Previous work established the link of ubiquilin proteins to several neurodegenerative diseases. The results from budding yeast indicate that ubiquilin proteins facilitate lysosome-dependent clearance of misfolded proteins by promoting inclusion body formation.
Abstract
Ubiquilin proteins contain a ubiquitin-like domain (UBL) and ubiquitin-associated domain(s) that interact with the proteasome and ubiquitinated substrates, respectively. Previous work established the link between ubiquilin mutations and neurodegenerative diseases, but the function of ubiquilin proteins remains elusive. Here we used a misfolded huntingtin exon I containing a 103-polyglutamine expansion (Htt103QP) as a model substrate for the functional study of ubiquilin proteins. We found that yeast ubiquilin mutant (dsk2Δ) is sensitive to Htt103QP overexpression and has a defect in the formation of Htt103QP inclusion bodies. Our evidence further suggests that the UBL domain of Dsk2 is critical for inclusion body formation. Of interest, Dsk2 is dispensable for Htt103QP degradation when Htt103QP is induced for a short time before noticeable inclusion body formation. However, when the inclusion body forms after a long Htt103QP induction, Dsk2 is required for efficient Htt103QP clearance, as well as for autophagy-dependent delivery of Htt103QP into vacuoles (lysosomes). Therefore our data indicate that Dsk2 facilitates vacuole-mediated clearance of misfolded proteins by promoting inclusion body formation. Of importance, the defect of inclusion body formation in dsk2 mutants can be rescued by human ubiquilin 1 or 2, suggesting functional conservation of ubiquilin proteins.
INTRODUCTION
Protein misfolding occurs spontaneously under normal physiological conditions, but conditions such as genetic mutations, environmental insults, and oxidative stress also stimulate it. Misfolded proteins are cytotoxic, but cells have developed protein quality control systems to restore correct protein folding or degrade misfolded proteins. The chaperone network prevents aggregation of misfolded proteins and assists correct refolding (Hartl et al., 2011). If refolding fails, misfolded proteins can be modified by ubiquitination, which leads to their degradation by the ubiquitin proteasome system (Varshavsky, 2012). Moreover, misfolded proteins can form inclusion bodies (IBs) that are cleared by lysosomes (Kroemer et al., 2010). For example, in patients with Huntington’s disease (HD), neuron cells have IBs that contain misfolded huntingtin (Chiti and Dobson, 2006). Increasing evidence suggests that compromised cellular capacity to dispose misfolded proteins directly contributes to the onset of numerous neurodegenerative diseases.
HD is a neurodegenerative disease with symptoms such as frontal cognitive deficits and involuntary abnormal movements (Walker, 2007). This disease is attributed to the expansion of CAG repeat (encoding glutamine) within the exon 1 of the huntingtin (HTT) gene, which causes terminal misfolding of the Htt protein. The HTT gene in normal individuals has 6–35 CAG repeats (polyQ), but expansions of >40 CAG repeats in the HTT gene cause HD (Andrew et al., 1993). The sequestration of misfolded proteins into IBs is believed to alleviate their cytotoxicity and facilitate their disposal by lysosomes (Chin et al., 2010; Tyedmers et al., 2010).
The huntingtin gene locus in the human genome spans 180 kb and consists of 67 exons. The transgenic R6/2 mouse is the most commonly used animal model of HD and expresses N-terminally truncated mutant human Htt with 125 polyQ repeats within exon 1. R6/2 mice develop HD-like symptoms, including motor and cognitive deficits (Mangiarini et al., 1996). Expression of truncated Htt with polyQ expansion and a proline-rich domain is sufficient to induce protein aggregation in vitro (Muchowski et al., 2000). Of interest, expression of this fragment in yeast and mammalian cells also results in IB (aggresome) formation, and this expression is well tolerated. However, expression of this Htt fragment lacking the proline-rich domain fails to form IBs and causes cytotoxicity in yeast cells, supporting the notion that IB formation alleviates the cytotoxicity of misfolded proteins (Krobitsch and Lindquist, 2000; Meriin et al., 2002; Wang et al., 2009). The capability of IB formation makes yeast an ideal model organism in which to study the biological processes of IB formation because of the availability of numerous genetic tools for yeast.
Previous studies indicate the ubiquitination of mutated Htt proteins. Striatum from HD brains showed elevated levels of ubiquitinated Htt N-terminal fragments (Mende-Mueller et al., 2001). The N-terminal domain of Htt with 44 polyQ repeats interacts with ubiquitination enzymes, which may contribute to Htt ubiquitination (Kalchman et al., 1996). The three lysine residues in the N-terminus of Htt are responsible for this modification, but the same residues are also subject to SUMOylation (Steffan et al., 2004). SUMOylation stabilizes Htt and reduces its ability to form aggregates, presumably by competing for ubiquitination. Recent evidence indicates that K48-linked ubiquitination of the N-terminal Htt fragments leads to proteasome-mediated degradation. However, in HD knock-in mice, aging decreases K48-linked ubiquitination and proteasome-mediated degradation of mutated Htt, whereas it increases K63-linked ubiquitination, indicating the complexity of the ubiquitination of misfolded proteins (Bhat et al., 2014).
Ubiquitinated proteins can be degraded through direct binding to proteasome receptors such as Rpn10 and Rpn13 (Husnjak et al., 2008). Ubiquitinated proteins can also interact with ubiquitin-like domain (UBL)–ubiquitin-associated (UBA) receptors, which subsequently transport the client proteins to proteasomes for degradation. UBL-UBA proteins contain an N-terminal UBL and either one or two UBA domains at the C-terminal (Kang et al., 2006). Previous work indicates that the human UBL-UBA proteins ubiquilin 1 and 2 are associated with various pathological inclusions, and mutations in ubiquilin 2 cause inherited amyotrophic lateral sclerosis (Daoud and Rouleau, 2011; Deng et al., 2011). Other studies suggest that ubiquilin 2 preferentially associates with Htt-polyQ aggregates compared with other protein inclusions (Doi et al., 2004; Wang and Monteiro, 2007; Rutherford et al., 2013). Some work indicated that ubiquilin proteins transport ubiquitinated substrates to the proteasome for degradation (Wang and Monteiro, 2007), and other research suggested that ubiquilin proteins facilitate IB formation, which may promote lysosome-medicated protein clearance (Heir et al., 2006). However, the molecular function of ubiquilin proteins in proteasome- or lysosome-mediated clearance of misfolded proteins remains largely unknown.
Here we used Htt exon 1 with 103-polyQ expansion and the proline-rich region (Htt103QP) as a model substrate to study the function of yeast ubiquilin, Dsk2, in the clearance of misfolded proteins. We found that dsk2Δ mutants exhibited slow growth and failed to form IB efficiently when Htt103QP was overexpressed. Our results indicate that the UBL domain of Dsk2 provides the functional specificity in Htt103QP IB formation. Surprisingly, Dsk2 is dispensable for proteasome-mediated protein degradation but is required for efficient delivery of Htt103QP into vacuoles (lysosomes), indicating that ubiquilin/Dsk2 facilitates lysosome-mediated clearance of mutated Htt by promoting IB formation. Furthermore, the rescue of the dsk2Δ mutant phenotype by human ubiquilin 1 and 2 indicates the functional conservation of this protein family.
