ability to rapidly stimulate substrate shedding (e.g., of ErbB ligands) provides a mechanism to reach a signaling threshold, or signaling pulse, that can produce a distinct stimulus-induced cellular response. However, stimulus-induced shedding can rapidly reduce the reservoir of substrate that is available to be shed from the cell surface. If substrate levels are not replenished at the cell surface, signaling activity is predicted to decrease (possibly below constitutive levels), and time may be required before sufficient substrate is available to repeat this process. Abbreviations: ADAM, A disintegrin and metalloproteinase; ECD, ADAM extracellular domain; ER, endoplasmic reticulum; GFs, growth factors; GPCRs, G-protein coupled receptors; ICD, ADAM intracellular domain; PDI, protein disulfide isomerase; LPS, lipopolysaccharide; RA, retinoic acid; ROS, reactive oxygen species; TIMP, tissue inhibitor of metalloproteinase.
Although mechanisms to maintain constitutive ADAM shedding are less defined, numerous stimuli are reported to rapidly activate shedding in an ADAM-specific and cell type–dependent manner. Many stimuli, such as reactive oxygen species (ROS), may act directly on the ADAM’s extracellular domain to alter its proteolytic activity without involving the cytoplasmic domain (22). Generally, stimulated ectodomain shedding provides a mechanism for cells to rapidly respond to changes in their extracellular environment. In the case of EGF-like ligands, stimulated ectodomain shedding rapidly enhances the production of active, soluble ligand. This pulse of active ligand may achieve a functional signaling threshold not obtainable under basal/constitutive shedding conditions (Figure 3b). A major challenge in the field is to accurately measure changes in the activity of a specific ADAM at the level of individual cells and in different developmental and pathological states in vivo.
Inhibition of ADAM Activity
TIMPs are small endogenous protein inhibitors of ADAMs, ADAMTSs, and matrix metalloproteinases. All four Timp genes (Timp1, -2, -3, and -4) encode small proteins that contain an N-terminal inhibitory domain that binds to the active site of metalloproteinase domains. TIMP3 can inhibit several ADAMs, although other TIMPs have more limited inhibitory activity toward ADAMs (23). TIMP3 is a secreted protein that binds to the extracellular matrix and is proposed to interact with a latent ADAM17 homodimer at the cell surface (24, 25). Detailed analysis of Timp3-deficient mice in lipopolysaccharide (LPS)-induced sepsis, hepatectomy, and Fas-induced hepatitis models has clearly demonstrated that TIMP3 is critical for regulating ADAM17 activity and limiting perturbations in TNFα signaling; this role impacts immune responses in vivo (6). Using either small-molecule inhibitors or recombinant prodomains, researchers have made extensive efforts to develop specific ADAM inhibitors besides endogenous TIMPs (26). These efforts have had limited success, however, because candidates’ poor inhibitor specificity and toxicity profiles have reduced their therapeutic applicability (27–29).
More recent approaches have focused on using antibody phage display to generate neutralizing antibodies to the ADAM extracellular domain (30, 31). The cross-domain and exosite specificity of the human monoclonal antibody ADAM17 inhibitor D1(A12) makes it a potent and specific ADAM17 inhibitor. A similar antibody phage display approach has been used to generate antibodies that bind and block substrate shedding. Neutralizing antibodies specific to NOTCH1 and NOTCH2 have been used to study Notch signaling in the crypt stem cell niche of the mouse small intestine (32–34) and in mouse models of liver cancer (35).
INTESTINAL DEVELOPMENT AND HOMEOSTASIS: A BRIEF OVERVIEW
The epithelium lining the gastrointestinal tract has multiple functions, including digestion, nutrient absorption, barrier function, and immunity. The intestinal epithelium is organized into proliferative crypts that undergo constant renewal to replenish differentiated cells along the crypt-villus axis; this replenishment is required to maintain intestinal homeostasis and tissue integrity. Many excellent reviews have recently been published on intestinal development and stem cell homeostasis (36–41). Here, we briefly summarize aspects of intestinal development, cell lineage specification, and intestinal stem niche homeostasis that ADAM signaling events are predicted to affect.
The primitive mouse gut tube is established at embryonic day (E)9.5 and is composed of a central lumen surrounded by pseudostratified, undifferentiated epithelium that progressively increases in circumference. At E14.5, the epithelium undergoes a wave of cytodifferentiation and villus morphogenesis, and, concurrently, intestinal stem cell (ISC)/progenitor cell populations are restricted to intervillus zones. The Wnt and Notch signaling pathways have essential roles in the proliferation and cytodifferentiation of the developing intestine. Functional differentiation of the intestine continues postnatally with the appearance of morphologically distinct Paneth cells at approximately postnatal day 7. The crypts become monoclonal, and the crypt number expands to match the rapid growth of the intestine. However, it is only after weaning that the intestine achieves full maturation (36–38, 40, 41).
Adult Intestinal Stem Cell Niche
In the adult small intestine, multipotent Lgr5+ (Leu-rich repeat–containing G protein–coupled receptor 5–expressing) crypt base columnar stem cells (CBCs), located in the bottom of the crypt, drive the replenishment of the epithelial cells lining the crypt-villus axis. Multiple extracellular signaling pathways, including Wnt, Notch, Eph, BMP, and ErbB, are important for maintaining and regulating the stem and progenitor compartments of the stem cell niche (Figure 4a). Importantly, Paneth cells, other epithelial cells, and stromal cells within the lamina propria contribute essential signals to maintain the stem cell niche (Figure 4b). Complex interactions between Wnt and Notch signaling are critical for ISC maintenance, stem cell/progenitor proliferation, and cell lineage specification within the crypt compartment. These signals are precisely integrated to maintain stem cell activity and to regulate the differentiation required for intestinal homeostasis. Several lines of evidence indicate that Notch signaling is active in multipotent ISCs (34, 42–44). Recent analysis of intestine-specific deletion of Notch receptors shows that NOTCH1 is the primary receptor regulating ISC function, but NOTCH1 and NOTCH2 regulate cell proliferation, cell fate specification, and postinjury regeneration (45, 46). Additional Notch inhibitor studies have demonstrated that Notch directly targets Lgr5+ CBCs and is required for stem cell proliferation and survival in an Atoh1-independent manner (32, 34, 45).
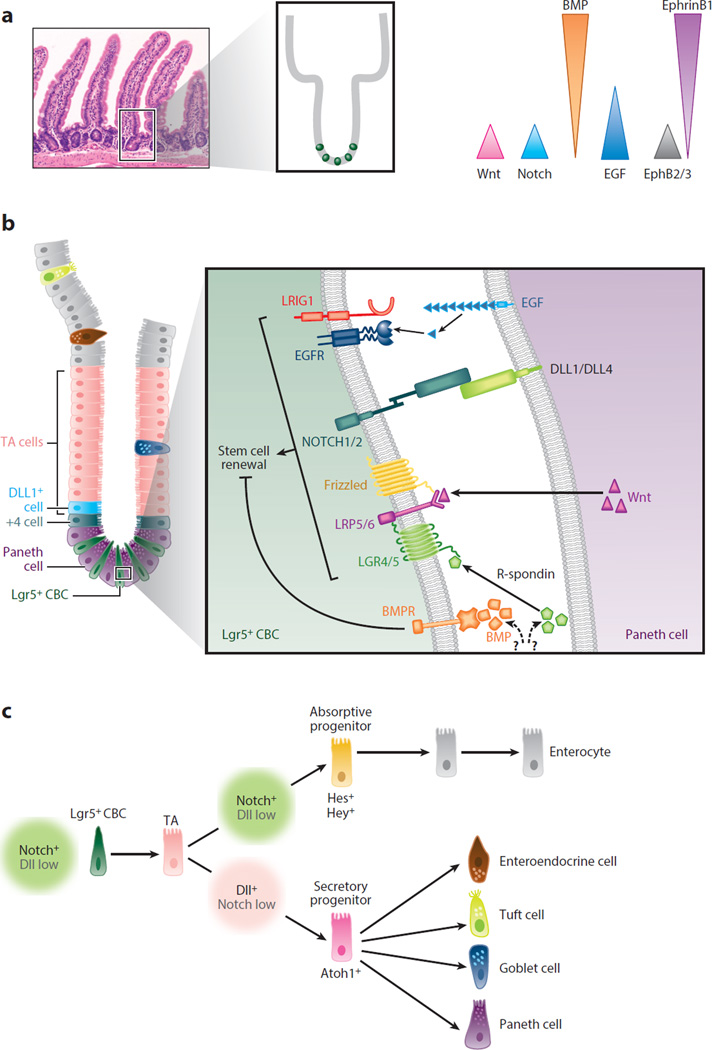
Figure 4.
Overview of intestinal homeostasis: regulation of the intestinal stem cell (ISC) niche and cell fate specification in the small intestine. (a) Crypt-villus architecture and signaling gradients involved in maintaining crypt homeostasis. The mouse small intestine is composed of repeating crypt-villus units. The replenishment of the entire epithelial lining of the intestine is a dynamic process that occurs every ~5–7 days. Complex interactions between multiple signaling pathways are precisely integrated to maintain epithelial cell renewal and crypt homeostasis. (Left) Hematoxylin and eosin staining of the mouse small intestine shows the repeating crypt-villus units (inset). (Right) Schematic showing the distribution of gradients for the Wnt, Notch, epidermal growth factor receptor (EGFR)/ErbB, BMP, and Eph/Ephrin signaling pathways along the crypt-villus axis. (b) Cell types of the intestine (left) and signaling within the stem cell niche (right). (Left) Schematic showing the cellular composition of the stem cell niche in the crypt compartment. Leu-rich repeat–containing G protein–coupled receptor 5–expressing (Lgr5+) crypt base columnar stem cells (CBCs) are positioned at the crypt base, intercalated between Paneth cells. Lgr5+ CBCs are an essential component of the ISC niche and give rise to rapidly proliferating transit-amplifying (TA) progenitor cells. +4 cells and DLL1+ cells are facultative reserve stem cells located at positions 4 and 5 from the crypt base. TA cells appear above the stem cell niche and rapidly migrate toward the crypt-villus junction. Before TA cells exit the crypts, they differentiate into distinct absorptive and secretory cell lineages. All differentiated postmitotic intestinal cells emerge from the crypts, migrate along the villus surface, and are eventually shed from the villus tips into the gut lumen. The one exception is Paneth cells, which first appear above the stem cell niche but are then retained at the crypt base. Paneth cells have a longer life span (>30 days) than other differentiated cell types (~5–7 days). (Right) Paneth cells provide signals required for regulating and maintaining Lgr5+ CBCs in the stem cell niche (inset). They produce Wnt ligands (e.g., Wnt3a), which bind to LRP5/6/Frizzled receptor complexes on Lgr5+ CBCs. The binding of R-spondin to LGR4/5 receptors enhances Wnt activity in Lgr5+ CBCs. Paneth cells release EGF (and other ErbB ligands) that bind to EGFR/ErbB receptors on Lgr5+ CBCs. LRIG1, a negative regulator of EGFR/ErbB signaling, can modulate stem cell/progenitor proliferation. DLL4 and DLL1 on the surface of Paneth cells bind to and activate Notch receptors on Lgr5+ CBCs. Wnt, EGFR/ErbB, and Notch signaling promotes stem cell survival, proliferation, and renewal. However, BMP ligands bind to BMPR receptors on Lgr5+ CBCs to limit cell proliferation and increase differentiation. BMPs are produced by myofibroblasts in the lamina propria, whereas the cellular sources of R-spondins are still under investigation. Other accessory cells within the lamina propria (e.g., pericryptal myofibroblasts, immune cells, and endothelial cells) contribute paracrine, and often redundant, signals (both agonistic and antagonistic) that can regulate the stem cell niche, particularly during tissue regeneration following injury and inflammation (not shown). (c) Notch regulates intestinal cell fate specification. Notch signaling is required for Lgr5+ CBC proliferation and survival. Notch also controls cell fate decisions of short-lived TA progenitors by regulating the key transcription factor Atoh1. NOTCH1 and NOTCH2 receptors and DLL1 and DLL4 control these events (see Figure 5 for more details). Notch+ (Dll-low) TA cells are fated toward the enterocyte lineage. Absorptive progenitors differentiate into enterocytes, the major cell type lining the villi. Dll+ (Notch-low) TA cells are fated toward the secretory lineage. Secretory progenitors undergo further specification and differentiate into goblet cells, enteroendocrine cells, tuft cells, and Paneth cells. Other specialized epithelial cell types (e.g., M cells associated with Peyer’s patches and cup cells) are found in the intestine, but their fate mapping is still poorly understood. Panels a and b adapted from Reference 149.
