Psychosine, a lysogalactosylceramide, induces cellular multiploidization. This psychosine-triggered multiploidization is modulated by cellular sphingolipids, with glycosphingolipid and sphingomyelin having opposite effects. Psychosine suppresses the sphingomyelin cluster and subsequent PIP2 production in cells undergoing mitosis.
Abstract
Endomitosis is a special type of mitosis in which only cytokinesis—the final step of the cell division cycle—is defective, resulting in polyploid cells. Although endomitosis is biologically important, its regulatory aspects remain elusive. Psychosine, a lysogalactosylceramide, prevents proper cytokinesis when supplemented to proliferating cells. Cytokinetic inhibition by psychosine does not inhibit genome duplication. Consequently cells undergo multiple rounds of endomitotic cell cycles, resulting in the formation of giant multiploid cells. Here we successfully quantified psychosine-triggered multiploid cell formation, showing that membrane sphingolipids ratios modulate psychosine-triggered polyploidy in Namalwa cells. Among enzymes that experimentally remodel cellular sphingolipids, overexpression of glucosylceramide synthase to biosynthesize glycosylsphingolipids (GSLs) and neutral sphingomyelinase 2 to hydrolyze sphingomyelin (SM) additively enhanced psychosine-triggered multiploidy; almost all of the cells became polyploid. In the presence of psychosine, Namalwa cells showed attenuated cell surface SM clustering and suppression of phosphatidylinositol 4,5-bisphosphate production at the cleavage furrow, both important processes for cytokinesis. Depending on the sphingolipid balance between GSLs and SM, Namalwa cells could be effectively converted to viable multiploid cells with psychosine.
INTRODUCTION
During somatic cell division, the mother cell replicates chromosomes and redistributes the intracellular contents to ensure the functional properties of the two daughter cells. Cytokinesis is the final step of mitosis, which divides daughter cells after appropriate segregation of the duplicated cellular contents (Carmena, 2008). In cytokinetic cells, the cleavage furrow—an indentation of the plasma membrane between two nascent daughter cells—further matures into a microtubule-derived midbody (Steigemann and Gerlich, 2009). Endomitosis is a special kind of cell cycle in which only cytokinesis is defective in the mitotic phase, enabling cells to increase cellular size and ploidy. However, the overall process of ensuring proper endomitosis has remained elusive, particularly regarding the membrane molecules involved and how this important mitotic event is regulated.
The cellular membrane is composed of lipids and embedded proteins, and various cell membrane activities are affected by lipids as constituents and/or signaling molecules. One class of membrane lipid constituents is made up of sphingolipids, biosynthesized from sphingosine and its acylated form, ceramide (Merrill and Sandhoff, 2002). Glycosphingolipids (GSLs), a glycosylated class of sphingolipids, comprise one of the major membrane components. GSLs are biosynthesized by glycosylation of ceramide, the lipid component of most GSLs. Psychosine is a galactosylsphingosine, also called a lysogalactosylceramide, that lacks the fatty acid amide bonded to sphingosine in ceramide. Psychosine exhibits various cellular activities when supplied to cell culture (Hannun and Bell, 1987, 1989; Suzuki, 1998; Lloyd-Evans et al., 2003a). Among them, psychosine induces polyploidization of cultured cells due to inhibition of cytokinesis. When the human monocytic cell line U937 was treated with psychosine, the DNA content of the cells increased to a ploidy of 32N (undergoing four rounds of failed cytokinesis in single cells) within 72 h of culture (Kanazawa et al., 2000). Time-lapse imaging indicated that polyploidization was achieved by a combination of cytokinetic prevention, lack of polyploidy-mediated apoptosis, and progression to S phase to duplicate chromosomes. Accordingly, the cell size was enlarged in multiploid cells arising from psychosine treatment.
It is noteworthy that, unlike chemical compounds that inhibit cytokinesis, psychosine is a naturally occurring mammalian lipid molecule, although knowledge about its natural expression is limited (Svennerholm et al., 1980; Suzuki, 1998; Kozutsumi et al., 2002). Psychosine is found in the brains of patients with the lysosomal storage disease globoid cell leukodystrophy (GLD). GLD patients have a genetic mutation of the lysosomal degradation enzyme β-galactosylceramidase, resulting in major accumulation of galactosylceramide (Miyatake and Suzuki, 1972; Wenger et al., 1974). Although its biosynthetic pathway is not established, psychosine causes the pathogenesis of GLD due to neuronal toxicity (Svennerholm et al., 1980; Shinoda et al., 1987; Suzuki, 1998). A giant polyploid cell, called a globoid cell, occurs in GLD patients, the presence of which is used as a diagnostic marker for the disease. A previous report indicated that similar giant multiploid cell formation is induced by psychosine (Kanazawa et al., 2000). Thus psychosine-mediated multiploid formation is regarded as a key event for globoid cell formation, although the molecular basis of formation is still elusive. When the intracellular events were examined, psychosine induced dispersal of TGN46-positive vesicles in COS-7 cells; thus vesicular intracellular trafficking event(s) downstream of the Golgi apparatus could be affected by psychosine to induce multiploid cells (Kanazawa et al., 2008). Attenuated vesicle transport caused by psychosine could affect delivery of both membrane proteins and lipids to the cleavage furrow.
In general, cellular membranes consist of glycerophospholipids, cholesterol, and sphingolipids. Sphingolipids consist of sphingosine with various modifications. Ceramides (Cers) are N-acylated sphingosine molecules that can be further modified with various polar head groups. The hydrophilic head group is provided by glycans or phosphocholine, which produce GSLs and sphingomyelin (SM) in the Golgi apparatus. A cytokinesis study in sea urchin eggs showed that the ganglioside Gal β1-3 GalNAc β1-4 (Sia α2-3) Gal β1-4 Glc β1-1 Cer (GM1), a GSL species, and cholesterol-enriched lipid microdomains accumulate during furrow progression (Ng et al., 2005). More recently, it was reported that the midbody of cytokinetic HeLa cells accumulates sphingolipid species such as C-24 hexosylceramide (Atilla-Gokcumen et al., 2014). Cellular SM clusters (here we define a SM cluster as a small aggregate composed of <10 SM molecules), which were stained using the earthworm toxin lysenin, were also suggested to be involved in phosphoinositide metabolism for cytokinesis completion (Abe et al., 2012). Hence sphingolipids appeared to play important roles in cytokinesis, but it is not clear how GSLs are involved in cytokinesis or related trafficking events.
In the present study, we examine the effects of cellular sphingolipids on the production of polyploid cells in the presence of psychosine. We quantitatively analyze psychosine-mediated polyploidization in the human B cell line Namalwa, which exhibits robust susceptibility to the effects of psychosine and is more responsive than U937 and COS-7 cells, which were shown in previous studies to form multiploid cells upon psychosine treatment (Kanazawa et al., 2000, 2008). Further sphingolipid perturbation studies show that cellular GSL and SM levels modulate cellular susceptibility to psychosine-induced polyploidization. We find evidence that membrane SM clustering is disrupted by psychosine. These data suggest that psychosine-triggered polyploidization may be sensitive to ratios of glycosphingolipids and sphingomyelin.
RESULTS
Lack of correlation between TDAG8 expression and psychosine-triggered cytokinetic defects
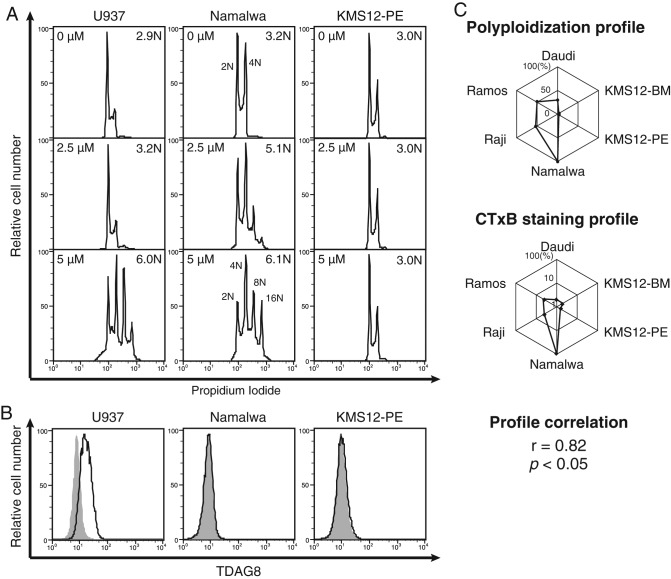
Psychosine is a lyso form of galactosylceramide. The biosynthetic pathway to formation of lyso-GSL is not known. Psychosine expression is observed in β-galactosylceramidase–deficient animals. Because psychosine elicits various cellular responses (Igisu and Suzuki, 1984; Hannun and Bell, 1987; Okajima and Kondo, 1995; Lloyd-Evans et al., 2003b), the mechanism of how cells respond to psychosine has been debated. It was previously suggested that the G protein–coupled receptor TDAG8 is a receptor for psychosine because TDAG8-expressing cells acquire psychosine sensitivity to induce multiploidy in RH7777 and HEK293 cells (Im et al., 2001). Subsequently TDAG8 was also proposed as a proton-sensing receptor (Wang et al., 2004). Furthermore, it was shown that macrophages from Tdag8-null mice undergo multiploidization upon psychosine treatment (Radu et al., 2006). The receptor responsible for psychosine-mediated polyploidization has yet to be identified. We compared psychosine-mediated polyploidization in U937, Namalwa, and KMS12-PE cell lines to examine the basis for induced cellular ploidy (Figure 1A). Among these cells, Namalwa B lymphoma cells were the most sensitive according to detection of 8/16N cells with 2.5 μM psychosine treatment. U937 cells were less sensitive than Namalwa cells, and myeloma KMS12-PE cells were not polyploidized with 5 μM psychosine. To determine whether TDAG8 expression correlates with psychosine-mediated multiploid cell nucleation, we examined its expression level in these cell lines (Figure 1B). TDAG8 was detected in U937 cells, whereas Namalwa and KMS12-PE cells were negative for staining. The finding that TDAG8-negative Namalwa cells had the highest sensitivity to psychosine is consistent with results in Tdag8-null macrophages, in which Radu et al. (2006) showed that TDAG8 does not seem to be involved in psychosine-induced multiploidy. Thus it is unlikely that TDAG8 functions as a specific receptor of psychosine to cause cytokinetic defects.