RESULTS
Dsk2 is required for mutated huntingtin inclusion body formation in budding yeast
IB formation is believed to be cytoprotective because it sequesters toxic misfolded proteins (Wang et al., 2009; Gong et al., 2012). In budding yeast, expression of mutated Htt with polyQ expansion leads to IB formation, but failure in this process causes toxicity to yeast cells (Duennwald et al., 2006; Wang et al., 2009). Therefore we speculated that yeast mutants with defective IB formation should exhibit slow growth when mutated Htt is expressed. In this context, we constructed a yeast strain harboring mfa1:PMFA1-Sphis5+ and an integrated plasmid that contains Htt with 103-polyQ expansion and the proline-rich region (Htt103QP). This Htt fragment was tagged with Flag at the N-terminus and green fluorescent protein (GFP) at the C-terminus and is under control of a galactose promoter (PGALFlag-Htt103QP-GFP; hereafter Htt103QP). This strain was crossed with all of the ∼4800 yeast deletion mutants from ATCC (Manassas, VA), and the haploid cells (MATa) containing a gene deletion and Htt103QP were selected (Tong et al., 2001; Daniel et al., 2006). The resulting ∼4800 mutants with Htt103QP were examined for their growth on glucose and galactose plates (Figure 1A). We also used a query strain with PGAL-CLB5 as a control to exclude the mutants that fail to grow on galactose plates. IB formation of the selected strains was examined after Htt103QP induction. From this genome-wide screen, we identified some yeast mutants that exhibited slow growth on galactose plates and an IB formation defect, including dsk2Δ mutants.
FIGURE 1:
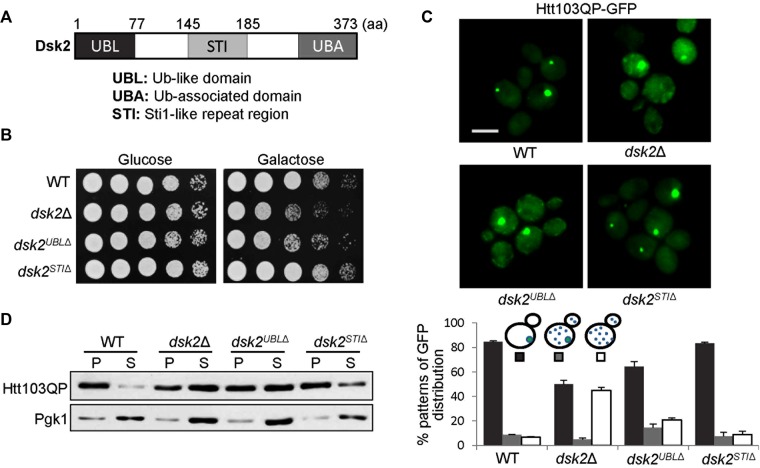
The absence of Dsk2 results in defects in Htt103QP IB formation. (A) The identification of yeast mutants sensitive to Htt103QP overexpression. About 4800 yeast deletion mutants with PGAL-Flag-Htt103QP-GFP (Htt103QP) were spotted onto glucose or galactose plates for growth assay. After 2 d of incubation at 30°C, the growth was examined. (B) dsk2Δ mutants are sensitive to Htt103QP overexpression. WT and dsk2Δ, rad23Δ, and ddi1Δ mutant cells were grown to saturation, 10-fold diluted, and spotted onto glucose and galactose plates for 2 d of incubation at 30°C. (C) dsk2Δ mutant cells exhibit defective Htt103QP IB formation. WT and dsk2∆, rad23∆, and ddi1∆ mutant cells with Htt103QP were grown in galactose medium at 30°C for 12 h to induce Htt103QP expression. Top, fluorescence microscopy, the GFP signal in representative cells. Bottom, percentage of cells with different patterns of Htt103QP-GFP signal (n > 100). The result is the average of three independent experiments. Scale bar, 5 μm. (D) dsk2∆ mutant cells show more soluble Htt103QP. Cells were grown in galactose medium at 30°C for 12 h to prepare cell lysates using a bead beater. The cell lysates were centrifuged and divided into detergent-soluble (supernatant) and detergent-insoluble (pellet) fractions. Htt103QP protein levels were determined by Western blotting with anti-GFP antibody. The Pgk1 levels are used as a loading control.
Dsk2, Rad23, and Ddi1 are the three UBL-UBA proteins in budding yeast that function as ubiquitin receptors to transport ubiquitinated proteins to the proteasome (Kang et al., 2006). To our surprise, we did not identify rad23Δ and ddi1Δ mutants from this screen. Therefore we first compared the growth of these three mutants and wild-type (WT) cells containing Htt103QP on glucose and galactose plates. Consistent with our screen, we noticed the slow growth of dsk2Δ mutants on galactose plates, but rad23Δ and ddi1Δ mutants grew similarly to WT cells (Figure 1B), indicating distinct roles of these three UBL-UBA proteins in response to the expression of Htt103QP. Next we examined the GFP signal in dsk2Δ, rad23Δ, and ddi1Δ mutants after induction of Htt103QP-GFP expression in galactose medium at 30°C for 12 h. Most of the WT cells (>80%) showed one bright GFP aggregate with variable sizes. rad23Δ and ddi1Δ mutant cells exhibited a similar phenotype as WT cells, but only 54% of dsk2Δ mutant cells showed a single GFP aggregate. Some dsk2Δ cells showed multiple tiny GFP aggregates or a mixture of tiny and big aggregates. Moreover, enhanced GFP background was observed in most dsk2∆ cells, indicating defective IB formation (Figure 1C).
Previous work demonstrated enriched distribution of mutated Htt in the detergent-insoluble fraction, presumably due to IB formation (Taylor et al., 2003; Gong et al., 2012). Moreover, the soluble mutated Htt oligomers before IB formation are cytotoxic (Takahashi et al., 2008; Lajoie and Snapp, 2010). Therefore we examined Htt103QP protein levels in supernatant (soluble fraction) and pellet (insoluble fraction) from WT, dsk2Δ, rad23Δ, and ddi1Δ mutant cells after induction of Htt103QP expression for 12 h. In WT cells, the majority of Htt103QP was detected in the pellet fraction, which is consistent with efficient IB formation. However, much less Htt103QP protein was detected in the pellet fraction from dsk2Δ mutant cells. In clear contrast, rad23Δ and ddi1Δ mutants showed similar distribution of Htt103QP as WT cells (Figure 1D). Taken together, these results indicate that Dsk2 promotes Htt103QP IB formation in budding yeast, and we speculate that the defect in Htt103QP IB formation causes slow growth of dsk2Δ mutants on galactose plates.
