Tgl4p and Tgl5p are two yeast lipid droplet proteins exhibiting triacylglycerol lipase and lysophospholipid acyltransferase activity. In the absence of lipid droplets, Tgl4p and Tgl5p are retained in the ER, where their properties change. Thus triacylglycerol substrate availability is an important regulator of Tgl4p and Tgl5p functionality.
Abstract
Tgl3p, Tgl4p, and Tgl5p are the major triacylglycerol lipases of the yeast Saccharomyces cerevisiae. Recently we demonstrated that properties of Tgl3p are regulated by the formation of nonpolar lipids. The present study extends these investigations to the two other yeast triacylglycerol lipases, Tgl4p and Tgl5p. We show that Tgl4p and Tgl5p, which are localized to lipid droplets in wild type, are partially retained in the endoplasmic reticulum in cells lacking triacylglycerols and localize exclusively to the endoplasmic reticulum in a mutant devoid of lipid droplets. In cells lacking steryl esters, the subcellular distribution of Tgl4p and Tgl5p is unaffected, but Tgl5p becomes unstable, whereas the stability of Tgl4p increases. In cells lacking nonpolar lipids, Tgl4p and Tgl5p lose their lipolytic activity but retain their side activity as lysophospholipid acyltransferases. To investigate the regulatory network of yeast triacylglycerol lipases in more detail, we also examined properties of Tgl3p, Tgl4p, and Tgl5p, respectively, in the absence of the other lipases. Surprisingly, lack of two lipases did not affect expression, localization, and stability of the remaining Tgl protein. These results suggest that Tgl3p, Tgl4p, and Tgl5p, although they exhibit similar functions, act as independent entities.
INTRODUCTION
Accumulation of free fatty acids and sterols can lead to lipotoxicity, resulting in defects in organelle membrane formation and cell integrity. Hence cells have developed mechanisms to store these lipids in their inert forms as triacylglycerols (TGs) and steryl esters (SEs), respectively, in defined organelles called lipid droplets (LDs; Zweytick et al., 2000; Czabany et al., 2007). Structurally, LDs are characterized by their hydrophobic core of nonpolar lipids, a phospholipid surface monolayer membrane, and a specific set of proteins associated with or embedded into the phospholipid monolayer. The ratio of the different nonpolar lipids stored in the hydrophobic core of LDs is determined by the cell type and by culture conditions. In the yeast Saccharomyces cerevisiae, which we used as our model system, LDs contain nearly equal amounts of TGs and SEs (Leber et al., 1994; Czabany et al., 2008; Grillitsch et al., 2011).
Formation of TGs provides an efficient mechanism by which to store excessive amounts of fatty acids, which can be used upon requirement. In the yeast, TGs are synthesized by two different pathways governed by acyl-CoA:diacyglycerol acyltransferase (Dga1p) and phospholipid:diacylglycerol acyltransferase (Lro1p), respectively (Dahlqvist et al., 2000; Oelkers et al., 2000, 2002; Sorger and Daum, 2002). The yeast enzyme Dga1p belongs to the diacylglycerol acyltransferase gene family DGAT2 and forms TG from diacylglycerol and acyl-CoA (Oelkers et al., 2002; Sorger and Daum, 2002). Like most enzymes contributing to lipid synthesis, Dga1p is located to the endoplasmic reticulum (ER), although small amounts of the protein are also detected at LDs (Sorger and Daum, 2002). The second major TG synthase, Lro1p, which is the yeast homologue of the human lecithin:cholesterol acyltransferase (Dahlqvist et al., 2000), resides exclusively at the ER. This enzyme catalyzes acyl-CoA–independent formation of TG by attaching an acyl chain from phosphatidylethanolamine or phosphatidylcholine, respectively, to diacylglycerol (Oelkers et al., 2000). During the exponential growth phase, TGs are mainly formed by Lro1p, whereas Dga1p is the major contributor to TG synthesis during the late exponential phase and in the stationary phase (Oelkers et al., 2002).
The second major component of yeast LDs is made up of SEs. These lipids are formed by two acyl-CoA:cholesterol acyltransferase–related enzymes, Are1p and Are2p, which are both located at the ER (Yang et al., 1996; Yu et al., 1996). Whereas Are2p prefers ergosterol as a substrate, Are1p uses ergosterol precursors as well. Both Are proteins catalyze acyl-CoA–dependent reactions.
Sandager et al. (2002) showed that LDs are completely absent in a dga1Δlro1Δare1Δare2Δ quadruple mutant (QM) lacking all four nonpolar lipid-synthesizing enzymes of the yeast. Of interest, the QM strain is viable and grows like wild type on rich medium. However, in the presence of exogenous fatty acids, the QM reveals a severe growth phenotype exhibiting extreme liposensitivity (Garbarino et al., 2009; Petschnigg et al. 2009; Connerth et al., 2010).
At the exit of the stationary phase, fatty acids and diacylglycerols derived from TG degradation serve as important building blocks for the synthesis of membrane lipids. Therefore, efficient TG hydrolysis accelerates this process (Kurat et al., 2009; Petschnigg et al., 2009). Degradation of TGs is mediated by TG lipases (Athenstaedt and Daum 2003, 2005). Athenstaedt and Daum (2003) showed that Tgl3p is the major TG lipase of the yeast S. cerevisiae. Tgl3p is mostly located at LDs, but was also found in the ER in minor quantities (Schmidt et al., 2013). In a mutant lacking TGL3, the residual lipase activity is catalyzed by two other lipases, called Tgl4p and Tgl5p (Athenstaedt and Daum, 2005; Kurat et al., 2006). In contrast to Tgl3p, which uses TG species with variable chain length as substrates, Tgl4p and Tgl5p exhibit a higher degree of specificity. Tgl4p hydrolyzes preferentially TG species with myristic acid (C14:0) and palmitic acid (C16:0), whereas Tgl5p also uses TGs with very long chain fatty acids, such as hexacosanoic acid (C26:0), as substrates. Differences in substrate selectivity affect the differential contribution of the three TG lipases to TG lipolysis. Under standard growth conditions, the TG level of a mutant lacking TGL3 increases ∼2.7-fold compared with wild type (Athenstaedt and Daum, 2003). In contrast, deletion of TGL4 causes only a moderate change in the total TG amount of the respective mutant. The lipolytic activity of Tgl5p, however, becomes apparent only in combination with a defect of one of the other major TG lipases or in vitro upon measurement of the enzymatic activity of purified Tgl5p (Athenstaedt and Daum, 2005).
Studies from our laboratory revealed that the three TG lipases, Tgl3p, Tgl4p, and Tgl5p, not only exhibit lipolytic activity but also might contribute to phospholipid synthesis as lysophospholipid acyltransferases (Rajakumari and Daum, 2010a, b). Whereas the preferred acyltransferase substrate of Tgl3p is lysophosphatidylethanolamine, Tgl4p and Tgl5p preferentially convert lysophosphatidic acid to phosphatidic acid in an acyl-CoA–dependent reaction. In addition, Tgl4p functions as SE hydrolase and also harbors an active center of a phospholipase (Kurat et al., 2009; Rajakumari and Daum, 2010b). These additional properties characterize the yeast orthologue of the mammalian adipocyte triglyceride lipase as a multifunctional enzyme. The lipolytic activity of Tgl4p is regulated via phosphorylation and linked to cell cycle progression (Kurat et al., 2009). This posttranslational modification, however, has no effect on the lysophospholipid acyltransferase activity of Tgl4p (Rajakumari and Daum, 2010b).
To shed some light on the regulation of TG lipases in the lipid metabolic network, we previously investigated the effect of the presence/absence of nonpolar lipid-forming enzymes on the major TG lipase, Tgl3p (Schmidt et al., 2013). These studies revealed that TGL3 is expressed irrespective of nonpolar lipid synthesis. In mutant cells lacking formation of nonpolar lipids, however, Tgl3p becomes highly unstable, resulting in a strongly reduced protein level. In a dga1Δlro1Δare1Δare2Δ QM that is devoid of LDs, Tgl3p was detected in the ER as nonfunctional enzyme. Hence both TG lipase activity and acyltransferase activity of Tgl3p seem to be exclusively attributed to LDs.
Although Tgl3p, Tgl4p, and Tgl5p share certain features, they differ from each other in other ways. To obtain a more complete picture of the nonpolar lipid regulatory network, we extended our previous studies (Schmidt et al., 2013) to the response of Tgl4p and Tgl5p to the presence/absence of nonpolar lipids. For this purpose, we studied gene expression, protein level, stability, intracellular localization, and function of Tgl4p and Tgl5p in strains compromised in nonpolar lipid formation. In addition, we studied the regulatory effects of Tgl3p, Tgl4p, and Tgl5p with regard to one another. Our studies revealed that the three TG lipases seem to act independently of each other, but that their function and cellular behavior is strongly influenced by the availability of their substrate.