In a separate signaling event, Notch controls cell fate decisions of short-lived, bipotent transit-amplifying (TA) progenitors by regulating the key transcription factor Atoh1 (34, 47). NOTCH1 and NOTCH2 receptors and DLL1 and DLL4 control these events (45, 46, 48). Upon Notch activation, Atoh1 expression is repressed in progenitors, driving differentiation toward the enterocyte lineage. In the absence of Notch signaling, progenitors express Atoh1 and are fated to the secretory lineage. ATOH1 target genes, such as Gfi1, Spdef, and Neurog3, are responsible for later specification events in the secretory lineage (Figure 4c). Some evidence suggests that goblet and Paneth cells belong to a shared lineage, but it is unclear how multipotent secretory progenitors are allocated and how they give rise to major secretory cell types (38, 41).
Intestinal Stem Cell Plasticity
Although Lgr5+ CBCs are required for normal intestinal homeostasis, analysis of novel stem cell markers and recent advances in lineage tracing suggest that, upon CBC injury/ablation, other facultative or reserve stem cell populations within the crypt can restore the Lgr5+ CBC stem cell pool and repopulate the stem cell niche. The exact identity of these facultative stem cell populations remains elusive, but secretory progenitors, including label-retaining Paneth cell progenitors, DLL1+ TA cells, and other +4 presumptive quiescent stem cell populations (Bmi1+, Lrig1+, Hopx+), are proposed to function in this role (38). Stem cell plasticity can be explained, in part, by broadly permissive chromatin found in different stem cell/progenitor cell populations (49), but the extracellular signals that regulate plasticity events are poorly defined.
ADAMs AND THE GASTROINTESTINAL TRACT
ADAM10 Function and the Intestine
ADAM10 is expressed in all intestinal cell types, including epithelial cells, lamina propria cells (pericryptal myofibroblasts, immune cells, and endothelial cells), smooth muscle cells, and enteric neurons. During development, ADAM10 is robustly expressed in E13.5 epithelium prior to cytodifferentiation, which indicates that it may have other, undefined role(s) in early versus late intestinal development (50). In the adult intestine, ADAM10 is abundantly expressed on the basolateral cell surface of all IECs (50). Under normal physiological conditions, ADAM10 specificity is therefore restricted to substrates expressed on the basolateral cell surface and can only interact with the contents of the gut lumen when barrier function is perturbed. As discussed below, the expression of ADAM10 in distinct IEC populations within the crypt compartment (e.g., Lgr5+ CBCs and TA progenitors) and in all differentiated, postmitotic IECs suggests that ADAM10may have unique functions in these different cell populations.
ADAM10 Acts Iteratively to Regulate the Notch Signaling Required to Maintain Intestinal Stem Cell Populations and Cell Lineage Specification
Phenotypic analysis of global Adam10-deficient mice provided the first evidence that ADAM10 is the α-secretase responsible for regulating Notch signaling. Specifically, Adam10-deficient embryos die at E9.5 as a result of developmental defects in somitogenesis, neurogenesis, and vasculogenesis; these features are similar to those observed in Notch-deficient mice (51). Data from conditional Adam10-deficient mice and biochemical studies have confirmed that ADAM10 is required for ligand-induced Notch activation in several different developmental settings (52–54). However, because global Adam10-deficient mice die prior to intestinal development, our current understanding of the role of ADAM10 in the developing and adult mouse intestine comes from analysis of different IEC-specific Adam10-deficient mouse models (Table 1). Analysis of Villin-Cre;Adam10 flox/flox mice (a model of constitutive ADAM10 inactivation in IECs that begins at approximately E15) and tamoxifen-inducible Villin-CreER;Adam10 flox/flox mice (a model that efficiently induces ADAM10 inactivation in adult IECs) has revealed that ADAM10 deficiency in immature and adult IECs reduces viability, decreases proliferation, and increases apoptosis; these effects lead to crypt degeneration. The conversion of the stem/progenitor compartments into postmitotic secretory cell populations points to an essential role of ADAM10 in regulating Notch and cell fate specification (50). In addition, decreased expression of the Notch target genes Hes1 and HeyL and a parallel increase in expression of genes encoding transcription factors involved in secretory fate specification (Atoh1, Gfi1, Spedef, Neurog3, and Sox9) was observed. Furthermore, lineage analysis using Atoh1LacZ reporter mice has revealed that stem cell/progenitor compartments in both the immature and adult intestine are completely converted to postmitotic ATOH1+ secretory cells. Genetic complementation studies using the Notch gain-of-function allele Rosa26NICD demonstrate that activated Notch can override Adam10 deficiency (50). This shows that Notch is the dominant pathway regulated by ADAM10 in the developing and adult intestine.
Table 1.
Effect of global and conditional Adam10 deletion on gastrointestinal tract function and pathophysiology
| ADAM10- deficient mouse models |
Tissue distribution | Experimental model |
Gastrointestinal phenotype |
Notch rescue |
Reference(s) |
|---|---|---|---|---|---|
| Null | |||||
| Adam10−/− | Global | Development | Embryonic lethal at E9.5; lethality prior to gastrointestinal development |
ND | 51 |
| IEC specific | |||||
|
Villin-Cre; Adam10flox/flox |
All IECs at ~ E15 | IEC development | Postnatal lethal atP1; ↓crypt cell proliferation, ↑secretory cell differentiation |
Yes | 50 |
|
Villin-CreER; Adam10flox/flox |
All IECs | Adult IEC homeostasis |
Lethality; ↓ISC markers, e.g., Olfm4, ↓crypt cell proliferation, ↑secretory cell differentiation |
Yes | 50 |
|
Cdx2P-CreER; Adam10flox/flox |
IECs in distal ileum, cecum, and colon |
APCflox/flox model | No adenoma initiation | Yes | 135 |
|
Lgr5-CreER; Adam10flox/flox |
Lgr5+ CBCs | ISC niche, crypt homeostasis |
Loss of Adam10-deficient Lgr5+ CBCs |
Yes | 50 |
| APCflox/flox model | No adenoma initiation | Yes | 135 | ||
|
Bmi1-CreER; Adam10flox/flox |
Bmi1+ cells | ISC niche, crypt homeostasis |
Loss of Adam10-deficient Bmi1+ ISCs |
Yes | P.J. Dempsey, unpublished observation |
|
Defensin4-Cre; Adam10flox/flox |
Paneth cells | Paneth cell biology | No overt phenotype | ND | P.J. Dempsey, M. Rajala &M.J. Myers Jr., unpublished observation |
|
Cryptidin2- dnAdam10 |
Paneth cells | Paneth cell biology | Mislocalization of Paneth cells |
ND | 63 |
| Leukocyte specific | |||||
|
PolyIC Mx-Cre; Adam10flox/flox |
Broad expression, hematopoietic cells |
Hematopoiesis | Enlarged liver with thrombocytosis and myeloid/granulocyte infiltration |
ND | 150, 151 |
| Increased mast cell infiltration in small intestine |
152 | ||||
|
CD19-Cre; Adam10flox/flox |
B cells | B cell development | Disturbed secondary lymphoid organ architecture, including Peyer’s patches, as a result of a compensatory increase in ADAM17/TNFα signaling axis |
ND | 21, 153, 154 |
|
LysM-CreER; Adam10flox/flox |
Neutrophils, monocytes, macrophages |
Inflammation models |
LPS TLR4-induced inflammatory polarization of macrophages |
ND | 128, 129 |
| DSS colitis | Severe, unremitting inflammation |
Yes | 129 | ||
Abbreviations: CBCs, crypt base columnar stem cells; DSS, dextran sodium sulfate; E, embryonic day; IEC, intestinal epithelial cell; ISC, intestinal stem cell; LPS, lipopolysaccharide; ND, not determined; P, postnatal day.
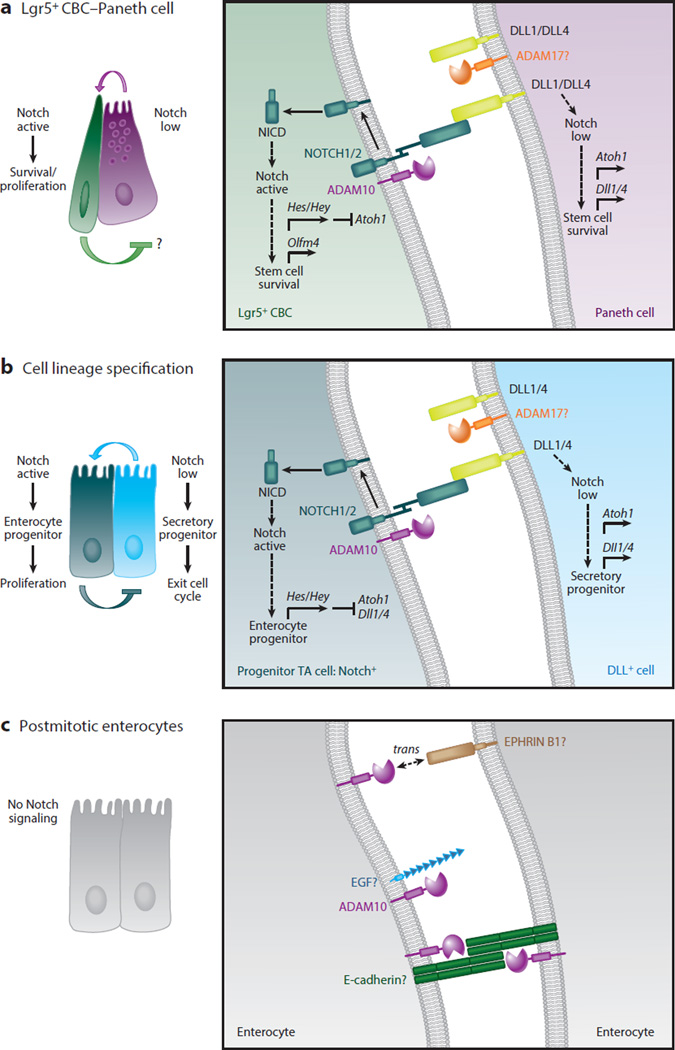
Active Notch signaling is present in Lgr5+ CBCs and is required for their maintenance (32, 34, 43–45, 55). Long-term lineage tracing of Adam10-deficient ISCs, performed under conditions of mosaic recombination in the Lgr5-EGFP-ires-CreER line or produced by reduced tamoxifen dosing in the Villin-CreER line, has shown that Adam10-deficient ISCs do not survive (50). However, activated Notch rescues Adam10-deficient ISCs and restores their capacity to populate the crypt-villus axis. Growth failure and the downregulation of the stem cell markers Lgr5 and Olfm4 in Adam10-deficient intestinal organoids is also rescued by activated Notch (50). Thus, cell autonomous, ADAM10-mediated Notch signaling is crucial for the survival and maintenance of Lgr5+ CBCs. Overall, ADAM10 acts iteratively to regulate Notch signaling associated with maintenance of Lgr5+ CBCs in the stem cell niche (Figure 5a) and cell lineage specification within the TA compartment (Figure 5b). No other ADAM can compensate for the loss of ADAM10, which is required for Notch signaling in development and normal adult crypt homeostasis (50).
Figure 5.
