Abstract
HIV infection is associated with an altered gut microbiome that is not consistently restored with effective antiretroviral therapy (ART). Interpretation of the specific microbiome changes observed during HIV infection is complicated by factors like population, sample type, and ART – each of which may have dramatic effects on gut bacteria. Understanding how these factors shape the microbiome during HIV infection (which we refer to as the HIV-associated microbiome) is critical for defining its role in HIV disease, and for developing therapies that restore gut health during infection.
Keywords: HIV, microbiome, dysbiosis, antiretroviral therapy, ART
Introduction
Antiretroviral therapy (ART) effectively halts replication of the Human Immunodeficiency Virus (HIV), however, infection is still associated with chronic inflammation and immune activation (1) as well as increased incidence of disorders including metabolic and cardiovascular disease (2). It is becoming increasingly clear that the collection of trillions of bacteria, fungi and viruses that colonize the intestine, the gut microbiome, plays a significant role in shaping health and disease including diseases that occur at increased incidence in individuals living with HIV (3). Recent findings of an altered, or “dysbiotic” microbiome in HIV-infected individuals supports this hypothesis. Since HIV infection results in massive depletion of CD4+ T cells in the gut-associated lymphoid tissue (GALT) and destruction of the gut epithelial barrier (4, 5), it is likely that HIV infection and the gut microbiome are intimately linked. Interestingly, ART has not consistently been found to restore a healthy gut microbiome in HIV-infected subjects, and the drugs themselves may in fact further promote dysbiosis. Thus, defining the mechanisms that cause dysbiosis during HIV infection and ART may reveal novel targets for therapies aimed at restoring a healthy gut microbiome and ameliorating HIV-associated diseases.
HIV disrupts gut immunity
The primary hallmark of HIV infection is depletion of CD4+ T cells. This is most evident in the GALT (5) which contains the majority of lymphocytes in the body (6). Although ART can restore CD4+ T cells in the periphery, GALT lymphocytes in most cases are slow to return to normal levels and restoration is most often incomplete (4). CD4+ lymphocytes in the GALT of healthy individuals consist primarily of effector T cells, such as IL-17-producing T-helper 17 (Th17) cells and regulatory T cells (Tregs). During HIV infection, the CD4+ T cell compartment in GALT suffers a significant loss of Th17 cells (7), which play critical roles in mitigating microbial invasion through the recruitment of neutrophils (8) and in maintaining the integrity of the gastrointestinal barrier (7). One potential driving factor of HIV-caused Th17 cell loss is that certain human-associated bacteria, such as Escherichia coli, can enhance HIV infection by inducing an activated state in these cells (9). Thus, HIV/bacterial-induced loss of Th17 cells in the gut may contribute to impaired mucosal T cell immunity, loss of barrier function, and microbial translocation (7).
Another hallmark of HIV infection is chronic immune activation, which could be caused by the loss of CD4+ Tregs – cells that can promote immune homeostasis and down-modulate effector activity of other immune cells by the production of IL-10 and TGF-beta (1). In contrast to the rapid loss of Th17 cells in blood, circulating Tregs are progressively lost during HIV infection and this decline has been associated with immune activation (10). In the gut compartment, Tregs have been shown to be more refractory to HIV infection than other CD4+ T cell subpopulations (11). However, the absolute number and function of Tregs in the gut is still decreased, suggesting HIV-driven Treg cell death and loss of Treg-mediated immune regulation (12). This is consistent with the observation of high levels of inflammation in both the periphery and in the gut of untreated HIV-infected individuals (13). Furthermore, Tregs have been shown to be important for colonization of some bacteria in the gut suggesting that loss of Treg function during HIV infection may contribute to dysbiosis (14).
Dysbiosis in HIV infection
The innate and adaptive immune systems are critical for maintaining a healthy gut microbiome. Knockout and transgenic mouse models have been particularly useful for demonstrating this; for example, loss of innate immune sensing in mice leads to alterations in bacterial species composition and increased gut inflammation, while B cell and T cell deficiencies in mice have been associated with reduced gut microbial diversity (15). Though precise mechanisms of immune interactions with gut microbes are still being elucidated, studies have shown that immunoglobulin A (IgA) binding to commensal bacteria may alter microbial gene expression (16), and that transfer of Tregs to T cell-knockout mice could restore gut microbial diversity (17). Antibody production by B cells and immune regulation by CD4+ T cells may therefore play essential roles in maintaining a healthy gut microbial environment. It would not be surprising then, that the rapid and profound breakdown in intestinal immunity caused by HIV infection, in particular the loss of CD4+ T cells, would have dramatic effects on the gut microbiome.
Initial studies of the effects of HIV infection on the gut microbiome were limited to qPCR-based methods for detection of only a few types of microbes. These studies found that feces from untreated HIV-infected subjects harbored higher amounts of bacterial DNA from the Proteobacteria phylum (18), and more frequently contained DNA from opportunistic pathogens such as Pseudomonas aeruginosa and Candida albicans (19). With the advent of next-generation sequencing (NGS) and informatics tools to analyze the millions of sequences generated by this technology, detailed microbiome profiling has offered a more descriptive picture of HIV-associated dysbiosis (Table 1). Measurements of alpha and beta diversity – which respectively describe the organismal richness and evenness within a single sample, and the similarity of microbiome composition between samples – have allowed recent NGS studies to consistently reveal an HIV-associated gut microbiome divergent from that of healthy control populations. Although some papers published by independent research groups show striking consistency in the nature of changes observed, other papers have produced unique findings. Comparing the results of these different studies to produce a comprehensive picture of the nature of community changes that occur with HIV and with ART is challenged by the complexity of the human microbiome, the diversity of populations examined and gastrointestinal sites surveyed, the complexity of HIV disease itself, and methodological differences between the studies (Fig. 1). Taken together, these papers have shed light on microbiome changes that occur with HIV infection throughout the gastrointestinal tract, but there are still open questions to be resolved.
Table 1.
Summary of HIV microbiome studies.