RESULTS
Protein levels of Tgl4p and Tgl5p in cells lacking nonpolar lipid-synthesizing enzymes
To investigate the fate of Tgl4p and Tgl5p in the absence of their substrate TGs, SEs, and/or their host organelle, the LD, we performed experiments with the double mutants dga1∆lro1∆ and are1∆are2∆, respectively, and the dga1∆lro1∆are1∆are2∆ QM. Whereas dga1∆lro1∆ is devoid of TGs, and are1∆are2∆ lacks SEs, both double mutants still contain LDs. In contrast, the QM lacks both nonpolar lipids and consequently LDs. To address whether TGL4 and TGL5 were expressed in cells bearing the different defects in nonpolar lipid formation, we first analyzed their mRNA levels in wild-type, are1∆are2∆, dga1∆lro1∆, and QM strains. As shown in Figure 1, A and B, the mRNA level of TGL4 increased in are1∆are2∆ compared with the other strains tested, whereas mRNA levels of TGL5 were similar in all four strains. This suggested that gene expression of TG lipases occurs independent of nonpolar lipid formation. Next we tested the effect of defects in nonpolar lipid formation on the level of TGL4 and TGL5 gene products. Despite differences in expression level, Tgl4p was present in similar amounts in all four strains examined. In addition, the protein level of Tgl5p was not affected by defects in nonpolar lipid synthesis (Figure 1C). These results demonstrate that regulation of lipolysis by Tgl4p and Tgl5p does not occur on the transcription or translation level.
FIGURE 1:
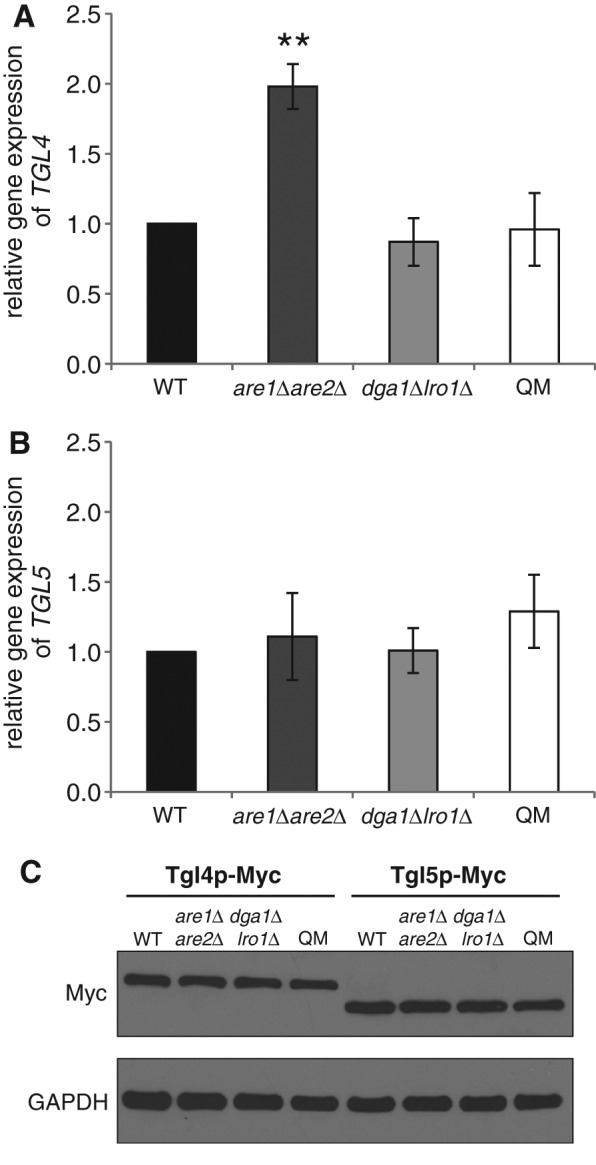
Gene expression and protein levels of Tgl4p and Tgl5p in cells defective in nonpolar lipid synthesis. Relative gene expression of (A) TGL4 and (B) TGL5 in wild-type (WT) (black bar), are1Δare2Δ (dark gray bar), dga1∆lro1∆ (gray bar), and dga1∆lro1∆are1∆are2∆ QM (white bar) strains was quantified by real-time PCR. Wild type was set at 1. Data are mean values from at least three independent experiments with the respective deviation. Significance was calculated by Student’s t-test (two tailed, unpaired). **p < 0.005, defined to be significant. (C) Protein analysis of Tgl4-Myc and Tgl5-Myc from total cell extracts of WT, are1Δare2Δ, dga1∆lro1∆, and QM strains grown to mid logarithmic phase. The primary antibody was directed against the Myc tag (Myc). The cytosolic marker GAPDH was used as loading control.
Protein stability of Tgl4p and Tgl5p in the presence/absence of nonpolar lipids
The finding of equal amounts of Tgl4p and Tgl5p in wild-type, are1∆are2∆, dga1∆lro1∆, and QM strains suggested similar protein stability of the TG lipases in the absence of nonpolar lipid formation. Nevertheless, we considered the possibility that the protein stability of Tgl4p and Tgl5p was affected in cells lacking TGs and/or SEs, as the major TG lipase Tgl3p became highly unstable in the absence of its substrate, TGs (Schmidt et al., 2013).
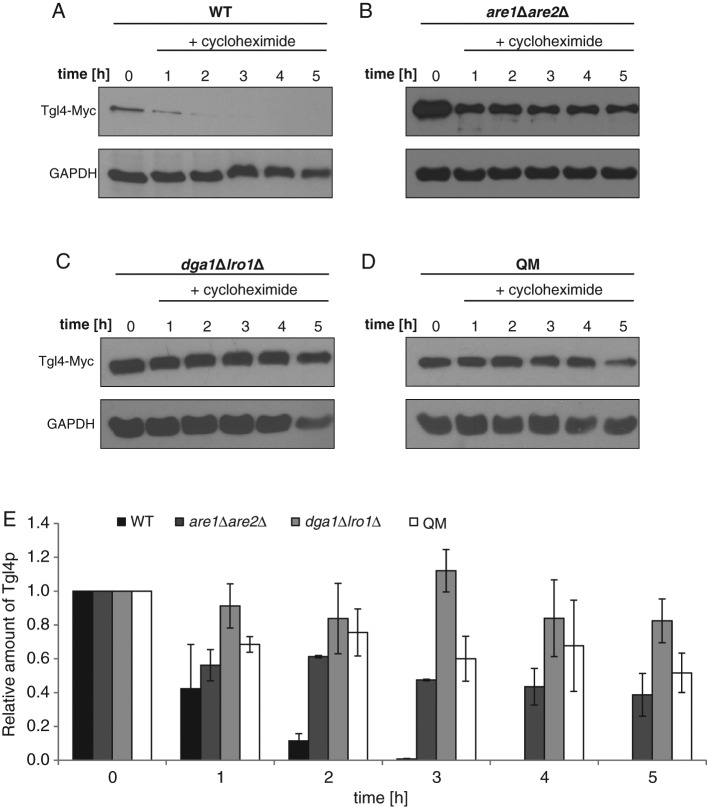
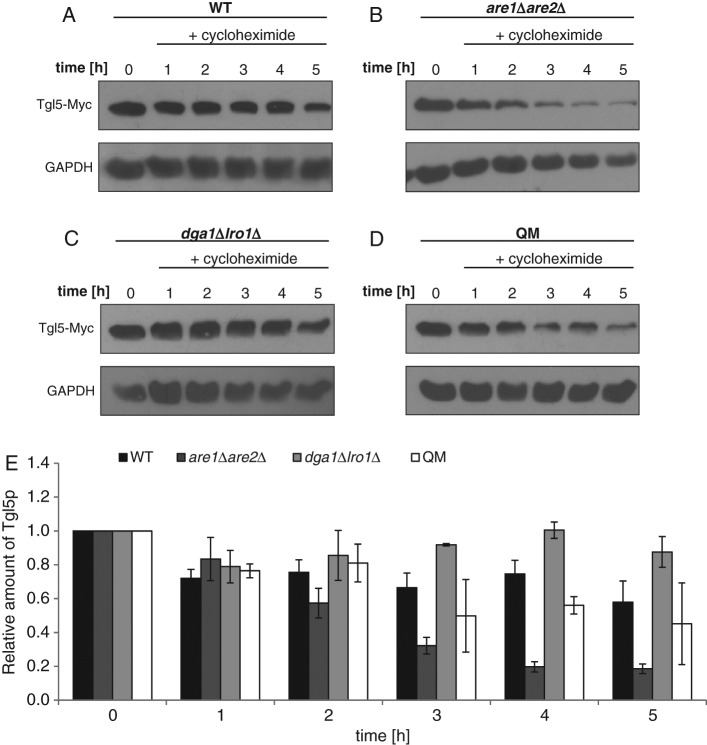
To examine the lifetime of Tgl4p and Tgl5p in wild-type, are1∆are2∆, dga1∆lro1∆, and QM strains, we blocked protein synthesis with cycloheximide (see Materials and Methods) and analyzed the amounts of polypeptides at different time points. In the dga1∆lro1∆ mutant poisoned with cycloheximide, the protein level of Tgl4p remained more or less constant for 5 h, and in are1∆are2∆, as well as in the QM, the protein level decreased only slightly (Figure 2, B–E). Surprisingly, in the wild type, Tgl4p was highly unstable (Figure 2, A and E). One hour after the addition of cycloheximide, the amount of Tgl4p dropped to 20% of the control, and after 3 h, this TG lipase was no longer detectable. Tgl5p exhibited stability in wild type and dga1∆lro1∆ over 5 h (Figure 3, A, C, and E). In the QM, the amount of Tgl5p decreased after 3 h and reached ∼50% of the wild-type level (Figure 3, D and E). However, the exclusive absence of SEs had an even stronger effect on the stability of Tgl5p. In the are1∆are2∆ mutant, the level of Tgl5p decreased already after 2 h and dropped to ∼25% of the wild-type level after 5 h (Figure 3, B and E). The surprising finding of these stability tests was the different behavior of Tgl4p and Tgl5p. Whereas Tgl4p gained stability in the absence of TGs or SEs, the opposite was seen with Tgl5p.