The UBL domain of Dsk2 is essential for inclusion body formation
Our results suggest that Dsk2, but not Rad23 and Ddi1, is required for Htt103QP IB formation in yeast cells. Next we examined which domain of Dsk2 contributes to this unique function. Dsk2, Rad23, and Ddi1 contain a UBL and a UBA domain that binds to the proteasome and ubiquitinated proteins, respectively (Verma et al., 2004). In addition, Dsk2 contains a Sti1-like repeat sequence (STI), which is found in proteins that bind to heat shock chaperones (Chang et al., 1997; Lassle et al., 1997). The STI domain in the human ubiquilin mediates the association with misfolded proteins (Stieren et al., 2011). We deleted the UBL or STI domain from the yeast genome to generate dsk2UBLΔ and dsk2STIΔ mutants, respectively, and then examined the sensitivity of these mutants to Htt103QP overexpression, as well as IB formation (Figure 2A). Because the UBA domain of Dsk2 has been shown to function as a stabilizing signal to protect Dsk2 from degradation (Heinen et al., 2011), we did not generate a dsk2 mutant with UBA deletion. Like dsk2Δ, dsk2UBLΔ mutant cells with Htt103QP exhibited similar slow-growth phenotype on galactose plates, but dsk2STIΔ mutant cells grew similarly to WT cells (Figure 2B). Consistently, dsk2UBLΔ mutant cells exhibited an obvious IB formation defect but one that was a little less dramatic than for dsk2Δ mutants. However, IB formation in dsk2STIΔ mutant cells was similar to that in WT cells (Figure 2C). We further examined the distribution of Htt103QP in the supernatant and pellet fractions in these dsk2 mutants after Htt103QP induction for 12 h. Most of the Htt103QP was detected in the insoluble pellet fraction in WT cells, but much less Htt103QP was detected in the pellet fraction in dsk2Δ and dsk2UBLΔ mutant cells. In contrast, dsk2STIΔ mutant cells showed similar distribution of Htt103QP in supernatant and pellet fractions as WT cells, which is consistent with efficient IB formation (Figure 2D). These data indicate that the UBL domain of Dsk2 is critical for its function in IB formation.
FIGURE 2:
Deletion of the DSK2 UBL domain causes defective Htt103QP IB formation. (A) Schematic illustration of the domains of the yeast Dsk2 protein. (B) dsk2UBL∆ and dsk2∆ mutants show sick growth when Htt103QP is overexpressed. Cells were grown to saturation, 10-fold diluted, and spotted onto glucose and galactose plates for 2 d of incubation at 30°C. (C) dsk2UBL∆ mutants show defective Htt103QP IB formation similar to dsk2∆ mutants. WT and dsk2∆, dsk2UBLΔ, and dskSTIΔ mutant cells with Htt103QP plasmid were grown in galactose medium for 12 h at 30°C to induce Htt103QP expression. Top, GFP signal in some representative cells by fluorescence microscopy. Bottom, percentage of cells with different patterns of GFP distribution (n > 100). The result is the average of three separate experiments. Scale bar, 5 μm. (D) dsk2UBLΔ and dsk2Δ mutants show more detergent-soluble Htt103QP. Cells with the indicated genotypes were grown in galactose medium for 12 h. Cell lysates were prepared and divided into detergent-soluble (supernatant) and -insoluble (pellet) fractions after centrifugation. Htt103QP protein levels were determined by Western blotting with anti-GFP antibody. Pgk1 levels were used as a loading control.
It is possible that the unique role of Dsk2 in IB formation is due to the specific binding of Htt103QP to Dsk2 but not to Rad23 and Ddi1. Thus we examined the interaction between Htt103QP and these three UBL-UBA proteins. For this purpose, we generated strains expressing Flag-Htt103QP-GFP and Myc-tagged Dsk2, Rad23, or Ddi1. After induction of Htt103QP in galactose medium for 4 h, when a majority of the cells did not form IBs, the cells were collected to prepare cell lysates. The coimmunoprecipitation results using anti-Myc antibody showed that Htt103QP binds to Dsk2, Rad23, and Ddi1 with similar affinity (Figure 3A). Therefore the function of Dsk2 in IB formation is unlikely to be attributed to its specific binding to Htt103QP.
FIGURE 3:
Cells expressing UBLDsk2-Rad23 or UBLDsk2-Ddi1 hybrid proteins show normal Htt103QP IB formation. (A) Dsk2, Rad23, and Ddi1 interact with Htt103QP. Cells expressing 13×Myc-tagged Dsk2, Rad23, or Ddi1, as well as Flag-Htt103QP, were grown in galactose medium at 30°C for 4 h. The cell lysates were pulled down with anti-Myc antibody. The protein levels of Myc-tagged Dsk2, Rad23, and Ddi1 and Flag-tagged Htt103QP in the cell lysates and immunoprecipitates were analyzed by Western blotting. (B) Schematic illustration of the domains in the constructed chimeras. To construct yeast strains expressing chimeras UBLDsk2-Rad23 and UBLDsk2-Ddi1, we replaced the C-terminal part of DSK2 gene (except the UBL domain) from the yeast genome with the corresponding sections of RAD23 and DDI1 genes using PCR-based homologous recombination. The resulting strains express the chimeras from the endogenous DSK2 promoter. (C) Cells expressing chimeras UBLDsk2-Rad23 and UBLDsk2-Ddi1 exhibit similar growth as WT cells when Htt103QP is overexpressed. WT, dsk2∆, UBLDsk2-RAD23, and UBLDsk2-DDI1 cells with Htt103QP plasmid were grown to saturation, 10-fold diluted, and spotted onto glucose and galactose plates, which were incubated at 30°C for 2 d. (D) Cells expressing UBLDsk2-Rad23 or UBLDsk2-Ddi1 chimera show similar Htt103QP distribution in soluble and insoluble factions as WT cells. WT, dsk2∆, UBLDsk2-RAD23, and UBLDsk2-DDI1 cells with Htt103QP plasmid were grown in galactose medium at 30°C for 12 h. Cells lysates from these cells were centrifuged and divided into detergent-soluble (supernatant) and -insoluble (pellet) fractions. Htt103QP protein levels were determined by Western blotting with anti-GFP antibody. Pgk1 levels are used as a loading control. (E) Cells expressing UBLDsk2-Rad23 or UBLDsk2-Ddi1 chimeras show similar Htt103QP IB formation as WT cells. WT, dsk2∆, UBLDsk2-RAD23, and UBLDsk2-DDI1 cells with Htt103QP plasmid were grown in galactose medium for 12 h at 30°C to induce Htt103QP expression. Top, GFP signal in these cells by fluorescence microscopy. Bottom, average percentage of cells with different GFP distribution patterns (n > 100). The results are the average of three separate experiments. Scale bar, 5 μm.
