Genetic deletion of one microtubule-binding domain in CENP-F results in multiple cellular changes, including changes in cell migration, adhesion, and cilia formation. All of these changes are related to an increase in microtubule stabilization in CENP-F mutants. This suggests that CENP-F plays a role in microtubule organization outside of mitosis.
Abstract
Microtubule (MT)-binding centromere protein F (CENP-F) was previously shown to play a role exclusively in chromosome segregation during cellular division. Many cell models of CENP-F depletion show a lag in the cell cycle and aneuploidy. Here, using our novel genetic deletion model, we show that CENP-F also regulates a broader range of cellular functions outside of cell division. We characterized CENP-F+/+ and CENP-F–/– mouse embryonic fibroblasts (MEFs) and found drastic differences in multiple cellular functions during interphase, including cell migration, focal adhesion dynamics, and primary cilia formation. We discovered that CENP-F–/– MEFs have severely diminished MT dynamics, which underlies the phenotypes we describe. These data, combined with recent biochemical research demonstrating the strong binding of CENP-F to the MT network, support the conclusion that CENP-F is a powerful regulator of MT dynamics during interphase and affects heterogeneous cell functions.
INTRODUCTION
As might be expected from its multifaceted domain structure and its complex and dynamic localization, centromere protein F (CENP-F) function at the cellular level is diverse. Previous in vitro studies showed that loss of CENP-F or dominant-negative CENP-F expression results in mitotic delay and misaligned chromosomes (Liao et al., 1995; Chan et al., 1998; Hussein and Taylor, 2002; Holt et al., 2005; Yang et al., 2005). Of interest, CENP-F was shown to have a number of functions during cellular interphase. CENP-F interacts with Hook2 at the centrosome, influencing cell shape (Moynihan et al., 2009), with soluble N-ethylmaleimide–sensitive factor attachment protein receptor proteins regulating the vesicular transport pathway (Pooley et al., 2006, 2008), and plays an important role in interphase, regulating primary cilium length (Jodoin et al., 2013). Recent findings show that CENP-F is required for redistribution of mitochondria (Kanfer et al., 2015). At the organismal level, loss of function leads to seemingly diverse neurological and muscular diseases (Khairallah et al., 2012; Holth et al., 2013; Waters et al., 2015; Filges et al., 2016). Human patients with CENP-F mutations present with ciliopathies and microcephaly, and CENP-F has been localized to the basal body of the cilia (Waters et al., 2015). Cardiac-specific loss of CENP-F in the mouse embryo results in adult-onset dilated cardiomyopathy and death (Dees et al., 2012). CENP-F overexpression has been used as a proliferation marker of various cancers (Clark et al., 1997; Rattner et al., 1997; Liu et al., 1998; Erlanson et al., 1999; Hui et al., 2005). Autoantibodies against CENP-F are hallmarks of specific cancers (Rattner et al., 1997; Tschernatsch et al., 2005), and CENP-F gene amplification is observed in squamous cell carcinomas (de la Guardia et al., 2001). Thus CENP-F function has a broad and varied effect at the level of organelles, cells, organs, and the organism itself.
Although multiple laboratories have conclusively demonstrated that CENP-F has wide-ranging control of diverse functions, the critical effect of CENP-F loss of function on its most fundamental relationship—that with the microtubule (MT) network—has not been resolved. Determining the role of CENP-F in regulation of the MT network is important for a mechanistic understanding of protein function in the diverse downstream events seen with loss and gain of gene function in development and disease.
Here we develop a novel genetic cell model to explore the role of this protein in regulation of the MT network and basic cell functions. Our data show that mutation of the CENP-F gene leads to an unexpected hyperstabilization of the MT network, with a unique loss of dynamic instability. With disruption of MT dynamics, CENP-F−/− cells exhibit dramatic loss of directionally persistent migration, defects in focal adhesion disassembly and lamellipodial formation/retraction, change in cilia frequency, and loss of regulation of cell shape. Of interest, changes in mitotic activity and development of aneuploidy were not observed. Taking the results together, examining this specific genetic variation provides a molecular mechanism for mutation of CENP-F function and the foundation for analysis and intervention in various developmental and pathological abnormalities seen with disruption of this complex gene product.
RESULTS AND DISCUSSION
Mutation of CENP-F in this model does not alter cell division rate or result in aneuploidy
Our CENP-F−/− mice were generated by Cre recombination of floxed exons 1–5 of CENP-F (Dees et al., 2012). It is therefore possible that exons 6–18 are still expressed and form a functional protein. To address this point, we first conducted serial PCR down the length of the CENP-F mRNA. We found that mRNA was still produced in CENP-F−/− mouse embryonic fibroblasts (MEFs) for exons 9–18 (Supplemental Figure S1A). Deletion of the floxed region of CENP-F was confirmed with real-time (RT) PCR using primers against CENP-F mRNA, as well as slot blotting with an anti–CENP-F mouse monoclonal antibody, ELDA6 (Supplemental Figure S1, A–D). To determine whether any CENP-F protein is generated from this truncated RNA, we completed immunofluorescence and Western blot analysis with antibodies generated against the domains other than the N-terminus of CENP-F (Supplemental Figure S1, E–G). Immunofluorescence with anti–CENP-F antibody ab5 demonstrates that there is protein in the cytoplasm of CENP-F+/+ MEFs that is absent in CENP-F−/− MEFs (Supplemental Figure S1, E and F). The ab5 was raised against the C-terminal portion of CENP-F. These data therefore suggest that no protein is made from the truncated mRNA. Western blot analysis of MEF lysates further supported this finding (Supplemental Figure S1G). Blotting with D10, another anti–CENP-F antibody specific to the C-terminus, reveals a clear band in CENP-F+/+ lysates. No band is visible, however in the entire CENP-F−/− lysate lane (Supplemental Figure S1G). We therefore conclude that no CENP-F protein is produced in our genetic knockout model.
Previous work reported that RNA interference (RNAi) knockdown of CENP-F results in mitotic delay and aneuploidy (Bomont et al., 2005; Holt et al., 2005; Yang et al., 2005; Feng et al., 2006). Therefore we first wanted to determine whether the present model of loss of CENP-F gene function altered the rate of cell division in MEFs. After following cell proliferation rates for 144 h, we found that the CENP-F+/+ MEFs had a doubling time of 57 h, whereas CENP-F−/− MEFs doubled in 59 h, suggesting that the rates of division are statistically unchanged with mutation of CENP-F (p = 0.06; Supplemental Figure S1H). Another predicted consequence of loss of CENP-F in MEFs is aneuploidy because CENP-F tethers chromosomes to kinetochore MTs (Bomont et al., 2005; Holt et al., 2005). To test this potential outcome, we conducted fluorescence-activated cell sorting on CENP-F+/+ and CENP-F−/− MEFs. These data demonstrated that there was no significant difference between CENP-F+/+ and CENP-F−/− MEF populations in the proportion of cells distributed across the cell cycle or in polyploidy (Supplemental Figure S1I). There were, however, statistically significant differences in the proportion of CENP-F−/− cells in subG0, G1, and G2 (Supplemental Figure S1I). Finally, we were able to visualize individual MEFs dividing to demonstrate successful chromosomal segregation in CENP-F−/− MEFs (Supplemental Figure S1, J–K′). Therefore, although cell division rate and polyploidy are unchanged with this specific model of loss of CENP-F function, there are changes within the cell cycle. Thus mutation of CENP-F did not result in cessation of the cell cycle or changes in DNA content, allowing us to further examine whether loss of CENP-F effects interphase cells.
This is the first study of nontransformed primary cells from a model of genetic loss of CENP-F. Therefore the differences we see with the loss of CENP-F could be attributed to a number of variables, including genetic versus RNAi deletion of CENP-F, primary versus transformed cell lines, and complete versus incomplete deletion of protein. Although we demonstrated the complete loss of CENP-F, other differences among these models are important avenues for future investigation.
