Abstract
Microbial eukaryotes encompass the majority of eukaryotic evolutionary and cytoskeletal diversity. The cytoskeletal complexity observed in multicellular organisms appears to be an expansion of components present in genomes of diverse microbial eukaryotes such as the basal lineage of flagellates, the Excavata. Excavate protists have complex and diverse cytoskeletal architectures and life cycles – essentially alternative cytoskeletal “landscapes” – yet still possess conserved microtubule- and actin-associated proteins. Comparative genomic analyses have revealed that a subset of excavates, however, lack many canonical actin-binding proteins central to actin cytoskeleton function in other eukaryotes. Overall, excavates possess numerous uncharacterized and “hypothetical” genes, and may represent an undiscovered reservoir of novel cytoskeletal genes and cytoskeletal mechanisms. The continued development of molecular genetic tools in these complex microbial eukaryotes will undoubtedly contribute to our overall understanding of cytoskeletal diversity and evolution.
Protists – not the more commonly studied multicellular eukaryotes such animals and plants – reflect the lion's share of eukaryotic genetic and cytoskeletal diversity [1,2]. These evolutionarily and morphologically diverse eukaryotes belong not to one specific evolutionary lineage, but instead are found in at least six main “supergroups”: Opisthokonts (animals, fungi, protists), Amoebozoa, Archaeplastida (e.g. plants and green algae), Rhizaria, Chromalveolata, and the Excavata [3-5]. Each of these supergroups contains microbial eukaryotes that have two morphological forms, amoeboid and flagellated, with conserved microtubule- and actin-associated proteins.
Because structural homologs of actin and tubulin are present in bacteria and archaea [6], it is clear that the cytoskeleton was present in the common ancestor of all cellular life. Elaborate cytoskeletal structures and dynamic regulation of the cytoskeleton arose during early eukaryotic evolution, however [7]. Comparisons of cytoskeletal structure and composition between diverse eukaryotes can thus inform our understanding of cytoskeletal evolution. While some functions of the cytoskeleton are conserved across eukaryotes in diverse supergroups (e.g., motility and mitosis), the molecular components and pathways underlying these mechanisms (particularly those involving the actin cytoskeleton) have extensive variation in less well studied eukaryotic supergroups like the Excavata [8].
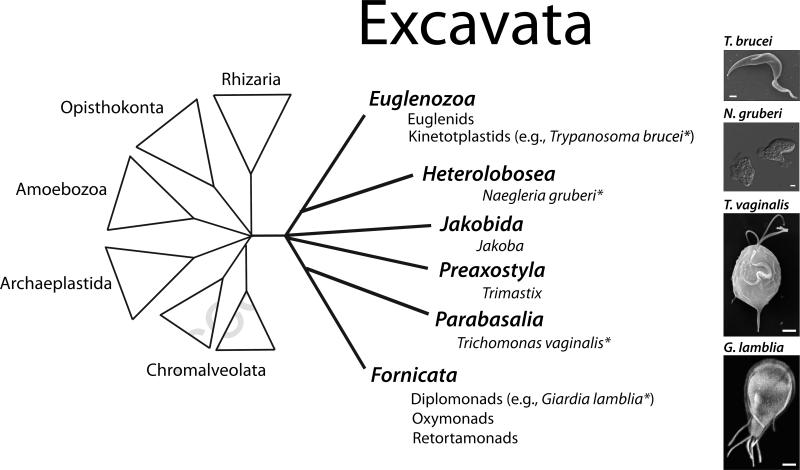
The Excavata is one of the primary evolutionary lineages of protists [3,9], and is proposed to be one of the deepest branching groups, closest to the common ancestor of all eukaryotes [10,11]. The evolutionary diversity within the Excavata represents genetic distances perhaps as large as those between plants and animals and fungi [3]. All excavates are flagellated or have flagellated stages, and, as a group, are defined by the presence of posteriorly directed flagella and flagellar root structures associated with the basal bodies [9]. Molecular phylogenetic support for this grouping is controversial, however [12]. Excavate biology is quite varied, and is represented by free-living, commensal, and common human parasitic forms of the following types of protists: Fornicata (diplomonads, oxymonads and retortamonads), Parabasalia, Euglenoza (both euglenids and kineoplastids), Heterolobosea, and the Jakobida and Preaxostyla. Based on their evolutionary distances and the complex composition of their diverse cytoskeletal structures, excavate protists may represent an undiscovered reservoir of novel cytoskeletal genes and cytoskeletal mechanisms.