FIGURE 1:
Cross-cell profiling of psychosine-mediated polyploidization and cellular factors. (A) Polyploidization of psychosine-treated cells. U937, Namalwa, and KMS12-PE cells were treated with 2.5 or 5 μM psychosine for 2 d before harvesting and measuring cellular DNA content by propidium iodide staining. Degree of multiploidy was expressed as average nuclear content value, where 2N represents normal diploid cells. (B) Expression of TDAG8. The same set of cell lines was assessed for TDAG8 expression. Cells were stained with anti-TDAG8 antibody and evaluated using FCM. (C) Positive correlation between the cross-cell profiles for GM1 level and psychosine-mediated polyploidization. Top, relative psychosine-mediated polyploidization profile among a set of six cell lines plotted in web-graph format. Relative PPIN values are expressed on the diagonal lines of a hexagon, with the plots located at the edge of the hexagon indicating stronger polyploidization. Cells with the strongest value were set to 100%. Middle, relative GM1 expression profile obtained by FCM staining using CTxB plotted in web-graph format. Owing to the use of fluorescence signals, data are plotted on a log scale. Bottom, Pearson’s r between these profiles and associated p value.
Quantitative determination and profiling of psychosine-mediated multiploidy
Psychosine susceptibility and resulting ploidy varied among cell types (Figure 1A). Therefore psychosine-induced multiploidy was quantified using six different B cell lines because quantitative profiling and correlation analyses of cellular phenotypes can be useful in uncovering genetic traits (Yamamoto et al., 2007). Lacking a standard procedure to quantitatively evaluate polyploidizing activity among different cell lines, we measured the nuclear status of the cell lines upon treatment with a graded dose of psychosine. Dose responses of each cell line were different for multiploidization. Therefore, to accurately quantify psychosine-mediated multiploidy, we determined the percentage of >4N cells with incremental doses of psychosine. For normalization, this value was divided by the concentration of psychosine used for each condition. The maximal value was used for each cell line to quantitatively express sensitivity for psychosine-mediated polyploidization. This value was called the psychosine-mediated ploidy index number (PPIN). When the six–cell line profile of PPIN was expressed as a web graph (Figure 1C), a similarity was found in the pattern with that of cell surface GM1 expression level, measured with the cholera toxin B subunit (CTxB), as in a previous study with the same set of cell lines (Takematsu et al., 2011). Pearson’s r between these profiles was positive (0.82). The presence of such a strong positive correlation suggested that the cell surface GM1 level can affect psychosine-mediated multiploid cell formation.
Requirement of glycosphingolipids in efficient psychosine-triggered multiploidization
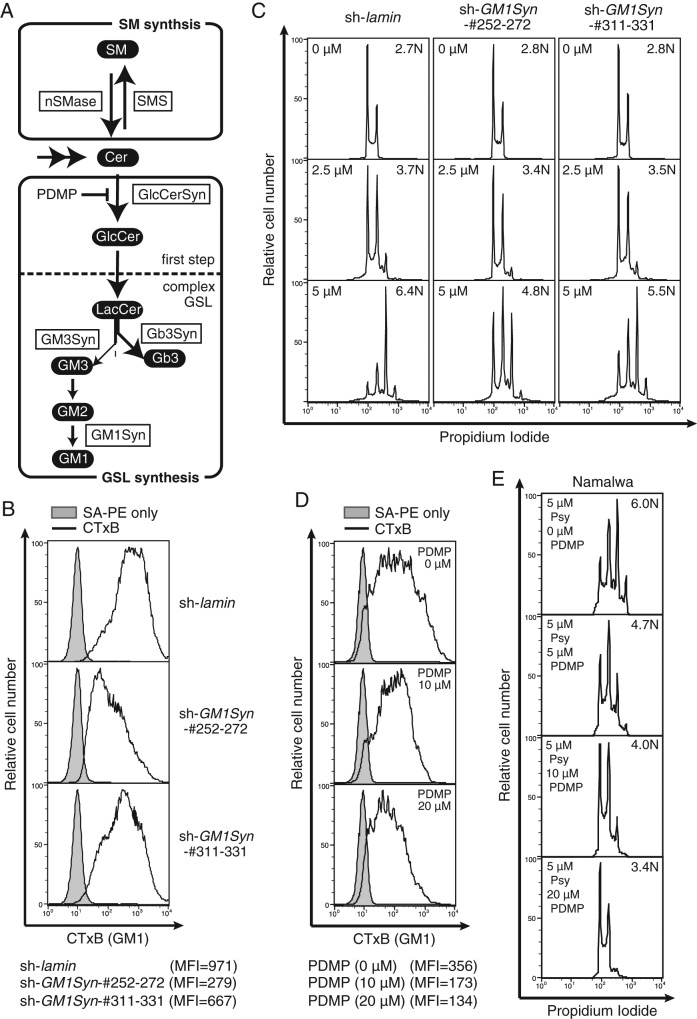
The GM1 level was hypothesized to be a cellular factor determining psychosine sensitivity. GSL expression has a propensity to be cell type and state specific (Kannagi et al., 1983; Hakomori and Zhang, 1997; Sonnino et al., 2007). To evaluate this positive correlation functionally, we chose Namalwa cells for the remainder of the study because they are sensitive to polyploidization. To examine the functional participation of cellular GSLs (the biosynthetic pathway is summarized in Figure 2A) in psychosine-triggered multiploidization, we examined the effect of GM1 knockdown by means of short hairpin RNA (shRNA)–treated Namalwa cells. Two shRNA species for GM1 synthase (GM1Syn; encoded by B3GALT4) exhibited a knockdown effect on CTxB staining, which probes cell surface GM1 expressed on Namalwa cells with high sensitivity (Takematsu et al., 2011). The shRNA corresponds to nucleotide residues 252–272 (Sh252-272) and 311–331 (Sh311-331), which exhibited roughly 70 and 30% reduction in flow cytometric CTxB staining relative to the control (ShLaminB), respectively (Figure 2B). The knockdown also resulted in loss of GM2 in liquid chromatography (LC)–mass spectrometry (MS) detection, whereas the Sia α2-3 Gal β1-4 Glc β1-1 Cer (GM3) level was not affected (Supplemental Figure S1). More prominent suppression in multiploidization was found in Sh252-272, as peak ploidy shifted to 4N in these cells. Although peak ploidy was 8N, similar to control, a relative increase in 2N/4N peaks was found in Sh311-331 (Figure 2C). These data indicate that cell surface GM1 levels can quantitatively modulate psychosine-triggered multiploidy.
FIGURE 2:
Reduced polyploidization of Namalwa cells with decreased GM1. (A) Biosynthetic pathway of sphingolipids in Namalwa cells. Lipids are indicated in white letters on black, and enzymes are indicated by boxed black letters. Inhibitors used in this study are depicted in black letters. (B) CTxB staining of GM1Syn-knockdown Namalwa cells. Namalwa cells were infected with lentivirus encoding shRNA for control (lamin) and GM1Syn (B3GALT4) and stained with CTxB. Gray indicates control staining. Bottom, mean fluorescence intensity (MFI) values of the CTxB staining. (C) GM1Syn-knockdown cells were treated with psychosine, and ploidy of the cells was analyzed as in Figure 1A. (D) Namalwa cells were treated with various doses of PDMP overnight. Cell surface GM1 level was examined using CTxB as in Figure 2B. (E) Namalwa cells were treated with a graded dose of PDMP and 5 μM psychosine (Psy) for 2 d, and ploidy of the cells was analyzed as in Figure 1A.
To examine whether psychosine-triggered multiploidy can be artificially manipulated by chemical inhibitors, we treated cells with (d-threo)-1-phenyl-2decanoylamino-3- morpholino-1-propanol (PDMP), a glucosylceramide synthase (GlcCerSyn) inhibitor that reduces cellular GSLs (Figure 2A; Inokuchi et al., 1989). The dosage of PDMP was carefully determined because its toxicity can cause cell cycle arrest. Addition of PDMP up to 20 μM did not attenuate proliferation of Namalwa cells (Supplemental Figure S2). PDMP treatment dose dependently reduced cell surface GM1 expression in Namalwa cells (Figure 2D). In the absence of PDMP, psychosine treatment resulted in the induction of 8N and 16N peaks. These multiploid peaks were suppressed with graded doses of PDMP (Figure 2E). These data show that levels of cell surface GM1, which is reduced by knockdown of GM1Syn or PDMP treatment, can affect psychosine-mediated polyploidization.