CENP-F–/– MEFs exhibit multiple altered cellular characteristics in interphase
In comparing CENP-F+/+ and CENP-F−/− MEFs, we immediately recognized numerous and distinct differences related to interphase cellular activities. We chose to focus on specific aspects of cell biology: focal adhesions, cell migration, primary cilia formation, and microtubule dynamics. These cell functions play critical roles in development, disease, and cancer. Prior loss of CENP-F phenotypes suggests that these key aspects of cell biology may be altered. Therefore it was pertinent to investigate these systems in our genetic loss-of-function MEF model.
Cell migration
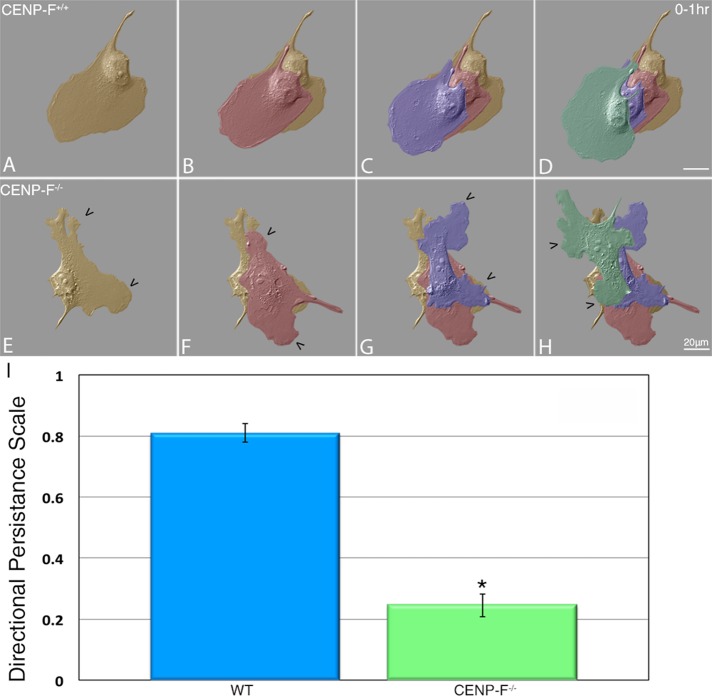
Dynamicity of MTs at the leading edge of migrating cells regulates directionally persistent migration (Petrie et al., 2009). If CENP-F plays a role in microtubule dynamics, then we would expect to see a difference in cell migration with loss of CENP-F. Live-cell differential interference contrast (DIC) microscopy clearly demonstrated that CENP-F+/+ cells exhibited directionally persistent migration over the 1-h time span tested (Figure 1, A–D). In contrast, CENP-F−/− MEFs were unable to maintain migration in a sustained direction (Figure 1, E–H), with quantification of directional persistence parameters showing a significant difference between CENP-F+/+ (0.81) and CENP-F−/− (0.24) MEFs (p < 0.001; Figure 1G). In addition, generation and retention of lamellipodial extensions varied greatly in these cells. Whereas lamellipodia were established along the axis of migration in CENP-F+/+ cells, they appeared randomly and transiently around the circumference of CENP-F−/− cells (Supplemental Movie S1).
FIGURE 1:
CENP-F−/− MEFs lack directionally persistent migration. Migration patterns of (A–D) CENP-F+/+ and (E–H) CENP-F−/− acquired by DIC microscopy over a period of 1 h. False-colored overlays of cells. Orange, time zero; red, 20 min; purple, 40 min; green, 60 min. CENP-F−/− MEFs fail to establish directionally persistent migration patterns as quantified in I. A value of 1.0 corresponds to movement in a straight line, and 0 equals random movement. CENP-F+/+ = 0.81 and CENP-F−/− = 0.24 (n = 20; *p < 0.001; error bars represent SEM).
Focal adhesion dynamics
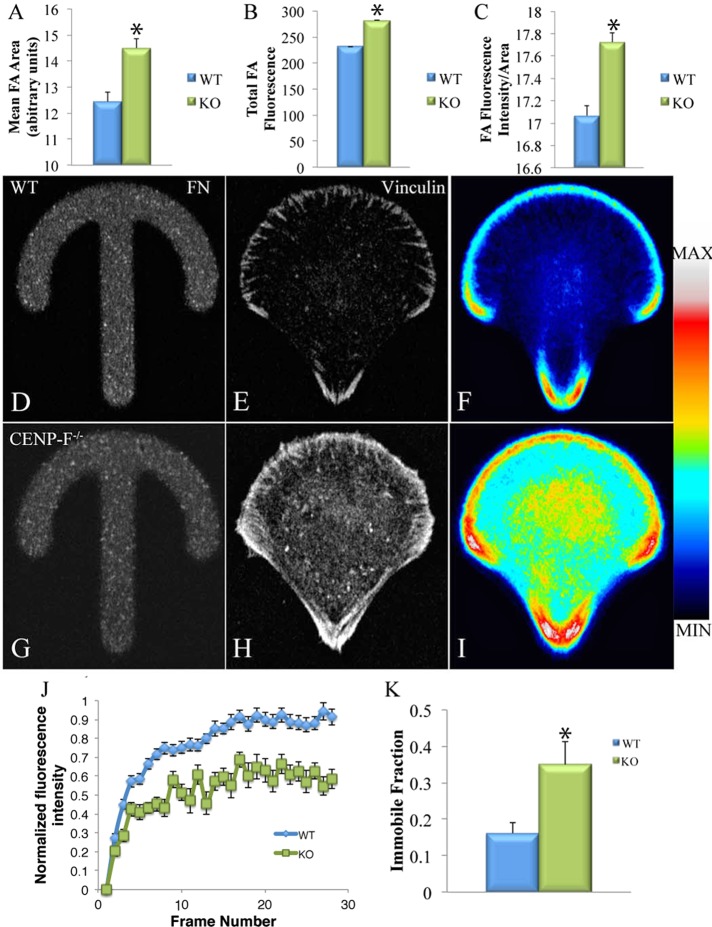
Directional migration is dependent in part on the ability of the cell to turn over integrin-based focal adhesions (FAs; Sastry and Burridge, 2000; Webb et al., 2002). Of interest, MTs and FAs are intimately related, with MT dynamics playing an integral role in FA disassembly (Kaverina et al., 1999; Small et al., 2002; Ezratty et al., 2005) and directional migration (Kaverina and Straube, 2011). We therefore predicted that loss of CENP-F would result in altered FA dynamics. To explore this concept, we stained CENP-F+/+ and CENP-F−/− MEFs for vinculin, a FA component, and imaged with confocal microscopy. We analyzed >3700 FAs in CENP-F+/+ and CENP-F−/− cells for area, mean fluorescence, and total fluorescence with ImageJ, using the Mann–Whitney U test for statistical significance. FAs in CENP-F−/− MEFs were significantly larger (p = 0.001) and had slightly higher mean fluorescence (p < 5.1E-05) and higher total fluorescence (p < 5.6E-05) (Figure 2, A–C).
FIGURE 2:
FAs of CENP-F−/− MEFs are larger and denser and disassemble more slowly than CENP-F+/+ MEFs. Immunofluorescence using α-vinculin antibodies visualizing FAs in CENP-F+/+ and CENP-F−/− MEFs. Quantification of (A) mean size (p = 0.001), (B) total fluorescence intensity (p < 5.1E-05), and (C) FA fluorescence intensity/area (p < 5.6E-05; n > 1500). Immunofluorescence using vinculin antibodies in MEFs plated on fibronectin crossbows. (D, G) Fluorescent fibronectin crossbows. (E, H) Vinculin staining in representative CENP-F+/+ and CENP-F−/− cells. When fluorescence of many cells is averaged, CENP-F−/− cells show greater FA and cytoplasmic vinculin fluorescence (D, I). n = 78 CENP-F+/+, n = 69 CENP-F−/−. (J) Fluorescence recovery curve from CENP-F+/+ and CENP-F−/− MEFs. Error bars represent SEM. (K) Quantification of immobile fraction from fluorescence recovery curve (p = 0.05; n = 10).