Unique interphase cytoskeletal arrays in excavate protists
The broad evolutionary diversity of free-living and parasitic Excavata is reflected in the diversity of their interphase cytoskeletal structures – many of which are associated with flagellar axonemes [13-16]. These unique interphase arrays are used in a variety of cellular functions: feeding, motility, and/or attachment to surfaces. Novel excavate cytoskeletal arrays include: the paraflagellar rod and subpellicular corset microtubules in trypanosomes like Trypanosoma brucei [16-18], the flagellar-associated axostyle and costa [19] and stable microtubule ribbons of the pelta in parabasalids such as Trichomonas vaginalis [20,21], and the ventral disc, caudal complex, marginal plates in the diplomonad parasite Giardia lamblia (see Figure 1 and recent reviews [13-16]). Many excavate protists can use these dynamic cytoskeletal arrays to facilitate cytological transformations between complex multi-stage life cycles [22,23]. The members of one group of excavates – the heteroloboseans – transform from actin-rich amoeboid cells to microtubule-rich flagellates upon environmental stress (see Naegleria gruberi, Figure 1 and [7]). This is an indication that the eukaryotic common ancestor likely had well-articulated actin and tubulin-based cytoskeletons [7]. T vaginalis also has the ability to transform from a flagellate to an amoeboid form upon interaction with host epithelial cells, although the molecular details of this transformation are unknown [21].
Figure 1. Evolutionary relationships of the Excavata.
Diagram highlighting major groups within the Excavata [3-5] as well as major primary eukaryotic lineages: Opisthokonts (e.g., metazoans and fungi), Archaeplastida (e.g., plants), and groups composed primarily of protists (Amoebozoa, Chromalveolata, and Rhizaria). Representative images (right) of excavates and asterisks indicate those excavates with genomes discussed in this review. Scale bars = 1μm.
This remarkable cytoskeletal complexity sometimes masks the unifying cytoskeletal features of the flagellated excavates –a feeding groove and flagellar roots associated with the basal bodies [9]. It remains unclear whether the flagellar roots and various flagellar associated structures of other excavates (e.g. the axostyle in parabasalids and paraflagellar rod in trypanosomes) have a common evolutionary origin. As more excavates are discovered and characterized, we may determine whether the molecular compositions of flagellar roots and the feeding groove have been retained across diverse excavates, providing additional and convincing phylogenomic support for excavates as a coherent group. The lack of such support would imply that these cytoskeletal structures are convergent, rather than homologous.
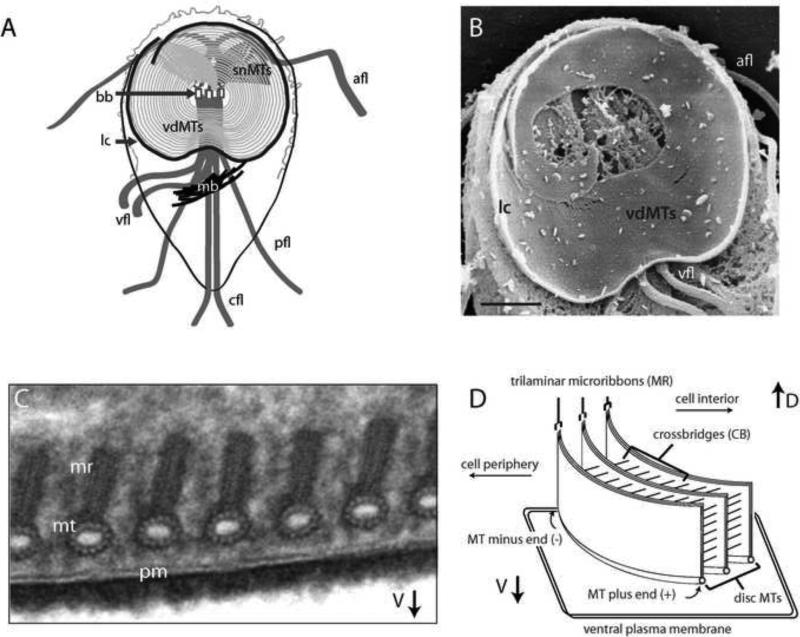
One example of a particularly elaborate and novel cytoskeletal organelle is the ventral disc in Giardia. While excavate trypanosomes, parabasalids, and some free-living diplomonads have retained the characteristic excavate feeding groove [24], the prevalent parasitic diplomonad Giardia lacks (and presumably has lost) this structure [24,25]. Giardia's cytology is instead dominated by the presence of a “suction-cup”-like microtubule organelle – the ventral disc – that enables strong extracellular attachment to the intestinal microvilli [8] to resist peristaltic flow. The ventral disc (Figure 2) is a left-handed spiral array of parallel microtubules and tightly associated trilaminar structures, or “microribbons”, connected by “crossbridge” structures [9]. Cryo-electron tomography of the ventral disc has revealed many novel protein complexes associated with the microtubules and the microribbons [26]. The composition of these densities remains unknown, but over thirty disc-associated proteins localize to the ventral disc or lateral crest (a fibrillar structure surrounding the ventral disc [27]). The disc likely evolved from the invention of novel cytoskeletal components, as most “disc-associated proteins”, or DAPs, lack any homology to known cytoskeletal proteins, or in fact to any protein, including proteins in other excavates.
Figure 2. Cytoskeletal complexity of the Giardia ventral disc.
