Cilia are cellular projections that can be motile to generate fluid flow or nonmotile to enable signaling. Both forms are based on shared components, and proteins involved in ciliary motility, like PACRG, may also function in ciliary signaling. Caenorhabditis elegans PACRG acts in a subset of nonmotile cilia to influence a learning behavior and promote longevity.
Abstract
Cilia are microtubule-based organelles that project from nearly all mammalian cell types. Motile cilia generate fluid flow, whereas nonmotile (primary) cilia are required for sensory physiology and modulate various signal transduction pathways. Here we investigate the nonmotile ciliary signaling roles of parkin coregulated gene (PACRG), a protein linked to ciliary motility. PACRG is associated with the protofilament ribbon, a structure believed to dictate the regular arrangement of motility-associated ciliary components. Roles for protofilament ribbon–associated proteins in nonmotile cilia and cellular signaling have not been investigated. We show that PACRG localizes to a small subset of nonmotile cilia in Caenorhabditis elegans, suggesting an evolutionary adaptation for mediating specific sensory/signaling functions. We find that it influences a learning behavior known as gustatory plasticity, in which it is functionally coupled to heterotrimeric G-protein signaling. We also demonstrate that PACRG promotes longevity in C. elegans by acting upstream of the lifespan-promoting FOXO transcription factor DAF-16 and likely upstream of insulin/IGF signaling. Our findings establish previously unrecognized sensory/signaling functions for PACRG and point to a role for this protein in promoting longevity. Furthermore, our work suggests additional ciliary motility-signaling connections, since EFHC1 (EF-hand containing 1), a potential PACRG interaction partner similarly associated with the protofilament ribbon and ciliary motility, also positively regulates lifespan.
INTRODUCTION
Cilia are organelles that emanate from the surface of most mammalian cell types and take on motile and nonmotile forms. Both forms are based on an axoneme of nine outer doublet microtubules nucleated from a centriolar structure termed the basal body and share a kinesin- and dynein-mediated intraflagellar transport (IFT) system for their biogenesis (Sung and Leroux, 2013). Motile cilia generate movement or flow using specialized axonemal structures that conform to a regular arrangement along microtubules believed to be dictated by the protofilament ribbon—a structure situated at the junction between the A- and B-tubules of outer doublet microtubules (Linck and Norrander, 2003; Linck et al., 2014). Loss of protofilament ribbon–associated proteins impairs ciliary motility in several organisms (Lorenzetti et al., 2004; Tanaka et al., 2004; Dawe et al., 2005; Suzuki et al., 2009; Wilson et al., 2010), but possible roles in nonmotile cilia have not been investigated.
Nonmotile (primary) cilia function as signaling centers specialized for the cells on which they are found (Oh and Katsanis, 2012), and several signaling pathways, including the highly conserved insulin/insulin-like growth factor (IGF) signaling pathway (Apfeld and Kenyon, 1999; Zhu et al., 2009; Yeh et al., 2013), depend on their proper functioning (Christensen et al., 2007; Eggenschwiler and Anderson, 2007; Satir et al., 2007; Lancaster and Gleeson, 2009; Johnson and Leroux, 2010). Nematode cilia are found exclusively in a subset of sensory neurons (60 neurons in total), where they modulate physiology, development, longevity, and most behaviors (reviewed in Apfeld and Kenyon, 1999; de Bono et al., 2002; Bargmann, 2006; Inglis et al., 2007; Bae and Barr, 2008). Of note, mutants with truncated cilia display increased longevity relative to wild-type animals, which has been attributed to anomalies in sensory perception (Apfeld and Kenyon, 1999). The current model for longevity control in Caenorhabditis elegans posits that activating the insulin/IGF-1 receptor (DAF-2) triggers a phosphorylation cascade that inhibits several transcription factors, including the forkhead box transcription factor (FOXO) DAF-16, which is required for lifespan extension (Kenyon, 2010).
C. elegans possesses only nonmotile cilia and therefore generally lacks motility-associated ciliary genes, but, of interest, it encodes orthologues of protofilament ribbon–associated proteins. In this work, we studied parkin coregulated gene (PACRG), a highly conserved ciliary protein (Avidor-Reiss et al., 2004; Li et al., 2004; Keller et al., 2005; Pazour, 2005; Stolc et al., 2005) that binds microtubules (Ikeda, 2008) and associates with the protofilament ribbon (Ikeda et al., 2007), with known ciliary roles restricted to motility (Lorenzetti et al., 2004; Dawe et al., 2005; Wilson et al., 2010; Thumberger et al., 2012). Given that a PACRG orthologue exists in C. elegans (PCRG-1) and that PACRG is present in mammalian neurons (Brody et al., 2008) that harbor only nonmotile cilia, we hypothesized that it has cilium-associated sensory/signaling functions previously masked by its role in ciliary motility. The absence of motile cilia in C. elegans makes it possible to test the role of PACRG in signaling independent of its function in ciliary motility.