To compare FAs more systematically, we used a reference cell approach. On plating of cells on a standardized fibronectin pattern, they take on identical shapes and can be compared using average fluorescence across many cells (Thery et al., 2006). The crossbow pattern has been used to simulate a polarized, migrating cell pattern (Theisen et al., 2012). MEFs were plated on a standardized crossbow pattern (Figure 2, D and G), fixed, stained with vinculin, and imaged (Figure 2, E and H). Approximately 70 CENP-F+/+ and CENP-F−/− cells were stacked and a visual average created (Figure 2, F and I). Based on this approach, it is clear that CENP-F−/− MEFs have a much higher average fluorescence in the distribution of FAs (Figure 2, F and I). Of interest, CENP-F−/− MEFs on average had increased diffuse vinculin staining throughout the cell, as shown by greater cytoplasmic fluorescence (Figure 2, H and I). These data suggest that although focal adhesions still form in the appropriate cellular locations, CENP-F−/− MEFs have significantly more vinculin.
To determine whether differences in FA size and density resulted from changes in FA dynamics, we used fluorescence recovery after photobleaching (FRAP) to calculate the dynamics of an FA-associated protein, Paxillin. CENP-F+/+ and CENP-F−/- MEFs were transfected with mCherry-Paxillin and imaged before and after a photobleaching event (Supplemental Movie S2). The fluorescence intensity of 30 FAs was measured in 10 cells for each condition. The resulting fluorescence recovery curve revealed that fluorescence of FAs in CENP-F−/− MEFs recovered to only ∼60%, whereas CENP-F+/+ MEFs recovered to 91% (Figure 2J). This difference in fluorescence recovery could be due to differences in either the rate of diffusion or the immobile fraction in the CENP-F+/+ and CENP-F−/− MEFs. The half-time of the recovery curves between CENP-F+/+ and CENP-F−/− were not statistically different, suggesting that the rate of diffusion of Paxillin is not different between MEFs. However, there was a significant difference in the immobile fraction, representing the proportion of bleached protein that fails to be turned over. The immobile fraction of the CENP-F−/− MEFs was twice that of the CENP-F+/+ MEFs (Figure 2K). Taken together, these data demonstrate altered focal adhesion dynamics in CENP-F−/− cells.
Ciliogenesis
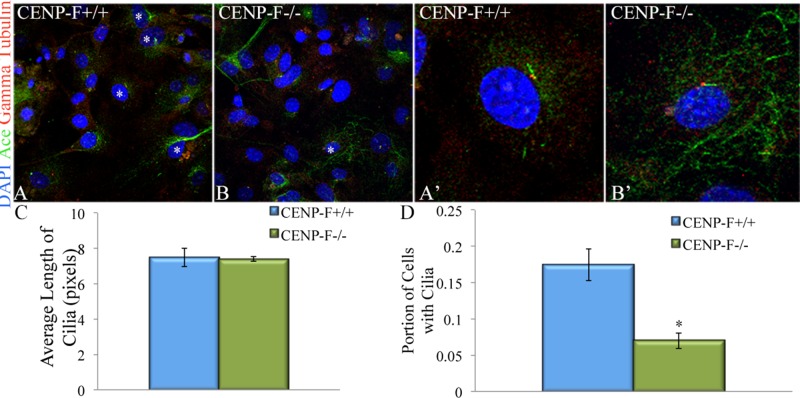
Although numerous studies have focused on CENP-F function in mitosis, a relationship between CENP-F and cilia during interphase has been identified. CENP-F has been found localized to the base of the cilium (Waters et al., 2015). Of importance, loss of CENP-F results in aberrant ciliary phenotypes (Jodoin et al., 2013; Waters et al., 2015; Filges et al., 2016). On the basis of this information, we looked closely at the primary cilia of CENP-F+/+ and CENP-F−/− MEFs. After culturing MEFs in serum starvation conditions, we were able to visualize the primary cilia with immunofluorescence labeling of acetylated tubulin and γ-tubulin (Figure 3, A–B′). Although we did not see a difference in the length of the primary cilia (Figure 3C), there was a significant decrease in cilia frequency in CENP-F−/− MEFs (Figure 3, A and B). On average, 17% of CENP-F+/+ MEFs had a primary cilium, compared with only ∼7% of CENP-F−/− MEFs (Figure 3D). These data suggest that with genetic loss of CENP-F before cilia formation, ciliogenesis fails to occur in a significant number of MEFs.
FIGURE 3:
Decrease in cilia formation in CENP-F−/− MEFs. (A, B) Fluorescence micrographs of MEFs after serum starvation to induce ciliogenesis. Cilia are labeled with antibodies raised against acetylated tubulin (green) and γ-tubulin (red). Nuclei are stained with DAPI. Images represent z-projections. Ciliated cells are labeled with an asterisk. (A′, B′) Increased magnification of ciliated MEFs. Cilia are the same length in both cells but occur less frequently in CENP-F−/− MEFs. (C) Quantification of cilia length in MEFs (p = 0.91; n = 25; error bars represent SEM). (D) Quantification of portion of ciliated cells (p < 5.0E-05; n = 30; error bars represent SEM).
MT dynamics
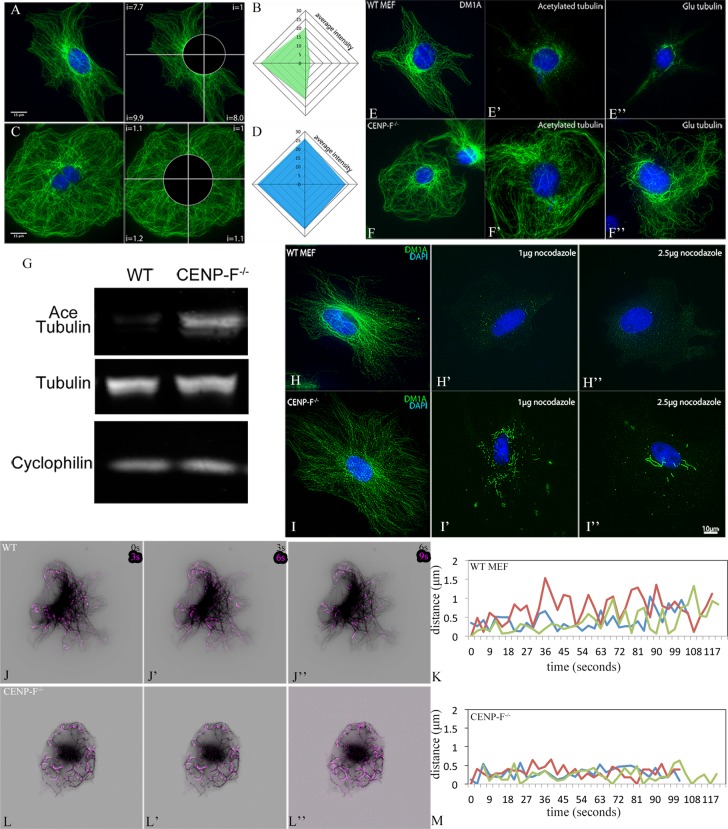
With all of the evidence suggesting a change in the MT network, we next examined the distribution of MTs in MEFs. Whereas CENP-F+/+ MEFs exhibited a typically asymmetric distribution of tubulin across the cell, CENP-F−/− cells lacked this asymmetric array (Figure 4; visualized with DM1A, an antibody against α-tubulin). Quantification of fluorescence intensity confirmed the lack of asymmetric distribution of MTs in the CENP-F−/− cells (Figure 4, B and D). In addition, we noted that CENP-F−/− MEFs have long, coiling MTs near the cell periphery, which are absent in their wild-type counterparts (Figure 4, A and C).