FIGURE 2:
Protein stability of Tgl4p in cells defective in nonpolar lipid synthesis. Western blot analysis of Tgl4-Myc was performed with total cell extracts from wild-type (WT) (A), are1Δare2Δ (B), dga1∆lro1∆ (C), and QM (D) strains grown for times as indicated after addition of 100 μg/ml cycloheximide to cells grown to mid logarithmic phase. The primary antibody was directed against the Myc tag (Tgl4-Myc). The cytosolic marker GAPDH was used as loading control. Western blot analysis shown is one representative experiment of at least two independent experiments. (E) Relative amounts of Tgl4-Myc analyzed by Western blotting were calculated using ImageJ with the respective deviations as shown. The amount of Tgl4-Myc at time point 0 (addition of cycloheximide) was set at 1. WT, black bar; are1Δare2Δ, dark gray bar; dga1∆lro1∆, gray bar; QM, white bar.
FIGURE 3:
Protein stability of Tgl5p in cells defective in nonpolar lipid synthesis. Western blot analysis of Tgl5-Myc was performed with total cell extracts from wild-type (WT) (A), are1Δare2Δ (B), dga1∆lro1∆ (C), and QM (D) strains grown for times as indicated after addition of 100 μg/ml cycloheximide to cells grown to the mid logarithmic phase. The primary antibody was directed against the Myc tag (Tgl5-Myc). The cytosolic marker GAPDH was used as loading control. Western blot analysis shown is one representative experiment of at least two independent experiments. (E) Relative amounts of Tgl5-Myc analyzed by Western blotting were calculated using ImageJ with the respective deviations as shown. The amount of Tgl5-Myc at time point 0 (addition of cycloheximide) was set at 1. WT, black bar; are1Δare2Δ, dark gray bar; dga1∆lro1∆, gray bar; QM, white bar.
In a mutant lacking lipid droplets, Tgl4p and Tgl5p are localized to the ER
Previous studies identified Tgl4p and Tgl5p as LD proteins in wild-type yeast cells (Athenstaedt and Daum, 2005; Kurat et al., 2006; Grillitsch et al., 2011; Currie et al., 2014). Hence the presence of considerable amounts of Tgl4p and Tgl5p in the QM, which is devoid of LDs, raised the question of the subcellular localization of the TG lipases in the absence of their original host organelle. In previous studies (Leber et al., 1998; Athenstaedt et al., 1999; Sorger and Daum, 2002), other proteins were found to be located to both LDs and the ER, suggesting a close relationship of these two organelles. Thus we examined the presence of the TG lipases in microsomal fractions (ER) from wild-type, are1∆are2∆, dga1∆lro1∆, and QM strains.
In wild type, Tgl4p was mostly present in LDs; only traces of the protein were detected in microsomal (ER) fractions (Figure 4A). A similar distribution of Tgl4p was observed in are1∆are2∆ cells devoid of SEs (Figure 4B). In the dga1∆lro1∆ double mutant lacking TGs, Tgl4p was still highly enriched in the LD fraction, but a substantial amount of the protein was also found in microsomes (Figure 4C). In the QM strain, the total amount of Tgl4p was detected in 30,000 × g and 40,000 × g microsomes (Figure 4D). Tgl5p was also highly enriched in the LD fraction from wild type and was also detected in microsomal fractions (Figure 5A). The absence of SEs in the are1∆are2∆ mutant did not affect the subcellular distribution of Tgl5p compared with wild type (Figure 5B). In contrast, in the double mutant dga1∆lro1∆, a larger portion of Tgl5p was shifted toward microsomes (Figure 5C), and in the QM lacking LDs, Tgl5p behaved like a true ER protein (Figure 5D). Further, in this case, the different behaviors of Tgl4p and Tgl5p are noteworthy. The tendency to exclude Tgl5p from LDs due to lack of TGs is much higher for this TG lipase than for Tgl4p.
FIGURE 4:
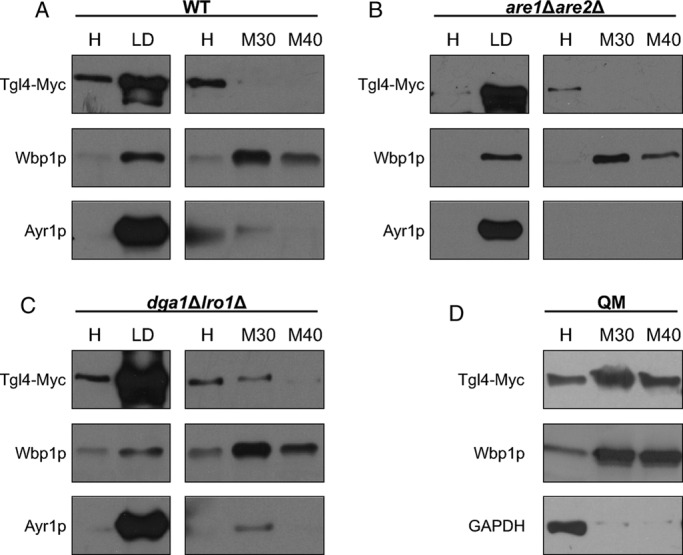
Localization of Tgl4p in wild-type (WT), are1Δare2Δ, dga1∆lro1∆, and dga1∆lro1∆are1∆are2∆ strains. Western blot analysis of Tgl4-Myc was performed with homogenate (H), 30,000 × g microsomal (M30), 40,000 × g microsomal (M40), and lipid droplet (LD) fractions from WT (A), are1Δare2Δ (B), dga1∆lro1∆ (C), and QM dga1∆lro1∆are1∆are2∆ (D) strains grown to the stationary phase. Primary antibodies were directed against the Myc tag (Tgl4-Myc), Wbp1p (ER marker), Ayr1p (LD marker), and GAPDH (cytosolic marker). Western blot analyses are representative of at least two independent experiments.
FIGURE 5:
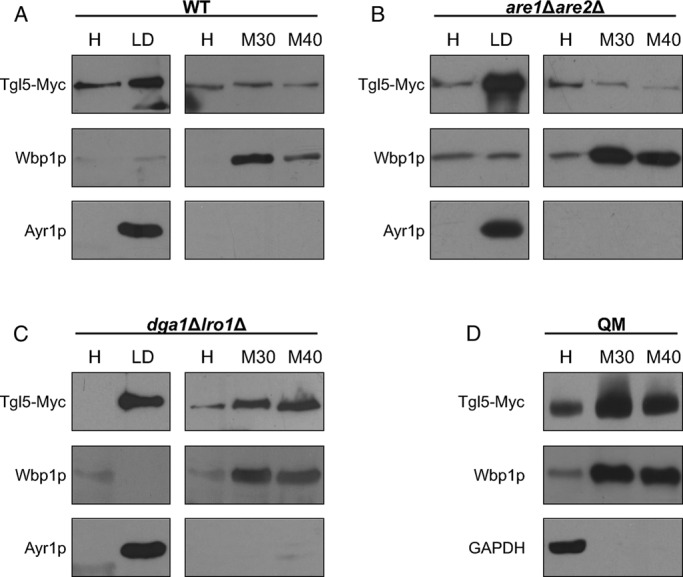
Localization of Tgl5p in wild-type (WT), are1Δare2Δ, dga1∆lro1∆, and dga1∆lro1∆are1∆are2∆ strains. Western blot analysis of Tgl5-Myc was performed with homogenate (H), 30,000 × g microsomal (M30), 40,000 × g microsomal (M40), and lipid droplet (LD) fractions from WT (A), are1Δare2Δ (B), dga1∆lro1∆ (C), and QM dga1∆lro1∆are1∆are2∆ (D) strains grown to the stationary phase. Primary antibodies were directed against the Myc tag (Tgl5-Myc), Wbp1p (ER marker), Ayr1p (LD marker), and GAPDH (cytosolic marker). Western blot analyses are representative of at least two independent experiments.