Cell-autonomous A disintegrin and metalloproteinase (ADAM)10 signaling acts iteratively to regulate Notch signaling in intestinal stem cells (ISCs) and transit-amplifying (TA) progenitors during crypt homeostasis. (a) ADAM10-mediated Notch signaling is required for the survival and maintenance of Leu-rich repeat–containing G protein–coupled receptor 5–expressing (Lgr5+) crypt base columnar stem cells (CBCs). Lgr5+ CBCs expressing NOTCH1/2 receptors are intercalated between Paneth cells expressing DLL1 and DLL4 ligands in the small intestinal stem cell niche. High Notch activity in Lgr5+ CBCs is required for the proliferation and survival of stem cells and maintenance of the stem cell pool. Notch signaling is activated in Lgr5+ CBCs when Dll ligand, found on the surface of Paneth cells, binds to Notch receptors expressed on the surface of Lgr5+ CBCs. Several studies have shown that the intestine tolerates Paneth cell depletion, suggesting that Notch ligand is likely provided by other cells as well. Notch is sequentially cleaved by ADAM10 and γ-secretase to generate the Notch intracellular domain (NICD), which translocates to the nucleus, where its forms an active transcriptional complex. In Notch-active Lgr5+ CBCs, Notch targets genes including those that encode the Hes/Hey transcription factors, which repress Atoh1 and Dll1/4 ligand transcription and enhance expression of the stem cell marker Olfm4. In DLL+ Paneth cells, low Notch activity, reinforced through Notch lateral inhibition, allows derepression of Atoh1 and Dll1/4 ligand expression. ADAM10 is not required in Paneth cells to maintain the Lgr5+ CBC stem cell pool, but it may be involved in other redundant signaling pathways (e.g., EGF) that contribute to the stem cell niche. (b) ADAM10-mediated Notch signaling is required for the fate specification of TA cells. Classical Notch lateral inhibition determines whether a TA cell becomes an absorptive or secretory progenitor. In Notch-active TA progenitors, Notch targets genes including those that encode the Hes/Hey transcription factors, which repress Atoh1 and Dll1/4 ligand transcription. These cells are fated to become absorptive progenitors, which undergo several rounds of proliferation before differentiating into postmitotic enterocytes. In DLL+ TA cells, low Notch activity allows derepression of Atoh1 and Dll1/4 ligand expression. These cells are fated to become secretory progenitors, which rapidly exit the cell cycle and differentiate into distinct secretory cell types. Under certain experimental conditions, Notch ligands can be subject to extracellular cleavage by ADAM proteases such as ADAM17 (as shown in panels a and b). (c) ADAM10 signaling in postmitotic differentiated intestinal epithelial cells (IECs). Although Notch signaling is restricted to the crypt compartment, ADAM10 is abundantly expressed on all differentiated IECs. This implies that ADAM10 is involved in other shedding events in these postmitotic IECs. Potential ADAM10 substrates include E-cadherin, EGF, and EPHRIN B1. The profound effects of ADAM10-deficiency in the ISC/progenitor compartment have hindered analysis of other ADAM10 substrates in vivo.
ADAM10-Mediated Notch Signaling and Insights into Intestinal Stem Cell Plasticity
Following injury or ablation of Lgr5+ CBCs, several reserve stem cell populations can repopulate the Lgr5+ stem cell compartment and reestablish intestinal homeostasis (38, 56–59). In an analogous manner, Adam10 deletion in Lgr5+ CBCs may lead to an imbalance within the stem cell niche that promotes permissive signals for stem cell plasticity, in which facultative stem cell populations can be mobilized to reestablish ISC homeostasis. Lineage tracing has shown that, in the absence of Adam10-deficient Lgr5+ CBCs, Notch signaling competent stem cells have a competitive advantage in replenishing the ISC compartment. Although the mechanism for this enhanced compensation has not been defined, it has been proposed that the reexpression of activated Notch permits the immediate progeny of Adam10-deficient Lgr5+ CBCs to revert to an undifferentiated stem cell phenotype capable of fulfilling the function of facultative stem cells (50). This model is consistent with the ability of secretory progenitors to revert to a stem cell state (56, 59). It will be interesting to test whether this compensatory mechanism works in other models of ISC injury/ablation. Recent lineage tracing experiments using Bmi1-CreER;Adam10 flox/flox mice have revealed that activated Notch can also rescue Adam10-deficient Bmi1+ ISCs (60; Y-H. Tsai & P.J. Dempsey, unpublished observation). In this case, activated Notch rescue occurred in a stochastic manner, highlighting the functional differences in Cre-mediated Adam10 deficiency between Bmi1+ cells and Lgr5+ CBCs. Further studies are needed to dissect out the precise roles of ADAM10-mediated Notch signaling associated with plasticity of the ISC niche.
Other ADAM10 Substrates in Intestinal Homeostasis
ADAM10 is probably involved in proteolytic processing of substrates other than Notch in the intestine, particularly in postmitotic IECs that do not possess functional Notch signaling. However, the severe and dominant Notch loss-of-function phenotype observed in Adam10-deficient mice has prevented direct analysis of other potential ADAM10 substrates in vivo. Nonetheless, several ADAM10 substrates (e.g., E-cadherin, EPHRIN B1, and EGF) implicated in crypt homeostasis have been identified with cell-based assays (Figure 5c). Previous experiments with global and conditional skin Adam10-deficient mice demonstrated the importance of ADAM10-mediated E-cadherin cleavage in the regulation of cell-cell adhesion, cell migration, and skin barrier function (54, 61, 62). A recent in vitro study showed that EphB signaling can regulate the formation of E-cadherin–based adhesions in polarized epithelial cells to control cell migration (63). A complex between ADAM10, EPHB2, and E-cadherin is proposed to initiate E-cadherin shedding in trans at sites of EphB/EPHRIN B1 interactions. In the crypt compartment, EphB signaling is also required for cell-cell repulsive signals that restrict Paneth cells to the crypt base. Deficiency in either Eph2/3 or ephrin B1 leads to mislocalization of Paneth cells (Figure 4). A similar, but milder, Paneth cell phenotype has been reported in mice constitutively expressing a mutant form of ADAM10 that lacks the prodomain and metalloproteinase domain and is controlled by the cryptdin-2 promoter (63). This is consistent with the results of in vitro experiments on other Eph/Ephrin interactions, in which ADAM10 cleaves Ephrin ligand in trans (17, 63). However, in Paneth cells, constitutive loss of ADAM10, under the control of the α-Defensin4 promoter-driven Cre line, does not result in a Paneth cell phenotype (Table 1) (P.J. Dempsey, M. Rajala & M.J. Myers Jr., unpublished observation). This discrepancy underscores the need for further investigation into ADAM10 signaling in postmitotic IECs in vivo. Such studies will require novel cell type–specific promoters that allow selective gene inactivation in different postmitotic cell types.
ADAM17 and the Intestine
Like ADAM10, ADAM17 is ubiquitously expressed throughout the gastrointestinal tract. However, the accumulation of immature ADAM17 in the ER, combined with the lack of specific antibodies for mature ADAM17 (especially mouse ADAM17), makes it difficult to study ADAM17 expression in vivo. One study has demonstrated that ADAM17 is restricted to the basolateral surface of polarized epithelial cells in human colon cancer cell lines and normal human colonic mucosa (64). This suggests that ADAM17, like ADAM10, is involved in selective signaling at the basolateral cell surface of IECs. In contrast to the detailed information we possess about the cellular distribution of ADAM10, it is not known how mature ADAM17 is distributed along the crypt-villus axis or whether epithelial cells express different levels of ADAM17 to nonepithelial cell types in the lamina propria. These questions await further investigation.
ADAM17 and Intestinal Development
ADAM17 was originally identified as the proteinase responsible for the ectodomain processing of membrane-anchored TNFα to generate soluble TNFα (65, 66). The ability of ADAM17 to regulate ectodomain processing of other substrates besides TNFα has been clearly demonstrated: Global Adam17-deficient (TaceΔZn/ΔZn) mice display perinatal lethality, whereas Tnfα-deficient mice and mice lacking Tnfr1/2 are viable and fertile (67, 68). The developmental defects observed in TaceΔZn/ΔZn mice are characteristic of epidermal growth factor receptor (Egfr)-deficient (Egfr−/−) mice (69–71). Defects in skin, heart, and mammary gland development are directly attributable to the inability of these mutants to process and activate specific EGFR ligands (72). Initial characterization of TaceΔZn/ΔZn mice, combined with genetic complementation studies using hypomorphic EgfrWa2 mice, confirms that ADAM17 is a critical upstream regulator of EGFR signaling (72, 73). This is a seminal observation in EGFR/ErbB biology, as it directly shows that many EGF-like ligand precursors (e.g., TGFα) must undergo ectodomain shedding to achieve full biological activity. Phenotypic variation and the timing of lethality in Egfr-deficient mice strongly depend on strain background (74). Strain background also influences the timing of lethality in TaceΔZn/ΔZn mice; this finding emphasizes that phenotypes observed upon inactivation of Adam17 or other genes in the EGFR/ErbB signaling axis should be compared with phenotypes in the same strain background (75).
Because of perinatal lethality (67, 76), a comprehensive analysis of the intestinal phenotype of TaceΔZn/ΔZn mice on a mixed background has not been performed. The intestine from E17.5 TaceΔZn/ΔZn embryos shows evidence of villus blunting and delayed epithelial maturation. Interestingly, no overt colonic phenotype has been observed in adult TaceΔZn/ΔZn mice on a different (S129S3) background (76; P.J. Dempsey, unpublished observation) (Table 2). This discrepancy is similar to the varying intestinal phenotypes reported in Egfr-null mice with different strain backgrounds. For example, Egfr-deficient mice on a CD-1 background survive past weaning, and overall gut architecture appears normal (70). However, Egfr-deficient mice on a mixed background display reduced postnatal viability and amore variable intestinal phenotype, and some mice exhibit features of necrotizing enterocolitis (71).
Table 2.
Effect of global and conditional Adam17 deletion on gastrointestinal tract function and pathophysiology
| ADAM17-deficient mouse models |
Tissue distribution |
Experimental model | Gastrointestinal phenotype | Reference(s) |
|---|---|---|---|---|
| Null | ||||
| TaceΔZn/ΔZn | Global | Development | Postnatal lethality dependent on genetic background. E17.5 intestine had variably blunted villi with delayed epithelial cell maturation |
51, 67 |
| Impaired development of Peyer’s patches |
76 | |||
| Hypomorphic | ||||
| Adam17Ex/Ex | Global | IEC development | Normal crypt-villus architecture, slightly heightened intestinal inflammation |
80 |
| DSS colitis | Increased susceptibility to injury, defective Stat3 signaling |
|||
| APCMin model | Decreased tumor burden | S. Schmidt &A. Chalaris, unpublished observation |
||
| Adam17wavedX/wavedX | IEC development | Normal crypt-villus architecture |
79 | |
| DSS colitis | Increased susceptibility to injury; defective AREG/EREG → EGFR signaling in nonhematopoietic cells of the GItract |
|||
| IEC specific | ||||
|
Villin-Cre; Adam17flox/flox |
All IECs at ~E15 |
IEC development | Normal crypt-villus architecture |
81, 82, 117 |
| DSS colitis | Increased susceptibility to injury |
81, 114 | ||
| Lactobacillus rhamnosus GG p40/DSS colitis |
p40-induced EGFR signaling is ADAM17 dependent |
81 | ||
| Total parenteral nutrition | Protection against mucosal atrophy as a result of ↓TNFα signaling |
82, 114, 117 | ||
|
Villin-CreER; Adam17flox/flox |
All IECs | Adult IEC homeostasis | Normal crypt-villus architecture |
114 |
| Adam17 deletion prior to DSS colitis |
Increased susceptibility to injury |
|||
| Adam17 deletion after DSS colitis |
No effect of epithelial cell restitution/regeneration |
|||
| Leukocyte specific | ||||
|
LysM-CreER; Adam17flox/flox |
Neutrophils, monocytes, macrophages |
Adult IEC homeostasis | Normal crypt-villus architecture |
P.J. Dempsey, unpublished observation |
| DSS colitis | No difference in acute intestinal injury |
|||
| Liver specific | ||||
|
Albumin-Cre; Adam17flox/flox |
Hepatocyte | LPS-induced TNFα/TNFR1 shedding |
↓TNFα and TNFR serum levels |
122 |
| Partial hepatectomy | ↓TNFα induction; ↓TNFα, TNFR, and AREG shedding in liver; no effect on liver generation |
|||
| CCl4-induced hepatotoxicity |
No effect on liver injury | |||
|
LysM-CreER; Adam17flox/flox |
Neutrophils, monocytes, macrophages |
LPS-induced TNFα/TNFR1 shedding |
↓TNFα and TNFR serum levels |
122 |
| Partial hepatectomy | ↓TNFα induction, no effect on liver regeneration |
|||
| CCl4-induced hepatotoxicity |
No effect on liver injury | |||
| Pancreas specific | ||||
|
Ptf1a-Cre; Adam17flox/flox |
All pancreatic cell lineages |
Pancreatic development | No gross pancreatic abnormalities |
141 |
| Mouse pancreatic intraepithelial neoplasia (mPanIN-KrasLSL- G12D) model |
Reduced tumorigenesis as a result of ↓ErbB ligand shedding leading to ↓EGFR/MEK signaling |
|||
| Cerulein-induced (KrasLSL-G12D) PDAC model |
Protected against metaplasia-mPanIN formation |
|||
| Cerulein-induced pancreatitis |
Protected against pancreatitis | |||
Abbreviations: DSS, dextran sodium sulfate; E, embryonic day; IEC, intestinal epithelial cell; LPS, lipopolysaccharide; mPanIN, murine pancreatic intraepithelial neoplasia; PDAC, pancreatic ductal adenocarcinoma.