FIGURE 4:
CENP-F is required for establishment of MT array asymmetry, and loss of CENP-F leads to an abundance of posttranslational modifications and resistance to depolymerization by NOC. Immunostaining of MTs with DM1A antibody (green) and DAPI (blue) in CENP-F+/+ (A) and CENP-F−/− (C) mouse embryonic fibroblasts. Scale bar, 15 μm. Schematic for fluorescence intensity quantification is shown on the right in A and C. Representative fluorescence intensity plots (30, each) of CENP-F+/+ (B) and CENP-F−/− (D) for each sample. Immunostaining of MTs (green) in wild-type MEFs (E–E′′) shows minimal amounts of acetylated (E′) and Glu (E′′) tubulin in relation to the typical staining of the complete MT network (E). In contrast, the MT network of CENP-F−/− MEFs (F) contains an abundance of posttranslational modifications, as indicated by immense acetylated (F′) and Glu (F′′) tubulin staining. Scale bar, 10 μm. (G) Western blot demonstrating the abundance of acetylated tubulin and tubulin in CENP-F+/+ and CENP-F−/− MEFs. Immunofluorescence analysis of the MT network (green) in CENP-F+/+ (H–H′′) and CENP-F−/− (I–I′′) MEFs treated for 2 h with no NOC (H, I), 1 μg/ml NOC (H′, I′), or 2.5 μg/ml NOC (H′′, I′′). Numerous short MTs persist in CENP-F−/- MEFs (I′, I′′), as compared with full depolymerization of MTs in CENP-F+/+ MEFs (H′, H′′). n = 25. Scale bar, 10 μm. Time-lapse imaging of CENP-F+/+ (J–J′′) and CENP-F−/- (L–L′′) MEFs expressing 3xEGFP-EMTB (black) to highlight the MT network. Images showing dynamic pixel differences were created by projecting pixel differences (purple) between sequential movie frames. CENP-F−/− MTs are very stable and move as an entire unit, as opposed to dynamic instability, exclusively at the tips of CENP-F+/+ MTs. Life history plots of MT dynamic instability can be observed in CENP-F+/+ MEFs (K) with periods of growth and shrinkage at the MT tips. In contrast, CENP-F−/- MEFs (M) experience very few periods of growth and catastrophe with long periods of pause.
Given that loss of CENP-F causes major changes in a number of interphase functions dependent on microtubule dynamics, we postulated that altered MT dynamics might underlie this phenotype. Although CENP-F has not been associated with regulation of MT posttranslational modifications, the overall structure of the MT network in CENP-F−/− cells, especially with “swirling” MT extensions at the cell periphery, is suggestive of MT hyperstabilization (Liu et al., 2005a, b). Posttranslational modifications, namely acetylation and proteolytic removal of the C-terminal tyrosine and glutamate residues, are hallmarks of long-lived, stable MTs (Webster et al., 1987; Infante et al., 2000). Acetylated and detyrosinated MTs are typically found in the perinuclear region, where the oldest MTs reside (Webster and Borisy, 1989; Li et al., 1996). Indeed, this concentration of modified tubulin is observed in the perinuclear region of the CENP-F+/+ MEFs (Figure 4, E′ and E′′). In sharp contrast, the entire length of the MT network, from perinuclear domain to cell periphery, was enriched in these posttranslational modifications of tubulin in CENP-F−/− MEFs (Figure 4, F′ and F′′). Western blotting demonstrated that whereas CENP-F+/+ and CENP-F−/− MEFs contain the same amount of tubulin, CENP-F−/− MEFs were enriched in acetylated tubulin (Figure 4G). This shows that CENP-F−/− MEFs contain an abundance of modified MTs compared with CENP-F+/+ MEFs.
To determine the relative stability of MTs in CENP-F+/+ and CENP-F−/− MEFs, we treated cells with varying concentrations of nocodazole (NOC) and assayed them with immunofluorescence for the presence of polymerized MTs. After a 2-h treatment with 1 μg/ml NOC, we observed a paucity of MTs in CENP-F+/+ cells, with only short remnants persisting in the perinuclear domain (Figure 4H′). Complete MT depolymerization was observed at 2.5 μg/ml NOC (Figure 4H′′). Conversely, the MT network of CENP-F−/− MEFs remained partially intact after 2 h of 1 and even 2.5 μg/ml NOC, with intact but shorter segments in the perinuclear area (Figure 4, I′ and I′′), demonstrating the stability of these MTs.
CENP-F–/– MTs have impaired dynamics
Hyperstabilization of the MT network most often results from reduction in MT dynamics, visualized as stunting of growth and regression at MT plus ends (Gardner et al., 2012). As shown here, loss of CENP-F function leads to hyperstabilization of the MT network. Alteration of MT dynamics would explain the phenotypes we have described.
To explore whether changes in MT dynamics occur with loss of CENP-F function, we imaged green fluorescent protein (GFP)–EMTB-transfected CENP-F+/+ and CENP-F−/− cells in real time. In agreement with previous reports in several cell systems (Altinok et al., 2007; Uchida et al., 2010; Garrison et al., 2012), CENP-F+/+ MEFs exhibited typical MT dynamics, with the majority of change occurring at the MT tips over time (Figure 4, J–J′′, and Supplemental Movie S3). Of interest, the MT network of CENP-F−/− MEFs demonstrated much greater movement than in CENP-F+/+ cells but in a completely different manner. In these cells, movement was greatest along the length of the MT and not associated the peripheral tips (Figure 4, L–L′′, and Supplemental Movie S3). MTs in CENP-F−/− MEFs exhibited a drastic increase in MT pause duration, with a subsequent decrease in the average growth and shrink rates (Table 1 and Figure 4, K and M). These data demonstrate the severe effect that loss of CENP-F has on the dynamics of the MT network.
TABLE 1:
MT dynamics is severely paused in CENP-F−/− MEFs.
| Dynamic parameter | WT (n = 15) | CENP-F–/– (n = 15) |
|---|---|---|
| Growth rate (μm/s) | 0.2796 ± 0.025 | 0.15165 ± 0.00374* |
| Shrink rate (μm/s) | 0.2714 ± 0.008 | 0.18591 ± 0.0191*** |
| Average pause duration (s) | 12.242 ± 1.09757 | 43.2889 ± 2.5005** |
Quantification of MT dynamics from MEFs transfected with 3xGFP-EMTB. Three individual microtubules were measured in 15 cells in each condition. n = 15. *p < 0.01, **p < 0.0005, ***p < 0.0002. Error = SEM.
Using this novel CENP-F model with the genetic loss of function, we were able to assay the function of CENP-F at the single-cell level. We found multiple disruptions in a variety of interphase activities; however, we did not find alteration in mitosis. The unifying feature behind all of the phenotypes we described, as well as in those in the literature, is the altered MT network. We are therefore presenting the first cell-biological evidence that CENP-F has a function tied to the destabilization of the MT network. With the identification of the effect of CENP-F on these novel yet basic cellular functions, we can now drill down to the molecular level and determine the mechanisms through which CENP-F functions in specific cellular processes in future studies.
Of importance, previous studies analyzing loss of CENP-F function clearly demonstrated delays in mitotic activity, misalignment of chromosomes, and multiple changes in functions related to the cytoplasm (Liao et al., 1995; Ashe et al., 2004; Holt et al., 2005; Soukoulis et al., 2005; Yang et al., 2005; Pooley et al., 2006, 2008; Varis et al., 2006; Vergnolle and Taylor, 2007; Moynihan et al., 2009), yet an underlying or unifying mechanism of disruption remains elusive. Here the dramatic changes in the overall structure of the cell and particularly in the MT network in interphase suggest broad and pervasive effects that influence all of these properties. Taken together, our data establish that loss of CENP-F function leads to increased presence of modified tubulin and hyperstabilization of the MT network. Given the broad importance of the MT network within cells, CENP-F has the potential to affect multiple cellular behaviors through its actions on the MT network.