Evaluation of the involvement of GSL species in polyploidy
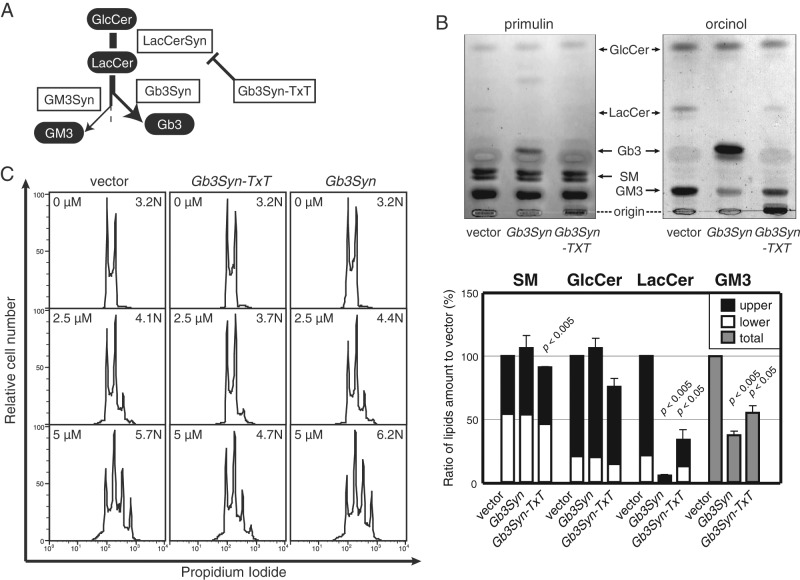
Cellular GSLs are present in multiple molecular species, and their balance varies among cells. Moreover, cellular metabolism of various classes of lipids is interconnected. Thus, in general, caution should be taken in evaluating experimental results from inhibitor treatment because these chemicals can simultaneously alter other, unexpected cellular metabolic pathway(s). To circumvent these potential pitfalls, we also examined cells with GSLs altered via modulation of glycosyltransferase gene expression. The major GSL species in Namalwa cells were previously shown to be in the ganglio series (Takematsu et al., 2011). Human B cells show drastic alteration in GSL species; activated germinal center B cells remodel major GSL species from the ganglio series (such as GM3, GM1) to the globo series (such as Gal α1-4 Gal β1-4 Glc β1-1 Cer [Gb3]). This was found to be due to the genetic dominance of Gb3 synthase (Gb3Syn, lactosylceramide α1-4 galactosyltransferase, encoded by A4GALT) at a pathway branch (Keusch et al., 2000; Kojima et al., 2000; Takematsu et al., 2011; Figure 3A). This dominant effect of Gb3Syn was exploited to modulate GSL expression. Of importance, Gb3Syn is a dual-function glycosyltransferase; in addition to α1-4 galactosyltransferase activity, Gb3Syn can form an intra-Golgi complex with Gal β1-4 Glc β1-1 Cer (LacCer) synthase (encoded by B4GALT6). Consequently mutant Gb3Syn (Gb3Syn-TxT, in which the DxD motif of the enzyme was converted to TxT) reduced LacCer and ganglio-series GSLs in a dominant-negative manner without biosynthesizing the globo series (Figure 3B, Supplemental Figure S3, and Supplemental Table S1; Takematsu et al., 2011). Therefore Gb3Syn-TxT cells were useful for evaluating the functional importance of overall GSL expression in psychosine-triggered polyploidy without using chemical inhibitors. Consistent with PDMP-mediated GSL inhibition (Figure 2E), Gb3Syn-TxT cells (exhibiting global GSL reduction downstream of LacCer) were also less susceptible to psychosine-mediated polyploidization, showing a prominent decrease in both 8N and 16N peaks and a consequent increase in the 2N peak in the presence of psychosine (Figure 3C). Unlike PDMP treatment, Gb3Syn-TxT cells did not alter the GlcCer level (Figure 3B and Supplemental Figure S3; Takematsu et al., 2011), and yet similar suppression of psychosine-mediated polyploidization was found in these cells (Figure 3C). This result was consistent with the suggestion that reduced GM1 can negatively modulate psychosine-triggered multiploidization. To examine the level effects of different glycolipid species, we introduced Gb3Syn into Namalwa cells, in which the major GSL species was shifted to Gb3 (Figure 3B; Takematsu et al., 2011). Unlike Gb3Syn-TxT cells, Gb3Syn cells exhibited comparable multiploidization upon psychosine addition. This result indicates that psychosine-mediated multiploidization is not altered by the change from ganglio- to globo-series GSL(s) (Figure 3C). These results suggest that specific GSL species such as GM1 and Gb3 can have similar effects in enhancing psychosine-triggered multiploidization.
FIGURE 3:
Effects of molecular GSL species. (A) Flowchart of GSL biosynthetic pathway branching in Namalwa cells. Gb3Syn dominantly regulates the biosynthetic pathway, and thus can effectively alter GSL profiles in Namalwa cells. (B) TLC analysis of sphingolipid species in vector, Gb3Syn, and Gb3Syn-TxT cells. Namalwa cells transfected with MSCV-IRES-EGFP virus (vector), MSCV-Gb3Syn-IRES-EGFP virus (Gb3Syn), and MSCV-Gb3Syn-TxT-IRES-EGFP virus (Gb3Syn-TxT) were polyclonally sorted by GFP positivity. Sphingolipids purified from these cells were analyzed by TLC. Lipids were separated in chloroform, methanol, and water (65:25:4). SM was visualized with primulin, and GSLs were visualized with orcinol-sulfate. The mobility of standard lipids is indicated. Relative mean band densities are plotted from three experimental replicates (bottom). Doublet bands on TLC were calculated separately when possible. (C) Effect of expression of Gb3Syn-TxT and Gb3Syn on psychosine-induced polyploidization. These cells were treated with psychosine, and polyploidization was measured as in Figure 1A.
Enhanced psychosine-induced multiploidization upon introduction of GlcCerSyn
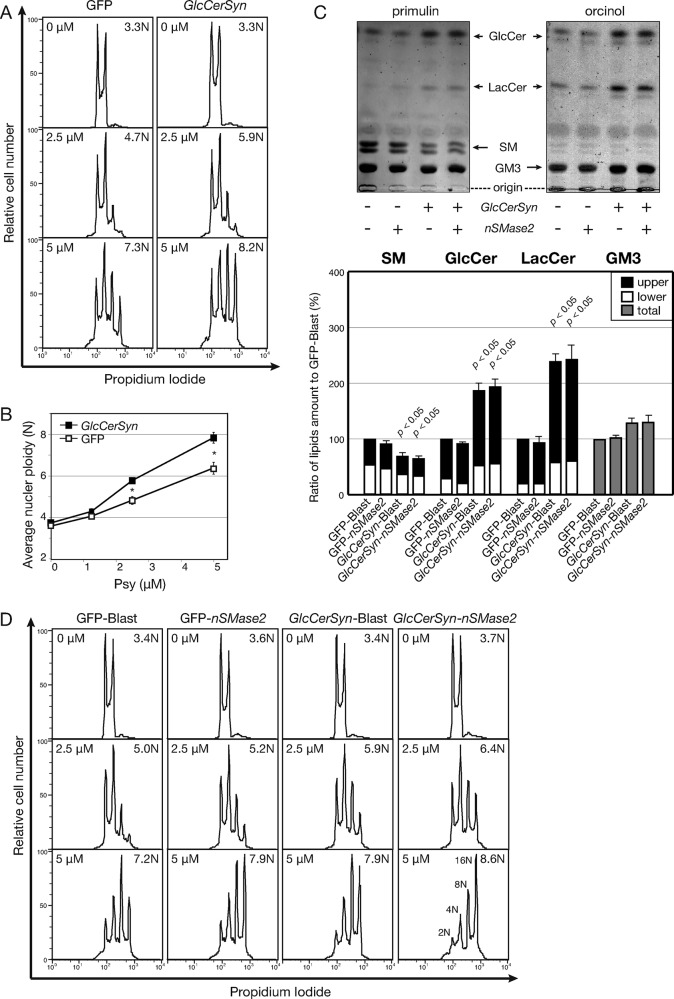
Reductions of GSL resulted in resistance to psychosine-triggered multiploidization (Figures 2D and 3C). Induction of GSL was used to examine enhanced cellular response to psychosine. GlcCerSyn (encoded by UGCG) responsible for cellular GSL levels was expressed to induce GSL levels. When GlcCerSyn was introduced into Namalwa cells, the ploidy value was not different from control cells. However, psychosine induced ploidy of the cells (Figure 4A). Because the difference in polyploidy was less than twofold, this experiment was repeated five times to confirm the reproducibility and statistical significance (Figure 4B). These data indicate that levels of GSLs quantitatively affect psychosine-triggered multiploidization.
FIGURE 4:
Enhancement of psychosine-induced polyploidization by cellular GSL. (A) Enhanced polyploidization of Namalwa cells infected with GlcCerSyn. Namalwa cells were transfected with MSCV-IRES-EGFP virus (GFP) and MSCV-GlcCerSyn-IRES-EGFP virus (GlcCerSyn) and polyclonally sorted by GFP positivity. Psychosine-triggered polyploidization was measured by FCM as in Figure 1A. (B) Quantification of GlcCerSyn-enhanced nuclear ploidy triggered by psychosine. The quantified mean nuclear ploidy from five independent experiments with graded psychosine (Psy) dose is plotted with SEM. Statistical significance was assessed using Student’s t test as indicated above the bars (*p < 0.05). (C) TLC analyses of cells transfected with GlcCerSyn and nSMase2. Namalwa cells used in Figure 3A were transfected with retrovirus MSCV-IRES-Blast virus (Blast) and MSCV-nSMase2-IRES-Blast virus (nSMase2) to produce four different types of cells. Sphingolipids were extracted from these cells and analyzed by TLC as in Figure 3B. (D) Additive effect of GlcCerSyn and nSMase2 expression on psychosine-triggered polyploidization. Psychosine-induced multiploidy was analyzed as in Figure 1A.
Additive effect of GlcCerSyn and sphingomyelinase
Modulation of GlcCerSyn expression reduces ceramide, as well as increases GSLs (Figure 2A and Supplemental Figure S3). Was the increase in GSLs or reduction in ceramide responsible for this phenotypic change? In addition to the de novo pathway, ceramide is also produced by sphingomyelinase (SMase). SM is strongly expressed on the plasma membrane and thus could serve as a source of ceramide. Among SMases, neutral SMase2 (nSMase2) functions to scramble ceramide from SM on the cell surface; thus, nSMase2 is a good candidate to alter the SM/ceramide balance (Hofmann et al., 2000; Marchesini et al., 2003; Tani and Hannun, 2007). Unlike GlcCerSyn expression, nSMase2 expression was expected to increase ceramide levels, although both of these enzymes favor the biosynthesis of GSLs downstream. When nSMase2 was expressed, ∼10% suppression in SM was detected by densitometry in TLC analyses (Figure 4C). By contrast, expression levels of Glc β1-1 Cer (GlcCer), LacCer, and GM3 were not affected. To account for the loss of SM, ceramide was increased threefold in nSMase2 cells in the lipidomic LC-MS analysis (Supplemental Figure S3). GlcCerSyn expression caused more apparent changes in GSL levels: approximate twofold increase in GlcCer and LacCer and ∼25% increase in GM3 on TLC and LC-MS (Figure 4C and Supplemental Figure S3). In both cases, we detected sensitization of Namalwa cells to psychosine-induced polyploidization (Figure 4D). These data indicate that an increase in GSLs rather than a reduction in ceramide sensitizes Namalwa cells to psychosine-mediated inhibition of cytokinesis to produce multiploid cells. The effects of GlcCerSyn and nSMase2 were additive because simultaneous expression of both enzymes resulted in the most sensitive phenotype to psychosine (Figure 4D). Of note, 2N cells were barely detectable under this condition, indicating that nearly the entire population of cells in culture was prevented from achieving cytokinesis by psychosine.