Schematic of the Giardia microtubule cytoskeleton (A) indicating the median body (mb), the ventral disc (vdMTs) and supernumerary (snMTs) microtubule spiral arrays, the overlap zone (oz) of the vdMTs, the lateral crest (lc), the eight basal bodies (bb), and the four flagellar pairs – ventral flagella, vfl; caudal flagella, cfl; posteriolateral flagella, pfl; and anterior flagella, afl. Actin localizes primarily to the axonemes and the cell periphery. The detergent extracted cytoskeleton SEM (B) highlights the spiral vdMT array, and the lateral crest (lc) as viewed from the ventral side. Associated with the ventral plasma membrane (pm), the ventral disc microtubules (mt), trilaminar microribbons (mr) are seen in cross-section in the TEM (C), provided by Cindi Schwartz (CU-Boulder). Schematic representation of the ventral disc (D) shows the architecture of the primary elements of the ventral disc including the curved MTs with the associated microribbons (MR) and linked crossbridges (CB).
Another unifying feature of excavates is the presence of one or more motile flagella, and excavate flagella generally retain the canonical “(9+2)” architecture. Trypanosomes have one recurrent flagellum, Trichmonas has four flagella, and all diplomonad protists, including Giardia lamblia, have eight flagella. Diplomonad flagella differ from those of other flagellates, however, in that each axoneme has an extended, non-membrane bound, cytoplasmic portion as well as a membrane-bound portion outside the cell body (Figure 1 and [13]). Intraflagellar transport (IFT) is required for the assembly of membrane-bound regions, but the cytoplasmic regions may be assembled by IFT-independent mechanisms [28]. In addition to the ventral disc and the eight flagella, Giardia also has another semi-organized interphase microtubule array of unknown function – the median body.
Discovering novel proteins associated with the microtubule cytoskeleton in the Excavata
Comparative genomic analysis is a powerful approach to identify conserved cytoskeleton proteins and to explore the genetic basis of structural and functional variation in cytoskeletal structures. A wide phylogenetic span of eukaryotic genomes in comparative genomic analyses enables deep studies of cytoskeletal conservation and evolution. Several excavate genomes have now been sequenced (see Table 1), and the availability of these diverse genomes, combined with the development of molecular genetic tools is facilitating better evolutionary and functional analyses of the cytoskeleton in excavates [7,11,29,30]. The number of sequenced excavate genomes is still a small fraction of the total number of sequenced eukaryotic genomes, however. Recent advances in sequencing technology and computational tools provide a fantastic opportunity to link excavate genomic information with classic morphological descriptions, highlighting cytoskeletal adaptations to various excavate lifestyles, such as parasitism.
Table 1.
Cytoskeletal comparative genomic analysis of the Excavata
| Euglenozoa | Heterolobosea | Parabasalia | Fornicata | Fungi | |
|---|---|---|---|---|---|
| T. brucei | N. gruberi | T. vaginalis | G. lamblia | S. cerevisae | |
| Genome size (Mb) | 26.1 | 41.0 | 176.4 | 11.0 | 12.5 |
| Protein-encoding genes | 10533 | 15727 | 99009 | 5901 | 6607 |
| Hypothetical proteins | 5827 | 5411 | 9801 | 1466 | 846 (2504) |
| Conserved Hypothetical proteins | 4642 | 5938 | 76496 | 2370 | n.a. |
| GO Term-annotated proteins | 5928 | n.a. | 13172 | 1777 | 6383 |
| MT-associated | 79 | 143 | 122 | 75 | 168 |
| Tubulin gene family | 11 | 24 | 16 | 8 | 4 |
| Kinesin gene family | 43 | 40 | 18 | 24 | 6 |
| Dynein (DHC) gene family | 12 | 15 | 36 | 13 | 1 |
| Actin-Associated | 16 | 137 | 73 | 4 | 42 |
| Actin | 1 | 29 | 12 | 1 | 1 |
| Actin Related Proteins (ARPs) | 7 | 49 | 11 | 3 | 10 |
| Myosins (MHC) gene family | 2 | 11 | 0 | 0 | 5 |
Comparison of four representative excavate genomes (Trypanosoma brucei strain 927 [30]; Naegleria gruberi strain NEG [7]; Trichomonas vaginalis isolate G3 [29]; Giardia lamblia isolate WBC6 [11]) with baker's yeast (Saccharomyces cervisae [70,71]). The amount of genetic novelty is estimated based on gene annotation as represented by protein-encoding genes lacking orthologs in other eukaryotes (hypothetical genes) and genes with a lack of structural or functional information based on Gene Ontology (GO) annotations. In yeast, both hypothetical and “dubious” annotations are noted with parentheses. Roughly 30-50% of protein-encoding genes in each genome have little annotation. Also shown are the relative complements of microtubule-associated and actin-associated genes. Note: yeast microtubule-associated proteins are obtained via Gene Ontology assignments, whereas those of the excavates are obtained via comparative genomic analyses). Although all excavates have a full complement of microtubule-associated proteins (as well as expanded kinesin and dynein motor families), there is a large variation in the numbers and types of actin-associated genes in excavate genomes.