| Study | Study location | N of HIV+ group (male/female) | Type of sample evaluated | Serum CD4+ T cells (cells/ul) | ART | Summary of observed dysbiosis (HIV+ vs controls) |
|---|---|---|---|---|---|---|
| Lozupone et al 2013, 2014 | Denver, USA | Untreated: 11/0 Treated: 7/1 |
Feces | Untreated: 551 (270–1095) Treated: 483 (204–876) |
Both +/− | Untreated: Bacteroides Bacteroides Prevotella Prevotella alpha-diversity alpha-diversityTreated:  Bacteroides Bacteroides Prevotella Prevotella |
| McHardy et al 2013 | Los Angeles, USA | Untreated: 20/0 Treated: 20/0 |
Anal washes | Untreated: 439 (271 SD) Treated: 534 (246 SD) |
Both +/− | Untreated: Coprococcus Coprococcus Alistipes Alistipes Fusobacteria Fusobacteria alpha-diversity alpha-diversityTreated: Intermediate dysbiosis |
| Vujkovic-Cvijin 2013 | San Francisco, USA | Untreated: 6/0 Treated: 16/0 |
Colon | Untreated: 356 (313–819) Treated: 374 (251–1110) |
Both +/− | Untreated: Bacteroides Bacteroides Pathogenic spp. Pathogenic spp.Treated: Intermediate dysbiosis |
| Dillon et al 2014 | Denver, USA | 13/5 | Feces, colon | 425 (238–782) | Untreated |
 Bacteroides Bacteroides Prevotella Prevotella Proteobacteria ProteobacteriaMucosa only:  alpha-diversity alpha-diversity |
| Mutlu et al 2014 | Chicago, USA | 16/5 | Feces, colon, ileum | 425 (106–948) | Treated |
 Bacteroides Bacteroides Prevotella Prevotella Proteobacteria Proteobacteria alpha-diversity alpha-diversity |
| Vasquez-Castellanos et al 2014 | Madrid, Spain | 12/3 | Feces | 584 (466–794 range) | Treated |
 Bacteroides Bacteroides Prevotella Prevotella Coprococcus Coprococcus |
| Yu et al 2014 | Washington, DC and New York City, USA | 76/0 | Anal swab | Swab 1: 580 (432–721) Swab 2: 232 (32–415) |
NRTI only, no combined therapy | Baseline: Fusobacteria Fusobacteria Firmicutes FirmicutesFollow-up 1–5 yrs:  Fusobacteria Fusobacteria alpha-diversity alpha-diversity |
| Dinh et al 2015 | Boston, USA | 17/4 | Feces | 668 (424–870) | Treated |
 Alistipes Alistipes Barnsiella Barnsiella Proteobacteria Proteobacteria |
| Nowak et al 2015 | Stockholm, Sweden | 16/15 | Feces | Before treatment: 355 (120–2470) |
Both +/− (longitudinal) | Before ART: Lachnobacterium Lachnobacterium Lactobacillus Lactobacillus alpha-diversity alpha-diversityFollowing ART:  Prevotella Prevotella alpha-diversity alpha-diversity |
| Yang et al 2016 | New York City, USA | 5/3 | Duodenum, proximal gut | 327 (12–708) | Untreated |
 Lactobacillus Lactobacillus Proteobacteria Proteobacteria Firmicutes FirmicutesLow CD4 count:  Burkholderia Burkholderia |
| Noguera-Julian et al 2016 | Barcelona, Spain and Stockholm, Sweden | Barcelona: 101/28 (15 untreated) Stockholm: 46/31 (all untreated) |
Feces | 700 (462–860 IQR) | Both +/− |
 alpha-diversity alpha-diversityUnique findings: Prevotella/Bacteroides abundance associated with sexual behavior |
 : increase,
: increase,
 : decrease; for CD4+ T cell count, numbers correspond to either mean or median (with either range, SD=standard deviation, or IQR=interquartile range); for ART, “+” corresponds to treated and “−” corresponds to untreated; NRTI = non-nucleoside reverse transcriptase inhibitor
: decrease; for CD4+ T cell count, numbers correspond to either mean or median (with either range, SD=standard deviation, or IQR=interquartile range); for ART, “+” corresponds to treated and “−” corresponds to untreated; NRTI = non-nucleoside reverse transcriptase inhibitor
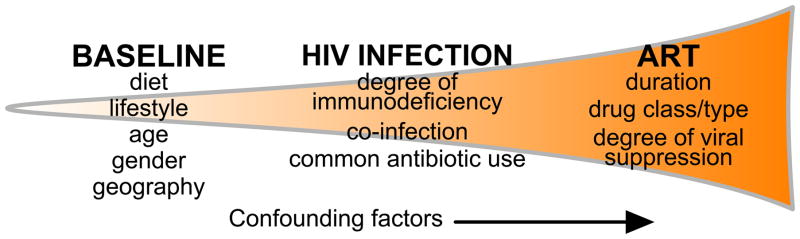
Figure 1. The myriad of variables in HIV microbiome studies may confound study results.
Many factors influence the intestinal microbiome throughout the course of HIV infection and treatment. An individual’s microbiome at baseline, before infection, will influence HIV-associated changes. These HIV-associated alterations themselves are influenced by the course of infection as well as any co-infection and treatment. In turn, these changes are further influenced by each patient’s course of ART. As a result, these confounding factors may hide ART-associated microbiome changes.
Effects of study population
A major confounding factor that has recently come to light is the effects of sexual behavior on fecal microbiome composition. One study that compared anal microbiome profiles between HIV-positive and HIV-negative men who have sex with men (MSM), found unprotected receptive anal intercourse to be significantly associated with alpha diversity (20). A recent study has shown that both HIV-positive and HIV-negative (MSM) have fecal microbiomes with increased alpha diversity and with high altered beta diversity associated with compositions enriched in the genus Prevotella and depleted in the genus Bacteroides (21). This microbiome profile was not evident in individuals who acquired HIV via heterosexual transmission or IV drug use. Studies of HIV-infected individuals conducted to date have most often used healthy control populations from the same geographic location that were matched for factors such as age and gender (22–26) but not for the practice of behaviors that lead to increased risk of contracting HIV. A Prevotella-rich/Bacteroides poor MSM-related microbiome type has been associated with HIV infection in multiple studies that did not control for sexual behavior (22–25). However, it was not observed in a study that had a cohort dominated by documented heterosexual transmission (27). This observation of MSM as a driving factor of large microbiome differences also potentially explains apparent discordant observations across studies with regards to alpha diversity, with one study showing an increase (23), others showing a decrease (22, 27) and others showing no change. When Nogera-Julian et al. examined alpha diversity while controlling for sexual behavior, they found MSM behavior leading to an increase in alpha diversity and within MSM, HIV-infection leading to a decrease (21). Controlling for sexual behavior will be crucial moving forward for determining which microbiome differences are related to HIV-induced immune dysfunction versus MSM sexual behavior.
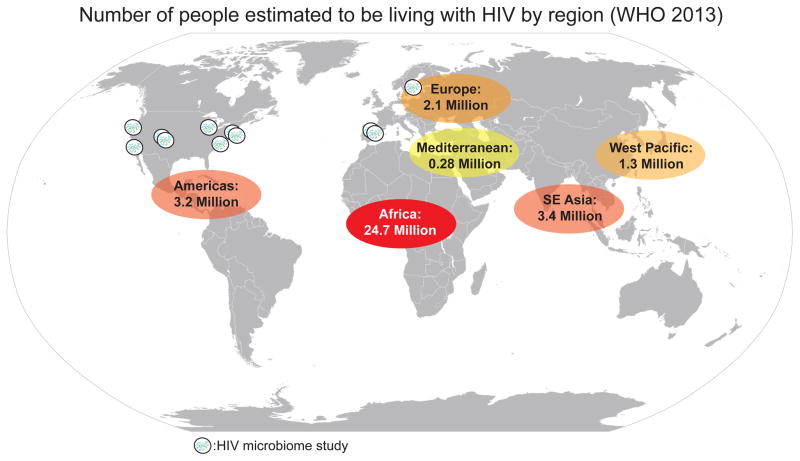
Another important complexity to consider is the baseline microbiome composition of the study population. Studies of healthy individuals have revealed that cultural differences between Western populations and agrarian societies in the developing world coincide with dramatic differences in gut microbiome composition, for instance with individuals in agrarian cultures having higher diversity microbiomes in health with relatively Prevotella-rich/Bacteroides-poor compositions that show a resemblance to that of MSM in Western cultures (28). The mode of transmission of HIV infection varies greatly across populations, with for instance transmission by MSM and IV drug use being more prevalent in the US and Europe and heterosexual and mother to child transmission being more important in the developing world (29). This leads to dramatic differences in demographic features of HIV-infected cohorts across populations that may also affect the microbiome at baseline or its response to disease, including age and sex. The studies of the HIV-associated gut microbiome published to date have largely focused on populations in the US and Europe and not the parts of the world where HIV is the most prevalent, including African populations (Fig. 2). Expanding HIV microbiome studies into the populations most affected by HIV is an important future direction.