Specificity of lysosphingolipid species on multiploidy
It was previously reported that both psychosine and glucopsychosine (GlcPsy) can induce U937 multiploidization (Kanazawa et al., 2000). Because the combination of Gb3Syn-TxT Namalwa cells and GlcCerSyn/nSMase2 Namalwa cells was a useful system for analyzing the effect of membrane GSLs, we used it to examine the effect of other lysosphingolipid species on multiploidization. Consistently, as with U937 cells, GlcPsy induced Namalwa cell multiploidization, whereas lyso-LacCer and sphingosylphosphorylcholine (SPC) did not (Supplemental Figure S4A). Similar to psychosine treatment (Figure 3C), Gb3Syn-TxT cells (reduced GSLs) were less polyploidized by GlcPsy. Unlike psychosine treatment (Figure 4D), GlcCerSyn/nSMase2 expression did not enhance GlcPsy-mediated polyploidization at a concentration of 5 μM (Supplemental Figure S4B) compared with control cells. In this condition, however, 2N cells were almost undetectable even in the controls. We titrated GlcPsy concentration to examine GlcCerSyn/nSMase2-mediated enhancement in GlcPsy-triggered multiploidization. At 1.25 μM, GlcPsy more efficiently induced GlcCerSyn/nSMase2 cell multiploidy than with controls (Supplemental Figure S4C). This enhancement was not increased at 2.5 μM GlcPsy probably due to the very efficient multiploidization-stimulating activity of GlcPsy.
Cell type–specific effect of GlcCerSyn expression
Given that membrane GSL can modulate psychosine susceptibility, GlcCerSyn elevation was examined to alter the psychosine sensitivity of unresponsive cells. Myeloma KMS12-PE cells were resistant to psychosine-induced multiploidy (Figure 1A). Introduction of GlcCerSyn in KMS12-PE cells did not alter this resistance (Supplemental Figure S5). Thus psychosine sensitivity can be modulated by GSL expression levels in susceptible cells (such as Namalwa cells), but cellular GSL levels alone do not sensitize cells to psychosine; instead, there may be a bona fide receptor affected by GSL levels.
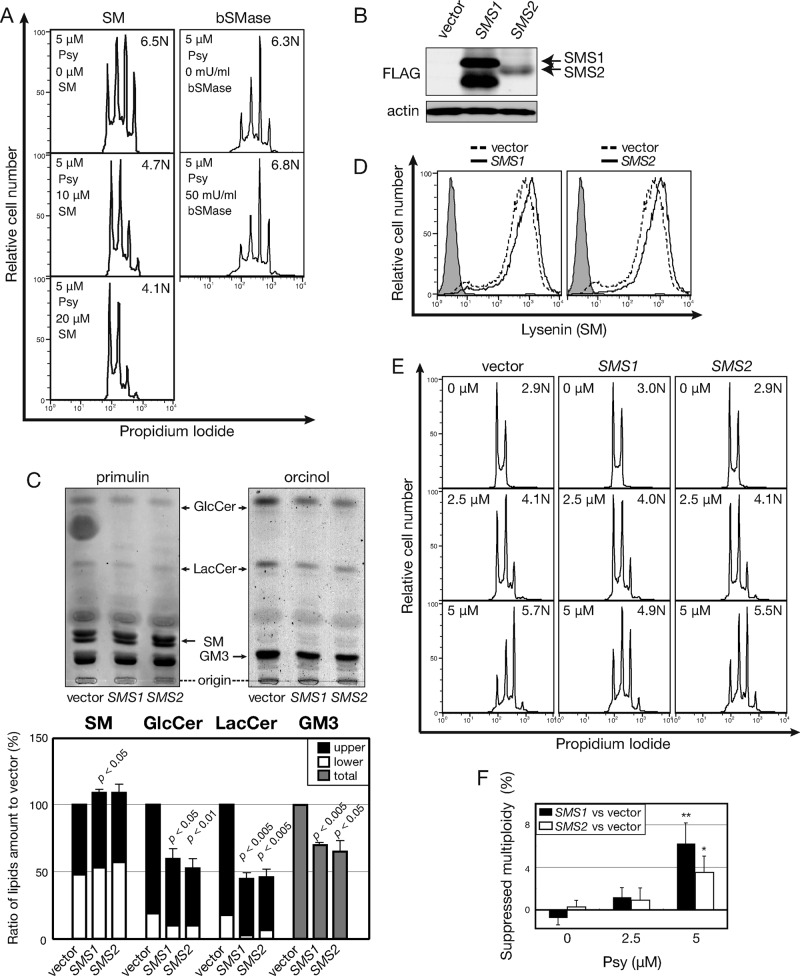
Suppression of psychosine susceptibility by sphingomyelin
Because nSMase2 cells exhibited enhanced psychosine-triggered polyploidization, we evaluated SM as a cellular factor controlling psychosine susceptibility (Figure 4D). SM was presumed to be independent of cellular GSLs because single nSMase2 introduction did not alter the GSL level (Figure 4C). We pretreated Namalwa cells with incremental doses of SM. Unlike GSL treatment of Namalwa cells, which had no effect (Supplemental Figure S6), SM treatment attenuated psychosine-mediated multiploidization in dose-dependent way (Figure 5A). When cell surface SM was digested by bacterial SMase (bSMase) treatment, which reduced SM levels and increased GSL levels (Supplemental Figure S7), enhanced psychosine-triggered multiploidy was detected (Figure 5A). Therefore SM levels can negatively modulate the psychosine effect. To perturb cellular GSL/SM biosynthesis with enzyme gene expression, we introduced SM synthase (SMS) cDNAs into Namalwa cells. SMS occurs in two isoforms: Golgi-specific SMS1 (Yamaoka et al., 2004) and plasma membrane/Golgi–specific SMS2 (Tafesse et al., 2007). In Namalwa cells, SMS1 was strongly expressed and SMS2 expression was weak, according to Western blotting of FLAG-tagged SMS proteins (Figure 5B). The apparent presence of the SM cycling pathway (Hannun, 1994) kept the cells from increasing SM levels >∼20% in SMS2 cells by TLC (Figure 5C). Lipidomic LC-MS showed a more prominent (∼30%) induction of SM in both SMS1 and SMS2 cells (Supplemental Figure S3). Therefore we used these cells for functional analyses of SM levels for psychosine-triggered multiploidization.
FIGURE 5:
Suppressed psychosine-induced polyploidization by induction of SM. (A) Effect of SM and bacterial SMase (bSMase) treatment on psychosine-induced polyploidization. Namalwa cells were treated with 10 or 20 μM SM or 50 mU/ml bSMase with 5 μM psychosine (Psy). Cellular DNA content was determined as in Figure 1A. (B) Expression of SMS1/2 as detected by Western blotting. Namalwa cells were transfected with pSP72-EF1-IRES-Blast vector (vector) and pSP72-EF1-SMS1/2-IRES-Blast vector (SMS1/2); stable transfectants were selected as polyclonal mixtures. Whole-cell lysates were prepared, and transgene-derived SMS1/2 was detected using anti-FLAG M2 antibody. (C) Effect of SMS1/2 on cellular levels of sphingolipids. Sphingolipids were isolated from these cells and analyzed by TLC as in Figure 3B. (D) Detection of cell surface SM by NT-lysenin. Induction of cell surface SM upon SMS introduction was detected using the GST-NT-lysenin probe by FCM. SM-bound lysenin was visualized as a complex with anti-GST and FITC-conjugated anti-goat IgG. Gray indicates control staining, and solid dashed black lines indicate Namalwa cells transfected with control vectors. (E) Effect of SMS1/2 expression on psychosine-induced polyploidization. SMS-transfected Namalwa cells were examined for psychosine-induced polyploidization as in Figure 1A. (F) Quantification of psychosine-mediated polyploidization in SMS-overexpressing cells. Psychosine-induced polyploidization was determined, and mean ploidy values of six independent experiments are expressed as percentage decrease from vector control. Statistical significance of the difference between SMS1 compared with vector is p < 0.05 and SMS2 compared with vector is p < 0.1.
Cell surface SM level and organization were monitored using NT-lysenin, an earthworm toxin specifically targeting SM clusters. Monomeric recombinant NT-lysenin was shown to be useful for SM detection (Ishitsuka and Kobayashi, 2004). When SMS1 and SMS2 cells were stained with NT-lysenin, consistent with the data from TLC analyses, a detectable increase in cell surface SM was observed (Figure 5D). Comparison of these cells with vector-transformed control cells showed suppressed psychosine-mediated polyploidization in both SMS1 and SMS2 cells (Figure 5E). Because the effect was subtle, the mean ploidy was calculated from six independent experiments, and results are expressed as the difference from control (Figure 5F). These data indicate that increased SM expression relative to GSL/GM1 can be a modulatory factor in psychosine sensitivity. A stronger effect in SMS1 cells than in SMS2 cells can be interpreted as a difference in the intracellular localization of these two isoforms. Collectively these results indicate that increased SM:GSL ratio reduces susceptibility to psychosine. When other parameters are fixed, SM expression negatively and GSL expression positively modulate psychosine-triggered multiploidization of Namalwa cells.