As with all eukaryotes, excavate protists have lost or gained conserved cytoskeletal genes and have retained cytoskeletal genes lost in other eukaryotic lineages. Comparative genomic analyses support that the majority of well-studied microtubule-associated cytoskeletal proteins occur in the excavates, including the structural components of flagella [31], kinesin and dynein microtubule motor protein families (Table 1 and [32,33]), and proteins involved in microtubule dynamics such as EB1, XMAP215, and katanin [7]. Generally, however, a large proportion of excavate genes lack homology to any gene in another eukaryotes (e.g. hypothetical genes); thus, there is still a need for more genomes and moleculr genetic tools in free-living excavates to ascertain the role of these novel proteins in excavate cytoskeletal structure and function (Table 1).
All lineages of eukaryotes are characterized by both expansions of specific cytoskeletal genes and gene families, as well as the loss of homologs of specific cytoskeletal genes and gene families (e.g., the loss of many kinesin families in yeasts [33]). The presence of a cytoskeletal gene family in multiple eukaryotic supergroups supports the idea that that cytoskeletal gene family was present in the common ancestor. Cytoskeletal gene families (e.g. kinesins, dyneins, and myosins) are conserved in most eukaryotes and are well represented in excavates, indicating these cytoskeletal gene families are ancient [32-34]. Myosins are lacking in Giardia and Trichomonas (see below), yet myosins are present some other excavates, however. Some novel types of cytoskeletal motors such as the kinesin-16 family may be ancestral, as they are conserved in diverse protists and not retained in fungal, metazoan or plant genomes [33]. Excavate protists also have novel types of dyneins and myosins [32,34], and the function of these novel classes of cytoskeletal genes remains understudied. Other novel motor gene families appear to have expanded in specific excavate lineages; for instance, both the diplomonads and Euglenozoa (e.g, euglenids and kinetoplastids) have expanded and novel kinesin and dynein family genes (Figure 1 and [32,33]). Members of the Euglenozoa and the Heterolobosea are proposed to be distantly related [24], and this relationship is reflected in the conservation of novel excavate-specific kinesin [7] and myosin subfamilies [34] in both groups. Homologs of the novel excavate kinesins and myosins might still be present in other eukaryotes that have not been sequenced, which would suggest that they have been lost in more well-studied eukaryotes.
Strong conservation of the protein components of a particular cellular structure (e.g. the ribosome [35]) across diverse organisms suggests a common evolutionary ancestry and evolutionary constraints on the function and assembly of that cellular structure. The eukaryotic flagellum is one such a cellular feature with conserved structure and conserved protein components [36-39] in diverse flagellated eukaryotes. Flagellated protists are found in the Excavata and every other eukaryotic supergroup, thus flagellar motility is undoubtedly an ancestral feature of eukaryotes [40]. The genome of the amoebo-flagellate excavate Naegleria gruberi retains an inventory of 173 flagellar structural genes including those required for basal body assembly, flagellar beating, and intraflagellar transport. In a comparative analysis of genomes primarily from free-living eukaryotes that included Naegleria and other excavates, 36 candidate flagella-associated genes were also identified [7]. In Giardia, genes encoding over eighty flagellar and basal body proteins that were previously identified through proteomic [37,38,41] and/or comparative genomic methods [42,43] are present in the genome [44]. Components of the BBSome and other basal body associated proteins (e.g., centrin, delta-tubulin and epsilon tubulin) are also present in excavates. Excavate homologs of flagellar genes involved in human ciliary-based genetic diseases have also been identified [45-47]. Thus comparative flagellar proteomics in diverse excavates could contribute to our overall understanding of flagellar structure and evolution in eukaryotes, and of the mechanisms of ciliary-based diseases.
Due to the conservation of flagellar structure, excavate flagellar biology informs general studies of eukaryotic flagellar structure and assembly. Investigations leading to one important discovery — how the kinesin-13 motor protein actively regulates flagellar length — were in fact initiated in excavate protists. The kinesin-13 family regulates microtubule dynamics in interphase and mitotic arrays, particularly the establishment of proper kinetochore-microtubule attachments and mitotic progression [48-50]. Recently kinesin-13 was reported to be present at the distal tips of the giardial axonemes [51]; this observation has been confirmed in trypanosomes [52] and in the green alga Chlamydmonas [53]. Active microtubule destabilization by kinesin-13 at the distal axonemal tip may promote microtubule disassembly and turnover of axonemal tubulin subunits. Because the ultrastructure as well as the structural and regulatory molecular components of flagellar axonemes are conserved in Giardia [28], trypanosomes and green algae [53], the active regulation of microtubule depolymerization at the flagellar tip by kinesin-13 appears to be a widespread and evolutionarily conserved mechanism important for flagellar length determination in many flagellates [52].