To determine further the role of the Dsk2 UBL domain in IB formation, we generated yeast strains expressing chimeric proteins UBLDsk2-Rad23 and UBLDsk2-Ddi1, in which the C-terminal part, but not the UBL domain of DSK2 gene, was replaced with the corresponding fractions of RAD23 and DDI1 genes (Figure 3B). We directly replaced the C-terminal fraction of DSK2 gene in the yeast genome with the PCR products of the corresponding RAD23 and DDI1 gene fragments that also contain a selectable marker. Therefore the resulting strains express chimeric genes UBLDsk2-RAD23 and UBLDsk2-DDI1 under the control of the endogenous DSK2 promoter. We first examined the growth on galactose plates of these strains containing Htt103QP. Yeast cells expressing UBLDsk2-Rad23 and UBLDsk2-Ddi1 chimeras grew similarly to WT cells on galactose plates (Figure 3C). Moreover, these cells showed efficient IB formation like WT cells (Figure 3E). Finally, these cells also showed enriched distribution of Htt103QP in the pellet fraction like WT cells (Figure 3D). Therefore our data support the conclusion that the UBL domain of Dsk2 contributes to its functional specificity in promoting Htt103QP IB formation.
Dsk2 is not required for proteasome-dependent degradation of Htt103QP
UBL-UBA proteins transport ubiquitinated proteins to proteasomes for degradation (Lowe et al., 2006). To test this possibility for Htt103QP, we first examined the protein stability of Htt103QP after a short period of induction, when no IB formation was noticed. We also examined the effect of the proteasome inhibitor MG132 on the stability of Htt103QP. To enhance MG132 permeability of yeast cells, we grew cells in medium containing 0.1% l-proline and added 0.003% SDS into the medium for 3 h before MG132 instillation as described (Liu et al., 2007). After Htt103QP induction in galactose medium for 1 h, glucose was added to the medium to shut off Htt103QP expression, and the Htt103QP protein level was analyzed over time. After 1 h of induction in galactose medium, the expression level of Htt103QP was high, but no IBs were observed. After addition of glucose in the medium, Htt103QP protein level declined over time, indicating efficient degradation. However, the presence of the proteasome inhibitor MG132 caused dramatic Htt103QP stabilization (Figure 4A). Using the same protocol, we found that MG132 treatment delayed the degradation of S-phase cyclin Clb5 (Figure 4B), indicating that MG132 inhibits proteasome activity under this experimental condition. Therefore Htt103QP is likely subject to proteasome-mediated degradation after a short period of induction.
FIGURE 4:
Htt103QP degradation after a short period of induction (1 h) is independent of Dsk2. (A) The degradation of Htt103QP after a short induction in the absence and presence of proteasome inhibitor. WT cells with Htt103QP plasmid growing in raffinose medium containing 0.1% l-proline at 30°C were pretreated with 0.003% SDS. After 3 h, dimethyl sulfoxide (DMSO) or 75 μM MG132 was added into the cultures. After incubation for 30 min, galactose (2%) was added into the medium to induce Htt103QP expression for 1 h. Finally, glucose (2%) was added into the cultures to shut off Htt103QP expression. The cells were collected over time to examine Htt103QP protein levels by Western blotting. Pgk1 levels were used as a loading control. Right, ratio change of Htt103QP to Pgk1 after Htt103QP expression shut-off. (B) Cell cycle protein Clb5 is stabilized in the presence of MG132. Yeast cells with Clb5-HA were treated as described. The Clb5 proteins levels were examined at the indicated time points after glucose addition. Right, ratio change. (C) dsk2Δ mutants show similar Htt103QP degradation kinetics as WT cells after a short induction. WT and dsk2Δ, rad23∆, and ddi1∆ mutant cells with Htt103QP plasmid were grown in 30°C raffinose medium to log phase, and then galactose (2%) was added to induce Htt103QP expression. After 1 h of induction, glucose (2%) was added into the medium to shut off Htt103QP expression. Cells were collected at the indicated times, and Htt103QP protein levels were determined by Western blotting with anti-GFP antibody. Pgk1, loading control. Right, ratio change of Htt103QP to Pgk1 after Htt103QP expression shut-off.
Next we assessed whether UBL-UBA proteins facilitate Htt103QP degradation after a short induction. For this purpose, we compared the Htt103QP degradation kinetics in WT, dsk2Δ, rad23Δ, and ddi1Δ mutants after 1 h of induction. The Htt103QP levels decreased over time in WT cells after the induction was shut off by glucose (Figure 4C). Of interest, rad23Δ but not dsk2Δ and ddi1Δ mutants showed compromised Htt103QP degradation (Figure 4C). This result suggests that Dsk2 and Ddi1 are dispensable for Htt103QP degradation after a short induction, but that cells need Rad23 for efficient Htt103QP degradation, presumably by transporting Htt103QP to proteasomes.
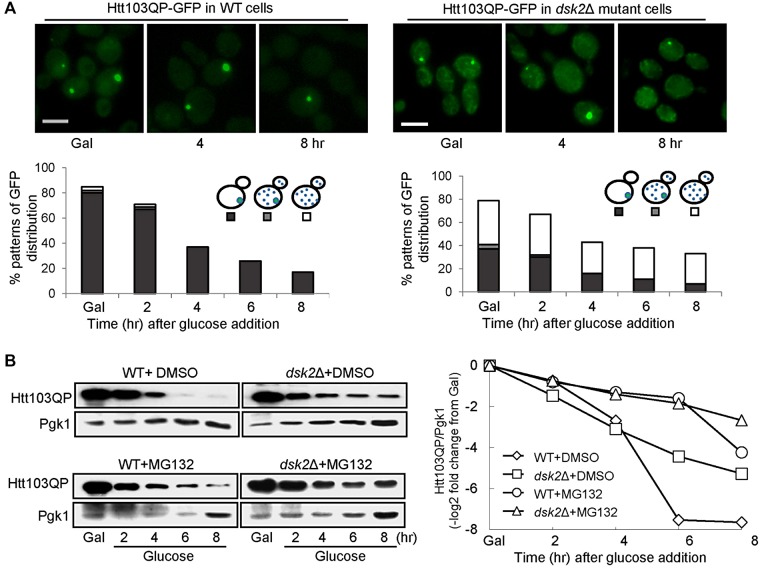
The efficient degradation of Htt103QP proteins before IB formation is likely through the proteasome. IB formation is observed in yeast cells after Htt103QP induction for ∼5 h, and Htt103QP proteins inside IBs might be degraded via distinct mechanisms, such as autophagy-dependent vacuolar degradation. If that is the case, the IB formation defect in dsk2Δ cells will impede Htt103QP clearance through the autophagy pathway. To test this possibility, we first induced Flag-Htt103QP-GFP expression in galactose medium for 12 h, which led to IB formation in WT cells. Then we added glucose and hydroxyurea (HU) into the cultures to shut off Htt103QP expression and block the cell cycle. We examined the GFP signal over time. Before glucose addition, 80% of WT cells showed IB structure, but IB structure was present in only 17% of cells after glucose addition for 8 h, and no obvious GFP signal was observed in other cells. Before glucose addition, dsk2∆ cells exhibited less efficient IB formation (37%), but 38% of cells showed multiple GFP dots. After glucose addition for 8 h, 26% of dsk2∆ cells still showed GFP aggregates (Figure 5A), indicating the less efficient clearance of Htt103QP-GFP in dsk2Δ mutants.