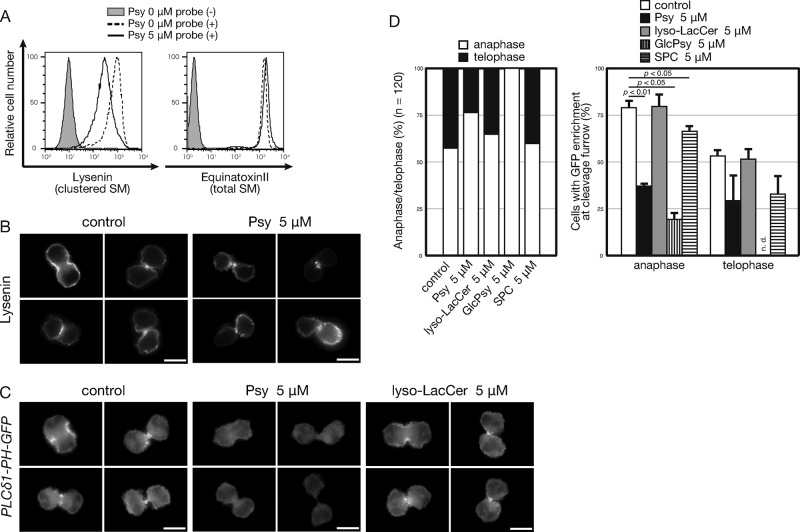
Suppressed cell surface SM clustering and phosphatidylinositol 4,5-bisphosphate production by psychosine
SM can be present on the cell membrane as clusters or domains. Here the phrase “cluster of lipids” is used to indicate small aggregates composed of <10 lipid molecules (Ishitsuka et al., 2004). By contrast, a “domain” is the specific area of the membrane with high labeling density of lipid-binding proteins. Here we examined the effect of psychosine on SM clusters. HeLa cells undergoing cytokinesis showed lysenin-positive SM cluster enrichment in the outer membrane of cleavage furrows (Abe et al., 2012). Subcellular localization of SM was examined in relation to the cleavage furrow using a monomeric NT-lysenin probe, which avoids formation of probe-mediated SM aggregation (Ishitsuka and Kobayashi, 2004). Psychosine treatment reduced NT-lysenin staining by flow cytometry in Namalwa cells (Figure 6A). Equinatoxin II is another SM-binding probe that preferentially binds to SM even when dispersed (Makino et al., 2015). In sharp contrast to lysenin, equinatoxin II stained both control and psychosine-treated cells with roughly equal intensity (Figure 6A), indicating that the abundance in cell surface SM per se was not attenuated. Thus psychosine attenuated SM clustering, which was presumably caused by cholesterol-mediated partitioning of the membrane. When psychosine-treated cells were examined under a fluorescence microscope, consistent overall reduction in lysenin staining was detected in both dividing (Figure 6B) and nondividing cells. Taken together, the data show that psychosine disrupts outer leaflet SM clustering, where phosphatidylinositol-4-phosphate 5-kinase is recruited to biosynthesize phosphatidylinositol 4,5-bisphosphate (PIP2) at the inner leaflet of the membrane (Abe et al., 2012). For further evaluation, we detected PIP2 production in the cleavage furrows of dividing cells using the PH–green fluorescent protein (GFP) probe, which contains the PH domain from PLCδ (Field et al., 2005). The cleavage furrow PIP2 level is important for furrow ingression at anaphase (Field et al., 2005). Therefore we examined psychosine-mediated loss of the cleavage furrow PH-GFP signal in cells during late mitosis. Cleavage furrows of psychosine-treated cells showed a reduced PH-GFP signal more prominently in anaphasic cells (Figure 6, C and D), indicating that psychosine disrupted SM clustering, which is associated with accumulation of PIP2 at the cleavage furrow. The number of telophasic cells (with enlongated cleavage furrow) was reduced upon psychosine treatment, probably caused by the defect in anaphase of psychosine-treated cells. Such results were consistent and more prominent in GlcPsy cells, whereas nonpolyploidizing lyso-LacCer and SPC did not exhibit any of these effects, despite similar physical properties shared by all of these lysosphingolipids (Figure 6, C and D). Therefore we propose that specific disruption of SM clusters by psychosine at the anaphase cleavage furrow causes compromised PIP2 production, which results in failed cytokinesis at a later stage.
FIGURE 6:
Alteration of cell surface SM expression upon psychosine treatment. (A) Cell surface expression of SM upon psychosine treatment. Namalwa cells were treated with 5 μM psychosine (Psy) overnight and stained with recombinant GST-NT-lysenin, which detects cluster SM as in Figure 5D, or with NT-equinatoxin II-GFP-His, for total SM. (B) Cell surface localization of the NT-lysenin signal upon psychosine treatment. Namalwa cells were treated with 5 μM psychosine overnight and stained with recombinant EGFP-NT-lysenin. EGFP fluorescence was detected using fluorescence microscopy. Four independent cells in late mitotic phases (two anaphasic and two telophasic) are shown for each condition. To represent the varied distribution found in the psychosine-treated samples, each of the four typical staining patterns is displayed. (C) Attenuated PIP2-binding PH-GFP probe signals in psychosine-treated Namalwa cells. Namalwa cells expressing PH-GFP (PLCδ1-PH-GFP) were treated with 5 μM each lysosphingolipid overnight, and the GFP signal was observed in mitotic cells. (D) Calculation of ratio of cells observed in Figure 6C. We counted 120 cells in late mitotic phases. Incidence of anaphasic and telophasic cells was determined from cell body shapes and DAPI signal. Both anaphasic and telophasic cells were assessed for enrichment of PH-GFP signal in the cleavage furrow.
DISCUSSION
In this study, we first quantified psychosine-triggered multiploid cell formation. This led to the realization that this phenomenon can be cell specific and quantitatively modulated by other cellular factor(s). Cellular sphingolipid species are modifier molecules. GSL can enhance and SM suppress multiploidy in sphingolipid-perturbed Namalwa cells. Unlike other studies on GSL-perturbation using small interfering RNAs (Atilla-Gokcumen et al., 2014), a significant increase in multiploid cells without psychosine was not detected (Figure 2C). At least in this system, altered levels of GSLs or SM per se did not trigger cytokinetic failure; instead, supplementation of psychosine triggered multiploid cell formation. Therefore GSLs and SM seemed to modulate a cytokinetic event that is inhibited by psychosine. Here almost all cells become multiploid when both GlcCerSyn and nSMase2 are overexpressed in Namalwa cells treated with psychosine (Figure 4D). In this condition, cytokinesis can be effectively disabled without disturbing genome duplication in the cell cycle. In other words, a specific balance among the three different sphingolipid classes seemed to allow cells to undergo successful endomitosis cycles rather than normal mitosis. Although this phenomenon is striking, and psychosine may be biosynthesized in certain situations, endogenous psychosine was not detected in Namalwa cells. Present data indicate that lack of psychosine in normal tissue is consistent with normal mitosis.
Relationship of sphingolipid effects to cleavage furrow regression and cytokinesis
Recent developments indicate that sphingolipids are functionally involved in cytokinesis. The present study showed that SM clustering was disturbed both at the cell surface and anaphase cleavage furrow in the presence of psychosine. By contrast, dispersed SM (stained by NT-equinatoxin) were not affected at the cell surface (Figure 6). The effect of psychosine clearly differs among these SM-binding probes. It is not clear how psychosine reduces SM clustering. A recent study showed that the presence of GSL dispersed SMs (Makino et al., 2015). Although psychosine has only one acyl chain, psychosine is a GSL and, as such, can physically disrupt SM clustering. In relation to this point, psychosine was shown to disrupt microdomains such as lipid rafts or detergent-insoluble membranes (White et al., 2009). However, it is notable that cholesterol depletion by methyl-β-cyclodextrin did not induce multiploidy in Namalwa cells (H.W. and H.T., unpublished data). Moreover, neither lyso-LacCer (Gal-Glc-sphingosine) nor N-acetyl-psychosine (cell-permeable Gal-Cer in which the sphingosine 2-amido group is the acetyl form) triggers polyploidization. It is reasonable to suppose that these nonpolyploidizing lyso/permeable-lipids disturb microdomains like psychosine. Therefore specific structural specificity seems to be responsible for the polyploidizing activity of psychosine (Kanazawa et al., 2000).
Involvement of particular sphingolipid species in cell division
GSL/GM1 enhances and SM suppresses polyploidization triggered by psychosine (Figures 4 and 5), most likely due to head group differences. This specificity may be better understood when some specific psychosine target molecules are considered. GSL and SM could modulate putative psychosine target(s). In fact, not only GSLs and/or SM participate in membrane microdomains to regulate cellular signaling (Hakomori and Igarashi, 1993; Iwabuchi et al., 2000); cellular (glyco)sphingolipids are also involved in membrane-mediated regulation of receptor(s) (Prinetti et al., 2009), cellular signaling components (Hakomori, 2000), and cellular trafficking events (Gkantiragas et al., 2001; Sprong et al., 2001). Expression of GSL regulates cell surface receptor behavior in the example of GM3 and insulin receptor signaling (Kabayama et al., 2007), although GM3 also may mediate cellular functions via GSL–GSL interactions (Kojima and Hakomori, 1991). More recently, it was shown that specific molecular species of hexosylceramides are enriched in the midbody of dividing cells (Atilla-Gokcumen et al., 2014). It was also shown that a specific molecular species of SM regulates transmembrane proteins through a specific recognition motif (Contreras et al., 2012). Regardless of the mechanisms, the present work represents a novel/rare cellular system in which GSL and SM compete in activity. This system could account for regulation of molecule(s) involved in cytokinesis. Such findings add to a further understanding of cytokinesis in which the involvement of sphingolipids is emerging (Ng et al., 2005; Abe et al., 2012; Atilla-Gokcumen et al., 2014). This finding may also contribute to understanding molecular mechanisms behind GLD pathogenesis in the formation of globoid cells, which are characteristic cells used for disease diagnosis.