ADAM17 is also implicated in regulating TNFα signaling during development. Analysis of TaceΔZn/ΔZn mice and RAG1−/− recipients of TaceΔZn/ΔZn bone marrow has demonstrated that nonlymphocyte expression of ADAM17 is required for normal T cell development, peripheral B cell maturation, and lymphoid organ structure formation (76). Impaired B cell follicle organization and germinal center formation in secondary lymphoid organs, including Peyer’s patches, indicates a loss of TNFα signaling. Phenotypically, this overlaps with Tnfα-deficient mice and the developmental role of TNFα signaling in nonepithelial cells during germinal center formation (68, 76).
Recently, two sibling pediatric patients lacking ADAM17 because of an autosomal recessive mutation in the Adam17 coding region were identified (77). Similar to TaceΔZn/ΔZn null mice, these patients displayed defects in hair, skin, and heart development. In addition, severe skin inflammation and persistent and recurrent skin infections have been linked to a loss of EGFR signaling, altered keratinocyte differentiation, and defective barrier function (77, 78). Early onset diarrhea and intestinal inflammation associated with villus blunting and increased mononuclear infiltrates were also observed, but these symptoms were variable and resolved over time. Although the mechanism underlying the intestinal inflammation has not been determined, the extent of intestinal inflammation parallels the severity of recurrent skin and gastrointestinal infections, which suggests the inflammation is systemic (77). Similar to the variations in postnatal lethality observed in Adam17-deficient mice and Egfr-deficient mice on different strain backgrounds, the differences in survival among patients lacking ADAM17 are most likely associated with differences in genetic background within the human population.
ADAM17 Is Not Essential for Maintenance of Normal Crypt Homeostasis
A more detailed analysis of ADAM17’s role in crypt homeostasis comes from studies using hypomorphic Adam17 mice and IEC-Adam17KO (Villin-Cre;Adam17flox/flox) mice (Table 2). Two independent groups have generated hypomorphic Adam17 mice (79, 80). These mice are viable but have developmental abnormalities of the eyes, skin, and heart that are reminiscent of TaceΔZn/ΔZn mice (79, 80). However, these mice have no overt intestinal phenotype; they possess normal crypt villus architecture and display normal crypt proliferation and differentiation. A slight increase in proinflammatory cytokine expression has been observed in the colonic mucosa of Adam17ex/ex mice, which suggests sensitization to inflammatory stimuli (79, 80). Similarly, IEC-Adam17KO mice on a C57BL/6J background do not have an intestinal phenotype (81, 82) (Table 2). No defects in the development and number of Peyer’s patches, in small intestinal barrier function, or in cytokine gene signatures have been observed (81, 82). The normal intestinal secretory differentiation in hypomorphic Adam17 mice and IEC-Adam17KO mice indicates that ADAM17 is not required for Notch signaling in the crypt compartment (50). Consistent with the lack of intestinal phenotype reported in different IEC-specific ErbB-deficient mice (83–86), the above observations indicate that ADAM17 signaling within IECs is not essential for maintaining intestinal homeostasis under normal physiological conditions.
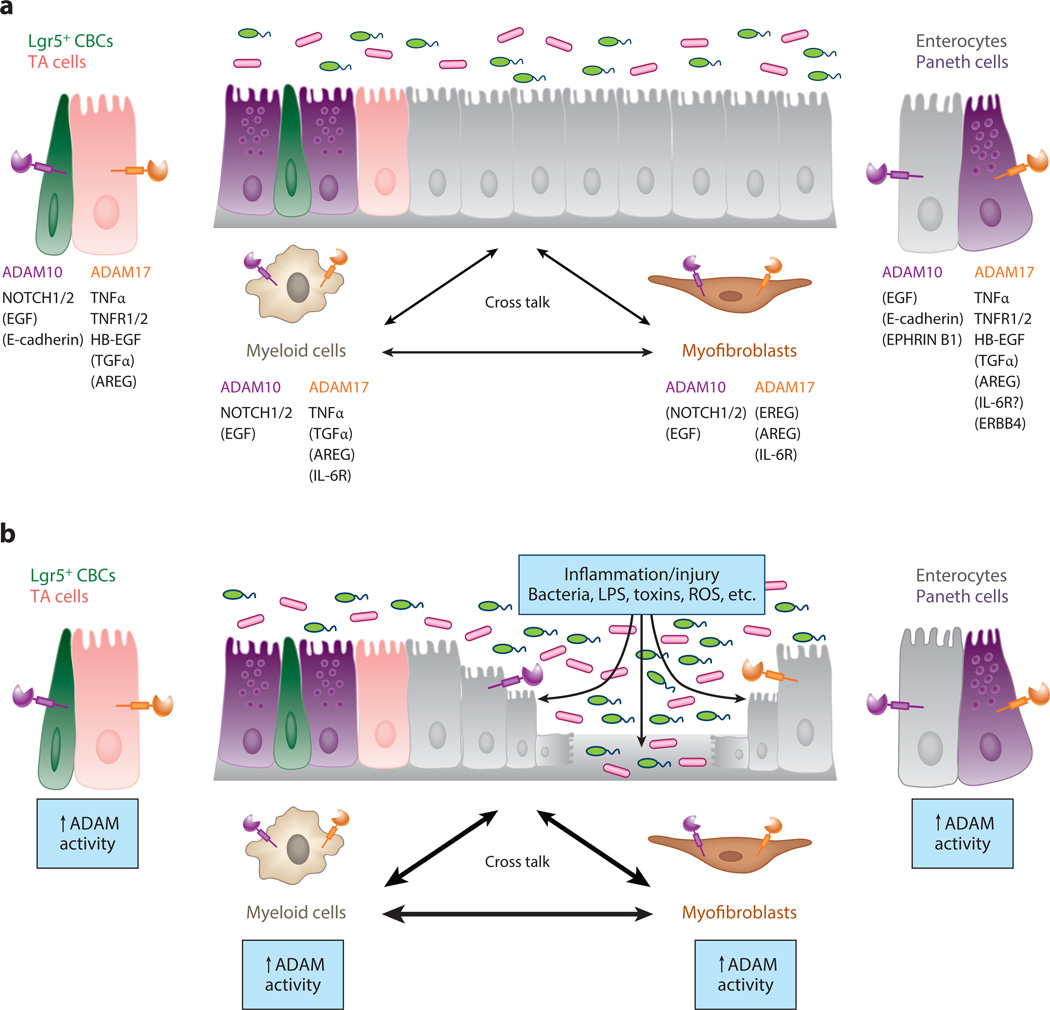
Why isn’t IEC-specific ADAM17 signaling essential for normal crypt homeostasis? Exogenous EGF can readily stimulate crypt proliferation in vivo and is essential for the growth of ISC organoids in culture (87). Recently, it was discovered that the stem cell marker LRIG1 negatively regulates EGFR/ErbB signaling and that its inactivation can increase crypt cell proliferation and tumor formation. This suggests that a significant reserve capacity of EGFR/ErbB signaling exists in the crypt compartment (88, 89). A degree of EGFR/ErbB signaling redundancy is implied by the overlapping and complementary functions of the four distinct EGFR/ErbB receptors (87) and the diverse range of ErbB ligands produced by different cell populations in the ISC niche. For example, Paneth cells express high levels of EGF (an ADAM10 substrate), although nonepithelial cells of the stem cell niche, such as pericryptal myofibroblasts, express AREG and EREG (ADAM17 substrates) (Figure 6). Thus, the lack of an intestinal phenotype, particularly in IEC-Adam17KO mice, is probably a result of redundant ligands from multiple sources; this redundancy can supplant the loss of autocrine ADAM17-dependent EGFR/ErbB signaling in IECs.
Figure 6.
A disintegrin and metalloproteinases (ADAMs) play a central role in intercellular communication during intestinal homeostasis and upon injury/inflammation. (a) A schematic diagram showing the specific roles of ADAM10 and ADAM17 signaling in cross talk between different cell types during normal intestinal homeostasis. For simplicity, only ADAM-mediated signaling between intestinal epithelial cells (IECs), myofibroblasts, and macrophages/myeloid cells is shown. ADAM activity is required in many other cell types of the gastrointestinal tract, including other immune cell populations (e.g., dendritic cells, T cells, and B cells), endothelial cells, and enteric neurons (not shown). For each cell type, ADAM substrate specificity and hierarchy may be different. For ADAM substrates, direct evidence exists for cleavage events in the gastrointestinal tract. For ADAM substrates listed in parentheses, in vitro data strongly suggest that ADAM-mediated cleavage events occur in the gastrointestinal tract. TA denotes transit amplifying. (b) Under conditions of intestinal inflammation and/or IEC injury, ADAM activity is upregulated. Loss of barrier integrity allows gut luminal contents to directly interact with ADAMs expressed on the basolateral surface of IECs. Numerous proinflammatory stimuli [e.g., lipopolysaccharide (LPS), reactive oxygen species (ROS), cytokines] will stimulate ADAM expression and proteolytic activity. Normally, enhanced ADAM activity and signaling cross talk are carefully integrated to resolve inflammation/injury and promote epithelial restitution and regeneration. However, under conditions of chronic and relapsing inflammation, these same signals, if sustained, may increase the risk of inflammation-associated cancer.
ADAMs AND THE GUT MICROBIOME
The epithelium lining the gut lumen plays an essential role in maintaining the integrity of the mucosal barrier between the host and its external environment. Mucosal defense and innate immune responses protect the host from commensal and pathogenic microbes in the intestinal lumen. ADAMs play an important role in the dynamic cross talk between luminal microorganisms and the host. Microbes and their byproducts (e.g., toxins, bacterial proteins) directly or indirectly interact with ADAMs to regulate signaling events in epithelial, stromal, and macrophage/dendritic cell populations of the intestine. In addition, ADAMs and their substrates can be intimately involved in the pathogenesis of various infectious agents.
LPS is a classic example of a Gram-negative bacterial product that can regulate ADAM activity. LPS-induced Toll-like receptor 4 (TLR4) signaling activates ADAM17-dependent TNFα shedding (62, 90), whereas nonepithelial TLR signaling confers protection against dextran sodium sulfate (DSS)-induced colitis via increased EGFR signaling (79). Recently, Yan et al. (81, 85) demonstrated that a Lactobacillus rhamnosus GG–derived soluble protein, p40, improves IEC homeostasis in IEC-Adam17KO mice by increasing ADAM17-mediated HB-EGF shedding and transactivation of EGFR (Table 2). Increased EGFR signaling is implicated in the marked gastric epithelial hyperplasia observed upon Helicobacter pylori infection. H. pylori can stimulate EGFR transactivation through multiple pathways, including rapid upregulation of ADAM17-mediated HB-EGF shedding (81, 91).
The virulence of specific microorganisms can depend upon direct binding to ADAMs or their substrates. For example, the Staphylococcus aureus α-hemolysin, a pore-forming cytotoxin, plays an important role in the virulence of this organism. ADAM10 is the functional cellular receptor for α-hemolysin. Direct binding of α-hemolysin to ADAM10 is proposed to upregulate ADAM10 activity, which results in increased E-cadherin cleavage and disruption of epithelial barrier function. This is critical for the pathogenesis of S. aureus (92, 93). In a different scenario, a protease toxin produced by enterotoxigenic Bacteroides fragilis binds to an unknown intestinal cell surface receptor to induce ADAM10-independent cleavage of E-cadherin (94). Alternatively, ADAM substrates can serve as host cell surface receptors for different microorganisms. E-cadherin is a functional cellular receptor for the InlA protein of the Gram-positive bacterium Listeria monocytogenes, and proHB-EGF is a receptor for the B fragment toxin of Corynebacterium diphtheria (95, 96). In the intestine, most of these interactions occur at the basolateral surface of IECs. This implies that barrier integrity must be compromised for these interactions to occur. More research is needed to further delineate the complex interactions between ADAMs, the microbiome, and mucosal defense.
ADAMs, INTESTINAL INJURY, AND INFLAMMATORY BOWEL DISEASE
Crohn’s disease and ulcerative colitis are the main forms of inflammatory bowel disease (IBD) in humans. IBD is a chronic relapsing condition in which high levels of inflammatory cytokines, such as TNFα, play a major role in tissue-damaging immune responses, loss of mucosal integrity, and barrier dysfunction. TNFα has pleiotropic effects during intestinal homeostasis and can induce both proinflammatory and anti-inflammatory responses in a dose- and cell context–dependent manner (97, 98). Although ADAM17 is a critical upstream regulator of TNFα signaling, limited data are currently available about its role in the pathogenesis of IBD. Previous studies have reported that Adam17 expression is upregulated in the intestinal mucosa of patients with IBD and in several mouse models of experimental colitis (99–103). Adam19 expression is also elevated in IBD (104). Apart from these reports, however, limited information exists on the role of these or other ADAMs in intestinal inflammation.