Figure 2.
Studies of the HIV-associated microbiome have not been published in the populations most impacted by HIV.
Effects of disease stage/severity
Gut microbiome dysbiosis has the potential to affect HIV disease in various ways throughout all phases in the natural history of HIV disease progression, from transmission through end-stage disease (Fig. 3). The degree and nature of immune dysfunction throughout HIV disease progression also varies substantially across individuals. Thus, important cohorts to consider include: 1) Those in the acute stage of HIV-infection, which occurs within 2–10 weeks of exposure and is characterized by flu-like symptoms and a marked decline in circulating CD4+ T cells, 2) Those with chronic (latent phase) infection, which typically lasts for 7–10 years in the absence of treatment and is characterized by recovered circulating CD4+ T cells that then slowly decline over time, 3) Those with severe immune deterioration (CD4+ T cells < 200 cells/ul) that leads to opportunistic infections and AIDS and 4) Elite controllers, individuals who have stable peripheral CD4+ T cell counts for many years in the absence of ART. Assessing microbiome composition in each of these different cohorts has unique challenges.
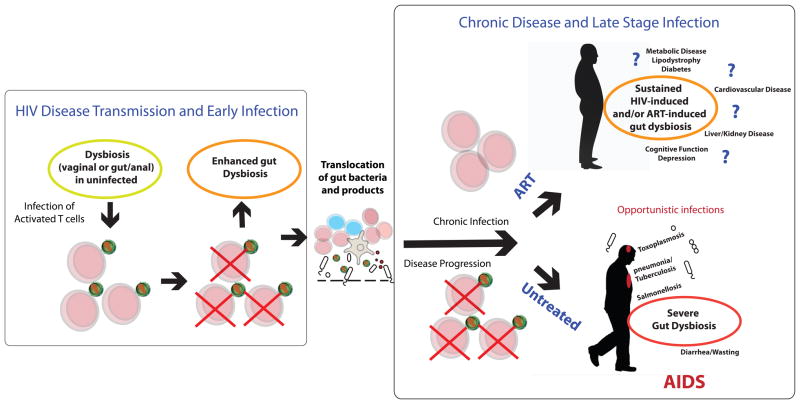
Figure 3. Microbiome influence through all stages of HIV infection.
The microbiome has the potential to have an influence throughout the entire natural history of HIV disease. Transmission: Since HIV preferentially infects activated T cells, dysbiotic microbiomes that activate CD4+ T cells can enhance disease transmission at mucosal sites. Disease Progression: The translocation of gut microbes through the intestinal barrier is thought to drive immune activation and disease progression. Chronic Infection: The gut microbiome differs from that of HIV-negative controls in chronic infection, and the myriad of health effects that may result from this dysbiotic microbiome are not understood. AIDS: Even more dramatic gut microbiome differences from healthy controls may occur with AIDS. Diarrhea, malabsorption, wasting and opportunistic infections that originate in the gut are common. ART: The gut microbiome of HIV-infected individuals on ART still differs from that of healthy controls. The effects of this dysbiotic microbiome on the health of individuals living a long time with HIV on ART are unknown, but many diseases that HIV-positive individuals on ART suffer from at increased incidence have been linked with gut microbiota compositional differences in HIV-negative individuals.
Gut microbiome changes that may occur in early/acute infection with HIV are very difficult to study, as individuals are usually past this stage of infection at the time of diagnosis. Longitudinal studies of high-risk cohorts have the most potential for catching this early stage of disease. In our cohort in Colorado, we characterized the fecal microbiome of 3 individuals with “recent” infection, characterized by likely acquisition within the prior year (23). Even these individuals, however, were most likely not within the acute phase of infection and data from only 3 individuals lacks statistical power to make definitive conclusions. The best existing data on what may be occurring in early infection comes from studies of the gut microbiome of non-human primates (NHPs) infected with Simian Immunodeficiency Virus (SIV). A spike in the abundance of Proteobacteria was observed in acutely infected macaques (30, 31), but this increase was temporary, with increased levels returning to normal levels at subsequent timepoints representing the chronic stage of infection. In general, fecal microbiome changes that occur in chronic infection have been more readily detectable in humans compared to NHPs. Multiple studies of SIV-infected macaques failed to detect any significant change in gut bacteria in animals with chronic infection (32, 33). This discrepancy may be in part due to MSM sexual behavior being a driving factor in humans. However human studies have uniquely used mucosal biopsies to assay microbiome differences in chronic infection and found a stronger microbiome change in that sample type (22, 24).
The majority of data produced thus far in HIV-infected humans not on ART are from individuals in the chronic phase of infection with mild to moderate immunodeficiency. However, a small number of individuals with a highly deteriorated immune system, indicative of AIDS, have been evaluated. Since advanced disease has been associated with higher incidences of opportunistic infections of many organs, including the gut, there is strong reason to believe that advanced disease will have a distinct microbiome signature compared to the chronic latent stage. In one study that included 6 individuals with a highly deteriorated immune status (CD4+ T cell counts between 120 and 150 cells/ul), decreased alpha diversity measured as the number of bacterial taxa in fecal samples was observed in this highly immune-deficient group compared to other individuals with chronic untreated infection (27), indicating a potential role for a breakdown in colonization resistance in opportunistic infections that occur with advanced disease. Consistent with this notion, in a comparison of the microbiome of the proximal gut (esophagus, stomach, and duodenum) of individuals with untreated HIV infection and controls, the environmental species Burkholdaria fungorum and Bradyrhizobium pachyrhizi colonized the duodenum of HIV-infected individuals with advanced disease but not those with normal CD4+ T cell counts (34). One study that compared anal microbiome profiles in HIV-positive and HIV-negative MSM surveyed many individuals with more advanced disease. These samples were collected at two time points from HIV-positive and HIV-negative MSM between 1989 and 1994, when clinicians collected swabs from the anal canal for evaluation of human papillomavirus (HPV) and anal cytology. Since these samples were collected before the advent of combination effective ART, the cohort was characterized by advancing disease between timepoint 1 and timepoint 2. At swab-1, the HIV-positive men had CD4+ T cell counts with a median of 580 (interquartile range (IQR): 432–721) cells/μl), and by the time of swab-2, 1–5 years later, CD4+ T cell counts in the HIV-positive men had fallen significantly [median (IQR) 232 (32–415) cells/μl] and 16 more HIV-positive men had developed AIDS. Despite this advancing disease between swab-1 and swab-2, microbiome differences between swabs were subtle, with some evidence for a decrease in diversity at swab-2 (20). Study of the gut microbiome in advanced AIDS is complicated by the high rates of antibiotic use in this population. Antibiotics can cause profound changes in gut microbiota composition so many studies of the HIV enteric microbiome have used recent antibiotic usage as an exclusion criteria. As a result few studies have examined HIV-infected subjects on antibiotics, which in turn has limited the numbers of studies of subjects with advance disease and co-infections. Further studies of cohorts that include individuals with advanced HIV disease will help to understand the effects of severe immunodeficiency on gut microbiome composition.