Cellular homeostatic regulation of SM levels
Perturbation of cellular SM levels was a rather difficult task. In the steady state in Namalwa cells, it is likely that the SM level is homeostatically controlled by the “SM cycle” when the SM level is increased (Figures 4C and 5C; Hannun, 1994; H.W. and H.T., unpublished data). In addition to accumulation in the form of ceramide detected in lipidomic analyses (Supplemental Figure S3), nSMase2-derived ceramide seemed to be efficiently converted to GlcCer when GlcCerSyn was introduced. These results suggest that GSL biosynthesis is remotely coupled to SM metabolism (Figure 5C and Supplemental Figure S3) and Namalwa cells might have a transport system that efficiently biosynthesizes GSLs (Tidhar and Futerman, 2013). Thus psychosine-mediated multiploidy cannot be understood only from SM cellular levels. In any case, it was very consistent that GSL level positively and SM level negatively affected the cytokinetic defect caused by psychosine. Thus the sphingolipid environment of the cellular membrane is an important factor controlling cytokinesis, at least in the presence of psychosine. To our knowledge, this is the first report to quantitatively identify cellular factor(s) altering the severity of multiploidization caused by psychosine. This information is useful for further understanding the mechanisms underlying psychosine-mediated multiploidization and the regulation of the elusive uncoupling of S phase and cytokinesis leading to successful endomitosis.
MATERIALS AND METHODS
Reagents and cell culture
Psychosine, GlcPsy, lyso-LacCer, and SPC were obtained from Matreya (State College, PA) and dissolved in ethanol as a 10 mM stock solution. Sphingomyelin (from bovine brain; Sigma-Aldrich, St. Louis, MO) used for pretreatment was dissolved in ethanol as a 10 mM stock solution. PDMP was dissolved in dimethyl sulfoxide as a 10 mM stock solution. Bacterial sphingomyelinase was obtained from Sigma-Aldrich. KMS-12 PE, Namalwa, U937, KMS-12 BM, Daudi, Raji, and Ramos cells were obtained from the Japanese Cell Research Bank and cultured in RPMI 1640 medium supplemented with nonessential amino acids, sodium pyruvate, 2-mercaptoethanol, and fetal bovine serum. A total of 1 × 105 cells in 24-well plates were treated with psychosine for 2 d before harvesting for DNA content assessment. The following probe/antibodies were used: biotin-conjugated cholera toxin B subunit (List Biological Laboratories, Campbell, CA); goat anti-human TDAG8 (N-19; Santa Cruz Biotechnology, Santa Cruz, CA); goat polyclonal anti–glutathione S-transferase (GST; Amersham Pharmacia Biotech, Uppsala, Sweden); mouse anti-FLAG (M2; Sigma-Aldrich); mouse anti-actin (C4; MP Biomedical, Santa Ana, CA); fluorescein isothiocyanate (FITC)–conjugated anti-goat immunoglobulin G (IgG; Vector Laboratories, Burlingame, CA); phycoerythrin (PE)-conjugated streptavidin (Caltag, Burlingame, CA); and horseradish peroxidase–conjugated anti-mouse IgG (Invitrogen, Carlsbad, CA).
Flow cytometry
Cells were stained with antibodies or recombinant toxin subunit in fluorescence-activated cell sorting buffer (1% bovine serum albumin, 0.1% NaN3 in phosphate-buffered saline [PBS]). After washing step(s), stained cells were analyzed by flow cytometry using FACScan or FACSCalibur (BD Biosciences, Franklin Lakes, NJ), and data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Quantitative determination of cellular ploidy
Multiploidy of the cells was examined as reported previously (Kanazawa et al., 2000). Briefly, cells treated with psychosine for 2 d were fixed in 70% ethanol overnight. After washing with PBS, the cells were incubated with RNase A (DNase-free; Nacalai Tesque, Kyoto, Japan) to remove RNA. The resultant cellular DNA was stained with propidium iodide (Nacalai Tesque) for subsequent flow cytometry (FCM) analyses using a FACScan or a FACSCalibur flow cytometer for FL-2 channel detection. To exclude doublets, cells were gated using the FL-2 Area and FL-2 Width. Average cell ploidy was calculated from the abundance ratio of each nuclear phase. In the multiploidization experiments, the comparison was made only using control cells cultured side by side. The PPIN value was developed to profile psychosine-induced multiploidy in six different B-cell lines, in which normalization of the cell-to-cell differences of psychosine sensitivity seemed appropriate. The concentration of psychosine to achieve maximum multiploidy was determined. Values of the maximum percentage of each cell with a ploidy >4N were divided by the concentration of psychosine to calculate the PPIN for normalization of cell variations in psychosine sensitivity and degree of multiploidy. When PPIN was not needed, average ploidy was calculated and is depicted on the upper right of the histograms.
shRNA-mediated knockdown of GM1 synthase
Target sequences for GM1Syn (B3GALT4) shRNA were chosen from the National Center for Biotechnology Information (NCBI) database (NCBI Reference Sequence NM_003782.3). Oligonucleotide corresponding to 252–272 and 311–331 was inserted into pENTR-U6 vector (Invitrogen) and transferred to pLenti6 (Invitrogen) lentivirus expression vector. Lentivirus plasmid vectors were transiently transfected into 293FT packaging cells, and lentivirus produced in the medium was used for subsequent infection. Infected cells were polyclonally selected in the culture medium containing 10 μg/ml blasticidin S for at least 3 wk.
Mass spectrometry analyses of gangliosides
For the extraction of total lipid, Namalwa cells with knockdown GM1Syn were suspended in chloroform:methanol (1:1), and the mixture was sonicated, incubated at 40°C for 1 h, and then centrifuged. The supernatant was retained, and the pellet was subjected to the same extraction by the same procedure using chloroform:methanol (1:2). The first and second supernatants were combined and evaporated. GM3(d18:1-[13C]16:0) was added for internal standards to the sample. The extracted lipids were subjected to mild alkaline hydrolysis and desalted with a Bond-elute C18 (Agilent Technology, Santa Clara, CA).
GM3 and GM2 molecular species were quantified using high-performance liquid chromatography (HPLC) coupled with electrospray ionization tandem mass spectrometry in multiple reaction monitoring negative-ionization mode. A triple-stage quadrupole Vantage AM instrument (Thermo Fisher, Waltham, MA) was calibrated by directly infusing a mixture of GM3 species extracted from milk; all ion source parameters and ionization conditions were optimized to improve sensitivity. Total lipid extracts from cells were dissolved in methanol and injected into an Accela 1250 HPLC pump (Thermo Fisher) and separated using a Develosil carbon 30 column (C30-UG-3-1 ×50 mm; Nomura Chemical Co., Seto, Japan). The gradient started with 100% solvent A (20% H2O/50% 2-propanol/30% methanol containing 0.1% acetic acid and 0.1% ammonia) for 5 min and then ramped to 100% solvent B (2% H2O/50% 2-propanol/48% methanol containing 0.1% acetic acid and 0.1% ammonia) over 30 min. One hundred percent solvent B was maintained for 4 min, and then the solvent was returned to 100% solvent A over 1 min and held there for 10 min. The flow rate throughout the chromatographic run was 50 μl/min.
The abundance of each molecular species was compared based on the relative percentage of the internal standard GM3(d18:1-[13C]16:0). Total GM3 and GM2 values were calculated by taking the sum of each molecular species detected, with the assumption that all species had similar ionization efficiency, comparable to that of the internal standard (Supplemental Figure S1). The signal strength for each lipid species is tabulated in Supplemental Table S2.
Lipidomic analyses
Genetically modified Namalwa cells were subjected to lipid isolation and analyzed for mass spectrometry as previously reported (Ogiso et al., 2014; Veillon et al., 2015). Protein amount in the residue after lipid extraction was determined using a BCA Protein Assay Kit (Thermo Fisher). Each level of measured lipids was normalized to the protein amount.
Retrovirus-mediated gene transfer
The preparation of Namalwa cells using modified mouse stem cell virus (MSCV) vectors (Clontech, Mountain View, CA) encoding human GlcCerSyn, Gb3Syn, and Gb3Syn-TxT combined with the internal ribosomal entry site (IRES)–controlled EGFP was reported previously (Takematsu et al., 2011). A similar vector with a blasticidin resistance gene as a selection marker was constructed to replace EGFP. The MSCV-IRES system was used throughout this study, such that bicistronic expression of the cDNA of interest is achieved with selective marker expression. The full open reading frame of human neutral sphingomyelinase 2 (nSMase2) cDNA was amplified and subcloned into pDONR221. The LR recombinase (Invitrogen) reaction was used to create the MSCV-nSMase2-IRES-Blast plasmid vector. MSCV plasmid vectors were transfected into Plat-A packaging cells (Morita et al., 2000) to prepare infectious retrovirus. The prepared retrovirus was spin infected into Namalwa cells as reported previously (Takematsu et al., 2011), and infectants were polyclonally sorted by GFP positivity or selected using 10 μg/ml blasticidin S for at least 2 or 3 wk, respectively.
Electroporation-mediated gene delivery
Because the retroviral vector lacks a strong promoter for transgene expression, plasmid vector was used to express SMS1/2 cDNA (Shakor et al., 2011). FLAG-tagged SMS1/2 cDNAs were subcloned into an expression vector downstream of the EF1a promoter, which bicistronically expresses the blasticidin resistance gene under the IRES. The blasticidin-selected stable polyclonal cells expressed SMS1 or SMS2 according to Western blotting.
Thin-layer chromatography
Cellular lipids were extracted as reported previously (Miyake et al., 1995). Briefly, after being washed with PBS, 6 × 107 cells were lysed with chloroform-methanol (2:1). The lipid fraction was prepared by Folch’s participation and saponified to remove phospholipids. Sphingolipid fractions were desalted and separated using TLC plates. SM was visualized using primulin under ultraviolet light, and GSLs were visualized using orcinol-sulfate. Data were digitally captured, and densities of each band from at least three experimental replicates were analyzed by densitometry.