Here we ascribe cilium-associated sensory/signaling roles to C. elegans PCRG-1. Instead of being a ubiquitous ciliary component, we find that it functions in a small subset of ciliated sensory neurons and takes on specific signaling roles. Specifically, loss of pcrg-1 results in altered gustatory plasticity, a learning behavior mediated by ASE neurons. We also find that PCRG-1 promotes longevity, acting upstream of the lifespan-promoting FOXO transcription factor DAF-16 and likely upstream of insulin/IGF signaling. Our findings provide the first evidence that the evolutionarily conserved ciliary protein PACRG linked to ciliary motility has sensory/signaling functions within the context of metazoan, nonmotile cilia. Our work also suggests further examples of ciliary motility–signaling connections. We show that the C. elegans orthologue of EF-hand containing 1 (EFHC1), a core protofilament ribbon protein also associated with ciliary motility and linked to juvenile myoclonic epilepsy (Ikeda et al., 2003, 2005; Suzuki et al., 2004, 2008, 2009), also localizes to a small subset of ciliated sensory neurons and promotes longevity, likely acting in the same genetic pathway as PCRG-1. More generally, our work extends our understanding of how signaling pathways are modulated by the ciliary organelle.
RESULTS
C. elegans PCRG-1 localizes to the cilia of ASE and outer labial quadrant neurons
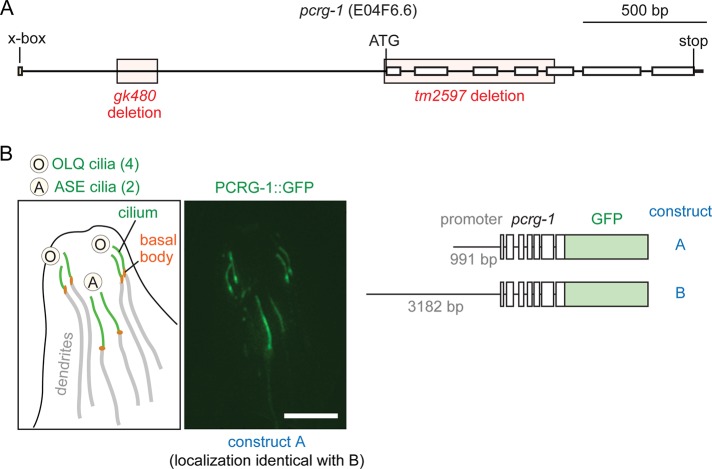
Given the established link between protist PACRG and motile cilia, we first sought to determine whether the uncharacterized C. elegans PCRG-1 protein also associates with (nonmotile) ciliary structures. We generated two constructs containing the complete pcrg-1 gene along with 991 and 3182 base pairs, respectively, of endogenous upstream promoter fused in-frame to green fluorescent protein (PCRG-1::GFP constructs A and B). We generated multiple transgenic lines for each construct and observed consistent localization of PCRG-1::GFP to cilia (Figure 1B). Surprisingly, PCRG-1::GFP was only observed in a subset of ciliated neurons, namely two ASE neurons and four outer labial quadrant (OLQ) neurons. Expression of PCRG-1::GFP in these neurons overlaps with the similarly limited expression pattern of EFHC-1, a core ciliary protofilament ribbon protein (see later discussion), suggesting that the observed expression pattern reflects that of the endogenous protein. ASE cilia are exposed to the external environment via openings in the cuticle (Perkins et al., 1986) and specifically mediate salt (NaCl) chemotaxis behaviors (Bargmann and Horvitz, 1991). Unlike ASE cilia, OLQ cilia are embedded in the cuticle and act as mechanosensors, modulating nose-touch avoidance and withdrawal responses, rate and bending angle of foraging (side-to-side head movements), and suppression of foraging upon nose touch or anterior touch to the body (Perkins et al., 1986; Kaplan and Horvitz, 1993; Hart et al., 1995). The restricted localization of C. elegans PCRG-1 to a subset of cilia provides evidence that the protein regulates specific sensory/signaling functions in nonmotile cilia.