FIGURE 5:
Htt103QP degradation is compromised in dsk2Δ mutants after IB formation. (A) Delayed clearance of Htt103QP-GFP signal in dsk2Δ mutants after a long period of induction. WT and dsk2Δ mutant cells were grown in galactose medium for 12 h at 30°C. After addition of glucose (2%) and HU (200 mM) into the cultures to shut off Htt103QP expression and block cell cycle progression, cells were collected at the indicated time points to examine the GFP signal by fluorescence microscopy. Top, GFP signal in representative cells at 0, 4, and 8 h after glucose addition. Bottom, percentage of cells with different GFP patterns (n > 100). Scale bar, 5 μm. (B) Htt103QP degradation kinetics in WT and dsk2Δ cells after a long induction. WT and dsk2Δ mutant cells with Htt103QP plasmid were grown in galactose medium for 12 h at 30°C. The cells were pretreated with 0.003% SDS for 3 h, and then DMSO or 75 μM MG132 was added into the cultures. After incubation for 30 min, glucose (2%) and HU (200 mM) were added into the medium to shut off Htt103QP expression and block cell cycle progression. Cells were collected at the indicated times, and Htt103QP protein levels were determined by Western blotting with anti-GFP antibody. Pgk1, loading control. Right, ratio change of Htt103QP to Pgk1 over time.
We further compared Htt103QP protein level in WT and dsk2Δ mutants. After cells were grown in galactose for 12 h, the cells were pretreated with 0.003% SDS, followed by the addition of proteasome inhibitor MG132 into some cultures for 30 min. Glucose and HU were then added into the medium, and cells were collected to determine Htt103QP levels. In WT cells, Htt103QP protein levels decreased over time, and MG132 treatment delayed but did not block Htt103QP clearance (Figure 5B), indicating that functional proteasomes may contribute partially to the clearance. dsk2∆ mutant cells exhibited delayed Htt103QP degradation even in the absence of MG132. The stabilization of Htt103QP was additive in dsk2Δ cells treated with MG132 (Figure 5B). These results support the possibility that Dsk2 may promote Htt103QP clearance through a proteasome-independent mechanism.
Dsk2 promotes Htt103QP clearance through the autophagy pathway
Misfolded proteins are prone to aggregation, and the aggregates are resistant to proteasome-dependent proteolysis, but they can be delivered to lysosomes/vacuoles for disposal (Ciechanover and Kwon, 2015). To test this possibility for Htt103QP, we constructed a VPH1-mApple strain, as Vph1 is a vacuolar membrane protein (Toulmay and Prinz, 2013). The fluorescence signal indicates the vacuole membrane localization of Vph1-mApple, but VPH1-mApple cells expressing Htt103QP showed a low frequency of mApple-GFP colocalization, which could be due to the vigorous degradation of Htt103QP-GFP inside the vacuole. Therefore we examined Htt103QP-GFP localization in pep4Δ VPH1-mApple cells, which lack the key vacuole protease Pep4 (Ammerer et al., 1986). The cells were grown in galactose medium for 12 h to induce Htt103QP expression, and the mApple and GFP signals were examined after addition of glucose and HU for 2 h. Strikingly, >90% of cells exhibited colocalization of GFP with the vacuole (mApple; Figure 6A). Vph1 exhibited the ring-like structure in WT cells described previously, but we found a homogeneous distribution of Vph1 inside the vacuole in pep4Δ cells. We speculate that vacuole membrane localization of Vph1 is dynamic, and Vph1 is degraded by the hydrolase Pep4 inside the vacuole; thus deletion of the PEP4 gene leads to vacuolar Vph1 accumulation as for other vacuolar membrane proteins (Li et al., 2015).
FIGURE 6:
The delivery of Htt103QP into vacuoles is autophagy dependent. (A) The vacuolar localization of Htt103QP-GFP in WT and atg1Δ and atg8Δ mutants. pep4Δ, atg1Δ pep4Δ, and atg8Δ pep4Δ cells expressing Vph1-mApple and Htt103QP were grown in galactose medium for 12 h, and then glucose (2%) and HU (200 mM) were added into the medium to shut off Htt103QP expression and block cell cycle progression. Two hours later, the cells were collected to examine GFP and mApple signals. Representative cell images are shown for the localization of Htt103QP-GFP and Vph1-mApple. Right, percentage of GFP-mApple colocalization (n > 100). (B) Quantitative intensity analysis. Statistical dot plot of the intensity of GFP and mApple within the vacuole of WT and atg1Δ, and atg8Δ mutant cells (n = 20), as well as the medians (black bars). The p values are from the statistical comparison between WT and each mutant.
The autophagy pathway is responsible for the clearance of misfolded proteins by delivering them to lysosomes/vacuoles for degradation. In this pathway, Atg8 is a ubiquitin-like protein that is anchored to the membrane of autophagosomes and likely mediates membrane fusion with the vacuole, whereas Atg1 is required for autophagy by promoting phagophore assembly (Wen and Klionsky, 2016). To test whether the vacuolar localization of Htt103QP is dependent on the autophagy pathway, we examined the vacuolar localization of Htt103QP-GFP in atg1Δ and atg8Δ cells. Strikingly, very few mutant cells showed obvious vacuolar GFP localization (Figure 6A). Quantitative analysis also indicated a much weaker vacuolar GFP signal in atg1Δ and atg8Δ cells than in WT cells (Figure 6B). Therefore the autophagy pathway is required for the delivery of Htt103QP proteins into the vacuole, supporting the conclusion that Htt103QP is subject to autophagy-dependent disposal after IB formation.
To determine the role of Dsk2 in vacuole-dependent disposal of Htt103QP, we compared the vacuolar localization of Htt103QP in pep4Δ and dsk2Δ pep4Δ cells. As described earlier, the cells were grown in galactose medium for 12 h. After addition of glucose and HU for 2 and 4 h, the cells were collected for fluorescence microscopy. At the 2-h time point, 95% of pep4Δ and 71% of dsk2Δ pep4Δ cells showed vacuolar GFP signal. At the 4-h time point, 83% of pep4Δ and 60% of dsk2Δ pep4Δ cells exhibited obvious vacuolar GFP signal (Figure 7A). Consistently, more dsk2Δ cells showed cytoplasmic GFP aggregates without noticeable vacuolar GFP signal compared with WT cells. In addition, the vacuolar GFP signal in dsk2∆ mutants was much weaker than that in WT cells, as evidenced by the quantitative intensity of GFP and mApple signals (Figure 7B). Therefore the delivery of Htt103QP-GFP into vacuoles is compromised in dsk2∆ mutant cells. We speculate that the IB formation defect in dsk2Δ mutants contributes to the failure of autophagy-dependent vacuole delivery of Htt103QP.
FIGURE 7:
The delivery of Htt103QP into vacuoles is compromised in dsk2Δ mutants. pep4Δ VPH1-mApple and dsk2Δ pep4Δ VPH1-mApple cells with Htt103QP plasmid were grown in galactose medium for 12 h, and then glucose (2%) and HU (200 mM) were added into the medium to shut off Htt103QP expression and block cell cycle progression. The cells were collected at 2 and 4 h after glucose addition to examine the GFP and mApple signals. (A) Representative cell images for the localization of Htt103QP-GFP and Vph1-mApple (2 h), as well as the percentage of the colocalization (n > 100). (B) Statistical dot plot of the intensity of GFP and mApple signals within the vacuole of WT and dsk2Δ mutant cells (n = 20) and medians (black bars). The p values are from the statistical comparison between WT and dsk2Δ cells.