Cellular surface staining using a nontoxic variant of lysenin and equinatoxin II probes
The preparation of GST-NT-lysenin and enhanced green fluorescent protein (EGFP)–NT-lysenin was reported previously (Abe et al., 2012). Briefly, GST-NT-lysenin was expressed in BL21 cells and purified using glutathione–Sepharose 4B resin. GST-NT-lysenin, anti-GST, and FITC-conjugated anti-mouse IgG were mixed and incubated overnight at 4˚C to form a staining complex. A total of 4 × 105 cells were stained using the “staining precomplex” for 30 min at room temperature and analyzed by FCM. The nontoxic variant of equinatoxin, NT-equinatoxin II-EGFP-His, was prepared as reported previously (Makino et al., 2015) and used for flow cytometric detection.
PH-GFP participation at cleavage furrows
The PH domain of PLCδ1 was modified from a previously reported plasmid, pEGFP-N1-PH, which was kindly provided by Tamas Balla (Varnai and Balla, 1998). The PH-GFP fragment was subcloned into the pENTR1A donor vector and subsequently cloned into the pSP72-EF1-DC-IRES-Blast-PolyA destination vector by an LR clonase reaction. The pSP72-EF1-PH-GFP-IRES-Blast-PolyA plasmid was introduced into Namalwa cells by electroporation and selected using 10 μg/ml blasticidin S. A selected clone (Namalwa-PH-GFP-1) with appropriate fluorescence intensity was used to monitor PIP2 concentration at the cleavage furrow. Briefly, 4 × 105 Namalwa-PH-GFP-1 cells/ml were cultured overnight with 5 μM psychosine. After washing, cells were fixed using fixation buffer (4% paraformaldehyde/0.2 M sucrose in PBS) for 15 min at room temperature. Cells were then suspended in mounting solution with 4′,6-diamidino-2-phenylindole (DAPI) for fluorescence microscopic observation as reported previously (Takematsu et al., 2011).
Experimental replication and statistical evaluation
Data are expressed as mean ± SE of the results of (at least) triplicate experiments. The statistical significance of the observed differences was evaluated using the Student’s t test. All experiments were performed at least twice, and representative results are shown. For profile correlation analyses among multiple cell panels, Pearson’s r (−1 ≤ r ≤ 1) was used, with a value of 1 indicating a perfect correlation and −1 a perfect negative correlation.
Supplementary Material
Acknowledgments
We thank Makoto Ogiso (Kanazawa Medical University, Uchinada, Japan) for LC-MS analyses, Takayuki Kanazawa (Shionogi & Co., Osaka, Japan) for the evaluation of PPIN values, and Toshio Kitamura (University of Tokyo, Tokyo, Japan) for Plat-A cells. We also thank Roger Laine (Louisiana State University, Baton Rouge, LA) for discussion. This work was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviations used:
- Cer
ceramide
- EGFP
enhanced green fluorescent protein
- FCM
flow cytometry
- Gb3
Gal α1-4 Gal β1-4 Glc β1-1 Cer
- GlcCer
Glc β1-1 Cer
- GM1
Gal β1-3 GalNAc β1-4 (Sia α2-3) Gal β1-4 Glc β1-1 Cer
- GM3
Sia α2-3 Gal β1-4 Glc β1-1 Cer
- GSL
glycosphingolipid
- IRES
internal ribosomal entry site
- LacCer
Gal β1-4 Glc β1-1 Cer
- MSCV
mouse stem cell virus
- MS
mass spectrometry
- PDMP
(d-threo)-1-phenyl-2decanoylamino-3-morpholino-1-propanol
- PIP2
phosphatidylinositol 4,5-bisphosphate
- SM
sphingomyelin
- SMS
SM synthase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-08-0555) on May 11, 2016.
REFERENCES
- Abe M, Makino A, Hullin-Matsuda F, Kamijo K, Ohno-Iwashita Y, Hanada K, Mizuno H, Miyawaki A, Kobayashi T. A role for sphingomyelin-rich lipid domains in the accumulation of PIP2 to the cleavage furrow during cytokinesis. Mol Cell Biol. 2012;32:1396–1407. doi: 10.1128/MCB.06113-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atilla-Gokcumen GE, Muro E, Relat-Goberna J, Sasse S, Bedigian A, Coughlin ML, Garcia-Manyes S, Eggert US. Dividing cells regulate their lipid composition and localization. Cell. 2014;156:428–439. doi: 10.1016/j.cell.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M. Cytokinesis: the final stop for the chromosomal passengers. Biochem Soc Trans. 2008;36:367–370. doi: 10.1042/BST0360367. [DOI] [PubMed] [Google Scholar]
- Contreras FX, Ernst AM, Haberkant P, Bjorkholm P, Lindahl E, Gonen B, Tischer C, Elofsson A, von Heijne G, Thiele C, et al. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature. 2012;481:525–529. doi: 10.1038/nature10742. [DOI] [PubMed] [Google Scholar]
- Field SJ, Madson N, Kerr ML, Galbraith KA, Kennedy CE, Tahiliani M, Wilkins A, Cantley LC. PtdIns(4,5)P2 functions at the cleavage furrow during cytokinesis. Curr Biol. 2005;15:1407–1412. doi: 10.1016/j.cub.2005.06.059. [DOI] [PubMed] [Google Scholar]
- Gkantiragas I, Brugger B, Stuven E, Kaloyanova D, Li XY, Lohr K, Lottspeich F, Wieland FT, Helms JB. Sphingomyelin-enriched microdomains at the Golgi complex. Mol Biol Cell. 2001;12:1819–1833. doi: 10.1091/mbc.12.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakomori SI. Cell adhesion/recognition and signal transduction through glycosphingolipid microdomain. Glycoconj J. 2000;17:143–151. doi: 10.1023/a:1026524820177. [DOI] [PubMed] [Google Scholar]
- Hakomori S, Igarashi Y. Gangliosides and glycosphingolipids as modulators of cell growth, adhesion, and transmembrane signaling. Adv Lipid Res. 1993;25:147–162. [PubMed] [Google Scholar]
- Hakomori S, Zhang Y. Glycosphingolipid antigens and cancer therapy. Chem Biol. 1997;4:97–104. doi: 10.1016/s1074-5521(97)90253-2. [DOI] [PubMed] [Google Scholar]
- Hannun YA. The sphingomyelin cycle and the second messenger function of ceramide. J Biol Chem. 1994;269:3125–3128. [PubMed] [Google Scholar]
- Hannun YA, Bell RM. Lysosphingolipids inhibit protein kinase C: implications for the sphingolipidoses. Science. 1987;235:670–674. doi: 10.1126/science.3101176. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Bell RM. Regulation of protein kinase C by sphingosine and lysosphingolipids. Clin Chim Acta. 1989;185:333–345. doi: 10.1016/0009-8981(89)90224-6. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Tomiuk S, Wolff G, Stoffel W. Cloning and characterization of the mammalian brain-specific, Mg2+-dependent neutral sphingomyelinase. Proc Natl Acad Sci USA. 2000;97:5895–5900. doi: 10.1073/pnas.97.11.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igisu H, Suzuki K. Progressive accumulation of toxic metabolite in a genetic leukodystrophy. Science. 1984;224:753–755. doi: 10.1126/science.6719111. [DOI] [PubMed] [Google Scholar]
- Im DS, Heise CE, Nguyen T, O’Dowd BF, Lynch KR. Identification of a molecular target of psychosine and its role in globoid cell formation. J Cell Biol. 2001;153:429–434. doi: 10.1083/jcb.153.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi J, Momosaki K, Shimeno H, Nagamatsu A, Radin NS. Effects of D-threo-PDMP, an inhibitor of glucosylceramide synthetase, on expression of cell surface glycolipid antigen and binding to adhesive proteins by B16 melanoma cells. J Cell Physiol. 1989;141:573–583. doi: 10.1002/jcp.1041410316. [DOI] [PubMed] [Google Scholar]
- Ishitsuka R, Kobayashi T. Lysenin: a new tool for investigating membrane lipid organization. Anat Sci Int. 2004;79:184–190. doi: 10.1111/j.1447-073x.2004.00086.x. [DOI] [PubMed] [Google Scholar]
- Ishitsuka R, Yamaji-Hasegawa A, Makino A, Hirabayashi Y, Kobayashi T. A lipid-specific toxin reveals heterogeneity of sphingomyelin-containing membranes. Biophys J. 2004;86:296–307. doi: 10.1016/S0006-3495(04)74105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi K, Zhang Y, Handa K, Withers DA, Sinay P, Hakomori S. Reconstitution of membranes simulating “glycosignaling domain” and their susceptibility to lyso-GM3. J Biol Chem. 2000;275:15174–15181. doi: 10.1074/jbc.275.20.15174. [DOI] [PubMed] [Google Scholar]
- Kabayama K, Sato T, Saito K, Loberto N, Prinetti A, Sonnino S, Kinjo M, Igarashi Y, Inokuchi J. Dissociation of the insulin receptor and caveolin-1 complex by ganglioside GM3 in the state of insulin resistance. Proc Natl Acad Sci USA. 2007;104:13678–13683. doi: 10.1073/pnas.0703650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T, Nakamura S, Momoi M, Yamaji T, Takematsu H, Yano H, Sabe H, Yamamoto A, Kawasaki T, Kozutsumi Y. Inhibition of cytokinesis by a lipid metabolite, psychosine. J Cell Biol. 2000;149:943–950. doi: 10.1083/jcb.149.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T, Takematsu H, Yamamoto A, Yamamoto H, Kozutsumi Y. Wheat germ agglutinin stains dispersed post-golgi vesicles after treatment with the cytokinesis inhibitor psychosine. J Cell Physiol. 2008;215:517–525. doi: 10.1002/jcp.21328. [DOI] [PubMed] [Google Scholar]
- Kannagi R, Papayannopoulou T, Nakamoto B, Cochran NA, Yokochi T, Stamatoyannopoulos G, Hakomori S. Carbohydrate antigen profiles of human erythroleukemia cell lines HEL and K562. Blood. 1983;62:1230–1241. [PubMed] [Google Scholar]