FIGURE 1:
PCRG-1::GFP fusion protein localization and pcrg-1 mutant alleles. (A) Gene model for E04F6.2 (pcrg-1) in the nematode. Positions and relative sizes of the two deletion alleles examined in this study (gk480, tm2597), as well as the position of the X-box sequence. (B) Localization of PCRG-1::GFP constructs A (991–base pair promoter) and B (3182–base pair promoter) to the four OLQ and the two ASE cilia of C. elegans. Left, schematic diagram outlining fusion protein localization in the nematode. Bases of cilia (basal bodies) are indicated by orange ovals, cilia by green lines, and dendrites by gray lines. Scale bar, 5 μm.
PCRG-1 is not essential for cilium biogenesis or intraflagellar transport
On the basis of the localization of PCRG-1 to cilia, we examined what ciliary defects, if any, resulted from disrupting its function. We tested two pcrg-1 mutant alleles, gk480 and tm2597 (Figure 1A). The gk480 allele disrupts part of the promoter region just downstream of a putative X-box transcriptional regulatory sequence commonly found in ciliary genes (Efimenko et al., 2005). The tm2597 allele completely removes exons 1–4 and part of exon 5 and is likely a null. Based on the consistent phenotypes displayed with both alleles (see later discussion), the gk480 allele likely prevents pcrg-1 expression by disrupting binding of the ciliary transcription factor DAF-19.
To test ciliary integrity of C. elegans cilia in pcrg-1 mutants, we conducted several standard assays for cilium structure and function. Of note, although most or all of GFP-tagged PCRG-1 appears to localize exclusively within ASE and OLQ cilia, we could not eliminate the possibility that it may be present (albeit at reduced levels) and function in other cilia. Many cilia in nematodes terminate at openings in the protective cuticle, exposing the sensory neurons to the external environment (reviewed in Bargmann, 2006; Inglis et al., 2007; Bae and Barr, 2008). This allows lipophilic dyes to permeate some ciliated sensory neurons, by which a dye-filling assay detects differences between normal cilia and structurally impaired (e.g., truncated) cilia. Both pcrg-1 alleles exhibit wild-type dye filling (Supplemental Figure S1A). This may not be unexpected, since ASE and OLQ cilia do not normally dye fill in wild-type animals (Perkins et al., 1986). pcrg-1 mutants were also wild type with respect to two other cilium-dependent behaviors, namely avoidance of high-osmolarity (Osm assay; Supplemental Figure S1B) and chemotaxis toward the volatile attractants isoamyl alcohol and acetone (Che assays; Supplemental Figure S1B). Furthermore, pcrg-1 mutants are superficially wild type with respect to development to adulthood (Supplemental Figure S1C) and egg-laying behavior (Supplemental Figure S1B), two physiological aspects regulated by sensory inputs (Fujiwara et al., 2002).
Given the apparent cell-specific expression of pcrg-1 in ASE and OLQ neurons, we specifically investigated cilia structure in these neurons, using gcy-5p::GFP to mark ASE cilia/neurons (Swoboda et al., 2000) and OSM-9::GFP to mark OLQ cilia/neurons (Tobin et al., 2002). pcrg-1 mutant ASE(R) cilium length is similar to wild type (Supplemental Figure S1, B and D), and we observed no difference in the OSM-9::GFP localization pattern between pcrg-1 mutant and wild-type animals (Supplemental Figure S1D). These data are consistent with PCRG-1 not playing a major role in cilium formation, even in neurons in which it is clearly present.
Given the absence of gross structural ciliary defects in the pcrg-1 mutant, we hypothesized that the IFT machinery required for ciliary protein delivery should still operate to build and maintain cilia. To test this, we monitored the behavior of a GFP-tagged core IFT protein (OSM-6; orthologue of mammalian IFT52) expressed specifically in the ASE(R) neurons (driven by the gcy-5 promoter) of pcrg-1 mutant animals. In wild-type C. elegans cilia, OSM-6 displays transport velocities of ∼0.7 μm/s in the middle segment and ∼1.1 μm/s in the distal segment (Snow et al., 2004). In pcrg-1 mutants, anterograde IFT is robust and largely normal, although velocities were minimally yet significantly slower than in wild-type animals (0.64 and 0.97 μm/s, respectively; Supplemental Figure S1E). Together these data show that PCRG-1 is not strictly required for general cilium structure/function, consistent with a largely normal axonemal ultrastructure (unpublished data) and a functioning IFT system. The absence of overt ciliary defects in pcrg-1 mutants may point to a more specific role for C. elegans PACRG in sensory reception and signaling.