The present data clearly demonstrate that loss of CENP-F results in major alterations in the interphase MT network. Knowing the key role of MTs in cell migration (Petrie et al., 2009), we postulated that CENP-F−/− cells would have migration defects. Our studies strongly support the hypothesis that loss of CENP-F function results in loss of directed cell migration. Directed migration is critical in development (Wei et al., 1996; Goodwin et al., 1999; Dees et al., 2012) and disease (Dees et al., 2012), and revealing a potential role for CENP-F in directed movement might bring insight into such events. Of interest, loss of CENP-F function in the embryonic heart leads to defects in trabeculation (Dees et al., 2012). Trabeculation—the inward growth of the ventricular wall—is dependent on myocyte migration through the burgeoning connective tissue space. Finally, CENP-F is a prognostic indicator for several cancers (Rattner et al., 1997; Tschernatsch et al., 2005; Dai et al., 2013; Aytes et al., 2014; Zhuo et al., 2015). Knockdown of CENP-F has been linked to decreases in cancer cell migration and invasion in vitro (Nishikawa et al., 2015). Thus understanding the essential role of CENP-F in regulation of directed cell movement through MT dynamics affects myriad issues in broad areas of embryogenesis, tissue repair, and cancer.
MTs affect cell migration not only directly through stabilization of the leading edge (Petrie et al., 2009) but also indirectly through their role in FA disassembly. Specifically, MTs go through lengthening and shortening cycles in order to target FAs (Kaverina et al., 1998). Based on our findings of MT stabilization, it is likely that altered FA disassembly could also play a role in impairing cell migration. Our findings of larger FAs and increased FA fluorescence in CENP-F−/− cells are consistent with the hypothesis that the altered MT dynamics in CENP-F−/− cells and the decrease in MT turnover would decrease the ability of a cell to break down FAs, a vital step in directional migration. Paxillin-transfected CENP-F−/− cells analyzed with FRAP had an immobile fraction twice that of CENP-F+/+ cells, suggesting that the difference in the fluorescence recovery curves is due to the increased immobility of Paxillin in the FAs of CENP-F−/− MEFs. These data support our hypothesis that hyperstabilization of MTs with loss of CENP-F disrupts interphase cellular functions.
Given our data and recent findings on the biochemical function of CENP-F, we propose that CENP-F acts to destabilize MTs. Multiple groups have found that CENP-F binds MTs with both carboxy- and amino-terminal MT-binding domains (Musinipally et al., 2013; Volkov et al., 2015). Of importance, CENP-F has even been shown to bind MTs as they are shrinking (Volkov et al., 2015). These studies, taken with our findings, strongly suggest that CENP-F plays a critical role in destabilizing microtubules. Future studies can now focus on determining the specific molecular mechanism by which CENP-F regulates MT dynamics in specific interphase cellular functions.
Implications
Disruption of CENP-F function leads to developmental abnormalities and disease outcomes in diverse cell types and models. Work from several groups has clearly demonstrated that CENP-F is critical in multiple processes influencing mitosis and interphase function. One common feature in all of these outcomes is the influence of CENP-F on the MT network in regulating these diverse functions. Here our data reveal a novel in vitro cellular function, namely that CENP-F is a powerful destabilizing element of the MT network in interphase. Discovery of these diverse CENP-F–dependent interphase traits has broad implications for the established roles of CENP-F in regulation of mitosis, chromosome pairing, migration, intracellular transport, and cell shape in development, cancer, and disease.
MATERIALS AND METHODS
Mouse lines
Mice were previously described (Moynihan et al., 2009). CENP-Ffl/−; CMV-CRE animals (The Jackson Laboratory, Bar Harbor, ME) were crossed to generate CENP-F−/−; CMV-CRE animals. CENP-F+/+; CMV-CRE animals were used to generate control MEFs. Dams were monitored daily for the presence of a vaginal plug, with noon on the day of the plug noted as 0.5 dpc. Mice were maintained in accordance with protocols approved by the Vanderbilt University Institutional Animal Care and Use Committee.
MEF isolation
Primary MEFs were isolated from embyronic day 13.5 embryos using standard procedures. MEFs were isolated as described in Moynihan et al. (2009). MEFs were used within three passages to avoid replicative senescence. Four independent yet interrelated methods were used to establish loss of N-terminal CENP-F DNA, RNA, and protein sequences. First, PCR analysis of genomic structure clearly demonstrated the predicted loss of CENP-F gene sequences (Dees et al., 2012; unpublished data). Next, quantitative RT-PCR of CENP-F+/+ and CENP-F−/− MEF RNA showed complete loss of these sequences within the totality of mRNA expressed in knockout MEFs (Supplemental Figure S1A). Finally, slot- (Supplemental Figure S1, B and C) and Western (Supplemental Figure S1G) blotting analyses of these MEFs synergistically demonstrated the loss of N-terminal protein structure in knockout cells. Taken together, these data conclusively demonstrate loss of DNA/RNA/protein structure in this cellular model test of CENP-F function.
Genotyping
PCR genotyping was based on three features to distinguish wild-type from Cre-mediated CENP-F exon 1–5 deletion: presence of the 5′ loxP site upstream of CENP-F, generation of a DNA band only possible with recombination after Cre-mediated excision, and the presence of the cTNT-Cre gene. The following primers were used:
Across the 5′ loxP site: 5′-AATAATGAAGCTGACACCAAAAACT,
3′-GAACCTACCGTCTGAGAACCACTG.
Recombination band: 5′-AATAATGAAGCTGACACCAAAAACT,
3′-GAGGAGCACAGGAGGGAAATG.
CMV-Cre: 5′-TCCGGGCTGCCACGACCAA,
3′-GGCGCGGCAACACCATTTTT.
PCR validation of CENP-F mRNA expression was completed with a series of primers specific to different exons of CENP-F. Total RNA extraction was conducted with TRIzol and with the use of a Qiagen (Hilden, Germany) RNeasy Mini Kit (50) for MEFs for the extraction. The RNA concentration was obtained by using an Eppendorf Biophotometer. cDNA was synthesized from the total RNA using an Applied Biosystems (Foster City, CA) 200 reaction High Capacity cDNA Reverse Transcription Kit. RT-PCR was used as the manufacturer instructed. The 25-μl reaction system contained 8.5 μl of double-distilled H2O, 7.5 μl of MgCl2, 5 μl of 5× buffer, 0.5 μl of dNTP mixtures, 0.5 μl of Taq DNA polymerase, 0.5 μl of both primers beginning at the 3′ and 5′ ends of the selected sequence, and 2 μl f cDNA template. Two thermal cycling protocols were followed: one for primers A, C, D, and F–H, and a second protocol for primers B and E. The following primary thermal cycling protocol was used: 95°C, 4 min; 16 cycles of 95°C for 30 s, 65°C decreasing by 0.5°C every cycle for 30 s, 74°C for 1 min, followed by 15 cycles of 94°C for 30 s, 59°C for 30 s, 74°C for 1 min, and a final extension cycle of 74°C for 5 min. Secondarily, the thermal cycling protocol for primers B and E proceeded as follows: 95°C for 4 min, 16 cycles of 95°C for 30 s, 63°C decreasing by 0.5°C every cycle for 30 s, 74°C for 1 min, followed by 15 cycles of 95°C for 30 s, 58°C for 30 s, 74°C for 1 min, and a final extension cycle of 74°C for 5 min. After thermal cycling was completed, an agarose gel was used to visualize the presence or absence of bands.
The RT-PCR primers were as follows:
Primer A, exons 1–3: forward, AGTTTGAATCGCTCGTGCTG; reverse, CGCTTAAGTTCCTGTTCCAGTT.