Enzymatic activities of Tgl4p and Tgl5p retained in the ER
Because considerable amounts of Tgl4p and Tgl5p were detected in ER fractions, especially in the QM, we investigated the functionality of the two enzymes in this compartment. For this purpose, we used the QM and the two quintuple mutants QM tgl4∆ and QM tgl5∆. First, we tested TG lipase activity in isolated microsomal fractions from these strains. As can be seen from Figure 6A, the lipolytic activity was not significantly changed by the additional deletion of TGL4 or TGL5, respectively. This result indicated that both gene products did not function as TG lipases in these samples.
FIGURE 6:
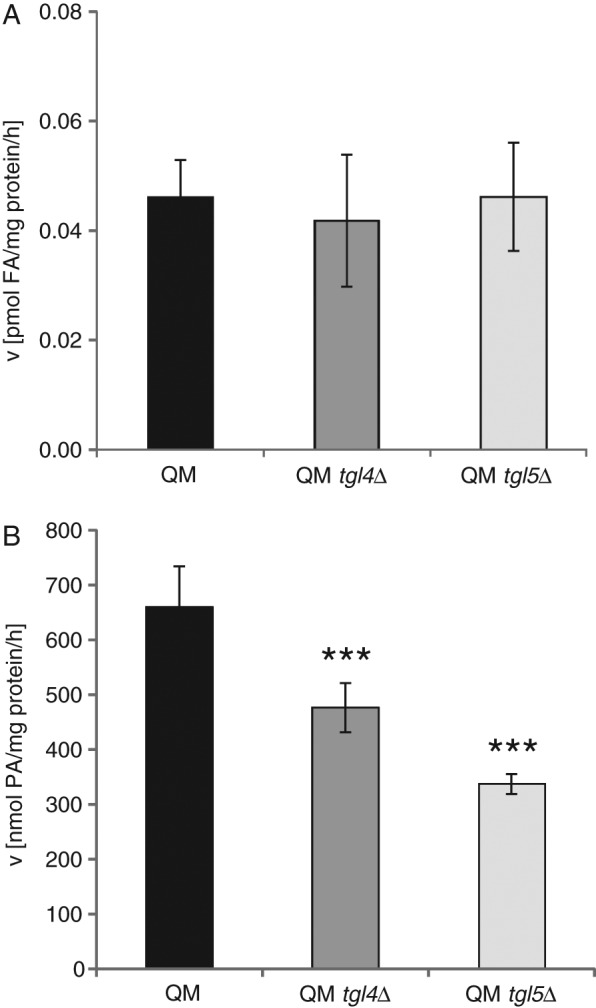
Triacylglycerol lipase activity and lysophosphatidic acid acyltransferase activity in microsomes from strains compromised in nonpolar lipid metabolism. For these experiments, microsomes from dga1∆lro1∆are1∆are2∆ (QM), dga1∆lro1∆are1∆are2∆tgl4∆ (QM tgl4∆), and dga1∆lro1∆are1∆are2∆tgl5∆ (QM tgl5∆) were isolated (for details, see Materials and Methods). TG lipase activity (A) and lysophosphatidic acid acyltransferase activity (B) in 30,000 × g ER fractions from QM (black bar), QM tgl4∆ (dark gray bar), and QM tgl5∆ (light gray bar) strains were analyzed. Assays were performed in triplicate from at least two independent biological samples. Data are mean values with the respective deviation. Significance was calculated by Student’s t-test (two tailed, unpaired). ***p < 0.001, defined to be significant.
In previous studies (Rajakumari and Daum, 2010a, b), we showed that Tgl4p and Tgl5p also harbor acyl-CoA–dependent lysophospholipid acyltransferase activity besides their ability to degrade TGs. Lysophosphatidic acid was the preferred substrate of Tgl4p and Tgl5p. We speculated that the two enzymes might have retained lysophospholipid acyltransferase activity when targeted to ER fractions. Indeed, comparison of this enzymatic activity measured in microsomes from QM, QM tgl4∆, and QM tgl5∆ showed that Tgl4p and Tgl5p may be active as acyltransferases in the ER. Figure 6B shows that acyl-CoA–dependent conversion of lysophosphatidic acid to phosphatidic acid decreased in microsomes from QM tgl4∆ to ∼75% and in microsomes from QM tgl5∆ to 50% of the control (QM). Thus both enzymes retained the potential to act as phospholipid biosynthetic enzymes in strains devoid of TG formation. We also quantified individual phospholipids from QM, QM tgl4∆, and QM tgl5∆ (Table 1). This pattern, however, was not significantly changed in the absence of TGL4 or TGL5, indicating that the lysophospholipid acyltransferase activity of both gene products in vivo was minor, at least under the conditions tested.
TABLE 1:
Phospholipid composition of cell-free homogenate from cells grown on YPD.
| Phospholipids in cell-free homogenate (mol%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Strain | CL | DMPE | LPLs | PA | PC | PE | PI | PS | Others |
| WT | 2.5 ± 0.5 | 3.2 ± 0.8 | 0.8 ± 0.3 | 2.4 ± 0.3 | 48.3 ± 1.5 | 25.3 ± 1.8 | 8.4 ± 0.9 | 5.8 ± 0.7 | 3.3 ± 0.8 |
| WT tgl4∆ | 2.3 ± 0.5 | 2.8 ± 0.5 | 0.7 ± 0.5 | 2.8 ± 0.3 | 49.1 ± 1.5 | 25.0 ± 1.0 | 8.7 ± 1.1 | 6.1 ± 0.5 | 2.5 ± 0.7 |
| WT tgl5∆ | 3.0 ± 0.2 | 3.2 ± 0.6 | 0.9 ± 0.2 | 2.9 ± 0.3 | 47.3 ± 1.3 | 23.9 ± 1.8 | 8.9 ± 0.9 | 5.7 ± 0.4 | 4.2 ± 0.9 |
| QM | 2.7 ± 1.0 | 3.7 ± 0.5 | 0.1 ± 0.1 | 2.0 ± 0.6 | 58.8 ± 3.5 | 20.4 ± 1.7 | 6.0 ± 1.9 | 4.2 ± 1.7 | 2.1 ± 0.6 |
| QM tgl4∆ | 2.8 ± 0.6 | 2.7 ± 1.4 | 0.4 ± 0.6 | 1.4 ± 1.2 | 55.7 ± 2.7 | 19.8 ± 1.3 | 8.1 ± 2.1 | 6.0 ± 3.0 | 3.1 ± 0.7 |
| QM tgl5∆ | 2.9 ± 0.9 | 4.4 ± 2.3 | 0.5 ± 0.7 | 1.7 ± 0.8 | 54.0 ± 1.0 | 19.5 ± 2.0 | 10.1 ± 2.0 | 6.8 ± 2.8 | 0.1 ± 0.1 |
WT, wild type; QM, quadruple mutant; CL, cardiolipin; DMPE, dimethylphosphatidylethanolamine; LPL, lysophospholipids; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine. Mean values of at least three measurements with SDs are shown.
Regulatory interaction of the three yeast triacylglycerol lipases Tgl3p, Tgl4p, and Tgl5p
Single deletions of TGL3, TGL4 and TGL5, respectively, have different effects on the accumulation of TGs (Athenstaedt and Daum, 2005; Schmidt et al., 2013). The highest accumulation of TGs was found in tgl3∆, a moderate increase in tgl4∆, and almost no change in tgl5∆. In our studies of the regulatory network of yeast TG lipases, we also investigated possible mutual regulatory effects of the three genes/gene products on each other. To study potential cross-talk of the yeast TG lipases, we used the double mutants tgl4∆tgl5∆, tgl3∆tgl5∆, and tgl3∆tgl4∆.