pcrg-1 is required for gustatory plasticity, a learning behavior dependent on ASE neurons
The expression of PCRG-1 in ASE neurons (Figure 1B) prompted us to analyze its potential role in chemotaxis to salts, which is critically dependent on these sensory neurons (Bargmann and Horvitz, 1991). We found that pcrg-1 mutants display wild-type chemotactic behaviors to various NaCl concentrations (Figure 2A). In addition to enabling chemotaxis, ASE neurons, along with ASI, ASH (Hukema et al., 2006), and possibly ADL and ADF neurons (Hukema et al., 2008), are essential for gustatory plasticity. This behavior represents the reduced attraction or avoidance of C. elegans to NaCl after prolonged exposure to this salt in the absence of food (Jansen et al., 2002; Bargmann, 2006). We discovered that pcrg-1 mutants show significant defects in gustatory plasticity, by which animals remain attracted to NaCl even after prolonged exposure to NaCl in the absence of food (Figure 2B). Of note, expression of PCRG-1::GFP in the pcrg-1 mutant did not rescue the gustatory plasticity defect, as overexpression of PCRG-1::GFP in wild-type animals results in a similar gustatory plasticity defect (Figure 2B). Together these data provide evidence that correct PCRG-1 levels are needed for proper signaling associated with gustatory plasticity. Of interest, both loss-of-function mutations and overexpression of GPC-1, a G-protein implicated in gustatory plasticity with a mutant phenotype similar to pcrg-1 mutants, also disrupts gustatory plasticity (Jansen et al., 2002). This observation led us to investigate a potential connection between PCRG-1 and G-proteins.
FIGURE 2:
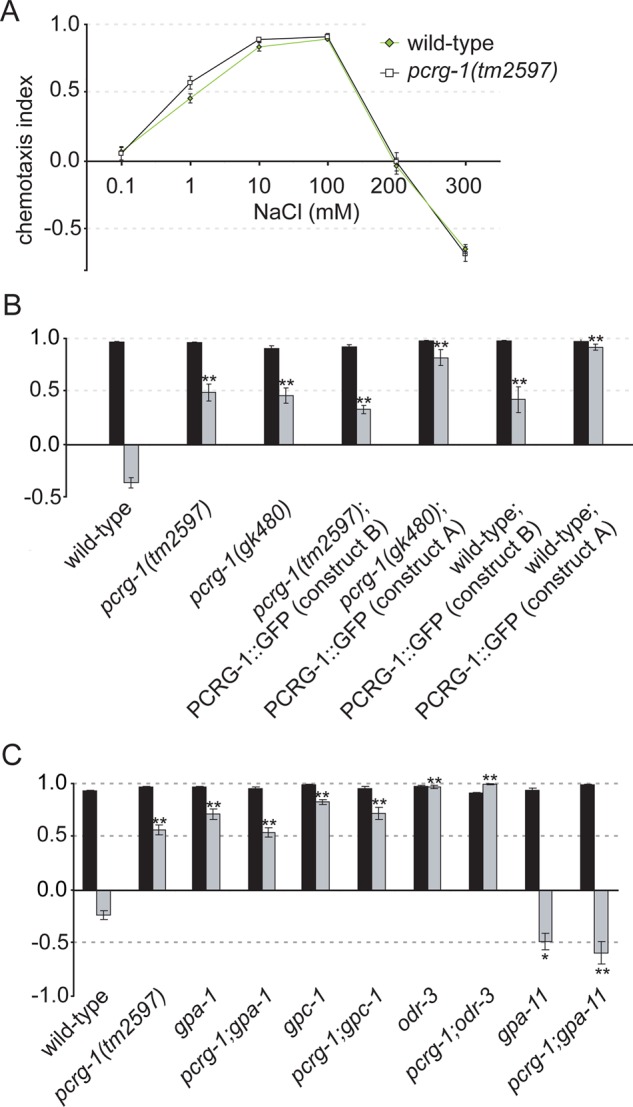
pcrg-1 animals show a defect in behavioral plasticity. (A) Naive responses to NaCl are not affected in pcrg-1 animals. Chemotaxis responses of wild-type (black line) and pcrg-1(tm2597) (green line) animals to various concentrations of NaCl. (B) Mutation of pcrg-1 affects gustatory plasticity. Worms are washed for 15 min in a low-salt buffer (black bars) or a buffer containing 100 mM NaCl (gray bars) and tested for chemotaxis to 25 mM NaCl. Wild-type animals show avoidance of NaCl after preexposure, whereas the two pcrg-1 mutants and the two transgenic strains carrying extra copies of pcrg-1 remain attracted to NaCl. **Statistically significantly different compared with wild type (p < 0.001). (C) pcrg-1 interacts with G-protein signal transduction in gustatory plasticity. Worms are washed for 15 min in a low-salt buffer (black bars) or a buffer containing 100 mM NaCl (gray bars) and tested for chemotaxis to 25 mM NaCl. *, Statistically significantly different compared to wild type (p < 0.05). **, Statistically significantly different compared to wild type (p < 0.001). Comparisons were also performed between double mutants and their corresponding single mutants. The only statistically significantly differences (p < 0.001) were between pcrg-1 and odr-3, pcrg-1 and pcrg-1;odr-3, pcrg-1, and gpa-11, and pcrg-1 and pcrg-1;gpa-11. Means + SEM; n > 100 for all assays.