On the other end of the spectrum, two studies have included small numbers of “elite-controllers” or “long-term non-progressors.” In one study, an HIV-positive individual who had a stable peripheral blood CD4+ count despite over 21 years of untreated infection, had a gut community similar to that of healthy subjects (26). Another study that surveyed the microbiome of 3 elite controllers, found that the microbiomes of the elite controllers were highly similar to one another but less similar to both viremic and HIV-negative groups. This study also reported that elite controllers had increased Bacteroidetes and decreased Actinobacteria and Proteobacteria compared to viremic patients (27). These results present the interesting possibility that non-progressive disease may be linked to host-microbiome homeostasis (26, 27).
Effects of Gastrointestinal site
Various studies have also targeted different regions of the gastrointestinal tract when surveying changes in the gut microbiome that occur with HIV. Fecal samples are commonly assayed (22, 23, 25), in part because of the ease in which they can be collected. Mucosal biopsies or brushings have been assayed in several studies (22, 26, 34), and have the potential to shed light on the bacteria that most intimately interact with the host immune system. Mucosal samples have been compared in HIV-infected individuals versus controls from various sites throughout the gastrointestinal tract, including from the duodenum (34), terminal ileum (24), and colon (22, 24, 26). The mucosa-associated microbiome can significantly differ down the length of the GI tract and from the microbiome of the feces (35). The anal microbiome of MSM may be of particular interest because of a potential role in HIV disease transmission (20). Not surprisingly, different sites of the GI tract had unique differences in the nature of microbiome changes that occurred with HIV, although these differences were also confounded by other varying factors between studies including underlying demographics of study populations, sexual behavior, disease stage/severity, or ART status.
The best studies for looking at the effects of sample type on observed differences with HIV infection are therefore those that assayed multiple sites from the same individuals. Dillon et al. observed enrichment of Prevotella and depletion of Bacteroides species in individuals with chronic untreated HIV infection in both colon biopsy and fecal samples but changes in abundance of various taxa from the Proteobacteria and Firmicutes phyla were only observed in the biopsies (22). Dillon et al. also observed a decrease in alpha diversity of individuals with chronic untreated infection in colon biopsy but not fecal samples. Another study that compared the microbiome of HIV-infected individuals on ART versus healthy controls using matched fecal, terminal ileum and left and right colonic biopsy samples found clustering by HIV status at all sites, but the most pronounced clustering was found in the terminal ileum and least pronounced difference in feces (24). Consistent with Dillon et al., decreased alpha diversity with HIV infection was found in biopsies but not in fecal samples. One further study used mucosal biopsy samples from the rectosigmoid region to compare the HIV-associated microbiome to healthy controls using Microarray (PhyloChip) analysis, and showed that the strongest changes with untreated HIV infection were increases in abundance of opportunistic pathogens (such as Staphylococcus and Pseudomonas species) and Proteobacteria, and decreases in abundance of bacteria from Bacteroides and Alistipes genera (26). Taken together, these results suggest that reduction in Bacteroides species is consistent across both mucosa and fecal samples in most studies, but that changes in Proteobacteria may occur more readily in specific mucosa-associated sites. Furthermore, decreases in alpha diversity with untreated HIV infection are evident in mucosal samples and not fecal samples in studies that are confounded by MSM sexual behavior (22, 24), except in the case of advanced disease, where decreased alpha diversity has been observed in feces (27). Decreased alpha diversity in feces was also observed in a study that controlled for MSM sexual behavior (21).
Studies that have examined the microbiome in the rectum yielded results that were in some respects distinct and in other respects consistent with studies that used feces and biopsies to assay gut microbiome composition. One study used an absorbent ophthalmic sponge applied under direct vision via anoscope to opposing sides of the rectal mucosa to compare the rectal mucosa of HIV-infected individuals to healthy controls (24). Some consistencies in HIV-associated alterations in fecal/biopsy samples of less prominent taxa were observed, such as depletion of Alistipes (23) and Coprococcus species (22) with untreated HIV infection. However, other commonly observed changes, such as those in abundance of Prevotella or Bacteroides organisms, did not occur (36). The observation of increased Fusobacterium and decreased Ruminococcus with HIV infection in the rectal mucosa was consistent with the results of a further study that compared anal swabs of HIV-positive and -negative MSM (20).
Effects of ART on the gut microbiota
Antiretroviral therapy (ART) has greatly decreased the morbidity and mortality of HIV infection (37), however, up to 40% of HIV-positive individuals receiving ART experience moderate to severe gastrointestinal (GI) symptoms (38). ART is also associated with potentially gut-linked diseases, such as elevated liver enzymes, diabetes, cardiovascular disease, alteration of fat deposits associated with metabolic disease, accelerated aging, and cognitive defects (39). Little is known about the role of the gut microbiome in GI disease during HIV infection, and why ART treatment may lead to an increased incidence of these diseases.
To date, eight papers have evaluated gut microbiome composition in HIV-infected individuals on ART, and all indicated that HIV-positive individuals on ART have a gut microbiome composition that differs from that of healthy control populations (21, 23, 24, 26, 27, 36, 40, 41). However, these studies differed in the magnitude of the observed changes, with some studies showing strong community level clustering between HIV-infected individuals on ART and HIV-negative controls (23, 24, 26) and others not (36). These studies varied in the degree to which the study design could differentiate whether microbiome differences observed in individuals on ART are driven by the drugs versus chronic HIV infection itself, and in the degree to which the observed results may have been confounded by MSM sexual behavior.
Studies that have compared untreated HIV-positive individuals, individuals on ART, and HIV-negative controls give more robust insights into ART-associated microbiome changes. These studies have indicated that at least some of the differences between HIV-positive individuals on ART and HIV-negative controls are consistent with those differentiating individuals with untreated HIV infection and HIV-negative controls. However, these studies largely did not control for MSM sexual behavior. For instance in our cohort, we observed no consistent movement towards the HIV-negative group at the community level with ART in individuals sampled before and after 6–12 months of ART and that most individuals on long-term (1–5 years) of ART clustered with individuals with untreated infection rather than healthy controls with a Bacteroides-poor/Prevotella-rich microbiome type (23). This result was initially interpreted to potentially indicate that HIV-associated microbiome changes were not completely ameliorated by ART. However, new data regarding the profound effects of MSM on the fecal microbiome indicates that the result may have more to do with the degree to which MSM sexual behavior persists in individuals on ART. However, we did find that certain taxa with increased relative abundance in individuals with untreated HIV infection compared to HIV-negative controls decreased strongly with ART, such as Peptococcus and Desulfovibrio (42). Robust response to ART suggests that these taxa are more likely differing because of HIV-associated immune dysfunction rather than MSM behavior.