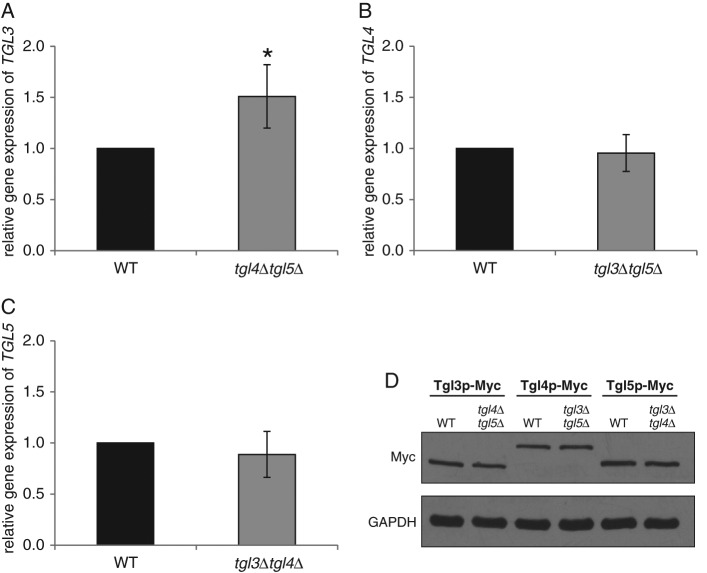
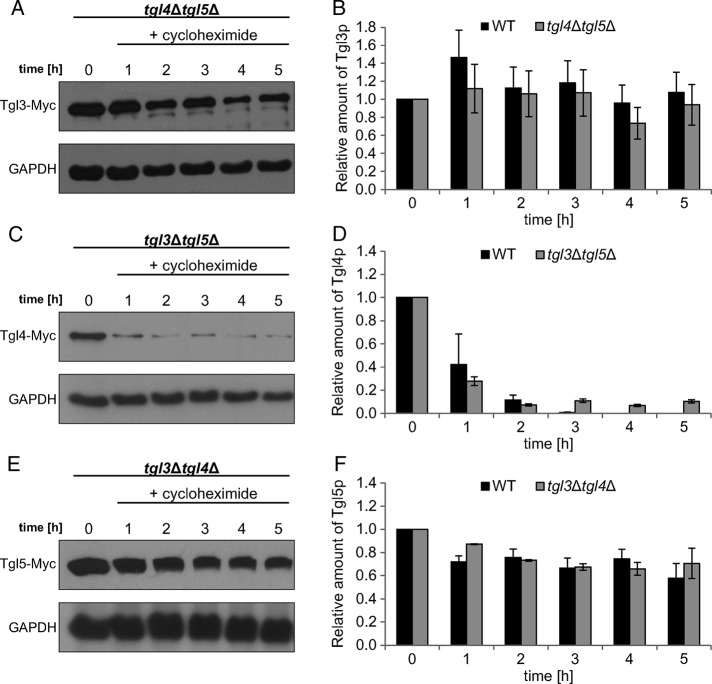
To our surprise, we found that there was very little regulatory interaction between TGL3, TGL4, and TGL5. Of the three TG lipase genes tested, only the expression level of TGL3 was slightly up-regulated in tgl4Δtgl5Δ (Figure 7A), but the protein level of Tgl3p remained the same as in the wild type (Figure 7D). Similarly, the lack of counterpart lipases did not affect the amount of Tgl4p and Tgl5p (Figure 7D). In terms of the protein stability, Tgl4p was slightly more stable in the tgl3∆tgl5∆ mutant background than in the wild type (Figure 8D), whereas the stability of Tgl3p and Tgl5p was not affected in tgl4∆tgl5∆ or tgl3∆tgl4∆ (Figure 8, B and F). Finally, we tested the intracellular distribution of Tgl3p, Tgl4p, and Tgl5p in the absence of the respective counterpart lipases. The major subcellular location of Tgl4p in the tgl3∆tgl5∆ mutant was still the LD fraction (Figure 9B). The minor amount of Tgl4p present in ER fractions seemed to be slightly increased compared with wild type (Figure 4). A more significant change was found for Tgl5p in the tgl3∆tgl4∆ mutant background (Figure 9C). Whereas in wild type (Figure 5), the dual localization of Tgl5p in LDs and ER was well expressed, only traces of Tgl5p were detected in microsomal fractions from tgl3∆tgl4∆. Similarly, Tgl3p was essentially restricted to LDs in the absence of TGL4 and TGL5 (Figure 9A). In the wild type, minor amounts of Tgl3p were also detected in the ER (Schmidt et al., 2013).
FIGURE 7:
Gene expression and protein levels of Tgl3p, Tgl4p, and Tgl5p in the absence of counterpart lipases. Relative gene expression of (A) TGL3 in wild type (WT) (black bar) and tgl4Δtgl5Δ (dark gray bar), (B) TGL4 in WT (black bar) and tgl3∆tgl5∆ (dark gray bar), and (C) TGL5 in WT (black bar) and tgl3∆tgl4∆ (dark gray bar) was measured by real-time PCR. Wild type was set at 1. Data are mean values from three independent experiments with the respective deviation. Significance was calculated by Student’s t-test (two tailed, unpaired). *p < 0.01, defined to be significant. (D) Protein analysis of Tgl3-Myc, Tgl4-Myc, and Tgl5-Myc from total cell extracts of WT, tgl4Δtgl5Δ, tgl3∆tgl5∆, and tgl3∆tgl4∆ strains grown to mid logarithmic phase. The primary antibody was directed against the Myc tag (Myc). Cytosolic marker GAPDH was used as loading control. Western blot analyses are representative of two independent experiments.
FIGURE 8:
Protein stability of Tgl3p, Tgl4p, and Tgl5pin the absence of counterpart lipases. Western blot analysis of (A) Tgl3-Myc in tgl4∆tgl5∆, (C) Tgl4-Myc in tgl3∆tgl5∆, and (E) Tgl5-Myc in tgl3∆tgl4∆ was performed with total extracts from cells grown for times as indicated after addition of 100 μg/ml cycloheximide to cells grown to mid logarithmic phase. The primary antibody was directed against the Myc tag (Tgl3-Myc, Tgl4-Myc, and Tgl5-Myc). The cytosolic marker GAPDH was used as loading control. Western blot analyses shown are one representative experiment of at least two independent experiments. Relative amounts of Tgl3-Myc (B), Tgl4-Myc (D), and Tgl5-Myc (F) obtained by three Western blots were calculated using ImageJ with the respective deviations as shown. Amounts of the respective protein at time point 0 (addition of cycloheximide) were set at 1. WT, wild type.
FIGURE 9:
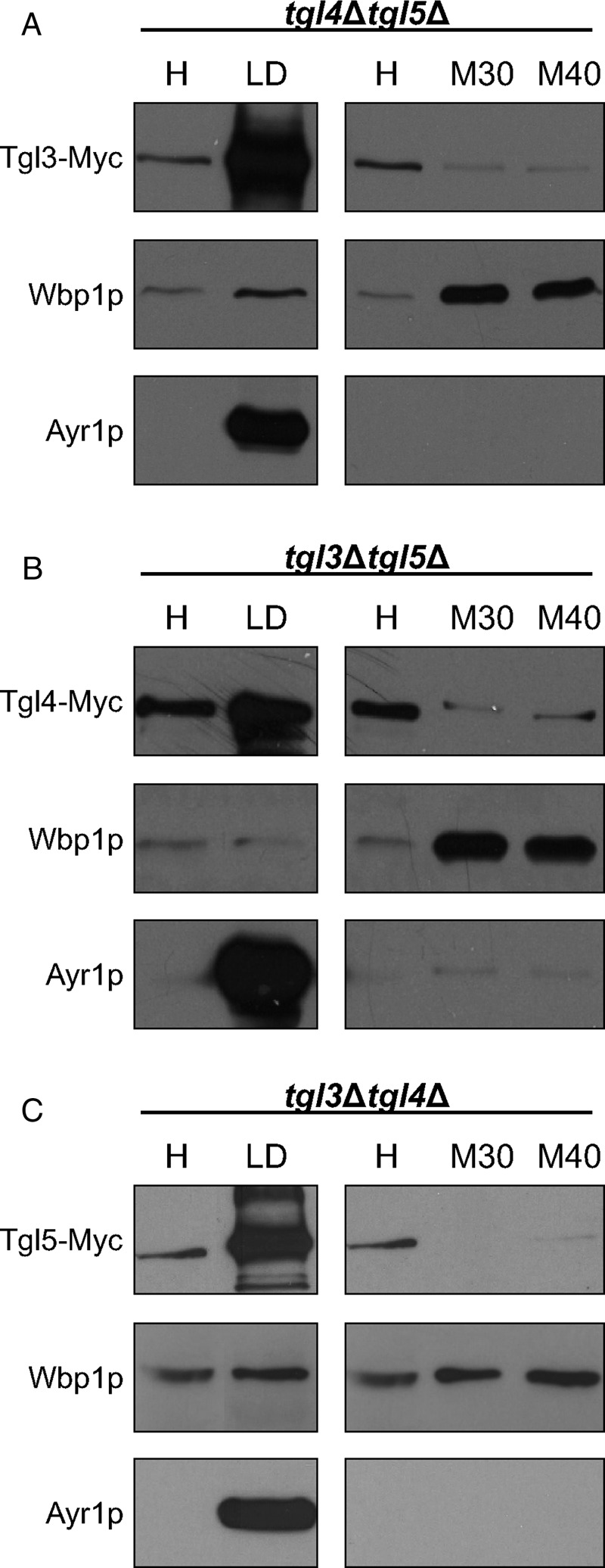
Localization of Tgl3p, Tgl4p, and Tgl5p in the absence of counterpart lipases. Western blot analysis of (A) Tgl3-Myc in tgl4∆tgl5∆, (B) Tgl4-Myc in tgl3∆tgl5∆, and (C) Tgl5-Myc in tgl3∆tgl4∆ using samples of homogenate (H), 30,000 × g microsomes (M30), 40,000 × g microsomes (M40), and LD fractions is shown. Primary antibodies were directed against the Myc tag (Tgl3-Myc, Tgl4-Myc, and Tgl5-Myc), Wbp1p (ER marker), and Ayr1p (LD marker). Western blot analyses are representative of at least two independent experiments.