pcrg-1 interacts genetically with G-proteins in gustatory plasticity
G-protein–coupled receptors represent an important interface through which cells respond to sensory stimuli. Their activation results in the dissociation of heterotrimeric G-proteins into α and βγ subunits, which regulate various downstream effectors. In C. elegans, at least 14 of the 21 known Gα subunits and one of the Gγ genes (gpc-1) are specifically expressed in sensory neurons (Zwaal et al., 1996; Roayaie et al., 1998; Jansen et al., 1999), making them promising candidates for contributing to the integration of several sensory cues in cooperation with PCRG-1 to enable gustatory plasticity. The pcrg-1 mutant phenotype is reminiscent of two G-protein mutant phenotypes: mutants of the Gα subunit GPA-1 and the Gγ subunit GPC-1 (Hukema et al., 2006). Like pcrg-1 mutants, these G-protein mutants show wild-type chemotaxis to NaCl but display defects in gustatory plasticity, with gpa-1 and gpc-1 working in the same pathway to modulate the behavior. It has been hypothesized that ASE neurons (which express pcrg-1) provide a signal to sensitize other ciliated cells expressing gpa-1 and/or gpc-1 to increase avoidance of normally attractive NaCl concentrations after prolonged exposure (Hukema et al., 2006).
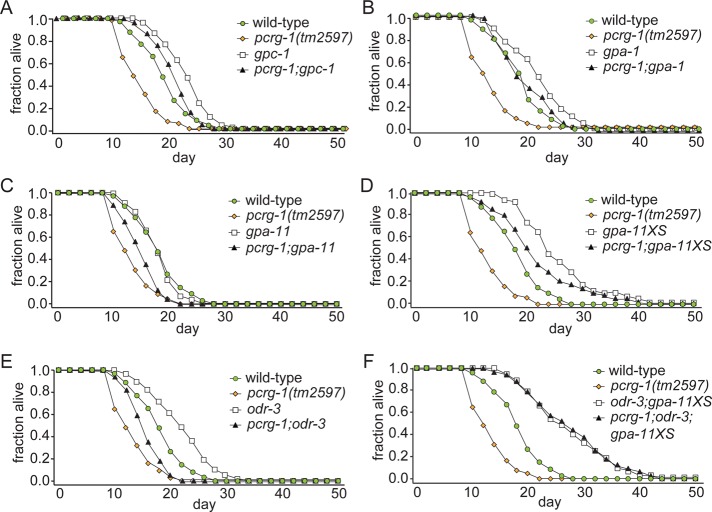
To analyze whether pcrg-1 genetically interacts with these G-protein genes in gustatory plasticity, we examined the behavior of double mutants. Mutant combinations of pcrg-1 with either gpc-1 or gpa-1 do not exacerbate the phenotype, suggesting that all three genes function in the same pathway in gustatory plasticity (Figure 2C). Mutants of another Gα subunit, ODR-3, also show gustatory plasticity defects in the absence of NaCl chemotaxis defects (Hukema et al., 2006). odr-3 mutants display a stronger gustatory plasticity defect than pcrg-1 mutants, by which they are always strongly attracted to NaCl. The pcrg-1;odr-3 double mutant also displays this phenotype (Figure 2C), but because odr-3 mutants display the maximum attraction to NaCl, it is possible that either the two genes function in parallel genetic pathways or odr-3 functions downstream of pcrg-1. Of interest, we also found that mutants of the Gα subunit GPA-11 show increased avoidance of NaCl after prolonged exposure to NaCl in the absence of food compared with wild-type animals and fully suppress the plasticity defect of pcrg-1 mutant animals (Figure 2C). This latter finding suggests that gpa-11 functions downstream of pcrg-1 in gustatory plasticity. Together our results provide evidence that PCRG-1 functionally interacts with several G-proteins to modulate the gustatory plasticity response.