A dearth of proteins associated with the actin cytoskeleton in the Excavata
The actin cytoskeleton not only enables amoeboid motility but also cytokinesis, endocytosis, and the maintenance of cell morphology and polarity. Comparative genomic analyses have revealed that a subset of excavates — Giardia, Trichomonas, and Trypanosoma —lack many proteins [35] considered central to actin cytoskeleton function in other eukaryotes [11,17,29] including myosin and regulators of the arp2/3 complex. The profound lack of canonical actin binding proteins is most profound in Giardia as homologs of canonical actin binding proteins are completely absent [11]. Despite the lack of known actin binding proteins, actin still has a role in fundamental cellular processes in Giardia such as membrane trafficking, cytokinesis, and cellular morphogenesis [8]. Actin also has an often overlooked structural role in flagella [54], and has been observed in the flagella of both Giardia [8] and trypanosomes [55]. Have these “core regulators” been lost from these organisms or did excavate protists diverge from the other eukaryotes prior to the evolution of these proteins? Comparative genomics of more free-living excavates could help to resolve this question, and more functional studies of the actin cytoskeleton in excavates like Giardia could reveal either novel or ancient mechanisms of actin cytoskeleton regulation.
Actin-based amoeboid locomotion is present in diverse eukaryotic lineages, and a phylogenomic approach associated with the Naegleria genome analysis identified 63 gene families conserved only in eukaryotes capable of amoeboid locomotion. This inventory does not include proteins that contribute to non-motile functions such as canonical actin, Arp2/3 (which nucleates actin filaments) or other general actin components. Nineteen candidate genes of unknown function that are strongly implicated in actin-based motility were also identified.
What is the composition and the extent of the complexity of actin regulatory mechanisms in the excavates? With sequenced excavate genomes and comparative proteomic methods, one can define potentially novel actin-associated proteins in excavates. Studies of highly divergent but conserved actin-associated proteins from protistan non-model organisms have revealed unanticipated functions and are providing a new understanding of actin regulation. For example ADF of Toxoplasma and Plasmodium does not bind and sever F-actin but rather binds G-actin to both sequester and perform nucleotide exchange [56,57]. This is a fascinating case where a conserved actin-associated protein is performing an important but unanticipated function that could be more widespread in other eukaryotes.
Conserved cellular processes with atypical mechanisms in the excavates
From comparative analyses, many protists lack what are commonly considered to be core components of conserved cellular processes such as mitosis, cytokinesis, and flagellar assembly. To ensure viability, cells must be able to divide the contents of their cytoplasm such that daughter cells receive both genetic and structural components (flagella and other organelles). Protists exhibit a diversity of mitoses with both internal and extranuclear spindles [58], and it remains unclear as to how conserved hallmark spindle-associated structures such as the kinetochore are in these organisms. A lack of conservation of such core cellular mechanisms could indicate a lack of constraints on these processes, and indicate that more variation in the mechanisms of mitosis and cytokinesis is tolerable to protistan cells.
Whether any excavate uses an actin-myosin contractile ring mechanism for cytokinesis remains unclear, as Trichomonas and the diplomonads lack myosin [11,29]. The actin-myosin contractile ring could be a more recent adaption in eukaryotes, however, as plants have myosin, but do not employ a contractile mechanism for cytokinesis. Constriction and abscission also appear as distinct cytokinetic stages in eukaryotes using an actin-myosin ring. Further cytokinesis can occur in the absence of myosin II, the motor that drives contraction, under certain conditions [59,60]. The phragmoplast – an interdigitated microtubule array in plants – is an alternative to the purse string mechanism that delivers cell plate forming vesicles precisely between daughter cells. The spindle remnant known as the midbody in animals has some proteins in common with the phragmoplast [61], and appears to have analogous functions in cytokinesis.
Have excavates evolved novel mechanism of cytokinesis, or have they retained and expanded an underlying mechanism found in all eukaryotes? Although there are several conserved midbody components in the excavates, the well-studied members (Giardia, Trichomonas and the Euglenozoa) all divide in a way that challenges the concept of a “midbody”. Mitosis and cytokinesis are uncoupled in the excavates as the spindle is dismantled before cytokinesis begins, thus there are no interdigitating microtubules to be found during cell scission. Rather it is thought that flagellar beating plays an important role in driving the final scission event [62]. A common feature of the excavate cleavage furrow is that it progresses from the anterior to the posterior and bisects cells on the anterior-posterior axis [63,64]. How the cleavage furrow is specified and subsequently driven toward the anterior remains unresolved. Despite this, the mechanism of cytokinesis in excavates likely involves membrane trafficking and the actin cytoskeleton [8] [65] [66]. Studying cytokinesis in eukaryotes such as excavates that lack a contractile ring and midbody/phragmoplast will lead to greater understanding of what underlies those prominent cytoskeletal arrays and perhaps which ancient cytokinetic mechanism that predates the divergence of plants and animals.
Future perspectives
Are the novel excavate cytoskeletal structures built from novel cytoskeletal proteins that are not present in other organisms, or do these structures demonstrate that there are alternative ways to build the cytoskeleton using conserved components? As molecular tools including affinity and localization tags are available for many excavate protists, we can expect the identification of novel cytoskeletal proteins. Our identification of over 20 novel proteins associated with the Giardia ventral disc [27] raises the possibility that the ventral disc evolved by the invention of novel cytoskeletal proteins, rather than the modification of an existing structure. Thus, there is a high likelihood of discovering novel cytoskeletal-associated genes, as well as uncovering novel functions of existing genes or expansions of existing gene families in diverse protists like excavates. The development of more functional genomic and reverse genetic approaches in more free-living excavates should provide an excellent opportunity to understand the composition, assembly and variation of cytoskeletal structures in these diverse eukaryotes across deep evolutionary time.