DISCUSSION
Redundancy in the enzymatic equipment of a cell raises the question of the reason for the occurrence of enzymes with overlapping functions. In many cases, different substrate specificities, expression levels at different growth phases/conditions, or intrinsic protein properties are the issue. Finally, differences in regulatory response might be a reason for the existence of such groups of enzymes.
As an example of enzymes with overlapping function, Tgl3p, Tgl4p, and Tgl5p were identified as the major TG lipases in the yeast S. cerevisiae (Athenstaedt and Daum, 2003, 2005; Kurat et al., 2006). These TG lipases exhibit similarities but also clear differences. First, Tgl3p, Tgl4p, and Tgl5p catalyze TG hydrolysis when assayed in vitro (Athenstaedt and Daum, 2003, 2005). However, enzymatic activities of isolated enzymes differ strongly, similar to the efficiency of the three enzymes in hydrolyzing TGs in vivo. Second, Tgl3p, Tgl4p, and Tgl5p are components of LDs but are also present in the ER. However, proportions of the proteins in the two compartments vary. Finally, Tgl3p, Tgl4p, and Tgl5p have a common side activity, namely as a lysophospholipid acyltransferase (Rajakumari and Daum, 2010a, b). However, differences in enzymatic activities and substrate specificity cause different effects on the lipid profile. This appears to be especially important for Tgl5p, which seems to be a more efficient lysophospholipid acyltransferase than a TG lipase.
To address regulatory aspects of the role of Tgl3p, Tgl4p, and Tgl5p in the yeast, we have been investigating reciprocal effects of TG biosynthetic and mobilizing machineries. The initial focus of our studies was on Tgl3p, the major TG lipase of the yeast (Schmidt et al., 2013, 2014). Here we extended our investigations to Tgl4p and Tgl5p to obtain a more complete picture of the nonpolar lipid metabolic network. First, we showed that transcriptional regulation of TGL3, TGL4, and TGL5 did not have a major effect on the lipolytic equipment of the cell. In the absence of TG synthesis and in cells accumulating aberrant amounts of TGs due to the lack of counterpart TG lipases, gene expression of TGL4 and TGL5 was unaffected (Figures 1 and 7). Similarly, gene expression of TGL3 exhibited only a slight decrease in cells defective in TG formation (Schmidt et al., 2013) and a marginal increase in the absence of counterpart lipases (Figure 7). Thus it is very unlikely that regulatory factors sense the cellular TG content and transmit the respective information to the transcription machinery. The second important level of regulation that we tested is regulation at the protein level in response to the presence/absence of TGs, SEs, or LDs. We observed clear differences among the three lipolytic enzymes. Whereas amounts of Tgl3p (Schmidt et al., 2013) were markedly reduced in strains with compromised TG synthesis, Tgl4p and Tgl5p levels seemed to be unaffected (Figure 1). However, experiments in vivo revealed that also the stability of Tgl4p and Tgl5p strongly depends on the capacity of a cell to form nonpolar lipids (Figures 2 and 3).
Observations concerning the stability of Tgl3p, Tgl4p, and Tgl5p upon depletion of cells of TGs, SEs, and LDs have to be discussed in connection with LD structure and composition, as well as the subcellular localization of the proteins. Structural analyses of Tgl3p localized to LDs revealed that the C-terminus of the protein, which determines the stability of Tgl3p, protrudes into the hydrophobic core of the droplet (Koch et al., 2014). This structural property may explain the different behavior of Tgl3p in cells lacking TGs or SEs. In the absence of SEs, Tgl3p remains associated with the droplet, perhaps by preferential interaction of its C-terminal part with TGs, resulting in stability of the TG lipase (Schmidt et al., 2013). In the absence of TGs, however, Tgl3p loses its affinity to the LDs and is degraded (Schmidt et al., 2013). Surprisingly, Tgl5p becomes unstable in the absence of SEs, whereas lack of TGs in dga1Δlro1Δ did not have a marked effect (Figure 3). In this mutant, a considerable amount of Tgl5p is retained in the ER, whereas in are1Δare2Δ, the TG lipase is associated with LDs (Figure 5). Of interest, stability of Tgl5p is slightly higher in the QM than in are1Δare2Δ, although both strains lack SEs. Thus it appears that not only the presence/absence of a nonpolar lipid determines the stability of Tgl5p, but so does the intracellular localization. In contrast to Tgl3p and Tgl5p, Tgl4p gained stability in cells lacking TGs and/or SEs (Figure 2). Further, in this case, the distribution of the enzyme between LDs and ER has to be taken into account. In dga1Δlro1Δ, a slight shift of the protein to the ER was observed (Figure 4C), but the vast majority of Tgl4p in dga1Δlro1Δ and are1Δare2Δ remained associated with the LD fraction. The molecular reason for this behavior of the enzyme is not known.
In the extreme situation of the QM lacking both nonpolar lipids and consequently LDs, all three TG lipases ended up in the ER (Figures 4 and 5; Schmidt et al., 2013). For Tgl3p residing in the ER, the C-terminus was exposed to the cytosol, resulting in rapid degradation (Koch et al., 2014). A similar conclusion can be drawn for Tgl5p in the QM. In this case too, the shift of the protein from LDs to the ER resulted in instability (Figures 3 and 5). Although we do not have molecular information about the structure of Tgl5p, we propose a similarity to Tgl3p. Because Tgl4p is rather stable in the ER (Figures 2 and 4), it appears that the bilayer environment of the ER is more favorable for this protein than the monolayer of LDs. In this case too, the molecular details of the protein remain to be elucidated.
The presence of TG lipases in the ER from wild type and especially under conditions of TG and LD depletion (Figures 4 and 5; Schmidt et al., 2013) raised the question of the enzymatic function(s) of these proteins in this compartment. At this point, differences between Tgl3p, Tgl4p, and Tgl5p have to be mentioned again. As discussed earlier, in terms of protein stability, Tgl5p located in the ER (Figures 3 and 5) behaved like Tgl3p, whereas stability of Tgl4p targeted to the ER was completely different (Figures 2 and 4). In terms of functionality, Tgl5p resembles Tgl4p as rather weak TG lipase, whereas Tgl3p catalyzes the highest lipolytic activity in the yeast. As another similarity, Tgl3p, Tgl4p, and Tgl5p lost their ability to hydrolyze TGs when retained in the ER (Figure 6; Schmidt et al., 2013). This makes sense from an economic viewpoint, as TGs are not present in the ER as substrate under regular conditions and is completely absent from strains like the QM. Of most interest, however, both Tgl4p and Tgl5p retained their lysophospholipid acyltransferase activity when located to the ER (Figure 6), in contrast to Tgl3p (Schmidt et al., 2013). Consistent with previous reports, the lysophosphatidic acid acyltransferase activity of Tgl5p exceeded that of Tgl4p (Figure 6; Rajakumari and Daum, 2010a, b). The fact that QM tgl4∆ and QM tgl5∆ did not exhibit marked changes in phospholipid pattern (Table 1) may be explained by the fact that other lysophospholipid acyltransferases, such as the major lysophosphatidic acid acyltransferases Slc1p and Ale1p (for review, see Henry et al., 2012), contribute more efficiently to lipid anabolism in vivo than do Tgl4p and Tgl5p.
The final point addressed in the present study was regulation of TGL3, TGL4, and TGL5 with regard to one another. No major effects were observed in that respect, but only a slight shift of Tgl4p from LDs to the ER (Figures 4 and 9), and an increase of Tgl3p and Tgl5p in LDs compared with the ER (Figures 5 and 9; Schmidt et al., 2013) in cells lacking counterpart lipases was seen. Taken together, previous results (Schmidt et al., 2013) and results from this study elucidate regulatory aspects of yeast TG lipases. Besides regulation at the protein level, changes in subcellular targeting of Tgl3p, Tgl4p, and Tgl5p have to be taken into account. Thus the organelle level of regulation must not be ignored as an important aspect affecting protein properties and probably also the functionality of these enzymes.
MATERIALS AND METHODS
Strains and culture conditions
Yeast strains used in this study are listed in Table 2. Cells were grown aerobically to the exponential growth phase or to the stationary phase at 30°C in yeast extract/peptone/dextrose (YPD) medium containing 1% yeast extract (Oxoid, Wesel, Germany), 2% peptone (Oxoid), and 2% glucose (Roth, Karlsruhe, Germany). The SD medium used for strain constructions consisted of 0.67% yeast nitrogen base (ForMedium, Hunstanton, UK), 2% glucose, and the respective amino acid supplements.