Evaluation of sigmoid biopsies (26) also has revealed that patients receiving ART had highly variable gut microbiomes, with the composition of some individuals more similar to a healthy control group and others more similar to the untreated HIV-positive group, however the results of this study that may also have been confounded because they did not control for sexual behavior. Nowak et al. also observed higher beta diversity with ART in a cohort dominated by heterosexual transmission, indicating high variability in microbiome composition with treatment that is likely not driven by behavior. A study that compared untreated HIV-positive individuals, individuals on ART, and HIV-negative controls using rectal sponge showed that individuals on ART exhibited similar compositional changes to those observed with untreated HIV infection but of intermediate magnitude (36).
There is also evidence that ART induces microbiome changes that are independent of those driven by HIV infection. For instance, one study that evaluated 19 individuals pre and post a median of 10 months of ART, described ART-associated compositional changes that were not related to differences observed between the untreated HIV-infected cohort and healthy controls (27). Three studies have now reported decreased alpha diversity in ART-treated individuals compared to untreated HIV-positive individuals (21, 27, 42), which may be indicative of further dysbiosis with ART, since low alpha diversity is often associated with disease states (43, 44). This result may be consistent with known GI side-effects such as diarrhea with particular ART drugs (38). Noguera-Julian et al observed lowest alpha diversity in individuals on unsuccessful ART, suggesting a compounding effect of HIV and ART on gut microbiome dysbiosis. However, McHardy et al. observed the opposite with regards to alpha diversity, with a slight but significant reduction in alpha diversity of rectal sponge samples in untreated HIV infection compared to healthy controls that was reversed to near equivalence to healthy controls in ART-treated patients (36). One potential confounding factor that may explain discrepancies with regards to alpha diversity between studies is the duration of ART. In fact, in an SIV model, ART initially increased gut dysbiosis, but this increase was ameliorated over time (31). This change in alpha diversity is likely due to the immune changes throughout ART duration as alpha diversity has been shown to be positively correlated with CD4+ T cell count (27). The discrepancies in alpha-diversity results may also be an artifact of sampling site, with different results possible in rectal sponge versus fecal samples.
Consistent with the notion that ART drugs can directly affect intestinal microbiota composition, there is evidence from early studies that proteinase inhibitors (PIs) can exert a direct effect on Candida albicans by reducing adherence of C. albicans to an epithelial cell layers in vitro (45). Both reduced adherence and immune reconstitution associated with ART could have contributed to the observed reduction of oral Candida infections in ART-treated, HIV-infected individuals. In a similar vein, a study investigating the link between the attachment and entry inhibitor (AEI), maraviroc, metabolic parameters, and the microbiome, observed a dramatic increase in intestinal Enterobacteriales in mice fed a high fat diet, which was significantly reduced when mice on the high fat diet were treated with a maraviroc regimen (46). Interestingly, mice fed a high fat diet and treated with maraviroc had significantly decreased body weight gains, liver weight, and alanine aminotransferase (ALT) levels compared to untreated high fat diet mice. Though direct antimicrobial effects cannot be determined from this study, it does suggest that even in the absence of HIV infection, ART influences the intestinal microbiome community composition, which in turn may affect metabolism.
The complex interaction between ART and microbiome composition may be even more confounded by direct drug metabolism by gut microbes. Although, there are currently no studies that specifically examine whether gut microbes can metabolically transform ART drugs, the microbiome has been shown to affect activity and toxicity of several other drugs. Gut microbiome composition influences the response to cancer therapy (47), the toxicity of acetaminophen (48), and the activity of cardiac drugs (49). As a result, baseline differences in the microbiome of HIV-infected individuals may affect both the effectiveness of therapy as well as the potential restoration of a healthy gut flora, contributing to the observed variation in microbiome community change in ART-treated patients. Studies which examine the effects of the gut microbiome on ART metabolism are needed.
Studies examining the impact of ART on intestinal microbiome composition are challenged by additional possible confounding factors including ART regimen (i.e. drug classes), duration of ART, and the degree to which the ART is successful in controlling viral load, in addition to all of the other factors already discussed (Fig. 1). For instance, even ART-treated cohorts can vary substantially in disease severity, with many studies restricting ART cohorts to individuals with negligible virus detected in plasma samples (23) and others including individuals with detectable viral loads and low CD4+ T cell counts despite being on ART (24). Further studies that include HIV-positive individuals with and without ART as well as HIV-negative controls, and those that data before and over time with treatment with specific drug combinations will further elucidate ART-associated microbiome differences. Examining the effects of ART in HIV-seronegative individuals also has the potential to produce insights into drug effects in the absence of HIV-induced immune dysfunction. These studies may also be relevant since treatment of high risk HIV-negative individuals with ART drugs (Pre-exposure prophylaxsis or PrEP) has been shown to be an effective HIV prophylactic (50), and thus understanding potential microbiome mediated side-effects is of increased importance.
The impact of HIV infection on the virome
The works that we have discussed thus far have all focused on gut bacteria. We know very little about how the composition of fungi, viruses and parasites living in the gut varies across the different stages of HIV disease and with ART, although opportunistic infections with all of these classes of organisms that occur in advanced disease suggest that subclinical manifestations may also happen. Of all of these groups, the most is known about viruses.
The majority of gut-associated virus is in the form of bacteriophage (or simply, phage), which are viruses that interact with microbes. Phage integrate into bacterial host genomes for varying periods of time and can mediate horizontal transfer of genes for antibiotic resistance, virulence, and host immune interaction (51). Given the co-dependent relationship between bacteria and phage, alterations in phage composition may have large effects on the gut microbiome and vice versa. Indeed, phage richness is greatly increased in IBD and is associated with an increase in gut bacterial diversity (52). Viruses that infect eukaryotic cells also inhabit the gut (53), and several have been linked to enteric disease. For example, diarrhea has been associated with multiple viruses in humans, including Picobirnaviruses (54), Adenoviruses, and Enteroviruses (55). Thus, viruses may impact gut health by altering the microbiome or by directly infecting human cells.
Recent evidence has suggested that HIV/SIV infection could induce alterations in the virome. RNA and DNA sequencing of feces from SIV-infected macaques showed an SIV-altered virome that was associated with enteric disease (32). SIV infection resulted in a significant increase in the abundance of vertebrate viral sequences in the feces, with the most prominent changes observed in viral sequences from the Picornaviridae and Adenoviridae families. Histology and viral antigen staining demonstrated an association between enteric disease and the presence of intestinal Adenovirus infection. These findings strongly suggest that virome changes may also play a role in HIV-associated enteric disease and microbial translocation.
Significant alterations of phage sequence abundance in the feces of SIV-infected macaques were also detected (32). Though changes in phage abundance did not associate with changes in gut bacteria or with enteric disease in SIV-infected monkeys (32), phage may still play a role in disease during HIV infection. In support of this notion, sequencing of DNA extracted from human plasma revealed a virome composed heavily of phage DNA in untreated HIV-infected subjects, and a virome completely lacking of phage sequence in healthy subjects (56). Though the disease impacts of increased phage in the blood were undetermined, it may be an indicator of bacterial translocation and therefore the severity of immune activation. Furthermore, there is evidence for the ability of phage to directly modulate immune cells (57), thus the direct contribution of phage to immune activation during HIV infection is an interesting possibility.