Comparative genomic and comparative cell biological analyses of excavate protists suffer from a lack of molecular genetic and genomic characterizations of free-living types; only one complete excavate genome is from a free-living protist, that of the heterolobosean Naegleria gruberi [7]. As is evident in the bacterial and archaeal domains, however, most microbes are free-living, and many phyla-level lineages of protists remain to be discovered [67-69]. Thus, a rich source of cytoskeletal gene novelty lies in the analysis of uncultivated microbial eukaryotic metagenomic data. With more environmental metagenomic and metatranscriptomic studies of microbial eukaryotes, we can expect to see environmental data from uncultured eukaryotes contributing to our understanding of cytoskeletal gene functional diversity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Patterson DJ. The diversity of eukaryotes. American Naturalist. 1999;154:S96–S124. doi: 10.1086/303287. [DOI] [PubMed] [Google Scholar]
- 2.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 3**.Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AG, Roger AJ. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci U S A. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [Phylogenomic classification of the diversity of eukaryotes supporting the monophyly of the Excavata using multiple concatenated protein-encoding genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Adl SM, Leander BS, Simpson AG, Archibald JM, Anderson OR, Bass D, Bowser SS, Brugerolle G, Farmer MA, Karpov S, et al. Diversity, nomenclature, and taxonomy of protists. Systematic biology. 2007;56:684–689. doi: 10.1080/10635150701494127. [Proposal to standardize eukaryotic taxonomy and develop future nomenclature guidelines to avoid confusion.] [DOI] [PubMed] [Google Scholar]
- 5.Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 6*.Ettema TJ, Lindas AC, Bernander R. An actin-based cytoskeleton in archaea. Molecular Microbiology. 2011;80:1052–1061. doi: 10.1111/j.1365-2958.2011.07635.x. [Discovery that the archaea have an actin homolog that is more similar to eukaryotic actin than to HSP70.] [DOI] [PubMed] [Google Scholar]
- 7**.Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [Comprehensive genomic analysis of primarily free-living eukaryotes with the excavate amoeboflagellate Naegleria. This study identified novel flagellar-associated and amoeboid motility genes in diverse eukaryotes.] [DOI] [PubMed] [Google Scholar]
- 8**.Paredez AR, Assaf ZJ, Sept D, Timofejeva L, Dawson SC, Wang CJ, Cande WZ. An actin cytoskeleton with evolutionarily conserved functions in the absence of canonical actin-binding proteins. Proc Natl Acad Sci U S A. 2011;108:6151–6156. doi: 10.1073/pnas.1018593108. [Functional and structural analysis of actin in Giardia using a Giardia-specific actin antibody. Giardia is a dipomonad protist lacking canonical actin-binding proteins.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Simpson AGB, Patterson DJ. The ultrastructure of Carpediemonas membranifera (Eukaryota) with reference to the “excavate hypothesis”. European Journal of Protistology. 1999;35:353–370. [Proposal to develop a morphological basis to group protists based on flagellar root and additonal cytoskeletal structures.] [Google Scholar]
- 10.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 11.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 12.Parfrey LW, Grant J, Tekle YI, Lasek-Nesselquist E, Morrison HG, Sogin ML, Patterson DJ, Katz LA. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol. 2010;59:518–533. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawson SC, House SA. Life with eight flagella: flagellar assembly and division in Giardia. Curr Opin Microbiol. 2010;13:480–490. doi: 10.1016/j.mib.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KE, Kim JH, Jung MK, Arii T, Ryu JS, Han SS. Three-dimensional structure of the cytoskeleton in Trichomonas vaginalis revealed new features. Journal of electron microscopy. 2009;58:305–313. doi: 10.1093/jmicro/dfp019. [DOI] [PubMed] [Google Scholar]
- 15.Fiori PL, Rappelli P, Addis MF. The flagellated parasite Trichomonas vaginalis: new insights into cytopathogenicity mechanisms. Microbes and infection / Institut Pasteur. 1999;1:149–156. doi: 10.1016/s1286-4579(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 16*.Hoog JL, Bouchet-Marquis C, McIntosh JR, Hoenger A, Gull K. Cryo-electron tomography and 3-D analysis of the intact flagellum in Trypanosoma brucei. Journal of structural biology. 2012;178:189–198. doi: 10.1016/j.jsb.2012.01.009. [Fine structural detail on the complex T. brucei cytoskeleton indicating novel components.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniels JP, Gull K, Wickstead B. Cell biology of the trypanosome genome. Microbiology and molecular biology reviews : MMBR. 2010;74:552–569. doi: 10.1128/MMBR.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohl L, Gull K. Molecular architecture of the trypanosome cytoskeleton. Molecular and Biochemical Parasitology. 1998;93:1–9. doi: 10.1016/s0166-6851(98)00014-0. [DOI] [PubMed] [Google Scholar]