Intestinal Epithelial Cell Responses Are Differentially Regulated by ADAM17 in Different Intestinal Injury/Repair Models
Although ADAM17 activity is not required in IECs during normal intestinal homeostasis, ADAM17 signaling is critical for IEC cytoprotection and restitution/regeneration in several experimental injury/inflammation models. IEC proliferation is crucial for adaptive growth after injury/inflammation, and EGFR/ErbB is a major mediator of these responses (87). In older studies of the acute DSS colitis model, hypomorphic Egfrwa2 mice and ErbB ligand–deficient mice displayed exacerbated intestinal inflammation (105, 106). More recent studies, using different IEC-Egfr/ErbBKO mice, show that all four ErbB receptors protect, in varying degrees, against DSS-induced colitis (85, 86, 107). Direct evidence that ADAM17 is an upstream regulator of EGFR/ErbB signaling in the DSS colitis model comes from an analysis of hypomorphic Adam17 mice under long-term exposure to low-dose DSS (79, 80). Hypomorphic Adam17 mice are more susceptible to DSS-induced colitis than wild-type mice, but treatment with exogenous ErbB ligands is protective (79, 80). Brandl et al. (79) have shown that DSS-induced colitis activates TLR-mediated MyD88 signaling, which dramatically increases expression of the genes encoding two ErbB ligands, Areg and Ereg. AREG and EREG are ADAM17 substrates (108), and it is postulated that ADAM17-dependent production of these soluble ligands is critical for activating ErbB signaling under these experimental conditions (79). Although the cellular source of these ErbB ligands was assigned to nonepithelial cell populations in these studies, more recent studies have highlighted the contribution of the ErbB ligand/EGFR signaling axis from specific nonepithelial cell types, including myeloid/immune cells and pericryptal myofibroblasts found in the lamina propria (87, 109, 110).
In an alternative pathway, TNFα-induced ErbB receptor expression and transactivation is reported to promote intestinal cell survival (97, 98). ERBB4 receptor expression is elevated in the mucosa of IBD patients, and TNFα-induced ERBB4 activation prevents colon cell apoptosis and is protective in mouse models of colitis. Mechanistically, Hilliard et al. (111) have demonstrated that TNFα-induced transactivation of ERBB4 depends on ADAM17-mediated ErbB ligand shedding in these colon cells. The ability of specific ERBB4 isoforms expressed in IECs to undergo ADAM17-mediated shedding may add another level of complexity to this event (112, 113). Our own preliminary experiments with IEC-Adam17KO mice demonstrate that ADAM17 signaling in IECs plays an important cytoprotective role in the acute DSS colitis model (114) (Table 2). In hypomorphic Adam17 mice and IEC-Adam17KO mice, loss of TNFα-induced ErbB receptor transactivation may, therefore, contribute to the exacerbated DSS-induced injury observed. These results illustrate the complex interactions between ADAM17 and the TNFR and EGFR/ErbB signaling pathways in acute colitis models (87, 97).
The mouse model of total parenteral nutrition (TPN) is an excellent system with which to study signal transduction pathways that contribute to mucosal atrophy in the absence of acute inflammatory changes or IEC cell destruction. Removal of enteral nutrition is associated with decreased crypt proliferation, increased IEC apoptosis, a loss of epithelial barrier function, and altered enteric microbiota; these changes result in a mild proinflammatory state and mucosal atrophy (115). Previous studies showed that upregulation of TNFα and its receptor, TNFR1, plays a central role in mediating TPN-induced mucosal atrophy. Improved IEC responses observed in TPN-treated Tnfr1KO mice depend, in part, on the maintenance of functional EGFR signaling in IECs. Exogenous EGF treatment can attenuate mucosal atrophy in TPN-treated mice (116). Ongoing studies with IEC-EgfrKO and IEC-ErbB4KO mice indicate that ErbB signaling in IECs is required for protection against TPN-induced mucosal atrophy (87, 114, 117; Y. Feng, D.H. Teitelbaum & P.J. Dempsey, unpublished observations). In addition, extensive analysis of TPN-treated Tnfr1KO, Tnfr2KO, and Tnfr1/2DKO mice has revealed that both TNFR1 and TNFR2 signaling contribute to TPN-induced epithelial barrier dysfunction (118).
Recent studies using TPN-treated IEC-Adam17KO mice have shown that ADAM17 deletion attenuates mucosal atrophy, with improved crypt proliferation, decreased IEC apoptosis, and reduced proinflammatory cytokine expression evident (82, 114, 117). Mechanistically, this protective effect results from a loss of IEC-specific TNFα signaling and the concomitant maintenance of functional EGFR signaling in IECs. In TPN-treated wild-type mice, the results of a TNFα blockade have confirmed that TNFα signaling is responsible for downregulation of EGFR signaling in IECs (82). Paradoxically, in light of the role of ADAM17 in activating different EGFR/ErbB ligands, TPN-treated IEC-Adam17KO mice show partial restoration of EGFR protein levels and signaling in IECs. EGFR kinase inhibitor studies have confirmed that the beneficial effects observed in TPN-treated IEC-Adam17KO mice depend on functional EGFR signaling (82, 114, 117). In contrast to the cytoprotective role of ADAM17 signaling in an acute DSS colitis model, ADAM17-mediated TNFα signaling within IECs has a significant and deleterious role in the development of TPN-induced mucosal atrophy.
IECs are critical for regulating intestinal immune responses to inflammation and injury, and recent studies have demonstrated the importance of cytokines/chemokines produced by IECs in inflammatory responses (98, 119–121). Roulis et al. (119) have shown that TNFα overexpression in IECs is responsible for Crohn’s-like symptoms in TnfΔARE/+ mice. Surprisingly, the intestinal inflammation observed in this model did not require direct autocrine TNFα/TNFR1 signaling within IECs but instead was triggered by paracrine TNFα signaling and myofibroblast activation. Similarly, in LPS-induced liver injury, ADAM17 expressed in hepatocytes and myeloid cells contributes to LPS-induced TNFα shedding (122). Overall, these observations implicate ADAM17 in complex intercellular communication involving TNFR and EGFR/ErbB signaling within multiple cell types; this communication allows cells to maintain intestinal homeostasis in a cell context–dependent manner when faced with intestinal stress and inflammation (87, 97). Further delineation of the cross talk between ADAM17-mediated TNFR and EGFR/ErbB signaling requires detailed analysis of conditional and cell type–specific knockouts in different injury/inflammation models.
Other ADAMs and Intestinal Inflammation
Information on other ADAMs in intestinal inflammation and IBD is limited. In previous studies, we showed that ADAM10 is rate limiting for Notch activation in crypt homeostasis and that ROS are potent stimulators of ADAM10 activity (50, 123). In the mouse intestine, deletion of NADPH oxidase 1, a major producer of ROS, decreases Notch signaling and increases secretory differentiation (124). Conversely, patients with ulcerative colitis display significantly decreased goblet cell differentiation, which is directly associated with increased Hes1 and decreased Atoh1 expression, indicative of increased NOTCH activity (125, 126). Assessing ADAM10 function in intestinal homeostasis is technically challenging, but these observations raise the possibility that ADAM10 is an important, and possibly rate-limiting, regulator of NOTCH signaling during intestinal inflammation and the development of colitis-associated cancer (CAC).
In macrophage populations, NOTCH is essential for macrophage polarization of inflammatory (M1) and regenerative (M2) macrophages and for the functional differentiation of dendritic cell populations in the intestine (127). Using myeloid-specific Adam10KO mice, researchers recently observed that ADAM10-dependent Notch signaling is required for LPS-induced M1-macrophage–associated gene expression (128). Our own preliminary studies of the acute DSS colitis model in myeloid-specific Adam10KO mice demonstrate that DSS causes profound, unremitting intestinal inflammation that can be rescued by active Notch signaling (129; Y-H. Tsai & P.J. Dempsey, unpublished observation). These results suggest a critical role for ADAM10 signaling in immune cell populations during intestinal inflammation, but they also caution against overinterpreting data from cell type–specific Adam10-deficient mice. In these mice, defective NOTCH signaling may alter cellular programming and function.
ADAMs AND GASTROINTESTINAL CANCER
Overexpression of proteolytically active ADAMs in human gastrointestinal tumors is correlated with cancer progression and invasiveness and poor clinical outcomes (130, 131). ADAMs can regulate many signaling pathways (Notch, EGFR/ErbB, IL-6/Stat3) involved in the pathogenesis of colorectal cancer (CRC), and they likely play a central role in cellular communication between cancer and stromal cells (e.g., fibroblasts, macrophages, and endothelial cells) within the tumor microenvironment. In conditions of chronic inflammation, such as ulcerative colitis, dysregulated ADAM signaling may increase the risk of CAC (130, 131). However, only recently have insights into the importance of ADAM10 and ADAM17 been obtained from experimental tumor models.
Activated Notch signaling is protumorigenic in CRC. Notch and Wnt signaling act cooperatively to control cell proliferation and tumorigenesis in the intestine, and many components of the Notch pathway are upregulated in CRC (132). Mechanistic insights into the intricate interactions between Notch and Wnt signaling in intestinal tumorigenesis come from studies using different APC mutant mouse models. Pharmacological inhibition of Notch with γ-secretase inhibitors blocks proliferation and induces secretory differentiation in established APCMin/+ adenomas, whereas overexpression of activated Notch increases adenoma formation and decreases survival in another APC+/− mutant mouse model (47, 133). However, in a model of biallelic APC loss, RBP-Jk deletion does not reduce short-term adenoma formation (134). By contrast, Adam10 deletion during biallelic APC loss in the Lgr5-CreER;APCflox/flox mouse tumor model dramatically reduces adenoma formation and improves survival. No ADAM10-deficient adenomas have been detected in this model, but activated Notch rescues ADAM10-deficient tumor formation (135; P.J. Dempsey, unpublished observations) (Table 1). One explanation for this result is that Adam10-deficient Lgr5+ CBCs do not survive to undergo APC-mediated transformation. It will be important to examine the effects of ADAM10 loss of function in both established mouse adenomas and colitis-associated tumor models.
The Notch pathway is also involved in CRC progression and metastasis. Concurrent Notch activation and p53 deletion triggers epithelial-to-mesenchymal transition and metastasis (136), whereas Notch-dependent local invasion and the intravasation of CRC cells can be triggered by Notch ligands expressed on endothelial cells (137). Although the importance of ADAMs has not been examined in the above models, a recent study defined a noncanonical role for JAG-1 expressed on endothelial cells in the CRC tumor microenvironment (138). In this experimental tumor model, ADAM17-dependent cleavage of JAG-1 from endothelial cells was responsible for the release of soluble JAG-1 ligand, which activated Notch on CRC cells and promoted a cancer stem cell phenotype.
Because the EGFR/ErbB signaling axis is an established therapeutic target in CRC (139), it is not surprising that ADAM-dependent ErbB signaling is involved in CRC tumorigenesis. Initial studies using autocrine EGFR-dependent colon cancer cell lines demonstrated that a combination of EGFR and ADAM17 inhibition resulted in cooperative growth inhibition, increased apoptosis, and reduced mitogen-activated protein kinase pathway activation (64, 140). The APCMin model has been extensively used to demonstrate the importance of EGFR/ErbB signaling in adenoma formation and intestinal tumorigenesis (139). In preliminary studies from the Chalaris lab (S. Schmidt & A. Chalaris, unpublished observation), hypomorphic Adam17ex/ex mice bred to the APCMin model show markedly reduced adenoma formation and tumor burden (Table 2). Another recent study has identified tumor-associated fibroblasts as key producers of EREG in experimental colitis–associated neoplasms and in patients with CAC (110). It is predicted that ADAM17 is responsible for EREG shedding in this CAC model. Comprehensive analysis of different cell type–specific Adam17-deficient mice is required to further delineate the cellular contributions of ADAM17 signaling to tumor formation in the APCMin and other CAC models. Direct evidence for the role of ADAM17/EGFR signaling in inflammation-associated tumorigenesis has been reported in an experimental pancreatic ductal adenocarcinoma (PDAC) mouse model. In KRAS-induced pancreatic tumorigenesis and pancreatitis-induced acinar-to-ductal metaplasia mouse models, Adam17 ablation blocks ligand-dependent EGFR activation, preventing acinar-to-ductal metaplasia and KRAS-driven tumorigenesis (141) (Table 2). Oncogenic KRAS mutations are also common in CRC but are resistant to EGFR/MEK-targeted therapies, as a result of JAK1/2-dependent STAT3 activation. MEK inhibitors block ADAM17-dependent shedding of the c-met receptor, which prevents production of the c-met decoy receptor that normally suppresses c-met–activated JAK1/2-STAT3 signaling (142).