- Keusch JJ, Manzella SM, Nyame KA, Cummings RD, Baenziger JU. Cloning of Gb3 synthase, the key enzyme in globo-series glycosphingolipid synthesis, predicts a family of alpha 1, 4-glycosyltransferases conserved in plants, insects, and mammals. J Biol Chem. 2000;275:25315–25321. doi: 10.1074/jbc.M002630200. [DOI] [PubMed] [Google Scholar]
- Kojima N, Hakomori S. Cell adhesion, spreading, and motility of GM3-expressing cells based on glycolipid-glycolipid interaction. J Biol Chem. 1991;266:17552–17558. [PubMed] [Google Scholar]
- Kojima Y, Fukumoto S, Furukawa K, Okajima T, Wiels J, Yokoyama K, Suzuki Y, Urano T, Ohta M. Molecular cloning of globotriaosylceramide/CD77 synthase, a glycosyltransferase that initiates the synthesis of globo series glycosphingolipids. J Biol Chem. 2000;275:15152–15156. doi: 10.1074/jbc.M909620199. [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y, Kanazawa T, Sun Y, Yamaji T, Yamamoto H, Takematsu H. Sphingolipids involved in the induction of multinuclear cell formation. Biochim Biophys Acta. 2002;1582:138–143. doi: 10.1016/s1388-1981(02)00148-8. [DOI] [PubMed] [Google Scholar]
- Lloyd-Evans E, Pelled D, Riebeling C, Bodennec J, de-Morgan A, Waller H, Schiffmann R, Futerman AH. Glucosylceramide and glucosylsphingosine modulate calcium mobilization from brain microsomes via different mechanisms. J Biol Chem. 2003a;278:23594–23599. doi: 10.1074/jbc.M300212200. [DOI] [PubMed] [Google Scholar]
- Lloyd-Evans E, Pelled D, Riebeling C, Futerman AH. Lyso-glycosphingolipids mobilize calcium from brain microsomes via multiple mechanisms. Biochem J. 2003b;375:561–565. doi: 10.1042/BJ20030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Abe M, Murate M, Inaba T, Yilmaz N, Hullin-Matsuda F, Kishimoto T, Schieber NL, Taguchi T, Arai H, et al. Visualization of the heterogeneous membrane distribution of sphingomyelin associated with cytokinesis, cell polarity, and sphingolipidosis. FASEB J. 2015;29:477–493. doi: 10.1096/fj.13-247585. [DOI] [PubMed] [Google Scholar]
- Marchesini N, Luberto C, Hannun YA. Biochemical properties of mammalian neutral sphingomyelinase 2 and its role in sphingolipid metabolism. J Biol Chem. 2003;278:13775–13783. doi: 10.1074/jbc.M212262200. [DOI] [PubMed] [Google Scholar]
- Merrill AH, Jr, Sandhoff K. Sphingolipis: metabolism and cell signaling. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoprotein and Membranes, 4th ed. Amsterdam: Elsevier Science; 2002. pp. 373–407. [Google Scholar]
- Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun. 1995;211:396–403. doi: 10.1006/bbrc.1995.1827. [DOI] [PubMed] [Google Scholar]
- Miyatake T, Suzuki K. Globoid cell leukodystrophy: additional deficiency of psychosine galactosidase. Biochem Biophys Res Commun. 1972;48:539–543. doi: 10.1016/0006-291x(72)90381-6. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Ng MM, Chang F, Burgess DR. Movement of membrane domains and requirement of membrane signaling molecules for cytokinesis. Dev Cell. 2005;9:781–790. doi: 10.1016/j.devcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Ogiso H, Taniguchi M, Araya S, Aoki S, Wardhani LO, Yamashita Y, Ueda Y, Okazaki T. Comparative analysis of biological sphingolipids with glycerophospholipids and diacylglycerol by LC-MS/MS. Metabolites. 2014;4:98–114. doi: 10.3390/metabo4010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima F, Kondo Y. Pertussis toxin inhibits phospholipase C activation and Ca2+ mobilization by sphingosylphosphorylcholine and galactosylsphingosine in HL60 leukemia cells. Implications of GTP-binding protein-coupled receptors for lysosphingolipids. J Biol Chem. 1995;270:26332–26340. doi: 10.1074/jbc.270.44.26332. [DOI] [PubMed] [Google Scholar]
- Prinetti A, Loberto N, Chigorno V, Sonnino S. Glycosphingolipid behaviour in complex membranes. Biochim Biophys Acta. 2009;1788:184–193. doi: 10.1016/j.bbamem.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Radu CG, Cheng D, Nijagal A, Riedinger M, McLaughlin J, Yang LV, Johnson J, Witte ON. Normal immune development and glucocorticoid-induced thymocyte apoptosis in mice deficient for the T-cell death-associated gene 8 receptor. Mol Cell Biol. 2006;26:668–677. doi: 10.1128/MCB.26.2.668-677.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakor AB, Taniguchi M, Kitatani K, Hashimoto M, Asano S, Hayashi A, Nomura K, Bielawski J, Bielawska A, Watanabe K, et al. Sphingomyelin synthase 1-generated sphingomyelin plays an important role in transferrin trafficking and cell proliferation. J Biol Chem. 2011;286:36053–36062. doi: 10.1074/jbc.M111.228593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda H, Kobayashi T, Katayama M, Goto I, Nagara H. Accumulation of galactosylsphingosine (psychosine) in the twitcher mouse: determination by HPLC. J Neurochem. 1987;49:92–99. doi: 10.1111/j.1471-4159.1987.tb03399.x. [DOI] [PubMed] [Google Scholar]
- Sonnino S, Mauri L, Chigorno V, Prinetti A. Gangliosides as components of lipid membrane domains. Glycobiology. 2007;17:1R–13R. doi: 10.1093/glycob/cwl052. [DOI] [PubMed] [Google Scholar]
- Sprong H, Degroote S, Claessens T, van Drunen J, Oorschot V, Westerink BH, Hirabayashi Y, Klumperman J, van der Sluijs P, van Meer G. Glycosphingolipids are required for sorting melanosomal proteins in the Golgi complex. J Cell Biol. 2001;155:369–380. doi: 10.1083/jcb.200106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigemann P, Gerlich DW. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 2009;19:606–616. doi: 10.1016/j.tcb.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Suzuki K. Twenty five years of the “psychosine hypothesis”: a personal perspective of its history and present status. Neurochem Res. 1998;23:251–259. doi: 10.1023/a:1022436928925. [DOI] [PubMed] [Google Scholar]
- Svennerholm L, Vanier MT, Mansson JE. Krabbe disease: a galactosylsphingosine (psychosine) lipidosis. J Lipid Res. 1980;21:53–64. [PubMed] [Google Scholar]
- Tafesse FG, Huitema K, Hermansson M, van der Poel S, van den Dikkenberg J, Uphoff A, Somerharju P, Holthuis JC. Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J Biol Chem. 2007;282:17537–17547. doi: 10.1074/jbc.M702423200. [DOI] [PubMed] [Google Scholar]
- Takematsu H, Yamamoto H, Naito-Matsui Y, Fujinawa R, Tanaka K, Okuno Y, Tanaka Y, Kyogashima M, Kannagi R, Kozutsumi Y. Quantitative transcriptomic profiling of branching in a glycosphingolipid biosynthetic pathway. J Biol Chem. 2011;286:27214–27224. doi: 10.1074/jbc.M111.234526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani M, Hannun YA. Neutral sphingomyelinase 2 is palmitoylated on multiple cysteine residues. Role of palmitoylation in subcellular localization. J Biol Chem. 2007;282:10047–10056. doi: 10.1074/jbc.M611249200. [DOI] [PubMed] [Google Scholar]
- Tidhar R, Futerman AH. The complexity of sphingolipid biosynthesis in the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833:2511–2518. doi: 10.1016/j.bbamcr.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillon L, Go S, Matsuyama W, Suzuki A, Nagasaki M, Yatomi Y, Inokuchi J. Identification of ganglioside GM3 molecular species in human serum associated with risk factors of metabolic syndrome. PLoS One. 2015;10:e0129645. doi: 10.1371/journal.pone.0129645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JQ, Kon J, Mogi C, Tobo M, Damirin A, Sato K, Komachi M, Malchinkhuu E, Murata N, Kimura T, et al. TDAG8 is a proton-sensing and psychosine-sensitive G-protein-coupled receptor. J Biol Chem. 2004;279:45626–45633. doi: 10.1074/jbc.M406966200. [DOI] [PubMed] [Google Scholar]
- Wenger DA, Sattler M, Hiatt W. Globoid cell leukodystrophy: deficiency of lactosyl ceramide beta-galactosidase. Proc Natl Acad Sci USA. 1974;71:854–857. doi: 10.1073/pnas.71.3.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AB, Givogri MI, Lopez-Rosas A, Cao H, van Breemen R, Thinakaran G, Bongarzone ER. Psychosine accumulates in membrane microdomains in the brain of krabbe patients, disrupting the raft architecture. J Neurosci. 2009;29:6068–6077. doi: 10.1523/JNEUROSCI.5597-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Takematsu H, Fujinawa R, Naito Y, Okuno Y, Tsujimoto G, Suzuki A, Kozutsumi Y. Correlation index-based responsible-enzyme gene screening (CIRES), a novel DNA microarray-based method for enzyme gene involved in glycan biosynthesis. PLoS One. 2007;2:e1232. doi: 10.1371/journal.pone.0001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka S, Miyaji M, Kitano T, Umehara H, Okazaki T. Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J Biol Chem. 2004;279:18688–18693. doi: 10.1074/jbc.M401205200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.