Disease implications of a dysbiotic gut
A dysbiotic gut has potential health implications through the entire life history of HIV infection, from transmission, early disease progression, chronic disease with and without ART, and end stage disease (Fig. 3). Transmission of HIV is a critical step in the viral life cycle, and early replication is essential for establishing viral reservoirs that allow for HIV to persist and cause immunodeficiency. Microbiome composition at mucosal sites where HIV is first encountered may have a significant impact on early HIV infection and therefore disease progression. Several studies have shown that dysbiosis of the vaginal microbiome in the form of Bacterial Vaginosis (BV), characterized by a depletion of Lactobacillus spp., can affect rates of heterosexual transmission of HIV both by increasing viral activity in the vagina of an infected woman so that the virus is more readily transmitted to a male partner (58), and by increasing the immune activation state in the HIV-negative woman such that the virus can more readily establish infection after exposure from her male partner (59). The degree to which gut dysbiosis may impact the transmission of HIV between couples engaging in anal intercourse is not known. However, interestingly, anal swabs collected from untreated HIV-infected subjects contained microbiomes dominated by Lactobacillales organisms, the abundance of which positively correlated with CD4+ T cell count and negatively correlated with inflammatory markers (41), suggesting the potential for Lactobacillus to play a protective role in the context of transmission via anal intercourse. The association of Lactobacillales bacteria with higher CD4+ T cell counts suggests a role for these organisms in controlling immunodeficiency; however, lack of a strong presence of Lactobacillales bacteria in colon biopsies and feces (22, 23) may indicate that they play a site-specific role in the anal mucosa.
Following transmission, HIV infection is associated with increased intestinal permeability, allowing for elevated microbial movement from the intestinal lumen into circulation, contributing to systemic, chronic inflammation which has been shown to drive HIV infection and disease progression (60) (Fig. 4). Increased serum levels of intestinal fatty acid binding protein (I-FABP) – which is indicative of enterocyte damage (61), serum levels of LPS (40, 62), sCD14 (40, 61, 62), and anti-flagellin antibodies (61, 62) – all markers of increased microbial translocation, are significantly increased in HIV-infected individuals compared to typical levels for healthy individuals. The degree to which changes in bacterial community composition that occur with HIV, such as the colonization of more pro-inflammatory types of microbes, are a driving factor of increased bacterial translocation, is not completely understood. However, preliminary studies have shown a negative correlation between sCD14 and Lactobacillales levels in anal swabs (41) and a positive correlation with Enterobacteriales levels in colonic biopsies (40). In principle, positive associations between T cell activation and the prevalence of particular bacteria indicate the potential to promote HIV infection, since HIV replicates most effectively in activated CD4+ T cells. Studies that examined the HIV-associated microbiome in biopsies found that particular bacteria (i.e. Staphylococcus and Prevotella) positively correlated with immune cell activation, including CD4+ T cells, in both the blood and the gut (22, 26). In contrast, Bacteroides abundance was negatively correlated with inflammatory markers in HIV infected individuals on and off of ART (24, 26), suggesting that Bacteroides organisms may offer a protective effect against HIV-associated inflammation. Controlling the abundance of pro-inflammatory bacteria or increasing the abundance of anti-inflammatory bacteria may therefore significantly limit inflammatory disease as well as HIV replication. Even with immune reconstitution following ART, patients still exhibit signs of intestinal damage and increased microbial translocation (40, 61, 62). In fact, sCD14 has been reported to be higher in subjects on ART compared to untreated subjects (61). This suggests that chronic inflammation that persists with ART can be related to gut dysbiosis.
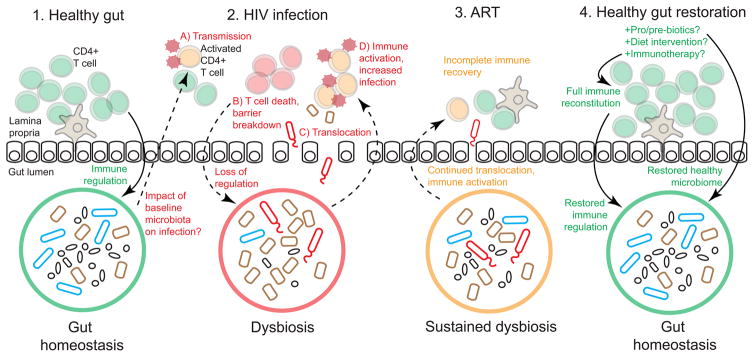
Figure 4. Gut health during HIV infection.
1) A healthy gut is characterized by homeostasis between the immune system and the microbiome. 2) Though the impact of the baseline microbiota on HIV infection is unknown, it is possible that gut microbes could impact (A) transmission, potentially by activating CD4+ T cells. (B) HIV infection leads to CD4+ T cell death and immune depletion, and loss of immune regulation of gut bacteria, resulting in a dysbiotic microbiome. (C) Translocation of dysbiotic bacteria leads to (D) immune activation and further HIV infection of activated CD4+ T cells. 3) ART suppresses viral replication but does not fully restore gut immunity; dysbiosis is sustained during ART and microbial translocation continues to cause chronic immune activation. 4) A healthy gut may be restored by supplementing ART with pro/prebiotics or diets that encourage growth of beneficial bacteria, and with immunotherapy that could help reconstitute gut immunity and restore immune regulation of the microbiome.
HIV infection has also been associated with a higher risk for other complications such as metabolic and cardiovascular diseases (2). These diseases have been associated with compositional differences in the gut microbiome in HIV-negative individuals (3), which would suggest that microbiome disturbances caused by HIV could be the source of their association with HIV infection. One of the earliest described metabolic diseases in HIV-infected patients was lipodystrophy, a syndrome characterized by fat gain at central body sites (i.e. abdomen) and fat loss in peripheral sites (i.e. face, arms), as well as insulin resistance (2) – diseases that are known to be affected by the gut microbiome (3). ART has also been suggested to play a role in metabolic disease during HIV infection (2), and since ART could have its own impact on microbiome composition as discussed above, it may worsen HIV-associated metabolic disease by independently altering the microbiome.
Increased risk of cardiovascular diseases such as atherosclerosis and myocardial infarction in HIV-infected individuals may also be linked to the microbiome (2). There is increasing evidence for the association of microbial metabolites like trimethylamine (TMA) and its derivative trimethylamine N-oxide (TMAO) with greater risk of cardiovascular disease in both humans and experimental mouse models (3). TMA has been associated with the severity of atherosclerosis in HIV-infected patients (63), and another study indicated that while plasma levels of TMAO did not differ between HIV-negative and HIV-positive individuals, levels increased after ART introduction and was associated with the use of PIs; furthermore, plasma TMAO was associated with myocardial perfusion defects in these HIV-infected individuals (64). Shifts in the microbial metabolome during HIV infection may therefore be drivers for cardiovascular disease. Integrative microbiome and metabolome analyses of HIV-infected cohorts will better define these links.
Microbiome-targeted therapy
Clinical trials evaluating the effects of certain probiotics (dietary supplements containing live bacteria) on HIV-infected individuals have yielded positive results. ART-treated patients with suppressed viral load responded to daily probiotic supplementation with decreases in immune cell activation (65), decreases in levels of serum inflammatory markers (66), and decreases in microbial translocation (67). Additionally, the use of prebiotics (dietary supplements containing nutrients that encourage growth of certain gut bacteria) by ART-naive patients resulted in reduced inflammation and CD4+ T cell activation (19). Thus, microbiome-targeted therapy in the form of pro- and prebiotics may be an effective way to reduce chronic inflammation and immune activation during HIV infection. An additional use for probiotics may be for the prevention of outgrowth of inflammatory or pathogenic bacteria that are increased with HIV infection. For example, there is evidence that probiotics could reduce growth of antibiotic-resistant Enterococci in humans and mice (68). Microbiome analyses of HIV-positive individuals treated with probiotics will help determine if a healthy gut microbiome can be recovered with probiotic therapy (Fig. 4).