Proinflammatory IL-6/STAT3 signaling is important in the pathogenesis of IBD and is directly involved in tumor growth and survival in human CRC and several murine CRC models (143). Multiple cell types produce IL-6 during intestinal inflammation and CAC development in a cell context–dependent manner. IL-6 has diverse protumorigenic actions, ranging from promoting tumorigenic immune responses to stimulating tumor cells directly, via STAT3 signaling. Soluble GP130-Fc protein, which suppresses the action of the IL-6/sIL-6R complex, is used to investigate IL-6 trans-signaling in vivo. Soluble GP130-Fc attenuates intestinal inflammation and tumor formation in several experimental models of colitis and CAC. Although ADAM17-dependent IL-6R shedding is responsible for the generation of sIL-6R, the cellular sites of sIL-6R production and IL-6 trans-signaling action (e.g., T cells, dendritic cells, macrophages, and IECs) diverge depending on the experimental model (144–147).
ADAM8 is an example of an ADAM that is expressed at very low levels in normal tissues but is aberrantly expressed in many solid tumors; overexpression is commonly associated with poor prognosis. In tumor cells, ADAM8 is implicated in interactions with β1-integrin and ADAM8- dependent FAK and ERK1/2 activation. In a mouse model of PDAC, recent studies using an ADAM8 inhibitor have demonstrated a marked reduction in tumor load, infiltration, and metastasis in vivo (148).
CONCLUDING REMARKS
ADAMs play a central role in the homeostasis of the gastrointestinal tract and in controlling cellular responses to intestinal injury and inflammation. Importantly, ADAM10 acts iteratively to regulate Notch signaling associated with the maintenance of ISCs and for cell lineage specification of the TA cells within the crypt compartment. Aberrant ADAM activity is associated with chronic inflammation, inflammation-associated cancer, and tumor progression. Further studies are needed to understand the role of ADAMs in intercellular communication and the hierarchy of ADAM-mediated signaling events in a context- and cell type–dependent manner. Identification of key ADAM substrates involved in different gastrointestinal disease states may lead to new therapeutic interventions. The development of novel and highly specific antibodies to target ADAMs and their substrates (e.g., Notch) is one such approach.
SUMMARY POINTS.
ADAM10 and ADAM17 are ubiquitously expressed in all cell types of the gastrointestinal tract. Other ADAMs, such as ADAM8, are expressed at low levels in normal tissues but are aberrantly expressed in gastrointestinal cancers.
ADAM10 is the α-secretase responsible for Notch activation in IECs. Notch is the dominant pathway regulated by ADAM10 in the intestine. ADAM10-dependent Notch signaling is required for maintenance and survival of Lgr5+ CBCs and specifies the fate of TA cells. ADAM10’s function in differentiated postmitotic IECs has not been defined.
ADAM17 signaling in IECs is not required for maintaining normal intestinal homeostasis.
In different experimental models of intestinal inflammation, ADAM17 signaling in IECs has essential but divergent functions. In an acute colitis model, for example, ADAM17 expression is protective because it maintains EGFR/ErbB signaling. In a TPN model, by contrast, ADAM17 exacerbates mucosal atrophy because of enhanced TNFα signaling from IECs.
ADAM10 and ADAM17 are implicated in tumor initiation and tumor progression in APC mutant mouse models.
ADAMs are responsible for controlling intercellular communication during normal intestinal homeostasis, injury/inflammation, and tumorigenesis in vivo. Detailed analysis of ADAM-mediated signaling events will be required at a cell type–specific level to further dissect out the complex cellular responses involved.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01-DK093697) and the University of Michigan Comprehensive Cancer Center (to P.J.D.). S.R. was supported by a fellowship program from the Digestive Health Institute, Children’s Hospital Colorado.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Jennifer C. Jones, Email: jennifer.2.jones@ucdenver.edu.
Shelly Rustagi, Email: shelly.rustagi@childrenscolorado.org.
Peter J. Dempsey, Email: peter.dempsey@ucdenver.edu.
LITERATURE CITED
- 1.Weber S, Saftig P. Ectodomain shedding and ADAMs in development. Development. 2012;139:3693–3709. doi: 10.1242/dev.076398. [DOI] [PubMed] [Google Scholar]
- 2.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol. Asp. Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endres K, Fahrenholz F. Regulation of α-secretase ADAM10 expression and activity. Exp. Brain Res. 2012;217:343–352. doi: 10.1007/s00221-011-2885-7. [DOI] [PubMed] [Google Scholar]
- 4.Reiss K, Saftig P. The “A disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin. Cell Dev. Biol. 2009;20:126–137. doi: 10.1016/j.semcdb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Scheller J, Chalaris A, Garbers C, Rose-John S. ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011;32:380–387. doi: 10.1016/j.it.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Khokha R, Murthy A, Weiss A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat. Rev. Immunol. 2013;13:649–665. doi: 10.1038/nri3499. [DOI] [PubMed] [Google Scholar]
- 7.Cho C. Testicular and epididymal ADAMs: expression and function during fertilization. Nat. Rev. Urol. 2012;9:550–560. doi: 10.1038/nrurol.2012.167. [DOI] [PubMed] [Google Scholar]
- 8.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, et al. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335:229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christova Y, Adrain C, Bambrough P, Ibrahim A, Freeman M. Mammalian iRhoms have distinct physiological functions including an essential role in TACE regulation. EMBO Rep. 2013;14:884–890. doi: 10.1038/embor.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maretzky T, McIlwain DR, Issuree PD, Li X, Malapeira J, et al. iRhom2 controls the substrate selectivity of stimulated ADAM17-dependent ectodomain shedding. PNAS. 2013;110:11433–11438. doi: 10.1073/pnas.1302553110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Maretzky T, Weskamp G, Monette S, Qing X, et al. iRhoms 1 and 2 are essential upstream regulators of ADAM17-dependent EGFR signaling. PNAS. 2015;112:6080–6085. doi: 10.1073/pnas.1505649112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dornier E, Coumailleau F, Ottavi JF, Moretti J, Boucheix C, et al. TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J. Cell Biol. 2012;199:481–496. doi: 10.1083/jcb.201201133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haining EJ, Yang J, Bailey RL, Khan K, Collier R, et al. The TspanC8 subgroup of tetraspanins interacts with A disintegrin and metalloprotease 10 (ADAM10) and regulates its maturation and cell surface expression. J. Biol. Chem. 2012;287:39753–39765. doi: 10.1074/jbc.M112.416503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prox J, Willenbrock M, Weber S, Lehmann T, Schmidt-Arras D, et al. Tetraspanin15 regulates cellular trafficking and activity of the ectodomain sheddase ADAM10. Cell Mol. Life Sci. 2012;69:2919–2932. doi: 10.1007/s00018-012-0960-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garbers C, Aparicio-Siegmund S, Rose-John S. The IL-6/gp130/STAT3 signaling axis: recent advances towards specific inhibition. Curr. Opin Immunol. 2015;34C:75–82. doi: 10.1016/j.coi.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, et al. Adam meets Eph: An ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans . Cell. 2005;123:291–304. doi: 10.1016/j.cell.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Janes PW, Wimmer-Kleikamp SH, Frangakis AS, Treble K, Griesshaber B, et al. Cytoplasmic relaxation of active Eph controls ephrin shedding by ADAM10. PLOS Biol. 2009;7:e1000215. doi: 10.1371/journal.pbio.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoeck A, Keller S, Riedle S, Sanderson MP, Runz S, et al. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem. J. 2006;393:609–618. doi: 10.1042/BJ20051013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimoda M, Principe S, Jackson HW, Luga V, Fang H, et al. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nat. Cell Biol. 2014;16:889–901. doi: 10.1038/ncb3021. [DOI] [PubMed] [Google Scholar]
- 21.Folgosa L, Zellner HB, El Shikh ME, Conrad DH. Disturbed follicular architecture in B cell A disintegrin and metalloproteinase (ADAM)10 knockouts is mediated by compensatory increases in ADAM17 and TNF-αshedding. J. Immunol. 2013;191:5951–5958. doi: 10.4049/jimmunol.1302042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willems SH, Tape CJ, Stanley PL, Taylor NA, Mills IG, et al. Thiol isomerases negatively regulate the cellular shedding of ADAM17. Biochem. J. 2010;428:439–450. doi: 10.1042/BJ20100179. [DOI] [PubMed] [Google Scholar]
- 23.Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim. Biophys. Acta. 2010;1803:55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu P, Derynck R. Direct activation of TACE-mediated ectodomain shedding by p38MAP kinase regulates EGF receptor–dependent cell proliferation. Mol. Cell. 2010;37:551–566. doi: 10.1016/j.molcel.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu P, Liu J, Sakaki-Yumoto M, Derynck R. TACE activation by MAPK-mediated regulation of cell surface dimerization and TIMP3 association. Sci. Signal. 2012;5:ra34. doi: 10.1126/scisignal.2002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moss ML, Bomar M, Liu Q, Sage H, Dempsey P, et al. The ADAM10 prodomain is a specific inhibitor of ADAM10 proteolytic activity and inhibits cellular shedding events. J. Biol. Chem. 2007;282:35712–35721. doi: 10.1074/jbc.M703231200. [DOI] [PubMed] [Google Scholar]
- 27.Fridman JS, Caulder E, Hansbury M, Liu X, Yang G, et al. Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clin. Cancer Res. 2007;13:1892–1902. doi: 10.1158/1078-0432.CCR-06-2116. [DOI] [PubMed] [Google Scholar]
- 28.Zhou BB, Peyton M, He B, Liu C, Girard L, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murumkar PR, DasGupta S, Chandani SR, Giridhar R, Yadav MR. Novel TACE inhibitors in drug discovery: a review of patented compounds. Expert Opin. Ther. Pat. 2010;20:31–57. doi: 10.1517/13543770903465157. [DOI] [PubMed] [Google Scholar]
- 30.Tape CJ, Willems SH, Dombernowsky SL, Stanley PL, Fogarasi M, et al. Cross-domain inhibition of TACE ectodomain. PNAS. 2011;108:5578–5583. doi: 10.1073/pnas.1017067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atapattu L, Saha N, Llerena C, Vail ME, Scott AM, et al. Antibodies binding the ADAM10 substrate recognition domain inhibit Eph function. J. Cell Sci. 2012;125:6084–6093. doi: 10.1242/jcs.112631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian H, Biehs B, Chiu C, Siebel CW, Wu Y, et al. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 2015;11:33–42. doi: 10.1016/j.celrep.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran IT, Sandy AR, Carulli AJ, Ebens C, Chung J, et al. Blockade of individual Notch ligands and receptors controls graft-versus-host disease. J. Clin. Investig. 2013;123:1590–1604. doi: 10.1172/JCI65477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huntzicker EG, Hotzel K, Choy L, Che L, Ross J, et al. Differential effects of targeting Notch receptors in a mouse model of liver cancer. Hepatology. 2015;61:942–952. doi: 10.1002/hep.27566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spence JR, Lauf R, Shroyer NF. Vertebrate intestinal endoderm development. Dev. Dyn. 2011;240:501–20. doi: 10.1002/dvdy.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 39.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Clevers HC, Bevins CL. Paneth cells: maestros of the small intestinal crypts. Annu. Rev. Physiol. 2013;75:289–311. doi: 10.1146/annurev-physiol-030212-183744. [DOI] [PubMed] [Google Scholar]