- 19.Honigberg BM, Vickerman K, Kulda J, Brugerolle G. Cytology and Taxonomy of Parasitic Flagellates. 1981:P205–227. [Google Scholar]
- 20.Benchimol M. Trichomonads under Microscopy. Microscopy and microanalysis : the official journal of Microscopy Society of America. Microbeam Analysis Society, Microscopical Society of Canada. 2004;10:528–550. doi: 10.1017/S1431927604040905. [DOI] [PubMed] [Google Scholar]
- 21.Ryan CM, de Miguel N, Johnson PJ. Trichomonas vaginalis: current understanding of host-parasite interactions. Essays in biochemistry. 2011;51:161–175. doi: 10.1042/bse0510161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adam RD. Biology of Giardia lamblia. Clin. Microbiol. Rev. 2001;14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fulton C, Dingle AD. Basal bodies, but not centrioles, in Naegleria. J Cell Biol. 1971;51:826–836. doi: 10.1083/jcb.51.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson AG. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int J Syst Evol Microbiol. 2003;53:1759–1777. doi: 10.1099/ijs.0.02578-0. [DOI] [PubMed] [Google Scholar]
- 25.Simpson AGB, Perley TA, Lara E. Lateral transfer of the gene for a widely used marker, alpha-tubulin, indicated by a multi-protein study of the phylogenetic position of Andalucia (Excavata). Molecular Phylogenetics and Evolution. 2008;47:366–377. doi: 10.1016/j.ympev.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 26*.Schwartz CL, Heumann JM, Dawson SC, Hoenger A. A Detailed, Hierarchical Study of Giardia lamblia's Ventral Disc Reveals Novel Microtubule-Associated Protein Complexes. PLoS One. 2012;7:e43783. doi: 10.1371/journal.pone.0043783. [Cryotomography study indicating novel supramolecular complexes associated with the microtubule spiral of the Giardia ventral disc.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Hagen KD, Hirakawa MP, House SA, Schwartz CL, Pham JK, Cipriano MJ, De La Torre MJ, Sek A,C, Du G, Forsythe BM, Dawson SC. Novel structural components of the ventral disc and lateral crest in Giardia intestinalis. PLoS NTD. 2011;5:e1442. doi: 10.1371/journal.pntd.0001442. [Identification of over 20 novel cytosketal proteins associated with the Giardia ventral disc.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Hoeng JC, Dawson SC, House SA, Sagolla MS, Pham JK, Mancuso JJ, Lowe J, Cande WZ. High-resolution crystal structure and in vivo function of a kinesin-2 homologue in Giardia intestinalis. Mol Biol Cell. 2008;19:3124–3137. doi: 10.1091/mbc.E07-11-1156. [Analysis of kinesin-2 function in Giardia indicating that intraflagellar transport is not likely required for assembly of cytoplasmic regions of axonemes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 31.Wickstead B, Gull K. The evolution of the cytoskeleton. The Journal of cell biology. 2011;194:513–525. doi: 10.1083/jcb.201102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickstead B, Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic. 2007;8:1708–1721. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Wickstead B, Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol Biol Cell. 2006;17:1734–1743. doi: 10.1091/mbc.E05-11-1090. [Identification of novel kinesin family conserved in protists but not present in plants, animals and fungi.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci U S A. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosometRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 36.Dutcher SK. Flagellar assembly in two hundred and fifty easy-to-follow steps. Trends Genet. 1995;11:398–404. doi: 10.1016/s0168-9525(00)89123-4. [DOI] [PubMed] [Google Scholar]
- 37.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- 39.Luck DJ. Genetic and biochemical dissection of the eucaryotic flagellum. J Cell Biol. 1984;98:789–794. doi: 10.1083/jcb.98.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 41.Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr., Culver BP, Yates JR, 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 43.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dawson SC. An insider's guide to the microtubule cytoskeleton of Giardia. Cell Microbiol. 2010;12:588–598. doi: 10.1111/j.1462-5822.2010.01458.x. [DOI] [PubMed] [Google Scholar]
- 45.Davy BE, Robinson ML. Congenital hydrocephalus in hy3 mice is caused by a frameshift mutation in Hydin, a large novel gene. Hum Mol Genet. 2003;12:1163–1170. doi: 10.1093/hmg/ddg122. [DOI] [PubMed] [Google Scholar]
- 46.Pfannenschmid F, Wimmer VC, Rios RM, Geimer S, Krockel U, Leiherer A, Haller K, Nemcova Y, Mages W. Chlamydomonas DIP13 and human NA14: a new class of proteins associated with microtubule structures is involved in cell division. J Cell Sci. 2003;116:1449–1462. doi: 10.1242/jcs.00337. [DOI] [PubMed] [Google Scholar]