Another approach to limit gut microbiome-mediated disease progression is treatments that directly restrict microbial translocation. A prophylactic antibiotic regimen of co-trimoxazole (TMP-SMX) has been shown to significantly reduce microbial translocation a year after starting ART (62). However, antibiotic treatment may lead to the loss of healthy gut flora and exacerbate a loss of diversity that is already observed with ART. Drugs that directly bind or neutralize LPS, such as sevelamer, could be another method for reducing immune activation caused by microbial translocation. Treatment of SIV-infected macaques with sevelamer was found to reduce systemic immune activation and inflammation (69), while a sevelamer trial in HIV-infected subjects found that the drug could improve cardiovascular health (70).
Combination therapy may be needed to fully restore gastrointestinal health during HIV infection. Studies using SIV-infected macaques showed that although pro- and prebiotics could aid in the recovery of CD4+ T cell numbers and functionality in ART-treated animals, it had no significant effect on microbial translocation (71). Supplementation with immunotherapy in addition to probiotics and ART was needed for full reconstitution of Th17 cells critical for gut immunity and reduction of microbial translocation in SIV-infected macaques (72). Full reconstitution of immune cells in the gut may also lead to restored immune regulation of the gut microbiome, which would in turn lead to recovery from HIV-associated dysbiosis. Immunotherapies aimed at boosting the function of monocytes/macrophages in the gut may also be effective for restoring gut health, particularly for reducing microbial translocation and subsequent inflammation. In support of this notion, elevated LPS levels in HIV-infected humanized mice was associated with poor clearance of translocated microbial products by macrophages (73). A multi-pronged approach of boosting both innate and adaptive immune function, along with probiotic therapy, may be needed for the full recovery of gut health in HIV-infected people (Fig. 4).
Conclusions
Although some studies have reported consistent observations of microbiome changes in HIV-infected cohorts, other studies have produced unique results. Understanding the impact of factors like population, disease state, ART, sexual behavior and even the virome on gut bacteria will be essential for interpreting the variation that is observed across studies. What is suggested from the current evidence is that HIV infection is associated with an altered gut microbiome, particularly with advanced disease and in mucosal tissue, and that ART is unable to consistently restore gut health. Given the increasing number of studies linking the microbiome to human disease, HIV-associated dysbiosis may be at the root of the cause for chronic diseases that persist during HIV infection, despite control of viral load by ART. Uncovering links between gut bacteria and HIV-associated disease will open the door for new therapies aimed at manipulating the microbiome. Probiotics have already been shown to have positive effects during HIV infection, including reducing immune activation and microbial translocation, and may even play a role in reducing mucosal HIV transmission. In conclusion, the microbiome has the potential to impact HIV disease at each step of the viral life cycle, from transmission to AIDS, which presents the opportunity for microbiome-targeted therapeutic intervention at each of these stages.
References
- 1.Paiardini M, Muller-Trutwin M. HIV-associated chronic immune activation. Immunol Rev. 2013;254:78–101. doi: 10.1111/imr.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nix LM, Tien PC. Metabolic syndrome, diabetes, and cardiovascular risk in HIV. Curr HIV/AIDS Rep. 2014;11:271–8. doi: 10.1007/s11904-014-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aron-Wisnewsky J, Clement K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol. 2015 doi: 10.1038/nrneph.2015.191. [DOI] [PubMed] [Google Scholar]
- 4.Kotler DP. HIV infection and the gastrointestinal tract. AIDS. 2005;19:107–17. doi: 10.1097/00002030-200501280-00002. [DOI] [PubMed] [Google Scholar]
- 5.Mehandru S, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–70. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mowat AM, Viney JL. The anatomical basis of intestinal immunity. Immunol Rev. 1997;156:145–66. doi: 10.1111/j.1600-065x.1997.tb00966.x. [DOI] [PubMed] [Google Scholar]
- 7.Brenchley JM, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–35. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang SC, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–9. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillon SM, et al. HIV-1 infection of human intestinal lamina propria CD4+ T cells in vitro is enhanced by exposure to commensal Escherichia coli. J Immunol. 2012;189:885–96. doi: 10.4049/jimmunol.1200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao Y, et al. The decrease of regulatory T cells correlates with excessive activation and apoptosis of CD8+ T cells in HIV-1-infected typical progressors, but not in long-term non-progressors. Immunology. 2009;128:e366–75. doi: 10.1111/j.1365-2567.2008.02978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allers K, et al. Gut mucosal FOXP3+ regulatory CD4+ T cells and Nonregulatory CD4+ T cells are differentially affected by simian immunodeficiency virus infection in rhesus macaques. J Virol. 2010;84:3259–69. doi: 10.1128/JVI.01715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno-Fernandez ME, Presicce P, Chougnet CA. Homeostasis and function of regulatory T cells in HIV/SIV infection. J Virol. 2012;86:10262–9. doi: 10.1128/JVI.00993-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity. 2013;39:633–45. doi: 10.1016/j.immuni.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato LM, Kawamoto S, Maruya M, Fagarasan S. The role of the adaptive immune system in regulation of gut microbiota. Immunol Rev. 2014;260:67–75. doi: 10.1111/imr.12185. [DOI] [PubMed] [Google Scholar]
- 16.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–39. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Kawamoto S, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–65. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Ellis CL, et al. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J Acquir Immune Defic Syndr. 2011;57:363–70. doi: 10.1097/QAI.0b013e31821a603c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gori A, et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the “COPA” pilot randomized trial. Mucosal Immunol. 2011;4:554–63. doi: 10.1038/mi.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G, Fadrosh D, Ma B, Ravel J, Goedert JJ. Anal microbiota profiles in HIV-positive and HIV-negative MSM. AIDS. 2014;28:753–60. doi: 10.1097/QAD.0000000000000154. [DOI] [PubMed] [Google Scholar]
- 21.Noguera-Julian M, et al. Gut Microbiota Linked to Sexual Preference and HIV Infection. eBioMedicine. 2016 doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dillon SM, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–94. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozupone CA, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–39. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutlu EA, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez-Castellanos JF, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 2015;8:760–72. doi: 10.1038/mi.2014.107. [DOI] [PubMed] [Google Scholar]
- 26.Vujkovic-Cvijin I, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 2013;5:193ra91. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowak P, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29:2409–18. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 28.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Cock KM, et al. Prevention of mother-to-child HIV transmission in resource-poor countries: translating research into policy and practice. JAMA. 2000;283:1175–82. doi: 10.1001/jama.283.9.1175. [DOI] [PubMed] [Google Scholar]