- 41.Sancho R, Cremona CA, Behrens A. Stem cell and progenitor fate in the mammalian intestine: Notch and lateral inhibition in homeostasis and disease. EMBO Rep. 2015;16:571–581. doi: 10.15252/embr.201540188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 43.Vooijs M, Ong CT, Hadland B, Huppert S, Liu Z, et al. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development. 2007;134:535–544. doi: 10.1242/dev.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopinke D, Brailsford M, Shea JE, Leavitt R, Scaife CL, Murtaugh LC. Lineage tracing reveals the dynamic contribution of Hes1 + cells to the developing and adult pancreas. Development. 2011;138:431–441. doi: 10.1242/dev.053843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carulli AJ, Keeley TM, Demitrack ES, Chung J, Maillard I, Samuelson LC. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev. Biol. 2015;402:98–108. doi: 10.1016/j.ydbio.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008;9:377–383. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fre S, Pallavi SK, Huyghe M, Lae M, Janssen KP, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. PNAS. 2009;106:6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pellegrinet L, Rodilla V, Liu Z, Chen S, Koch U, et al. Dll1- and Dll4-mediated Notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230–1240.e7. doi: 10.1053/j.gastro.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim TH, Li F, Ferreiro-Neira I, Ho LL, Luyten A, et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai YH, VanDussen KL, Sawey ET, Wade AW, Kasper C, et al. ADAM10 regulates Notch function in intestinal stem cells of mice. Gastroenterology. 2014;147:822–834.e13. doi: 10.1053/j.gastro.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, et al. The disintegrin/ metalloprotease ADAM 10 is essential for Notch signalling but not for α-secretase activity in fibroblasts. Hum. Mol. Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- 52.Groot AJ, Habets R, Yahyanejad S, Hodin CM, Reiss K, et al. Regulated proteolysis ofNOTCH2 and NOTCH3 receptors by ADAM10 and presenilins. Mol. Cell. Biol. 2014;34:2822–2832. doi: 10.1128/MCB.00206-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J. Biol. Chem. 2009;284:31018–31027. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber S, Niessen MT, Prox J, Lullmann-Rauch R, Schmitz A, et al. The disintegrin/ metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development. 2011;138:495–505. doi: 10.1242/dev.055210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fre S, Hannezo E, Sale S, Huyghe M, Lafkas D, et al. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLOS ONE. 2011;6:e25785. doi: 10.1371/journal.pone.0025785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 57.Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. PNAS. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian H, Biehs B, Warming S, Leong KG, Rangell L, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van EsJH, Sato T, van deWetering M, Lyubimova A, Nee AN, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, et al. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and β-catenin translocation. PNAS. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maretzky T, Scholz F, Koten B, Proksch E, Saftig P, Reiss K. ADAM10-mediated E-cadherin release is regulated by proinflammatory cytokines and modulates keratinocyte cohesion in eczematous dermatitis. J. Investig. Dermatol. 2008;128:1737–1746. doi: 10.1038/sj.jid.5701242. [DOI] [PubMed] [Google Scholar]
- 63.Solanas G, Cortina C, Sevillano M, Batlle E. Cleavage of E-cadherin by ADAM10 mediates epithelial cell sorting downstream of EphB signalling. Nat. Cell Biol. 2011;13:1100–1107. doi: 10.1038/ncb2298. [DOI] [PubMed] [Google Scholar]
- 64.Merchant NB, Voskresensky I, Rogers CM, Lafleur B, Dempsey PJ, et al. TACE/ADAM-17: a component of the epidermal growth factor receptor axis and a promising therapeutic target in colorectal cancer. Clin. Cancer Res. 2008;14:1182–1191. doi: 10.1158/1078-0432.CCR-07-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-α. Nature. 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 66.Peschon JJ, Torrance DS, Stocking KL, Glaccum MB, Otten C, et al. TNF receptor–deficient mice reveal divergent roles for p55 and p75 in several models of inflammation. J. Immunol. 1998;160:943–952. [PubMed] [Google Scholar]
- 67.Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 68.Winsauer C, Kruglov AA, Chashchina AA, Drutskaya MS, Nedospasov SA. Cellular sources of pathogenic and protective TNF and experimental strategies based on utilization of TNF humanized mice. Cytokine Growth Factor Rev. 2014;25:115–123. doi: 10.1016/j.cytogfr.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 69.Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- 70.Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 71.Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, et al. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- 72.Blobel CP, Carpenter G, Freeman M. The role of protease activity in ErbB biology. Exp. Cell Res. 2009;315:671–682. doi: 10.1016/j.yexcr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sunnarborg SW, Hinkle CL, Stevenson M, Russell WE, Raska CS, et al. Tumor necrosis factor-α converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J. Biol. Chem. 2002;277:12838–12845. doi: 10.1074/jbc.M112050200. [DOI] [PubMed] [Google Scholar]
- 74.Strunk KE, Amann V, Threadgill DW. Phenotypic variation resulting from a deficiency of epidermal growth factor receptor in mice is caused by extensive genetic heterogeneity that can be genetically and molecularly partitioned. Genetics. 2004;167:1821–1832. doi: 10.1534/genetics.103.020495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gelling RW, Yan W, Al-Noori S, Pardini A, Morton GJ, et al. Deficiency of TNFαconverting enzyme (TACE/ADAM17) causes a lean, hypermetabolic phenotype in mice. Endocrinology. 2008;149:6053–6064. doi: 10.1210/en.2008-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li N, Boyd K, Dempsey PJ, Vignali DA. Non–cell autonomous expression of TNF-α–converting enzyme ADAM17 is required for normal lymphocyte development. J. Immunol. 2007;178:4214–4221. doi: 10.4049/jimmunol.178.7.4214. [DOI] [PubMed] [Google Scholar]
- 77.Blaydon DC, Biancheri P, Di WL, Plagnol V, Cabral RM, et al. Inflammatory skin and bowel disease linked to ADAM17 deletion. N. EnglJMed. 2011;365:1502–1508. doi: 10.1056/NEJMoa1100721. [DOI] [PubMed] [Google Scholar]
- 78.Franzke CW, Cobzaru C, Triantafyllopoulou A, Loffek S, Horiuchi K, et al. Epidermal ADAM17 maintains the skin barrier by regulating EGFR ligand–dependent terminal keratinocyte differentiation. J. Exp. Med. 2012;209:1105–1119. doi: 10.1084/jem.20112258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brandl K, Sun L, Neppl C, Siggs OM, Le Gall SM, et al. MyD88 signaling in nonhematopoietic cells protects mice against induced colitis by regulating specific EGF receptor ligands. PNAS. 2010;107:19967–19972. doi: 10.1073/pnas.1014669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chalaris A, Adam N, Sina C, Rosenstiel P, Lehmann-Koch J, et al. Critical role of the disintegrin metalloprotease ADAM17 for intestinal inflammation and regeneration in mice. J. Exp. Med. 2010;207:1617–1624. doi: 10.1084/jem.20092366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yan F, Liu L, Dempsey PJ, Tsai YH, Raines EW, et al. A Lactobacillus rhamnosus GG–derived soluble protein, p40, stimulates ligand release from intestinal epithelial cells to transactivate epidermal growth factor receptor. J. Biol. Chem. 2013;288:30742–30751. doi: 10.1074/jbc.M113.492397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feng Y, Tsai YH, Xiao W, Ralls MW, Stoeck A, et al. Loss of ADAM17-mediatedTNFαsignaling in intestinal cells attenuates mucosal atrophy in a mouse model of parenteral nutrition. Mol. Cell. Biol. 2015;35:3604–3621. doi: 10.1128/MCB.00143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee D, Yu M, Lee E, Kim H, Yang Y, et al. Tumor-specific apoptosis caused by deletion of the ERBB3 pseudo-kinase in mouse intestinal epithelium. J. Clin. Investig. 2009;119:2702–2713. doi: 10.1172/JCI36435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rowland KJ, McMellen ME, Wakeman D, Wandu WS, Erwin CR, Warner BW. Enterocyte expression of epidermal growth factor receptor is not required for intestinal adaptation in response to massive small bowel resection. J. Pediatr. Surg. 2012;47:1748–1753. doi: 10.1016/j.jpedsurg.2012.03.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yan F, Cao H, Cover TL, Washington MK, Shi Y, et al. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Investig. 2011;121:2242–2253. doi: 10.1172/JCI44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Y, Dube PE, Washington MK, Yan F, Polk DB. ErbB2 and ErbB3 regulate recovery from dextran sulfate sodium–induced colitis by promoting mouse colon epithelial cell survival. Lab. Investig. 2012;92:437–450. doi: 10.1038/labinvest.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frey MR, Polk DB. ErbB receptors and their growth factor ligands in pediatric intestinal inflammation. Pediatr. Res. 2014;75:127–132. doi: 10.1038/pr.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wong VW, Stange DE, Page ME, Buczacki S, Wabik A, et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 2012;14:401–408. doi: 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Horiuchi K, Kimura T, Miyamoto T, Takaishi H, Okada Y, et al. Cutting edge: TNF-α–converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J. Immunol. 2007;179:2686–2689. doi: 10.4049/jimmunol.179.5.2686. [DOI] [PubMed] [Google Scholar]
- 91.Saha A, Backert S, Hammond CE, Gooz M, Smolka AJ. Helicobacter pylori CagL activates ADAM17 to induce repression of the gastric H, K-ATPase αsubunit. Gastroenterology. 2010;139:239–248. doi: 10.1053/j.gastro.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Inoshima I, Inoshima N, Wilke GA, Powers ME, Frank KM, et al. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat. Med. 2011;17:1310–1314. doi: 10.1038/nm.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wilke GA, Wardenburg JB. Role of a disintegrin and metalloprotease 10 in Staphylococcus aureus α-hemolysin–mediated cellular injury. PNAS. 2010;107:13473–13478. doi: 10.1073/pnas.1001815107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J. Clin. Investig. 2014;124:4166–4172. doi: 10.1172/JCI72334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bonazzi M, Lecuit M, Cossart P. Listeria monocytogenes internalin and E-cadherin: from bench to bedside. Cold Spring Harb. Perspect. Biol. 2009;1:a003087. doi: 10.1101/cshperspect.a003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Naglich JG, Metherall JE, Russell DW, Eidels L. Expression cloning of a diphtheria toxin receptor: identity with a heparin-binding EGF-like growth factor precursor. Cell. 1992;69:1051–1061. doi: 10.1016/0092-8674(92)90623-k. [DOI] [PubMed] [Google Scholar]
- 97.Dube P, Punit S, Polk DB. Redeeming an old foe: protective as well as pathophysiological roles for tumor necrosis factor in inflammatory bowel disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2014;308:G161–G170. doi: 10.1152/ajpgi.00142.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leppkes M, Roulis M, Neurath MF, Kollias G, Becker C. Pleiotropic functions of TNF-αin the regulation of the intestinal epithelial response to inflammation. Int. Immunol. 2014;26:509–515. doi: 10.1093/intimm/dxu051. [DOI] [PubMed] [Google Scholar]
- 99.Brynskov J, Foegh P, Pedersen G, Ellervik C, Kirkegaard T, et al. Tumour necrosis factor α converting enzyme (TACE) activity in the colonic mucosa of patients with inflammatory bowel disease. Gut. 2002;51:37–43. doi: 10.1136/gut.51.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cesaro A, Abakar-Mahamat A, Brest P, Lassalle S, Selva E, et al. Differential expression and regulation of ADAM17 and TIMP3 in acute inflamed intestinal epithelia. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296:G1332–G1343. doi: 10.1152/ajpgi.90641.2008. [DOI] [PubMed] [Google Scholar]
- 101.Colon AL, Menchen LA, Hurtado O, De Cristobal J, Lizasoain I, et al. Implication of TNF- α convertase (TACE/ADAM17) in inducible nitric oxide synthase expression and inflammation in an experimental model of colitis. Cytokine. 2001;16:220–226. doi: 10.1006/cyto.2001.0969. [DOI] [PubMed] [Google Scholar]
- 102.Forsyth CB, Banan A, Farhadi A, Fields JZ, Tang Y, et al. Regulation of oxidant-induced intestinal permeability by metalloprotease-dependent epidermal growth factor receptor signaling. J. Pharmacol. Exp. Ther. 2007;321:84–97. doi: 10.1124/jpet.106.113019. [DOI] [PubMed] [Google Scholar]