PCRG-1 positively regulates longevity in C. elegans, potentially independently of G-proteins implicated in gustatory plasticity
Of interest, the G-proteins believed to function in the same pathway as PCRG-1 in gustatory plasticity also modulate lifespan. Increased lifespan is seen in gpa-1, gpc-1, and odr-3 mutants and in animals overexpressing gpa-11 (Lans and Jansen, 2007). This prompted us to analyze longevity in pcrg-1 mutants. Of interest, the two pcrg-1 alleles show significantly reduced lifespan compared with wild type (Figure 3A and Supplemental Table S1), both of which are fully rescued by GFP-tagged PCRG-1 (Figure 3, B and C, and Supplemental Table S1). This confirms that decreased longevity specifically arises from disrupting the pcrg-1 gene and that PCRG-1::GFP localization to cilia is biologically relevant. Unlike previously identified longevity-promoting proteins, PCRG-1::GFP expression in wild-type animals does not increase lifespan (Figure 3D and Supplemental Table S1). This is likely because many longevity-promoting proteins are kinases involved in signaling pathways like the insulin signaling pathway. Consequently, increased signaling via increased kinase activity may further extend longevity in a wild-type background. In contrast, PCRG-1 is believed to associate with the protofilament ribbon, a structural component of ciliary axonemes, where it is suspected to anchor signaling molecules. The amount of PACRG associated with the protofilament is likely fixed, so that an increase of PACRG may not lead to increased signaling in this context.
FIGURE 3:
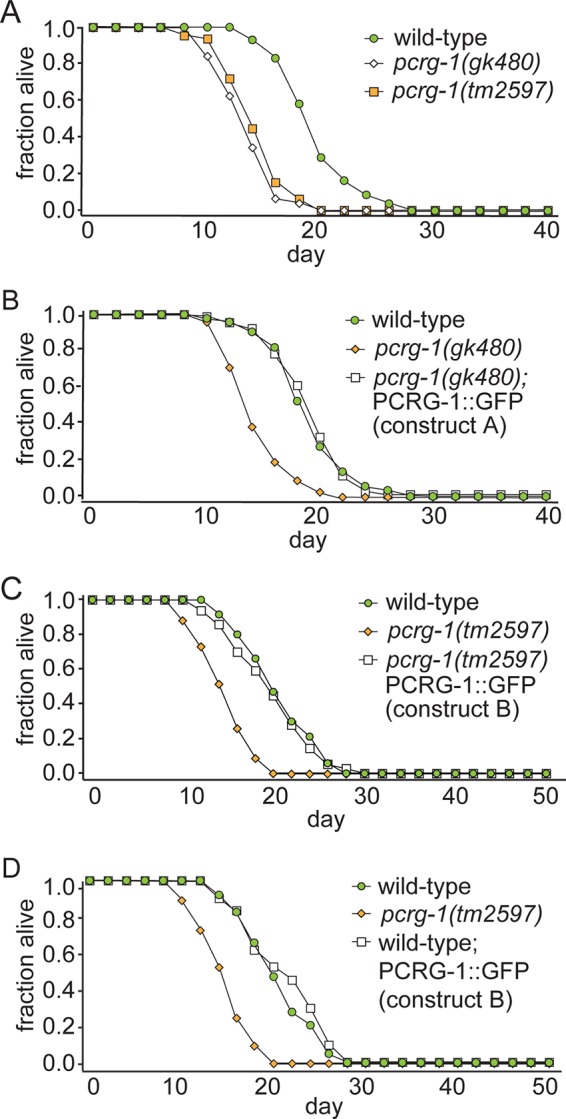
C. elegans pcrg-1 animals show a significant decrease in lifespan compared with wild type. (A) Both alleles of pcrg-1 (gk480, tm2597) show a significant decrease in lifespan compared with the wild-type N2 strain. (B) Expression of PCRG-1::GFP (construct A with 991– base pair promoter) rescues the lifespan defect shown in the gk480 allele. (C) Expression of PCRG-1::GFP (construct B with 3182–base pair promoter) rescues the lifespan defect shown in the tm2597 allele. (D) Expression of PCRG-1::GFP (construct B) in the wild-type (N2) background does not affect the lifespan of the worm. For number of animals and p values comparing differences between strains in lifespan experiments, see Supplemental Table S1.