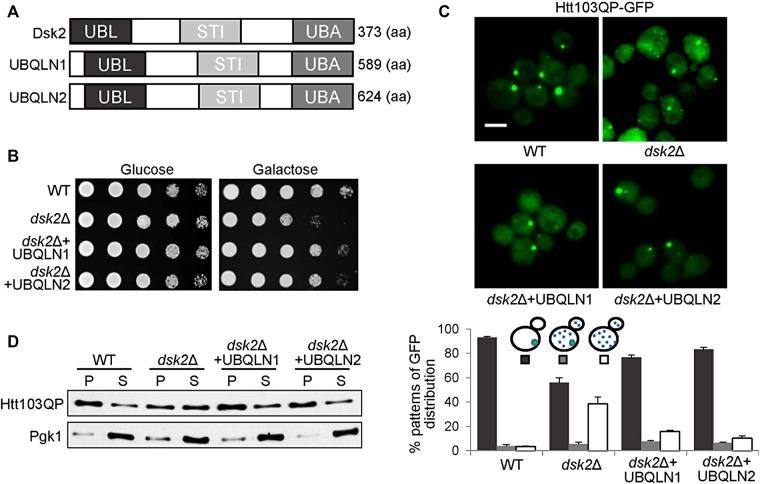
Expression of human ubiquilin 1 and 2 restores Htt103QP inclusion body formation in dsk2Δ mutants
Human ubiquilin 1 and 2 are the yeast Dsk2 homologues and also contain UBL, STI, and UBA domains (Elsasser and Finley, 2005; Lowe et al., 2006; Figure 8A). Mutations in the two ubiquilin genes are linked to neurodegenerative diseases (Takalo et al., 2013). To determine whether ubiquilin 1 and 2 have a conserved function like Dsk2 in IB formation, we constructed yeast-expressing plasmids for human ubiquilin 1 and 2 and then introduced them into dsk2Δ mutants. We found that expression of ubiquilin 1 and 2 suppressed the growth and IB formation defects in dsk2Δ mutants overexpressing Htt103QP (Figure 8, B and C). In addition, more Htt103QP proteins were detected in the pellet fraction in dsk2Δ mutant cells with the ubiquilin plasmids than in dsk2Δ mutant cells (Figure 8D). These results indicate that the function of ubiquilin proteins in IB formation is conserved from yeast to human.
FIGURE 8:
Human ubiquilin 1 and 2 suppress the phenotype of dsk2Δ mutants. (A) Schematic illustration of the domains in yeast Dsk2 and human ubiquilin (UBQLN) proteins. (B) Human ubiquilin 1 or ubiquilin 2 rescues the growth defect of dsk2Δ mutants expressing Htt103QP. WT and dsk2Δ and dsk2Δ mutants with plasmids containing human ubiquilin genes were grown to saturation, 10-fold diluted, and spotted onto synthetic glucose or galactose plates. The plates were incubated at 30°C for 2 d. (C) Human ubiquilin 1 or 2 restores Htt103QP IB formation in dsk2Δ mutants. The same strains were grown in synthetic medium containing galactose for 12 h at 30°C to induce Htt103QP expression. Top, GFP signal in representative cells. Bottom, average percentage of cells with different patterns of GFP distribution from three separate experiments (n > 100). Scale bar, 5 μm. (D) Human ubiquilin 1 or 2 restores Htt103QP distribution in detergent-soluble/insoluble factions in dsk2Δ mutants. The same yeast strains were grown in galactose medium for 12 h at 30°C, and the cell lysates were centrifuged and separated into supernatant and pellet fractions. Htt103QP protein levels were determined by Western blotting with anti-GFP antibody. Pgk1, loading control.
DISCUSSION
In addition to proteasome-mediated protein degradation, lysosomes/vacuoles play a critical role in the clearance of misfolded proteins. However, it is not fully understood how misfolded proteins are packaged and delivered into lysosomes. Using misfolded Htt103QP as a model substrate, we demonstrated the role of yeast ubiquilin Dsk2 in IB formation. Surprisingly, Dsk2 is not required for Htt103QP degradation after a short induction before noticeable IB formation, but the clearance of Htt103QP after a long induction is impaired in dsk2Δ mutants, presumably due to less efficient IB formation. We observed vacuolar localization of Htt103QP that is dependent on the autophagy pathway, but dsk2∆ mutant cells showed significantly compromised vacuolar localization of Htt103QP. Therefore our data from budding yeast suggest that ubiquilin proteins promote autophagy-mediated clearance of misfolded proteins by facilitating IB formation. The suppression of the IB formation defect in dsk2Δ mutants by the expression of human ubiquilin proteins indicates the functional conservation of this protein family.
Ubiquilin proteins have been implicated in the pathogenesis of numerous neurodegenerative diseases (Takalo et al., 2013). It appears that ubiquilin proteins play a critical role in maintaining proteostasis, but the mechanism remains elusive. The observation of delayed degradation of a proteasome substrate in ubiquilin 2 mutant cells supports the possibility that ubiquilin delivers ubiquitinated substrates to proteasome for degradation (Deng et al., 2011). However, we found that, after a short induction, the degradation of Htt103QP in dsk2Δ mutants is as efficient as in WT cells. We reason that ubiquilin/Dsk2 is dispensable for proteasome-mediated degradation of Htt103QP. In contrast, deletion of another UBL-UBA–encoding gene, RAD23, led to delayed degradation of Htt103QP after a short induction, suggesting that Rad23 likely transports ubiquitinated Htt103QP to proteasomes for degradation.
The UBA domain of Dsk2 protein is likely responsible for the interaction with ubiquitinated proteins (Ohno et al., 2005). The interaction of Htt103QP with the three UBL-UBA proteins indicates that Htt103QP is likely ubiquitinated in budding yeast. Recent work found the ubiquitination of Htt103QP in yeast cells (Yang et al., 2016). Previous data show that huntingtin is phosphorylated at serine 13 and 16 by an inflammatory kinase IKK in mammalian cells, and this phosphorylation is essential for huntingtin ubiquitination (Thompson et al., 2009). Because there is no IKK homologue in budding yeast, it is unclear how Htt103QP is ubiquitinated. One possibility is that a yeast kinase phosphorylates Htt103QP to allow its ubiquitination. Alternatively, Htt103QP could be ubiquitinated in budding yeast without phosphorylation, but further experiments are needed to test these possibilities.