- 103.Kirkegaard T, Pedersen G, Saermark T, Brynskov J. Tumour necrosis factor-αconverting enzyme (TACE) activity in human colonic epithelial cells. Clin. Exp. Immunol. 2004;135:146–153. doi: 10.1111/j.1365-2249.2004.02348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Franze E, Caruso R, Stolfi C, Sarra M, Cupi ML, et al. High expression of the “A Disintegrin And Metalloprotease” 19 (ADAM19), a sheddase for TNF-α in the mucosa of patients with inflammatory bowel diseases. Inflamm. Bowel Dis. 2013;19:501–511. doi: 10.1097/MIB.0b013e31828028e8. [DOI] [PubMed] [Google Scholar]
- 105.Egger B, Procaccino F, Lakshmanan J, Reinshagen M, Hoffmann P, et al. Mice lacking transforming growth factor α have an increased susceptibility to dextran sulfate-induced colitis. Gastroenterology. 1997;113:825–832. doi: 10.1016/s0016-5085(97)70177-x. [DOI] [PubMed] [Google Scholar]
- 106.Egger B, Schmid SW, Naef M, Wildi S, Buchler MW. Efficacy and safety of weight-adapted nadroparin calcium versus heparin sodium in prevention of clinically evident thromboembolic complications in 1,190 general surgical patients. Dig. Surg. 2000;17:602–609. doi: 10.1159/000051969. [DOI] [PubMed] [Google Scholar]
- 107.Frey MR, Edelblum KL, Mullane MT, Liang D, Polk DB. The ErbB4 growth factor receptor is required for colon epithelial cell survival in the presence of TNF. Gastroenterology. 2009;136:217–226. doi: 10.1053/j.gastro.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, et al. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 2004;164:769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lu N, Wang L, Cao H, Liu L, Van Kaer L, et al. Activation of the epidermal growth factor receptor in macrophages regulates cytokine production and experimental colitis. J. Immunol. 2014;192:1013–1023. doi: 10.4049/jimmunol.1300133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Neufert C, Becker C, Tureci O, Waldner MJ, Backert I, et al. Tumor fibroblast–derived epiregulin promotes growth of colitis-associated neoplasms through ERK. J. Clin. Investig. 2013;123:1428–1443. doi: 10.1172/JCI63748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hilliard VC, Frey MR, Dempsey PJ, Peek RM, Jr, Polk DB. TNF-αconverting enzyme-mediated ErbB4 transactivation by TNF promotes colonic epithelial cell survival. Am. J. Physiol. Gastrointest. Liver Physiol. 2011;301:G338–G346. doi: 10.1152/ajpgi.00057.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rio C, Buxbaum JD, Peschon JJ, Corfas G. Tumornecrosis factor-α–converting enzyme is required for cleavage of erbB4/HER4. J. Biol. Chem. 2000;275:10379–10387. doi: 10.1074/jbc.275.14.10379. [DOI] [PubMed] [Google Scholar]
- 113.Vecchi M, Rudolph-Owen LA, Brown CL, Dempsey PJ, Carpenter G. Tyrosine phosphorylation and proteolysis: pervanadate-induced, metalloprotease-dependent cleavage of the ErbB-4 receptor and amphiregulin. J. Biol. Chem. 1998;273:20589–20595. doi: 10.1074/jbc.273.32.20589. [DOI] [PubMed] [Google Scholar]
- 114.Tsai Y-H, Feng Y, Wade AW, Teitelbaum DH, Dempsey PJ. Intestinal cell responses in different injury/regeneration models are differentially dependent on ADAM17. Gastroenterology. 2012;142:S61. [Google Scholar]
- 115.Demehri FR, Barrett M, Ralls MW, Miyasaka EA, Feng Y, Teitelbaum DH. Intestinal epithelial cell apoptosis and loss of barrier function in the setting of altered microbiota with enteral nutrient deprivation. Front. Cell Infect. Microbiol. 2013;3:105. doi: 10.3389/fcimb.2013.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Feng Y, Teitelbaum DH. Epidermal growth factor/TNF-α transactivation modulates epithelial cell proliferation and apoptosis in a mouse model of parenteral nutrition. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G236–G249. doi: 10.1152/ajpgi.00142.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Feng Y, Tsai YH, Teitelbaum DH, Dempsey PJ. Intestinal epithelial cell responses to total parenteral nutrition (TPN) are under complex regulation by ADAM17. Gastroenterology. 2012;142:S167–S168. [Google Scholar]
- 118.Feng Y, Teitelbaum DH. Tumour necrosis factor-α–induced loss of intestinal barrier function requires TNFR1 and TNFR2 signalling in a mouse model of total parenteral nutrition. J. Physiol. 2013;591:3709–3723. doi: 10.1113/jphysiol.2013.253518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roulis M, Armaka M, Manoloukos M, Apostolaki M, Kollias G. Intestinal epithelial cells as producers but not targets of chronic TNF suffice to cause murine Crohn-like pathology. PNAS. 2011;108:5396–5401. doi: 10.1073/pnas.1007811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stadnyk AW. Intestinal epithelial cells as a source of inflammatory cytokines and chemokines. Can. J. Gastroenterol. 2002;16:241–246. doi: 10.1155/2002/941087. [DOI] [PubMed] [Google Scholar]
- 121.Xue X, Ramakrishnan S, Anderson E, Taylor M, Zimmermann EM, et al. Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology. 2013;145:831–841. doi: 10.1053/j.gastro.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McMahan RS, Riehle KJ, Fausto N, Campbell JS. A disintegrin and metalloproteinase 17 regulates TNF and TNFR1 levels in inflammation and liver regeneration in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2013;305:G25–G34. doi: 10.1152/ajpgi.00326.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sanderson MP, Erickson SN, Gough PJ, Garton KJ, Wille PT, et al. ADAM10 mediates ectodomain shedding of the betacellulin precursor activated by p-aminophenylmercuric acetate and extracellular calcium influx. J. Biol. Chem. 2005;280:1826–1837. doi: 10.1074/jbc.M408804200. [DOI] [PubMed] [Google Scholar]
- 124.Coant N, Ben Mkaddem S, Pedruzzi E, Guichard C, Treton X, et al. NADPH oxidase 1 modulates WNT and NOTCH1 signaling to control the fate of proliferative progenitor cells in the colon. Mol. Cell. Biol. 2010;30:2636–2650. doi: 10.1128/MCB.01194-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zheng X, Tsuchiya K, Okamoto R, Iwasaki M, Kano Y, et al. Suppression of hath1 gene expression directly regulated by hes1 via Notch signaling is associated with goblet cell depletion in ulcerative colitis. Inflamm. Bowel. Dis. 2011;17:2251–2260. doi: 10.1002/ibd.21611. [DOI] [PubMed] [Google Scholar]
- 126.Gersemann M, Stange EF, Wehkamp J. From intestinal stem cells to inflammatory bowel diseases. World J. Gastroenterol. 2011;17:3198–3203. doi: 10.3748/wjg.v17.i27.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat. Rev. Immunol. 2013;13:427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- 128.Xu H, Zhu J, Smith S, Foldi J, Zhao B, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat. Immunol. 2012;13:642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tsai YH, Rakshit R, Chung J, Maillard I, Dempsey PJ. Myeloid-specific deletion of ADAM10 dramatically increases susceptibility to DSS-induced colitis. Gastroenterology. 2013;144:S308. [Google Scholar]
- 130.Duffy MJ, McKiernan E, O’Donovan N, McGowan PM. Role of ADAMs in cancer formation and progression. Clin. Cancer Res. 2009;15:1140–1144. doi: 10.1158/1078-0432.CCR-08-1585. [DOI] [PubMed] [Google Scholar]
- 131.Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer. 2008;8:929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 132.Noah TK, Shroyer NF. Notch in the intestine: regulation of homeostasis and pathogenesis. Annu. Rev. Physiol. 2013;75:263–288. doi: 10.1146/annurev-physiol-030212-183741. [DOI] [PubMed] [Google Scholar]
- 133.van EsJH, van Gijn ME, Riccio O, van den Born M, Vooijs M, et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 134.Peignon G, Durand A, Cacheux W, Ayrault O, Terris B, et al. Complex interplay between β- catenin signalling and Notch effectors in intestinal tumorigenesis. Gut. 2011;60:166–176. doi: 10.1136/gut.2009.204719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tsai YH, Feng Y, Rakshit R, Fearon ER, Dempsey PJ. Cell-autonomous ADAM10 signaling is required for adenoma initiation after APC mutation. Gastroenterology. 2013;144:S51–S52. [Google Scholar]
- 136.Chanrion M, Kuperstein I, Barriere C, ElMarjou F, Cohen D, et al. Concomitant Notch activation and p53 deletion trigger epithelial-to-mesenchymal transition and metastasis in mouse gut. Nat.Commun. 2014;5:5005. doi: 10.1038/ncomms6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sonoshita M, Itatani Y, Kakizaki F, Sakimura K, Terashima T, et al. Promotion of colorectal cancer invasion and metastasis through activation of NOTCH-DAB1-ABL-RHOGEF protein TRIO. Cancer Discov. 2015;5:198–211. doi: 10.1158/2159-8290.CD-14-0595. [DOI] [PubMed] [Google Scholar]
- 138.Li J, Ye X, Fan F, Xia L, Bhattacharya R, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell. 2013;23:171–185. doi: 10.1016/j.ccr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fiske WH, Threadgill D, Coffey RJ. ERBBs in the gastrointestinal tract: recent progress and new perspectives. Exp. Cell Res. 2009;315:583–601. doi: 10.1016/j.yexcr.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dong J, Opresko LK, Dempsey PJ, Lauffenburger DA, Coffey RJ, Wiley HS. Metalloprotease-mediated ligand release regulates autocrine signaling through the epidermal growth factor receptor. PNAS. 1999;96:6235–6240. doi: 10.1073/pnas.96.11.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ardito CM, Gruner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Van Schaeybroeck S, Kalimutho M, Dunne PD, Carson R, Allen W, et al. ADAM17-dependent c-MET-STAT3 signaling mediates resistance to MEK inhibitors in KRAS mutant colorectal cancer. Cell Rep. 2014;7:1940–1955. doi: 10.1016/j.celrep.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 143.Waldner MJ, Neurath MF. Master regulator of intestinal disease: IL-6 in chronic inflammation and cancer development. Semin. Immunol. 2014;26:75–79. doi: 10.1016/j.smim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 144.Atreya R, Mudter J, Finotto S, Mullberg J, Jostock T, et al. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in Crohn disease and experimental colitis in vivo. Nat. Med. 2000;6:583–588. doi: 10.1038/75068. [DOI] [PubMed] [Google Scholar]
- 145.Becker C, Fantini MC, Schramm C, Lehr HA, Wirtz S, et al. TGF-βsuppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity. 2004;21:491–501. doi: 10.1016/j.immuni.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 146.Matsumoto S, Hara T, Mitsuyama K, Yamamoto M, Tsuruta O, et al. Essential roles of IL-6 trans-signaling in colonic epithelial cells, induced by the IL-6/soluble-IL-6 receptor derived from lamina propria macrophages, on the development of colitis-associated premalignant cancer in a murine model. J. Immunol. 2010;184:1543–1551. doi: 10.4049/jimmunol.0801217. [DOI] [PubMed] [Google Scholar]
- 147.Mitsuyama K, Matsumoto S, Rose-John S, Suzuki A, Hara T, et al. STAT3activation via interleukin-6 trans-signalling contributes to ileitis in SAMP1/Yit mice. Gut. 2006;55:1263–1269. doi: 10.1136/gut.2005.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schlomann U, Koller G, Conrad C, Ferdous T, Golfi P, et al. ADAM8as a drug target in pancreatic cancer. Nat. Commun. 2015;6:6175. doi: 10.1038/ncomms7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 150.Weber S, Wetzel S, Prox J, Lehmann T, Schneppenheim J, et al. Regulation of adult hematopoiesis by the a disintegrin and metalloproteinase 10 (ADAM10) Biochem. Biophys. Res. Commun. 2013;442:234–241. doi: 10.1016/j.bbrc.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 151.Yoda M, Kimura T, Tohmonda T, Uchikawa S, Koba T, et al. Dual functions of cell-autonomous and non-cell-autonomous ADAM10 activity in granulopoiesis. Blood. 2011;118:6939–6942. doi: 10.1182/blood-2011-06-357210. [DOI] [PubMed] [Google Scholar]
- 152.Faber TW, Pullen NA, Fernando JF, Kolawole EM, McLeod JJ, et al. ADAM10 is required for SCF-induced mast cell migration. Cell. Immunol. 2014;290:80–88. doi: 10.1016/j.cellimm.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gibb DR, El Shikh M, Kang DJ, Rowe WJ, El Sayed R, et al. ADAM10 is essential for Notch2- dependent marginal zone B cell development and CD23 cleavage in vivo. J. Exp. Med. 2010;207:623–635. doi: 10.1084/jem.20091990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chaimowitz NS, Martin RK, Cichy J, Gibb DR, Patil P, et al. A disintegrin and metalloproteinase 10 regulates antibody production and maintenance of lymphoid architecture. J. Immunol. 2011;187:5114–5122. doi: 10.4049/jimmunol.1102172. [DOI] [PMC free article] [PubMed] [Google Scholar]