- 47.Keller LC, Romijn EP, Zamora I, Yates JR, 3rd, Marshall WF. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 48.Mennella V, Rogers GC, Rogers SL, Buster DW, Vale RD, Sharp DJ. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat Cell Biol. 2005;7:235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- 49.Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- 50.Kline-Smith SL, Walczak CE. The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol Biol Cell. 2002;13:2718–2731. doi: 10.1091/mbc.E01-12-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dawson SC, Sagolla MS, Mancuso JJ, Woessner DJ, House SA, Fritz-Laylin L, Cande WZ. Kinesin-13 regulates flagellar, interphase, and mitotic microtubule dynamics in Giardia intestinalis. Eukaryot Cell. 2007;6:2354–2364. doi: 10.1128/EC.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blaineau C, Tessier M, Dubessay P, Tasse L, Crobu L, Pages M, Bastien P. A novel microtubule-depolymerizing kinesin involved in length control of a eukaryotic flagellum. Curr Biol. 2007;17:778–782. doi: 10.1016/j.cub.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 53.Piao T, Luo M, Wang L, Guo Y, Li D, Li P, Snell WJ, Pan J. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4713–4718. doi: 10.1073/pnas.0808671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato-Minoura T, Hirono M, Kamiya R. Chlamydomonas inner-arm dynein mutant, ida5, has a mutation in an actin-encoding gene. The Journal of cell biology. 1997;137:649–656. doi: 10.1083/jcb.137.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sahasrabuddhe AA, Bajpai VK, Gupta CM. A novel form of actin in Leishmania: molecular characterisation, subcellular localisation and association with subpellicular microtubules. Molecular and Biochemical Parasitology. 2004;134:105–114. doi: 10.1016/j.molbiopara.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 56.Schuler H, Mueller AK, Matuschewski K. A Plasmodium actin-depolymerizing factor that binds exclusively to actin monomers. Molecular Biology of the Cell. 2005;16:4013–4023. doi: 10.1091/mbc.E05-02-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mehta S, Sibley LD. Toxoplasma gondii actin depolymerizing factor acts primarily to sequester G-actin. The Journal of biological chemistry. 2010;285:6835–6847. doi: 10.1074/jbc.M109.068155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raikov IB. The diversity of forms of mitosis in protozoa: A comparative review. European Journal of Protistology. 1994;30:252–269. [Google Scholar]
- 59.Kanada M, Nagasaki A, Uyeda TQ. Adhesion-dependent and contractile ring-independent equatorial furrowing during cytokinesis in mammalian cells. Mol Biol Cell. 2005;16:3865–3872. doi: 10.1091/mbc.E05-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uyeda TQ, Kitayama C, Yumura S. Myosin II-independent cytokinesis in Dictyostelium: its mechanism and implications. Cell Structure and Function. 2000;25:1–10. doi: 10.1247/csf.25.1. [DOI] [PubMed] [Google Scholar]
- 61.Eme L, Moreira D, Talla E, Brochier-Armanet C. A complex cell division machinery was present in the last common ancestor of eukaryotes. PLoS One. 2009;4:e5021. doi: 10.1371/journal.pone.0005021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ralston KS, Lerner AG, Diener DR, Hill KL. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot Cell. 2006;5:696–711. doi: 10.1128/EC.5.4.696-711.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sagolla MS, Dawson SC, Mancuso JJ, Cande WZ. Three-dimensional analysis of mitosis and cytokinesis in the binucleate parasite Giardia intestinalis. J Cell Sci. 2006;119:4889–4900. doi: 10.1242/jcs.03276. [DOI] [PubMed] [Google Scholar]
- 64.Ribeiro KC, Coutinho L, Pereira AN, Benchimol M. Understanding the extranuclear spindle of the closed mitosis in trichomonads. Memorias do Instituto Oswaldo Cruz. 2000;95:105–106. [Google Scholar]
- 65.Barquilla A, Crespo JL, Navarro M. Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14579–14584. doi: 10.1073/pnas.0802668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garcia-Salcedo JA, Perez-Morga D, Gijon P, Dilbeck V, Pays E, Nolan DP. A differential role for actin during the life cycle of Trypanosoma brucei. The EMBO journal. 2004;23:780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moon-van der Staay SY, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- 68.Stoeck T, Hayward B, Taylor GT, Varela R, Epstein SS. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist. 2006;157:31–43. doi: 10.1016/j.protis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 69.Dawson SC, Pace NR. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc Natl Acad Sci U S A. 2002;99:8324–8329. doi: 10.1073/pnas.062169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Issel-Tarver L, Christie KR, Dolinski K, Andrada R, Balakrishnan R, Ball CA, Binkley G, Dong S, Dwight SS, Fisk DG, et al. Saccharomyces Genome Database. Methods in enzymology. 2002;350:329–346. doi: 10.1016/s0076-6879(02)50972-1. [DOI] [PubMed] [Google Scholar]
- 71.Dwight SS, Harris MA, Dolinski K, Ball CA, Binkley G, Christie KR, Fisk DG, Issel-Tarver L, Schroeder M, Sherlock G, et al. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic acids research. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]