We tested for genetic interactions between pcrg-1 and the G-protein signaling mutants implicated in gustatory plasticity. Of interest, mutant combinations of pcrg-1 with gpc-1 or gpa-1 show lifespans more similar to those of the G-protein single mutants (p > 0.05; Figure 4, A and B, and Supplemental Table S1). The statistical analyses are consistent with all three genes functioning in the same pathway, with PCRG-1 acting upstream of the two G-proteins (Lans and Jansen, 2007). However, since the p value for the pcrg-1;gpc-1 double mutant compared with the gpc-1 single mutant is only slightly above the 0.05 cutoff for significance (0.0825), the possibility that pcrg-1 functions in a parallel pathway to gpc-1 could not be ruled out. In contrast, pcrg-1 clearly acts in a pathway parallel to that of the two other G-protein genes tested, gpa-11 and odr-3, as double mutants display a lifespan significantly different from that of either mutant alone (Figure 4, C and E), and overexpression of gpa-11 increases the short lifespan of the pcrg-1 mutant (Figure 4, D and F). These data, together with evidence that gpa-1, gpc-1, gpa-11, and odr-3 are believed to all act in the same genetic pathway to regulate longevity (Lans and Jansen, 2007), suggest that pcrg-1 may promote longevity independently of these G-proteins.
FIGURE 4:
PCRG-1 does not appear to interact genetically with G-proteins to affect lifespan. (A) The lifespan of a pcrg-1;gpc-1 double mutant is not statistically different from that of gpc-1. (B) The lifespan of a pcrg-1;gpa-1 double mutant is not statistically different from that of gpa-1. (C) The lifespan of a pcrg-1;gpa-11 double mutant is significantly different from that of either single mutant. (D) The lifespan of a pcrg-1;gpa-11XS mutant strain is significantly different from that of gpa-11XS and pcrg-1 mutants. (E) The lifespan of a pcrg-1;odr-3 double mutant is significantly different from that of either single mutant. (F) The lifespan of a pcrg-1;odr-3; gpa-11XS mutant strain is indistinguishable from that of odr-3;gpa-11XS. For number of animals and p values comparing differences between strains in lifespan experiments, see Supplemental Table S1.
PCRG-1 regulates lifespan upstream of the longevity-promoting FOXO transcription factor DAF-16
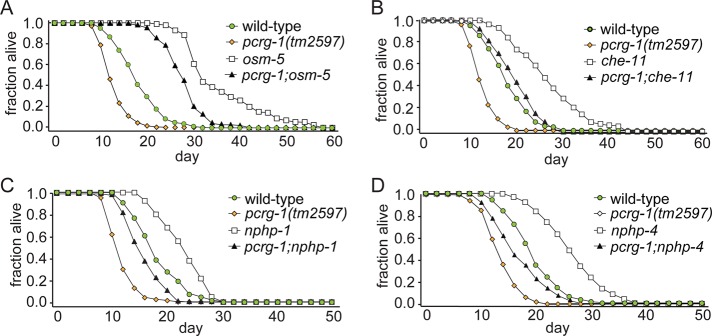
Mutations in insulin signaling genes, including the insulin/IGF-1 receptor homologue daf-2, greatly extend lifespan (Kenyon et al., 1993). This lifespan extension depends on the FOXO family transcription factor DAF-16, which translocates to the nucleus to promote longevity in daf-2 mutants (Lee et al., 2001). A seminal study by Apfeld and Kenyon (1999) established that loss of ciliogenic genes increased lifespan in C. elegans, where cilia are believed to influence longevity by controlling the insulin/IGF-1 pathway (Apfeld and Kenyon, 1999). It is notable that disruption of pcrg-1 (confirmed in two mutants) results in decreased lifespan; in contrast, ablation of IFT (Apfeld and Kenyon, 1999; Schafer et al., 2006; Lans and Jansen, 2007), the transition zone (membrane diffusion barrier at the ciliary base; Winkelbauer et al., 2005; Bialas et al., 2009), or most sensory G-proteins (except GPA-2; Lans and Jansen, 2007) all cause increased lifespan. To test for interactions between PCRG-1 and other ciliary proteins, we queried for genetic interactions between pcrg-1 (both alleles) and IFT mutants (osm-5 and che-11) or transition zone mutants (nphp-1 and nphp-4), which exhibit increased lifespan to varying degrees (Apfeld and Kenyon, 1999; Winkelbauer et al., 2005; Wolf et al., 2005). In all cases, double mutants show intermediate lifespans compared with individual mutants (Figure 5, A–D, Supplemental Figure S2, A–D, and Supplemental Table S1). This intermediate phenotype is interesting, as it suggests that PCRG-1 functions parallel to other ciliary (ciliogenic) proteins that affect lifespan. Hence the different (opposite) phenotypes of pcrg-1 and ciliogenic (IFT/transition zone) mutants on lifespan are supported by these genetic epistasis results. Furthermore, the results may not be surprising, given that PCRG-1 is restricted to a small number of ciliated neurons and likely contributes to specific ciliary signaling functions as opposed to affecting all cilia in the animal.