Primer B, exons 2–5: forward, GCTCAGTTTACTTCTAAATGCTTTA; reverse, TTTTGCATGCAAAGCTTTAA.
Primer C, exons 6–8: forward, AAAGTGAGCCTGCCTGTTTC; reverse, CTCTTGAAGCTCCTTTTCTTTCTC.
Primer D, exons 9–11: forward, GAGCTGTCCCGACAGCATC; reverse, CCTTCATTTCTTCTGCAAAATTCT.
Primer E, exon 12: forward, CTAAGAATACCTCTCAGGAAATC; reverse, CTTATTTGCTTTCAAGCACA.
Primer F, exon 13: forward, GTTGTTCGGTGCCAAAACGAT; reverse, GGTTACATGGAAGGGATTCCAACT.
Primer G, exons 14–16: forward, GTTAGCACAATATCTGGGAAGGAA; reverse, AACTCTGGAGATCCTGTCTGGAC.
Primer H, exons 17 and 18: forward, GGTTTGCTGACATCCCAACTG; reverse, TACATATGGGAGACCTGAGCACC.
Western and slot blotting
Slot blotting was conducted according to Moynihan et al. (2009) using appropriate Odyssey secondary antibodies (Licor, Lincoln, NE) and imaging (Pierce).
For Western blot, pelleted CENP-F+/+ and CENP-F−/− MEFs were solubilized in Laemmli sample buffer in the presence of protease inhibitors. Equivalent amounts of extracts were run on 3–8% SDS gels using standard methodologies (Lim et al., 2004) and transferred to nitrocellulose paper in the presence of 20% methanol, 10 mM β-mercaptoethanol, and 0.1% SDS. Blots were washed with phosphate-buffered saline (PBS) and blocked with 5% bovine serum albumin (BSA) in PBS for 1 h, followed by a 1-h incubation with anti–CENP-F antibody D10 (kindly provided by Tim Yen, Fox Chase Cancer Center, Philadelphia, PA) diluted 1:3000 in 5% BSA/PBS. Blots were washed in PBS with 0.1% Triton X-100, followed by reaction with goat anti-mouse (IRDye 800CW; Li-Cor) and similar washing for imaging on an Odyssey system. Controls for loading equivalence were conducted under similar conditions with anti-tubulin antibody DM1A (Abcam, Cambridge, UK).
Flow sorting
MEFs were fixed in 70% ethanol in a single-cell suspension of 500,000 cells in 500 μl, washed in PBS, and stained with propidium iodide/Triton X-100 solution with RNase at 4°C overnight. Samples were then analyzed for nuclear content at the Vanderbilt University Flow Cytometry Core. Replicates were run in triplicate.
Quantification of proliferation ((3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay)
For the quantification of cell proliferation, we used the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI). CENP-F+/+ and CENP-F−/− cells were plated at a density of 5000 cells per 200 μl of medium in three 96-well plates. Cells were incubated at 37°C overnight. At 24, 96, and 144 h, one 96-well plate was removed from the incubator, and 20 μl of CellTiter Reagent was added and incubated for 2 h. Absorbance was read at 490 nm using a plate reader. Data were analyzed against a standard curve to determine the rate of cell proliferation.
Imaging
To investigate migration movies, cells were plated on 1 μg/ml fibronectin on MatTek dishes, and DIC images were acquired at 1-min intervals on a DeltaVision system (Applied Precision) at the Vanderbilt University Epithelial Biology core. These movies were analyzed for distance and velocity using ImageJ Cell Tracker software. Directional persistence was then calculated by dividing the total displacement by the distance traveled and is presented on a scale of 0–1, where 0 is random movement and 1 is movement in a straight line (Benesh et al., 2013).
MT dynamics movies frames were acquired every 3 s for 2 min on the DeltaVision system at 100× magnification. CENP-F+/+ and CENPF−/− cells were transfected with 0.5 μg of 3xGFP-EMTB (plasmid 26741; Addgene) using FuGENE 6 (Promega) according to standard protocols. Images showing dynamic movie pixels were created by projecting pixel differences between movie frames using an ImageJ macro available at rsb.info.nih.gov/ij/macros/Slice-to-Slice%20Difference.txt.
For quantification of FAs, fixed MEFs on glass coverslips were blocked in 5% goat serum and 1% BSA for 1 h at room temperature. FAs were stained with α-vinculin antibody (1:200) for 1 h at room temperature and with anti-mouse immunoglobulin G 488 nm (1:500) and TOPRO (1:1000) for 45 min at room temperature. Cells were imaged on Zeiss LSM 510 Meta inverted confocal microscope in the Vanderbilt Cell Imaging Shared Resource Core at 40×. Projections were created from 0.3-μm slices through the full thickness of FAs. In ImageJ, background fluorescence and diffuse perinuclear labeling were removed using the threshold function. FAs were selected and analyzed using the Analyze Particle function. FAs were measured for total and average fluorescence in arbitrary units and area in pixels for both CENP-F+/+ and CENP-F−/− MEFs and were statistically analyzed with the Mann–Whitney U test.
In reference cell migration assays, cells were plated and fixed on fibronectin crossbow micropatterns (CYTOO, Grenoble, France) and stained with vinculin antibody as described. Single images were taken as described. Cells that completely occupied micropatterns were stacked in ImageJ, and a reference cell was created for CENP-F+/+ and CENP-F−/− MEFs by averaging vinculin fluorescence in each stack and applying an ImageJ LUT.
All statistics were completed with a Student’s t test with a 95% confidence interval unless otherwise specified.
FRAP
Cells were transfected with mCherry-Paxillin using FuGENE 6 at 48 h before imaging. Frames were acquired every 10 s for 10 min. A region of interest was set in a region containing FAs. A 1-s, 405-nm laser event was scheduled on the third frame. Movies were generated on the DeltaVision system at 40× magnification. Fluorescence intensity in bleached and nonbleached FAs was measured using ImageJ software. Bleached areas were normalized to nonbleached areas both prebleach and postbleach in order to obtain fluorescence recovery values. Data represent the fraction of fluorescence recovery in the bleached region. Immobile fraction, fi, was calculated from fi = 1 − f(f)/f(p), where f(f) is final fluorescence intensity and f(p) is prebleach fluorescence.
Cilia quantification
Cilia growth was established by culturing MEFs for 48 h in medium containing 0.5% FBS (Plotnikova et al., 2009). Cilia number and length were quantified with ImageJ. Cilia frequency was determined by dividing the number of cells that contain a primary cilium by total number of cells.
Nocodazole treatment
NOC was diluted into dimethyl sulfoxide. Cells were treated with 0, 1, and 2.5 μg/ml for 2 h before analysis (Moynihan et al., 2009).
Quantification of MT array asymmetry
MT array asymmetry was quantified as previously described (Miller et al., 2009). Representative images are shown.
Antibodies
Cells were fixed for 10 min at room temperature in 2% paraformaldehyde with 0.25% Triton X-100 before antibody labeling:
DM1A: mouse monoclonal (ab7291; Abcam).
Acetylated tubulin: mouse monoclonal (ab24610; Abcam).
Anti-tubulin detyrosinated (Glu): rabbit polyclonal (AB3201; Millipore, Billerica, MA).
Cyclophilin B (ab16045; Abcam).
γ-Tubulin (ab16504; Abcam).
Vinculin (ab18058; Abcam).
Alexa Fluor 488–phalloidin (A12379; Invitrogen, Carlsbad, CA).
Anti–CENP-F (ab5; Abcam): rabbit polyclonal antibody raised against the carboxy terminus of CENP-F.
Anti-CENP-F antibody D10 (from Tim J. Yen). The epitope for this antibody is within the C-terminal domain of CENP-F (Liao et al., 1995).
ELDA6: new mouse monoclonal antibody generated and characterized in our laboratory. The epitope for this antibody resides within amino acids 134–187 in the N-terminal domain of murine CENP-F (E. Pfaltzgraff, E. Mace, J. Schultz, K. Compton, Z. Shancer, and D. Bader, unpublished data).