The three UBL-UBA proteins in budding yeast are Dsk2, Rad23, and Ddi1. Although all three proteins show similar binding affinity to Htt103QP, only Dsk2 is required for Htt103QP IB formation. Moreover, expression of chimeric proteins containing the Dsk2 UBL domain and other domains of Rad23 or Ddi1 is able to promote IB formation. Therefore the UBL but not the UBA domain of Dsk2 contributes to its functional specificity in IB formation. It is possible that a unique interaction between the Dsk2 UBL domain and other proteins is responsible for this specificity. One candidate is the proteasome subunit Rpn10, an intrinsic ubiquitin receptor. The UBL domain of Dsk2 shows specific interaction with Rpn10. Unlike other proteasome proteins, Rpn10 is present in proteasome-bound and free forms. Previous work shows that overexpression of Dsk2 is toxic to yeast cells and results in increased accumulation of ubiquitin conjugates, but the effects of Dsk2 overexpression are alleviated by Rpn10 overexpression (Matiuhin et al., 2008), most likely due to extraproteasomal Rpn10, which restricts Dsk2’s access to the proteasome (Walters and Zhang, 2008). Therefore the nonproteasomal Rpn10 may interact with Dsk2 to facilitate IB formation. However, we cannot exclude the possibility that the interaction of Dsk2 with proteasomal Rpn10 facilitates the degradation of other protein substrates but does not play a role in Htt103QP IB formation.
The colocalization of ubiquilin with autophagosome cargo protein p62/SQSTM indicates the potential role of ubiquilin in autophagy-mediated clearance of misfolded proteins (Ceballos-Diaz et al., 2015; Osaka et al., 2015). Using budding yeast as a model organism, we show that ubiquilin Dsk2 promotes the formation of Htt103QP IBs. Moreover, we demonstrate the essential role of the autophagy pathway in the delivery of Htt103QP into the vacuole, and this delivery process is significantly impaired in dsk2Δ cells. These results support the conclusion that Dsk2 promotes the clearance of Htt103QP through the autophagy pathway. However, the link between ubiquilin proteins and the autophagy pathway remains unclear. The ubiquitin-binding protein Cue5 was shown to mediate the clearance of polyQ proteins through autophagy (Lu et al., 2014a, b). It will be of interest to test the Cue5-Dsk2 interaction and investigate the role of ubiquilin/Dsk2 in the recruitment of misfolded proteins to the autophagy machinery. Together our results demonstrate that ubiquilin proteins promote IB formation and facilitate the clearance of mutated Htt through the autophagy pathway. Moreover, this mechanism is conserved from yeast to human cells.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions
All of the yeast strains used in this study are isogenic to Y300, a W303 derivative. The relevant genotypes are listed in Supplemental Table S1. Gene deletions and epitope tagging were performed using a PCR-based protocol (Longtine et al., 1998). The 13×Myc-tagged Dsk2, Rad23, and Ddi1, dsk2UBL∆ and dsk2STI1∆ domain-deletion mutants, and UBLDsk2-RAD23, UBLDsk2-DDI1 chimeras were confirmed with PCR. The Flag- and GFP-tagged Htt103QP fragment with galactose-inducible promoter (PGALFLAG-Htt103QP-GFP) was originally from the Lindquist lab (Duennwald et al., 2006) and integrated into yeast genome. Ubiquilin 1 and 2–expressing plasmids were constructed by inserting the gene fragment of human ubiquilin 1 and 2 into a pRS415 vector. The plasmids p4458 FLAG-hPLIC-1 (ubiquilin 1) and p4455 FLAG-hPLIC-2 (ubiquilin 2), were originally from the Howley lab (Harvard Medical School; plasmids 8663 and 8661; Addgene, Cambridge, MA). Yeast extract/peptone medium supplied with raffinose, glucose, or galactose was used for the growth of yeast strains, except for those carrying plasmids.
Fluorescence image analysis
The analysis of Htt103QP IB formation in fixed cells was carried out using a fluorescence microscope (EVOS; Thermo Fisher Scientific, Waltham, MA). The cells were collected and fixed with 4% paraformaldehyde for 10 min at room temperature. Fluorescence signals from these cells were examined under a fluorescence microscope with a 60× objective.
Western blotting
Protein samples were prepared using an alkaline method and resolved by 10% SDS–PAGE. Anti-Myc antibody was purchased from Covance (Madison, WI); anti-Flag antibody was from Sigma-Aldrich (St. Louis, MO); anti-GFP antibody was from Santa Cruz Biotechnology (Santa Cruz, CA); and anti-Pgk1 antibody was from Molecular Probes (Eugene, OR). The horseradish peroxidase–conjugated goat anti-mouse immunoglobulin G secondary antibody was purchased from Jackson ImmunoResearch (West Grove, PA).
Protein fractionation assay
Cells expressing Htt103QP were collected and resuspended in RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 25 mM EDTA, 0.2% [vol/vol] Triton X-100) supplemented with protease inhibitors and phenylmethylsulfonyl fluoride and then broken using a bead beater. The lysates were centrifuged at 14,000 rpm for 30 min at 4°C to separate supernatant and pellet fractions, and the pellet was resuspended in 1× loading buffer (equal volume as the supernatant). Equal volumes of supernatant and pellet fractions were loaded and subject to Western blot analysis.
Coimmunoprecipitation assay
Cell cultures growing in galactose at 30°C for 4 h (to induce Htt103QP expression) were collected and washed once with water. After being resuspended in RIPA buffer (25 mM Tris, pH 7.5, 10 mM EDTA, 150 mM NaCl, and 0.05% Tween-20) supplied with protease inhibitors, cells were broken with a bead beater. The resulting cell extracts were incubated with primary antibody overnight at 4°C. The cell extracts were then incubated with protein A/G PLUS agarose beads (Santa Cruz Biotechnology) for 2 h at room temperature. After incubation, the beads were collected by centrifugation and washed three times with RIPA buffer supplied with protease inhibitors. After removal of RIPA buffer, protein loading buffer was added, and the protein samples were boiled for 5 min for Western blotting.
Statistical analysis
Experimental data are expressed as mean ± SEM. The distribution of fluorescence intensity is compared between WT cells and dsk2Δ mutants by using the Mann–Whitney test (two tailed) and between WT cells and atg1Δ and atg8Δ mutants by using one-way analysis of variance, followed by a Dunnett’s multiple comparison test to compare the difference between each of the mutants and WT cells. Differences with p < 0.05 are considered statistically significant.
Supplementary Material
Acknowledgments
We thank the yeast community at Florida State University for comments and suggestions. We thank Hong-Guo Yu for providing the template plasmid for the construction of the VPH1-mApple strain and Fengzhi Jin for advice on yeast strain construction. This work was supported in part by Grant RO1GM102115 from the National Institutes of Health/National Institute of General Medical Sciences to Y.W.
Abbreviations used:
- Htt
huntingtin
- IB
inclusion body
- polyQ
poly-glutamine
- UBL-UBA
ubiquitin-like and ubiquitin-associated domains
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-01-0026) on May 11, 2016.