TABLE 2:
Yeast strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| Wild type | BY4741 Mat a; his3∆1; leu2∆0; met15∆0; ura3∆0 | EUROSCARF (Frankfurt, Germany) |
| Tgl4-Myc | BY4741; TGL4-13Myc::HIS3MX6 | This study |
| Tgl5-Myc | BY4741; TGL5-13Myc::HIS3MX6 | This study |
| are1∆are2∆ | BY4742; are1::kanMX4, are2::URA3 | Kindly provided by M. Spanova (this laboratory) |
| are1∆are2∆ Tgl4-Myc | BY4742; are1::kanMX4, are2::URA3 TGL4-MYC13::HIS3MX6 | This study |
| are1∆are2∆ Tgl5-Myc | BY4742; are1::kanMX4, are2::URA3 TGL5-MYC13::HIS3MX6 | This study |
| dga1∆lro1∆ | BY4741; lro1∆::URA3KL dga1∆::kanMX4 | This study |
| dga1∆lro1∆ Tgl4-Myc | dga1∆lro1∆; TGL4-13Myc::HIS3MX6 | This study |
| dga1∆lro1∆ Tgl5-Myc | dga1∆lro1∆; TGL5-13Myc::HIS3MX6 | This study |
| QM | BY4741; dga1∆::kanMX4 lro1∆::kanMX4 are1∆::kanMX4 are2∆::kanMX4 | Athenstaedt (2011) |
| QM Tgl4-Myc | QM; TGL4-13Myc::HIS3MX6 | This study |
| QM Tgl5-Myc | QM; TGL5-13Myc::HIS3MX6 | This study |
| QM tgl4∆ | QM; tgl4∆::URA3KL | This study |
| QM tgl5∆ | QM; tgl5∆::URA3KL | This study |
| tgl4∆tgl5∆ | BY4741; tgl4∆::kanMX4 tgl5∆::kanMX4 | Athenstaedt and Daum (2005) |
| tgl3∆tgl4∆ | BY4741; tgl3∆::kanMX4 tgl4∆::kanMX4 | Athenstaedt and Daum (2005) |
| tgl3∆tgl5∆ | BY4741; tgl3∆::kanMX4 tgl5∆::kanMX4 | Athenstaedt and Daum (2005) |
| tgl4∆tgl5∆ Tgl3-Myc | tgl4∆tgl5∆; TGL3-13Myc::HIS3MX6 | This study |
| tgl3∆tgl5∆ Tgl4-Myc | tgl3∆tgl5∆; TGL4-13Myc::HIS3MX6 | This study |
| tgl3∆tgl4∆ Tgl5-Myc | tgl3∆tgl4∆; TGL5-13Myc::HIS3MX6 | This study |
Genetic techniques
Chromosomal tagging and deletions were performed by homologous recombination using the PCR-mediated method described by Longtine et al. (1998). In brief, the inserts for the construction of Tgl3-Myc, Tgl4-Myc, Tgl5-Myc, tgl4∆, or tgl5∆ strains were obtained by PCR from plasmids pFA6a-13Myc-HIS3MX6, pFA6a-HIS3MX6, or pFA6a-URA3KL. Primers used for DNA amplification are listed in Table 3. For transformation of yeast strains, 400–700 ng of DNA were used, employing the high-efficiency lithium acetate transformation protocol (Gietz et al., 1995). After transformation, cells were plated on SD medium lacking the respective amino acid. Positive transformants were verified for correct integration of the fusion cassette by analytical PCR of whole yeast cell extracts.
TABLE 3:
Primers used in this study.
| Primer | Sequence (5′ → 3′) |
| Tgl3F1 | GTGCAGTCGAATTTAAATTAGACGACATAATAAGAGCAAGACGGAGTAGGCGGATCCCCGGGTTAATTAA |
| Tgl3S2 | ATCGAGCTCTATCAATAAAAAAAATAAGACAGAAAAAAGTGGAAACGATAATCGATGAATTCGAGCTCG |
| Tgl4F2 | CGAGGCCTTCTTCTTCAACGCAGCACAAAAGCACCACCAGTTTTACTCAACGGATCCCCGGGTTAATTAA |
| Tgl4S2 | CATAGATGAAAAAGAATATCTAGAGGATATATAAGCAAGCCCGTGTTTTCATCGATGAATTCGAGCTCG |
| Tgl4S1 | TGTAATAATTATTGAAGGGAGTACAGGTATATGTAATAAAAGTCTGAATGCGTACGCTGCAGGTCGAC |
| Tgl5F2 | CGGCTGCCACAAACGACAATTTCATGAACAATTCAGACATTTTTCAAAATCGGATCCCCGGGTTAATTAA |
| Tgl5S2 | TGAGAATATAGAAAGCTTTTTATATAAAAATGTACTTATTGTCTTTCATTTCAATCGATGAATTCGAGCTCG |
| Tgl5S1 | AAAAGACATCATAAACAGCACAAGGAAGACGGTTCTGTTTCGTTGCTATGCGTACGCTGCAGGTCGAC |
| RT-Act1-fwd | CAGAGAAAAGATGACTCAAATTATG |
| RT-Act1-rev | TTCTACCGGAAGAGTACAAGG |
| RT-Tgl3-fwd | GCCAACAATCCGAGCATAAC |
| RT-Tgl3-rev | GGTGCCAAGTATGGTCTCG |
| RT-Tgl4-fwd | TGCCCGACATGTGTATGCTTTTTAGAAT |
| RT-Tgl4-rev | CTTGGGCCACGTAGCTTTTGCAC |
| RT-Tgl5-fwd | CCGGGAGTTGACTTGGAAGAATCC |
| RT-Tgl5-rev | GGAGAAGGCAATGGCTGAAGAGGA |
RNA isolation and real-time PCR
Total RNA from cells grown to the early logarithmic phase at 30°C in YPD was isolated using the RNeasy kit from Qiagen (Hilden, Germany) following the manufacturer’s instructions. After DNaseI digestion, real-time PCR was performed using the SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen, Carlsbad, CA) as described by the manufacturer. Reactions were performed in sealed MicroAmp Optical 96-well reaction plates, and amplification was measured using an ABI 7500 instrument (Applied Biosystems). Samples were quantified using the ∆∆Ct method described by Livak and Schmittgen (2001) with ACT1 serving as internal control. With this method, differences in mRNA expression relative to control were calculated. Primers used for real-time PCR are listed in Table 3.
Isolation and characterization of subcellular fractions
LDs and microsomal fractions were isolated from cells grown to the early stationary phase in YPD medium following published procedures (Zinser et al., 1991; Zinser and Daum, 1995; Leber et al., 1994). The protein concentration of samples was analyzed by the method of Lowry et al. (1951) using bovine serum albumin as a standard. Before protein determination, LD samples were delipidated with two to three volumes of diethyl ether. After withdrawal of the organic phase, residual diethyl ether was removed under a stream of nitrogen. Proteins were precipitated with trichloroacetic acid at a final concentration of 10% and solubilized in 0.1% SDS and 0.1 M NaOH.
SDS–PAGE was performed as described by Laemmli (1970) using 12.5% separation gels. Western blot analysis was carried out according to Haid and Suissa (1983). Proteins were detected by using rabbit or mouse antisera as primary antibodies and peroxidase-conjugated goat anti-rabbit or anti-mouse as secondary antibodies. Primary antibodies were directed against the Myc-tag, Wbp1p (ER marker), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; cytosolic marker), and Ayr1p (LD marker). Protein amounts of 10–20 μg from each fraction were loaded onto SDS gels for Western blot analysis. Quantification of immunoreactive bands detected by Western blot analysis was performed by measuring the relative intensities of bands using ImageJ.
Determination of protein stability
To block protein synthesis in yeast, cycloheximide was added at a final concentration of 100 μg/ml to cells grown to the mid logarithmic growth phase in YPD at 30°C. At time points indicated, aliquots of cells were harvested by centrifugation at 13,000 × g for 10 min at 4°C. The cell pellet was suspended in lysis buffer (1.85 M NaOH, 7.5% β-mercaptoethanol) and incubated for 10 min on ice. Subsequently, trichloroacetic acid was added at a final concentration of 25%, and proteins were precipitated on ice for at least 10 min. Precipitated proteins were sedimented by centrifugation at 13,000 × g at 4°C for 10 min, washed once with ice-cold distilled water, and dissolved in Laemmli buffer (Laemmli, 1970) at 37°C for 30 min. Gel electrophoresis, Western blot analysis, and quantification of immunoreactive bands were performed as described.