DAPI or TOPRO was used to visualize DNA.
Supplementary Material
Acknowledgments
We thank the members of the Bader laboratory for many constructive and insightful discussions concerning this work. We also thank Joseph Roland and the Vanderbilt Epithelial Biology Center for assistance with DeltaVision imaging and Sean Schaffer of the Vanderbilt Cell Imaging Shared Resource for assistance with quantification and imaging of focal adhesions. We thank Tim J. Yen for the generous gift of the anti-CENP-F antibody. This work is supported by American Heart Association Grant 12PRE10950005 to E.R.P. and National Institutes of Health Grants NHLBI 5T32HL007751 to P.M.M., R01GM086610 to R.O., and R01 HL037675 to D.M.B. R.O. is a scholar of the Leukemia and Lymphoma Society.
Abbreviations used:
- CENP-F
centromere protein F
- DIC microscopy
differential interference microscopy
- FA
focal adhesion
- FRAP
fluorescence recovery after photobleaching
- MEF
mouse embryonic fibroblast
- MT
microtubule
- RNAi
RNA interference.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-12-0848) on May 4, 2016.
REFERENCES
- Altinok A, Kiris E, Peck AJ, Feinstein SC, Wilson L, Manjunath BS, Rose K. Model based dynamics analysis in live cell microtubule images. BMC Cell Biol. 2007;8(Suppl 1):S4. doi: 10.1186/1471-2121-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M, Pabon-Pena L, Dees E, Price KL, Bader D. LEK1 is a potential inhibitor of pocket protein-mediated cellular processes. J Biol Chem. 2004;279:664–676. doi: 10.1074/jbc.M308810200. [DOI] [PubMed] [Google Scholar]
- Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A, Pienta KJ, Shen MM, et al. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesh EC, Miller PM, Pfaltzgraff ER, Grega-Larson NE, Hager HA, Sung BH, Qu X, Baldwin HS, Weaver AM, Bader DM. Bves and NDRG4 regulate directional epicardial cell migration through autocrine extracellular matrix deposition. Mol Biol Cell. 2013;24:3496–3510. doi: 10.1091/mbc.E12-07-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomont P, Maddox P, Shah JV, Desai AB, Cleveland DW. Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F. EMBO J. 2005;24:3927–3939. doi: 10.1038/sj.emboj.7600848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GM, Allred DC, Hilsenbeck SG, Chamness GC, Osborne CK, Jones D, Lee WH. Mitosin (a new proliferation marker) correlates with clinical outcome in node-negative breast cancer. Cancer Res. 1997;57:5505–5508. [PubMed] [Google Scholar]
- Dai L, Lei N, Liu M, Zhang JY. Autoantibodies to tumor-associated antigens as biomarkers in human hepatocellular carcinoma (HCC) Exp Hematol Oncol. 2013;2:15. doi: 10.1186/2162-3619-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees E, Miller PM, Moynihan KL, Pooley RD, Hunt RP, Galindo CL, Rottman JN, Bader DM. Cardiac-specific deletion of the microtubule-binding protein CENP-F causes dilated cardiomyopathy. Dis Model Mech. 2012;5:468–480. doi: 10.1242/dmm.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Guardia C, Casiano CA, Trinidad-Pinedo J, Baez A. CENP-F gene amplification and overexpression in head and neck squamous cell carcinomas. Head Neck. 2001;23:104–112. doi: 10.1002/1097-0347(200102)23:2<104::aid-hed1005>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Erlanson M, Casiano CA, Tan EM, Lindh J, Roos G, Landberg G. Immunohistochemical analysis of the proliferation associated nuclear antigen CENP-F in non-Hodgkin’s lymphoma. Mod Pathol. 1999;12:69–74. [PubMed] [Google Scholar]
- Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- Feng J, Huang H, Yen TJ. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma. 2006;115:320–329. doi: 10.1007/s00412-006-0049-5. [DOI] [PubMed] [Google Scholar]
- Filges I, Bruder E, Brandal K, Meier S, Undlien DE, Waage TR, Hoesli I, Schubach M, de Beer T, Sheng Y, et al. Stromme syndrome is a ciliary disorder caused by mutations in CENPF. Hum Mutat. 2016;37:359–363. doi: 10.1002/humu.22960. [DOI] [PubMed] [Google Scholar]
- Gardner MK, Zanic M, Howard J. Microtubule catastrophe and rescue. Curr Opin Cell Biol. 2012;25:14–22. doi: 10.1016/j.ceb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison AK, Shanmugam M, Leung HC, Xia C, Wang Z, Ma L. Visualization and analysis of microtubule dynamics using dual color-coded display of plus-end labels. PLoS One. 2012;7:e50421. doi: 10.1371/journal.pone.0050421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin RL, Pabon-Pena LM, Foster GC, Bader D. The cloning and analysis of LEK1 identifies variations in the LEK/centromere protein F/mitosin gene family. J Biol Chem. 1999;274:18597–18604. doi: 10.1074/jbc.274.26.18597. [DOI] [PubMed] [Google Scholar]
- Holt SV, Vergnolle MA, Hussein D, Wozniak MJ, Allan VJ, Taylor SS. Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. J Cell Sci. 2005;118:4889–4900. doi: 10.1242/jcs.02614. [DOI] [PubMed] [Google Scholar]
- Holth JK, Bomben VC, Reed JG, Inoue T, Younkin L, Younkin SG, Pautler RG, Botas J, Noebels JL. Tau loss attenuates neuronal network hyperexcitability in mouse and Drosophila genetic models of epilepsy. J Neurosci. 2013;33:1651–1659. doi: 10.1523/JNEUROSCI.3191-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D, Reiman T, Hanson J, Linford R, Wong W, Belch A, Lai R. Immunohistochemical detection of cdc2 is useful in predicting survival in patients with mantle cell lymphoma. Mod Pathol. 2005;18:1223–1231. doi: 10.1038/modpathol.3800409. [DOI] [PubMed] [Google Scholar]
- Hussein D, Taylor SS. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J Cell Sci. 2002;115:3403–3414. doi: 10.1242/jcs.115.17.3403. [DOI] [PubMed] [Google Scholar]
- Infante AS, Stein MS, Zhai Y, Borisy GG, Gundersen GG. Detyrosinated (Glu) microtubules are stabilized by an ATP-sensitive plus-end cap. J Cell Sci. 2000;113:3907–3919. doi: 10.1242/jcs.113.22.3907. [DOI] [PubMed] [Google Scholar]
- Jodoin JN, Shboul M, Albrecht TR, Lee E, Wagner EJ, Reversade B, Lee LA. The snRNA-processing complex, Integrator, is required for ciliogenesis and dynein recruitment to the nuclear envelope via distinct mechanisms. Biol Open. 2013;2:1390–1396. doi: 10.1242/bio.20136981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfer G, Courtheoux T, Peterka M, Meier S, Soste M, Melnik A, Reis K, Aspenstrom P, Peter M, Picotti P, et al. Mitotic redistribution of the mitochondrial network by Miro and Cenp-F. Nat Commun. 2015;6:8015. doi: 10.1038/ncomms9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I, Krylyshkina O, Small JV. Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J Cell Biol. 1999;146:1033–1044. doi: 10.1083/jcb.146.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I, Rottner K, Small JV. Targeting, capture, and stabilization of microtubules at early focal adhesions. J Cell Biol. 1998;142:181–190. doi: 10.1083/jcb.142.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I, Straube A. Regulation of cell migration by dynamic microtubules. Semin Cell Dev Biol. 2011;22:968–974. doi: 10.1016/j.semcdb.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairallah RJ, Shi G, Sbrana F, Prosser BL, Borroto C, Mazaitis MJ, Hoffman EP, Mahurkar A, Sachs F, Sun Y, et al. Microtubules underlie dysfunction in duchenne muscular dystrophy. Sci Signal. 2012;5:ra56. doi: 10.1126/scisignal.2002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhao Y, Chou IN. Nickel (Ni2+) enhancement of alpha-tubulin acetylation in cultured 3T3 cells. Toxicol Appl Pharmacol. 1996;140:461–470. doi: 10.1006/taap.1996.0243. [DOI] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CC, Zuppinger C, Guo X, Kuster GM, Helmes M, Eppenberger HM, Suter TM, Liao R, Sawyer DB. Anthracyclines induce calpain-dependent titin proteolysis and necrosis in cardiomyocytes. J Biol Chem. 2004;279:8290–8299. doi: 10.1074/jbc.M308033200. [DOI] [PubMed] [Google Scholar]