REFERENCES
- Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- Bhat KP, Yan S, Wang CE, Li S, Li XJ. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc Natl Acad Sci USA. 2014;111:5706–5711. doi: 10.1073/pnas.1402215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos-Diaz C, Rosario AM, Park HJ, Chakrabarty P, Sacino A, Cruz PE, Siemienski Z, Lara N, Moran C, Ravelo N, et al. Viral expression of ALS-linked ubiquilin-2 mutants causes inclusion pathology and behavioral deficits in mice. Mol Neurodegener. 2015;10:25. doi: 10.1186/s13024-015-0026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Nathan DF, Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol Cell Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin LS, Olzmann JA, Li L. Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem Soc Trans. 2010;38:144–149. doi: 10.1042/BST0380144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Kwon YT. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Exp Mol Med. 2015;47:e147. doi: 10.1038/emm.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Yoo J, Bettinger BT, Amberg DC, Burke DJ. Eliminating gene conversion improves high-throughput genetics in Saccharomyces cerevisiae. Genetics. 2006;172:709–711. doi: 10.1534/genetics.105.047662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud H, Rouleau GA. A role for ubiquilin 2 mutations in neurodegeneration. Nat Rev Neurol. 2011;7:599–600. doi: 10.1038/nrneurol.2011.163. [DOI] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H, Mitsui K, Kurosawa M, Machida Y, Kuroiwa Y, Nukina N. Identification of ubiquitin-interacting proteins in purified polyglutamine aggregates. FEBS Lett. 2004;571:171–176. doi: 10.1016/j.febslet.2004.06.077. [DOI] [PubMed] [Google Scholar]
- Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Gong H, Romanova NV, Allen KD, Chandramowlishwaran P, Gokhale K, Newnam GP, Mieczkowski P, Sherman MY, Chernoff YO. Polyglutamine toxicity is controlled by prion composition and gene dosage in yeast. PLoS Genet. 2012;8:e1002634. doi: 10.1371/journal.pgen.1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Heinen C, Acs K, Hoogstraten D, Dantuma NP. C-terminal UBA domains protect ubiquitin receptors by preventing initiation of protein degradation. Nat Commun. 2011;2:191. doi: 10.1038/ncomms1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heir R, Ablasou C, Dumontier E, Elliott M, Fagotto-Kaufmann C, Bedford FK. The UBL domain of PLIC-1 regulates aggresome formation. EMBO Rep. 2006;7:1252–1258. doi: 10.1038/sj.embor.7400823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalchman MA, Graham RK, Xia G, Koide HB, Hodgson JG, Graham KC, Goldberg YP, Gietz RD, Pickart CM, Hayden MR. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J Biol Chem. 1996;271:19385–19394. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J Mol Biol. 2006;356:1027–1035. doi: 10.1016/j.jmb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie P, Snapp EL. Formation and toxicity of soluble polyglutamine oligomers in living cells. PLoS One. 2010;5:e15245. doi: 10.1371/journal.pone.0015245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassle M, Blatch GL, Kundra V, Takatori T, Zetter BR. Stress-inducible, murine protein mSTI1. Characterization of binding domains for heat shock proteins and in vitro phosphorylation by different kinases. J Biol Chem. 1997;272:1876–1884. doi: 10.1074/jbc.272.3.1876. [DOI] [PubMed] [Google Scholar]
- Li M, Rong Y, Chuang YS, Peng D, Emr SD. Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol Cell. 2015;57:467–478. doi: 10.1016/j.molcel.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Liu C, Apodaca J, Davis LE, Rao H. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques. 2007;42:158. doi: 10.2144/000112389. 160, 162. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lowe ED, Hasan N, Trempe JF, Fonso L, Noble ME, Endicott JA, Johnson LN, Brown NR. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr D Biol Crystallogr. 2006;62:177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]
- Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014a;158:549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- Lu K, Psakhye I, Jentsch S. A new class of ubiquitin-Atg8 receptors involved in selective autophagy and polyQ protein clearance. Autophagy. 2014b;10:2381–2382. doi: 10.4161/15548627.2014.981919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- Matiuhin Y, Kirkpatrick DS, Ziv I, Kim W, Dakshinamurthy A, Kleifeld O, Gygi SP, Reis N, Glickman MH. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende-Mueller LM, Toneff T, Hwang SR, Chesselet MF, Hook VY. Tissue-specific proteolysis of Huntingtin (htt) in human brain: evidence of enhanced levels of N- and C-terminal htt fragments in Huntington’s disease striatum. J Neurosci. 2001;21:1830–1837. doi: 10.1523/JNEUROSCI.21-06-01830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno A, Jee J, Fujiwara K, Tenno T, Goda N, Tochio H, Kobayashi H, Hiroaki H, Shirakawa M. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure. 2005;13:521–532. doi: 10.1016/j.str.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Osaka M, Ito D, Yagi T, Nihei Y, Suzuki N. Evidence of a link between ubiquilin 2 and optineurin in amyotrophic lateral sclerosis. Hum Mol Genet. 2015;24:1617–1629. doi: 10.1093/hmg/ddu575. [DOI] [PubMed] [Google Scholar]
- Rutherford NJ, Lewis J, Clippinger AK, Thomas MA, Adamson J, Cruz PE, Cannon A, Xu G, Golde TE, Shaw G, et al. Unbiased screen reveals ubiquilin-1 and -2 highly associated with huntingtin inclusions. Brain Res. 2013;1524:62–73. doi: 10.1016/j.brainres.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- Stieren ES, El Ayadi A, Xiao Y, Siller E, Landsverk ML, Oberhauser AF, Barral JM, Boehning D. Ubiquilin-1 is a molecular chaperone for the amyloid precursor protein. J Biol Chem. 2011;286:35689–35698. doi: 10.1074/jbc.M111.243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Kikuchi S, Katada S, Nagai Y, Nishizawa M, Onodera O. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet. 2008;17:345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- Takalo M, Salminen A, Soininen H, Hiltunen M, Haapasalo A. Protein aggregation and degradation mechanisms in neurodegenerative diseases. Am J Neurodegener Dis. 2013;2:1–14. [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Tanaka F, Robitschek J, Sandoval CM, Taye A, Markovic-Plese S, Fischbeck KH. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet. 2003;12:749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, Martinez-Vincente M, Arrasate M, O’Rourke JG, Khashwji H, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Toulmay A, Prinz WA. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J Cell Biol. 2013;202:35–44. doi: 10.1083/jcb.201301039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Walker FO. Huntington’s disease. Semin Neurol. 2007;27:143–150. doi: 10.1055/s-2007-971176. [DOI] [PubMed] [Google Scholar]
- Walters KJ, Zhang N. Rpn10 protects the proteasome from Dsk2. Mol Cell. 2008;32:459–460. doi: 10.1016/j.molcel.2008.10.020. [DOI] [PubMed] [Google Scholar]
- Wang H, Monteiro MJ. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun. 2007;360:423–427. doi: 10.1016/j.bbrc.2007.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Meriin AB, Zaarur N, Romanova NV, Chernoff YO, Costello CE, Sherman MY. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J. 2009;23:451–463. doi: 10.1096/fj.08-117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Klionsky DJ. An overview of macroautophagy in yeast. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Hao X, Cao X, Liu B, Nystrom T. Spatial sequestration and detoxification of Huntingtin by the ribosome quality control complex. Elife. 2016;5:e11792. doi: 10.7554/eLife.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.