- 30.Glavan TW, Gaulke CA, Hirao LA, Sankaran-Walters S, Dandekar S. SIV-infection-driven changes of pattern recognition receptor expression in mesenteric lymph nodes and gut microbiota dysbiosis. J Med Primatol. 2015;44:241–52. doi: 10.1111/jmp.12187. [DOI] [PubMed] [Google Scholar]
- 31.Klase Z, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol. 2015;8:1009–20. doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Handley SA, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–66. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna P, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L, et al. HIV-induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS. 2016;30:19–29. doi: 10.1097/QAD.0000000000000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McHardy IH, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dieffenbach CW, Fauci AS. Thirty years of HIV and AIDS: future challenges and opportunities. Ann Intern Med. 2011;154:766–71. doi: 10.7326/0003-4819-154-11-201106070-00345. [DOI] [PubMed] [Google Scholar]
- 38.Kartalija M, Sande MA. Diarrhea and AIDS in the era of highly active antiretroviral therapy. Clin Infect Dis. 1999;28:701–5. doi: 10.1086/515191. quiz 6–7. [DOI] [PubMed] [Google Scholar]
- 39.Montessori V, Press N, Harris M, Akagi L, Montaner JS. Adverse effects of antiretroviral therapy for HIV infection. CMAJ. 2004;170:229–38. [PMC free article] [PubMed] [Google Scholar]
- 40.Dinh DM, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Santiago J, et al. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS. 2013;27:1921–31. doi: 10.1097/qad.0b013e3283611816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes. 2014;5:562–70. doi: 10.4161/gmic.32132. [DOI] [PubMed] [Google Scholar]
- 43.Weingarden A, et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome. 2015;3:10. doi: 10.1186/s40168-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willing BP, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–54 e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 45.Borg-von Zepelin M, et al. HIV-Protease inhibitors reduce cell adherence of Candida albicans strains by inhibition of yeast secreted aspartic proteases. J Invest Dermatol. 1999;113:747–51. doi: 10.1046/j.1523-1747.1999.00747.x. [DOI] [PubMed] [Google Scholar]
- 46.Perez-Matute P, Perez-Martinez L, Aguilera-Lizarraga J, Blanco JR, Oteo JA. Maraviroc modifies gut microbiota composition in a mouse model of obesity: a plausible therapeutic option to prevent metabolic disorders in HIV-infected patients. Rev Esp Quimioter. 2015;28:200–6. [PubMed] [Google Scholar]
- 47.Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–70. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106:14728–33. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–8. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caceres CF, O’Reilly KR, Mayer KH, Baggaley R. PrEP implementation: moving from trials to policy and practice. J Int AIDS Soc. 2015;18:20222. doi: 10.7448/IAS.18.4.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breitbart M, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci U S A. 2002;99:14250–5. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norman JM, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–60. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang T, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banyai K, et al. Sequence heterogeneity among human picobirnaviruses detected in a gastroenteritis outbreak. Arch Virol. 2003;148:2281–91. doi: 10.1007/s00705-003-0200-z. [DOI] [PubMed] [Google Scholar]
- 55.Holtz LR, et al. Geographic variation in the eukaryotic virome of human diarrhea. Virology. 2014;468–470:556–64. doi: 10.1016/j.virol.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li SK, et al. Detection and identification of plasma bacterial and viral elements in HIV/AIDS patients in comparison to healthy adults. Clin Microbiol Infect. 2012;18:1126–33. doi: 10.1111/j.1469-0691.2011.03690.x. [DOI] [PubMed] [Google Scholar]
- 57.Foca A, Liberto MC, Quirino A, Marascio N, Zicca E, Pavia G. Gut inflammation and immunity: what is the role of the human gut virome? Mediators Inflamm. 2015;2015:326032. doi: 10.1155/2015/326032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen CR, et al. Bacterial vaginosis associated with increased risk of female-to-male HIV-1 transmission: a prospective cohort analysis among African couples. PLoS Med. 2012;9:e1001251. doi: 10.1371/journal.pmed.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmid G, Markowitz L, Joesoef R, Koumans E. Bacterial vaginosis and HIV infection. Sex Transm Infect. 2000;76:3–4. doi: 10.1136/sti.76.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–71. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 61.Sandler NG, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203:780–90. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vesterbacka J, et al. Kinetics of microbial translocation markers in patients on efavirenz or lopinavir/r based antiretroviral therapy. PLoS One. 2013;8:e55038. doi: 10.1371/journal.pone.0055038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Srinivasa S, et al. Plaque burden in HIV-infected patients is associated with serum intestinal microbiota-generated trimethylamine. AIDS. 2015;29:443–52. doi: 10.1097/QAD.0000000000000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haissman JM, et al. Microbiota-dependent marker TMAO is elevated in silent ischemia but is not associated with first-time myocardial infarction in HIV infection. J Acquir Immune Defic Syndr. 2015 doi: 10.1097/QAI.0000000000000843. [DOI] [PubMed] [Google Scholar]
- 65.d’Ettorre G, et al. Probiotics Reduce Inflammation in Antiretroviral Treated, HIV-Infected Individuals: Results of the “Probio-HIV” Clinical Trial. PLoS One. 2015;10:e0137200. doi: 10.1371/journal.pone.0137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Falasca K, Vecchiet J, Ucciferri C, Di Nicola M, D’Angelo C, Reale M. Effect of Probiotic Supplement on Cytokine Levels in HIV-Infected Individuals: A Preliminary Study. Nutrients. 2015;7:8335–47. doi: 10.3390/nu7105396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Villar-Garcia J, et al. Effect of probiotics (Saccharomyces boulardii) on microbial translocation and inflammation in HIV-treated patients: a double-blind, randomized, placebo-controlled trial. J Acquir Immune Defic Syndr. 2015;68:256–63. doi: 10.1097/QAI.0000000000000468. [DOI] [PubMed] [Google Scholar]
- 68.Crouzet L, Rigottier-Gois L, Serror P. Potential use of probiotic and commensal bacteria as non-antibiotic strategies against vancomycin-resistant enterococci. FEMS Microbiol Lett. 2015;362:fnv012. doi: 10.1093/femsle/fnv012. [DOI] [PubMed] [Google Scholar]
- 69.Kristoff J, et al. Early microbial translocation blockade reduces SIV-mediated inflammation and viral replication. J Clin Invest. 2014;124:2802–6. doi: 10.1172/JCI75090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandler NG, et al. Sevelamer does not decrease lipopolysaccharide or soluble CD14 levels but decreases soluble tissue factor, low-density lipoprotein (LDL) cholesterol, and oxidized LDL cholesterol levels in individuals with untreated HIV infection. J Infect Dis. 2014;210:1549–54. doi: 10.1093/infdis/jiu305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klatt NR, et al. Probiotic/prebiotic supplementation of antiretrovirals improves gastrointestinal immunity in SIV-infected macaques. J Clin Invest. 2013;123:903–7. doi: 10.1172/JCI66227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ortiz AM, et al. IL-21 and probiotic therapy improve Th17 frequencies, microbial translocation, and microbiome in ARV-treated, SIV-infected macaques. Mucosal Immunol. 2015 doi: 10.1038/mi.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hofer U, et al. Inadequate clearance of translocated bacterial products in HIV-infected humanized mice. PLoS Pathog. 2010;6:e1000867. doi: 10.1371/journal.ppat.1000867. [DOI] [PMC free article] [PubMed] [Google Scholar]