- Liu L, Vo A, Liu G, McKeehan WL. Distinct structural domains within C19ORF5 support association with stabilized microtubules and mitochondrial aggregation and genome destruction. Cancer Res. 2005a;65:4191–4201. doi: 10.1158/0008-5472.CAN-04-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Vo A, McKeehan WL. Specificity of the methylation-suppressed A isoform of candidate tumor suppressor RASSF1 for microtubule hyperstabilization is determined by cell death inducer C19ORF5. Cancer Res. 2005b;65:1830–1838. doi: 10.1158/0008-5472.CAN-04-3896. [DOI] [PubMed] [Google Scholar]
- Liu SC, Sauter ER, Clapper ML, Feldman RS, Levin L, Chen SY, Yen TJ, Ross E, Engstrom PF, Klein-Szanto AJ. Markers of cell proliferation in normal epithelia and dysplastic leukoplakias of the oral cavity. Cancer Epidemiol Biomarkers Prev. 1998;7:597–603. [PubMed] [Google Scholar]
- Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan KL, Pooley R, Miller PM, Kaverina I, Bader DM. Murine CENP-F regulates centrosomal microtubule nucleation and interacts with Hook2 at the centrosome. Mol Biol Cell. 2009;20:4790–4803. doi: 10.1091/mbc.E09-07-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musinipally V, Howes S, Alushin GM, Nogales E. The microtubule binding properties of CENP-E’s C-terminus and CENP-F. J Mol Biol. 2013;425:4427–4441. doi: 10.1016/j.jmb.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R, Goto Y, Kurozumi A, Matsushita R, Enokida H, Kojima S, Naya Y, Nakagawa M, Ichikawa T, Seki N. MicroRNA-205 inhibits cancer cell migration and invasion via modulation of centromere protein F regulating pathways in prostate cancer. Int J Urol. 2015;22:867–877. doi: 10.1111/iju.12829. [DOI] [PubMed] [Google Scholar]
- Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova OV, Pugacheva EN, Golemis EA. Primary cilia and the cell cycle. Methods Cell Biol. 2009;94:137–160. doi: 10.1016/S0091-679X(08)94007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooley RD, Moynihan KL, Soukoulis V, Reddy S, Francis R, Lo C, Ma LJ, Bader DM. Murine CENPF interacts with syntaxin 4 in the regulation of vesicular transport. J Cell Sci. 2008;121:3413–3421. doi: 10.1242/jcs.032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooley RD, Reddy S, Soukoulis V, Roland JT, Goldenring JR, Bader DM. CytLEK1 is a regulator of plasma membrane recycling through its interaction with SNAP-25. Mol Biol Cell. 2006;17:3176–3186. doi: 10.1091/mbc.E05-12-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner JB, Rees J, Whitehead CM, Casiano CA, Tan EM, Humbel RL, Conrad K, Fritzler MJ. High frequency of neoplasia in patients with autoantibodies to centromere protein CENP-F. Clin Invest Med. 1997;20:308–319. [PubMed] [Google Scholar]
- Sastry SK, Burridge K. Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Exp Cell Res. 2000;261:25–36. doi: 10.1006/excr.2000.5043. [DOI] [PubMed] [Google Scholar]
- Small JV, Geiger B, Kaverina I, Bershadsky A. How do microtubules guide migrating cells. Nat Rev Mol Cell Biol. 2002;3:957–964. doi: 10.1038/nrm971. [DOI] [PubMed] [Google Scholar]
- Soukoulis V, Reddy S, Pooley RD, Feng Y, Walsh CA, Bader DM. Cytoplasmic LEK1 is a regulator of microtubule function through its interaction with the LIS1 pathway. Proc Natl Acad Sci USA. 2005;102:8549–8554. doi: 10.1073/pnas.0502303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen U, Straube E, Straube A. Directional persistence of migrating cells requires Kif1C-mediated stabilization of trailing adhesions. Dev Cell. 2012;23:1153–1166. doi: 10.1016/j.devcel.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Thery M, Racine V, Piel M, Pepin A, Dimitrov A, Chen Y, Sibarita JB, Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc Natl Acad Sci USA. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschernatsch M, Stolz E, Strittmatter M, Kaps M, Blaes F. Antinuclear antibodies define a subgroup of paraneoplastic neuropathies: clinical and immunological data. J Neurol Neurosurg Psychiatry. 2005;76:1702–1706. doi: 10.1136/jnnp.2003.033225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida M, Mourino-Perez RR, Roberson RW. Live-cell imaging of microtubule dynamics in hyphae of Neurospora crassa. Methods Mol Biol. 2010;638:259–268. doi: 10.1007/978-1-60761-611-5_19. [DOI] [PubMed] [Google Scholar]
- Varis A, Salmela AL, Kallio MJ. Cenp-F (mitosin) is more than a mitotic marker. Chromosoma. 2006;115:288–295. doi: 10.1007/s00412-005-0046-0. [DOI] [PubMed] [Google Scholar]
- Vergnolle MA, Taylor SS. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr Biol. 2007;17:1173–1179. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- Volkov VA, Grissom PM, Arzhanik VK, Zaytsev AV, Renganathan K, McClure-Begley T, Old WM, Ahn N, McIntosh JR. Centromere protein F includes two sites that couple efficiently to depolymerizing microtubules. J Cell Biol. 2015;209:813–828. doi: 10.1083/jcb.201408083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AM, Asfahani R, Carroll P, Bicknell L, Lescai F, Bright A, Chanudet E, Brooks A, Christou-Savina S, Osman G, et al. The kinetochore protein, CENPF, is mutated in human ciliopathy and microcephaly phenotypes. J Med Genet. 2015;52:147–156. doi: 10.1136/jmedgenet-2014-102691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells—over and over and over again. Nat Cell Biol. 2002;4:E97–100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- Webster DR, Borisy GG. Microtubules are acetylated in domains that turn over slowly. J Cell Sci. 1989;92:57–65. doi: 10.1242/jcs.92.1.57. [DOI] [PubMed] [Google Scholar]
- Webster DR, Gundersen GG, Bulinski JC, Borisy GG. Assembly and turnover of detyrosinated tubulin in vivo. J Cell Biol. 1987;105:265–276. doi: 10.1083/jcb.105.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Bader D, Litvin J. Identification of a novel cardiac-specific transcript critical for cardiac myocyte differentiation. Development. 1996;122:2779–2789. doi: 10.1242/dev.122.9.2779. [DOI] [PubMed] [Google Scholar]
- Yang Z, Guo J, Chen Q, Ding C, Du J, Zhu X. Silencing mitosin induces misaligned chromosomes, premature chromosome decondensation before anaphase onset, and mitotic cell death. Mol Cell Biol. 2005;25:4062–4074. doi: 10.1128/MCB.25.10.4062-4074.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo YJ, Xi M, Wan YP, Hua W, Liu YL, Wan S, Zhou YL, Luo HW, Wu SL, Zhong WD, et al. Enhanced expression of centromere protein F predicts clinical progression and prognosis in patients with prostate cancer. Int J Mol Med. 2015;35:966–972. doi: 10.3892/ijmm.2015.2086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.