Preparation of total cell extracts for lipid analysis
For lipid analysis, total cell extracts were prepared from yeast cells grown to the stationary phase in YPD. Cells were harvested by centrifugation at 3000 × g for 5 min at room temperature and suspended in breaking buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl) containing 1 mM phenylmethylsulfonyl fluoride. Then yeast cells were disintegrated by vigorous shaking in the presence of glass beads for 10 min at 4°C. After cell disruption, residual cell debris were removed by centrifugation at 3000 × g for 5 min. The supernatant was used for protein determination and lipid analysis.
Lipid analysis
Lipids from total cell extracts were isolated by the method of Folch et al. (1957) using chloroform/methanol (2:1 [vol/vol]) as solvent. Individual phospholipids from total cell extracts (2 mg of protein) were separated by two-dimensional (2D) thin-layer chromatography (TLC) using chloroform/methanol/25% ammonia (65:35:5 per volume) as solvent system for the first dimension and chloroform/acetone/methanol/acetic acid/water (50:20:10:10:5 per volume) for the second dimension. Spots of individual phospholipids were visualized by staining with iodine vapor, scraped off the plate, and quantified by the method of Broekhuyse (1968).
Enzyme analysis
TG lipase activity was measured as described previously (Schmidt et al., 2013) using 30,000 × g fractions of the ER (300–400 μg of protein) as enzyme sources.
Lysophospholipid acyltransferase activity was measured in a final volume of 200 μl using 30,000 × g microsomes (ER; ∼20 μg of protein) as enzyme source. Reactions were carried out at 30°C in a mixture containing 100 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 100 μM lysophosphatidic acid (Sigma-Aldrich, Vienna, Austria), and 20 μM [14C]oleoyl-CoA (41.9 mCi/mmol; New England Nuclear, Waltham, MA). After 2 min, the reaction was stopped by adding 3 ml of chloroform/methanol (1:2 [vol/vol]) and 0.7 ml 1% perchloric acid, and lipids were extracted by vortexing. After centrifugation, the upper phase was removed, and the organic phase containing the lipids was washed twice with 1% perchloric acid before drying under a stream of nitrogen. Lipids were dissolved in a small volume of chloroform/methanol (2:1 [vol/vol]), unlabeled lipid extract from total cells (2 mg of protein) was added, and the combined extracts were spotted onto a TLC plate (Merck, Darmstadt, Germany). Individual phospholipids were separated by 2D TLC as described. After visualization of the individual spots of phospholipids by staining with iodine vapor, the spot corresponding to phosphatidic acid was scraped off the plate, and radioactivity was measured by liquid scintillation counting using LSC Safety (Baker, Deventer, Netherlands) with 5% water as scintillation cocktail.
Acknowledgments
We thank the Austrian Centre of Industrial Biotechnology Graz for providing the ABI 7500 instrument. This research was supported by Austrian Science Fund (FWF) Projects W901 DK Molecular Enzymology and P23029 to G.D. and P26308 to K.A.
Abbreviations used:
- ER
endoplasmic reticulum
- LD
lipid droplet
- QM
quadruple mutant
- SE
steryl ester
- TG
triacylglycerol.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-09-0633) on May 11, 2016.
REFERENCES
- Athenstaedt K. YALI0E32769g (DGA1) and YALI0E16797g (LRO1) encode major triacylglycerol synthases of the oleaginous yeast Yarrowia lipolytica. Biochim Biophys Acta. 2011;1811:587–596. doi: 10.1016/j.bbalip.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K, Daum G. YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J Biol Chem. 2003;278:23317–23323. doi: 10.1074/jbc.M302577200. [DOI] [PubMed] [Google Scholar]
- Athenstaedt K, Daum G. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J Biol Chem. 2005;280:37301–37309. doi: 10.1074/jbc.M507261200. [DOI] [PubMed] [Google Scholar]
- Athenstaedt K, Weys S, Paltauf F, Daum G. Redundant systems of phosphatidic acid biosynthesis via acylation of glycerol-3-phosphate or dihydroxyacetone phosphate in the yeast Saccharomyces cerevisisae. J Bacteriol. 1999;181:1458–1463. doi: 10.1128/jb.181.5.1458-1463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekhuyse RM. Phospholipids in tissues of the eye. I. Isolation, characterization and quantitative analysis by two-dimensional thin-layer chromatography of diacyl and vinyl-ether phospholipids. Biochim Biophys Acta. 1968;152:307–315. doi: 10.1016/0005-2760(68)90038-6. [DOI] [PubMed] [Google Scholar]
- Connerth M, Czabany T, Wagner A, Zellnig G, Leitner E, Steyrer E, Daum G. Oleate inhibits steryl ester synthesis and causes liposensitivity in yeast. J Biol Chem. 2010;285:26832–26841. doi: 10.1074/jbc.M110.122085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E, Guo X, Christiano R, Chitraju C, Kory N, Harrison K, Haas J, Walther TC, Farese RV., Jr High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J Lipid Res. 2014;55:1465–1477. doi: 10.1194/jlr.M050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabany T, Athenstaedt K, Daum G. Synthesis, storage and degradation of neutral lipids in yeast. Biochim Biophys Acta. 2007;1771:299–309. doi: 10.1016/j.bbalip.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Czabany T, Wagner A, Zweytick D, Lohner K, Leitner E, Ingolic E, Daum G. Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J Biol Chem. 2008;283:17065–17074. doi: 10.1074/jbc.M800401200. [DOI] [PubMed] [Google Scholar]
- Dahlqvist A, Ståhl U, Lenman M, Banàs A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA. 2000;97:6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Garbarino J, Padamsee M, Wilcox L, Oelkers PM, D’Ambrosio D, Ruggles KV, Ramsey N, Jabado O, Turkish A, Sturley SL. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J Biol Chem. 2009;284:30994–31005. doi: 10.1074/jbc.M109.050443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Grillitsch K, Connerth M, Köfeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim Biophys Acta. 2011;1811:1165–1176. doi: 10.1016/j.bbalip.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–324. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch B, Schmidt C, Ploier B, Daum G. Modifications of the C terminus affect functionality and stability of yeast triacylglycerol lipase Tgl3p. J Biol Chem. 2014;389:19306–19316. doi: 10.1074/jbc.M114.556944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein SD. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leber R, Landl K, Zinser E, Ahorn H, Spök A, Kohlwein SD, Turnowsky F, Daum G. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol Biol Cell. 1998;9:375–386. doi: 10.1091/mbc.9.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber R, Zinser E, Zellnig G, Paltauf F, Daum G. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast. 1994;10:1421–1428. doi: 10.1002/yea.320101105. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem. 2002;277:8877–8881. doi: 10.1074/jbc.M111646200. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J Biol Chem. 2000;275:15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]
- Petschnigg J, Wolinski H, Kolb D, Zellnig G, Kurat CF, Natter K, Kohlwein SD. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J Biol Chem. 2009;284:30981–30993. doi: 10.1074/jbc.M109.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Daum G. Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol Biol Cell. 2010a;21:501–510. doi: 10.1091/mbc.E09-09-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Daum G. Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J Biol Chem. 2010b;285:15769–15776. doi: 10.1074/jbc.M109.076331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandager L, Gustavsson MH, Ståhl U, Dahlqvist A, Wiberg E, Banàs A, Lenman M, Ronne H, Stymne S. Storage lipid synthesis is non-essential in yeast. J Biol Chem. 2002;277:6478–6482. doi: 10.1074/jbc.M109109200. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Athenstaedt K, Koch B, Ploier B, Daum G. Regulation of the yeast triacylglycerol lipase Tgl3p by formation of nonpolar lipids. J Biol Chem. 2013;288:19939–19948. doi: 10.1074/jbc.M113.459610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Athenstaedt K, Koch B, Ploier B, Korber M, Zellnig G, Daum G. Defects in triacylglycerol lipolysis affect synthesis of triacylglycerols and steryl esters in the yeast. Biochim Biophys Acta. 2014;1842:1393–1402. doi: 10.1016/j.bbalip.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Sorger D, Daum G. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J Bacteriol. 2002;184:519–524. doi: 10.1128/JB.184.2.519-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bard M, Bruner DA, Gleeson A, Deckelbaum RJ, Aljinovic G, Pohl TM, Rothenstein R, Sturley SL. Sterol esterification in yeast: a two-gene process. Science. 1996;272:1353–1356. doi: 10.1126/science.272.5266.1353. [DOI] [PubMed] [Google Scholar]
- Yu C, Kennedy NJ, Chang CC, Rothblatt JA. Molecular cloning and characterization of two isoforms of Saccharomyces cerevisiae acyl-CoA:sterol acyltransferase. J Biol Chem. 1996;271:24157–24163. doi: 10.1074/jbc.271.39.24157. [DOI] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweytick D, Athenstaedt K, Daum G. Intracellular lipid particles of eukaryotic cells. Biochim Biophys Acta. 2000;1469:101–120. doi: 10.1016/s0005-2736(00)00294-7. [DOI] [PubMed] [Google Scholar]