FIGURE 5:
PCRG-1 functions in a parallel pathway to ciliary proteins to regulate longevity. (A) The lifespan of a pcrg-1;osm-5 double mutant is significantly different from that of either single mutant. (B) The lifespan of a pcrg-1;che-11 double mutant is significantly different from that of either single mutant. (C) The lifespan of a pcrg-1;nphp-1 double mutant is significantly different from that of either single mutant. (D) The lifespan of a pcrg-1;nphp-4 double mutant is significantly different from that of either single mutant. For number of animals and p values comparing differences between strains in lifespan experiments, see Supplemental Table S1.
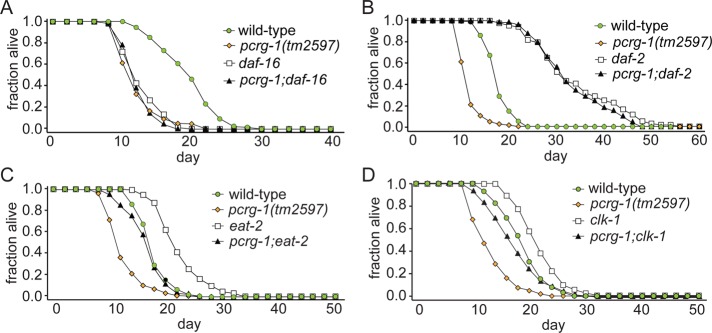
Many pathways that regulate longevity converge on the lifespan-promoting transcription factor DAF-16, including the insulin-signaling pathway. We tested double mutants with either pcrg-1 allele and the short-lived daf-16(mu86) null allele. If PCRG-1 extends lifespan independently of DAF-16 function, double mutants would be predicted to show decreased lifespans compared with the two single mutants. For example, mutants of the AMP-activated protein kinase AAK-2 promote longevity independent of DAF-16 function, and double mutants show reduced lifespan compared with both aak-2 and daf-16 single mutants (Apfeld et al., 2004). In contrast, we find that loss of pcrg-1 does not reduce the lifespan of daf-16 mutants (Figures 6A and Supplemental Table S1), indicating that pcrg-1 modulates longevity in cooperation with daf-16. Next, we wanted to determine whether pcrg-1 and daf-16 modulate longevity as part of the insulin-signaling pathway. The hypomorphic, nonnull nature of the daf-2(e1370) allele prevents definitive epistasis experiments, but removing pcrg-1 activity does not reduce the lifespan of daf-2 mutants; indeed, the daf-2;pcrg-1 double mutant is as long-lived as the daf-2 mutant (Figures 6B and Supplemental Table S1). This finding strongly supports the notion that the pcrg-1 mutant is not simply “sick” since it can live much longer than wild type by simply reducing insulin signaling. In addition, our data are consistent with both proteins functioning in the same pathway, with PCRG-1 functioning upstream of DAF-2 as part of the insulin pathway and DAF-16 functioning downstream of both proteins.
FIGURE 6:
PCRG-1 functions upstream of DAF-16 and likely the insulin signaling pathway to regulate longevity. (A) The lifespan of a pcrg-1(tm2597);daf-16 double mutant is indistinguishable from that of either single mutant. (B) The lifespan of a pcrg-1(tm2597);daf-2 double mutant is indistinguishable from that of daf-2. (C) The lifespan of a pcrg-1;eat-2 double mutant is significantly different from that of either single mutant. (D) The lifespan of a pcrg-1;clk-1 double mutant is significantly different from that of either single mutant. For number of animals and p values comparing differences between strains in lifespan experiments, see Supplemental Table S1.
To provide further evidence that PCRG-1 functions in the insulin-signaling pathway and not another longevity-related pathway we investigated dietary restriction and mitochondrial function (Klass, 1977; Ewbank et al., 1997). We tested for genetic interactions between the short-lived pcrg-1 mutant and 1) long-lived eat-2 mutant (impaired in feeding) and 2) long-lived clk-1 mutant (impaired in mitochondrial activity). In contrast to the extremely long-lived pcrg-1;daf-2 double mutants, pcrg-1;eat-2 and the pcrg-1;clk-1 double mutants exhibit intermediate lifespans (Figure 6, C and D, and Supplemental Table S1). These results suggest that PCRG-1 does not directly act within the diet restriction or mitochondrial pathways but instead functions in a parallel pathway, consistent with a specific role for PACRG in the insulin/IGF-1 pathway.
C. elegans EFHC-1 shows restricted, partially overlapping localization to PCRG-1
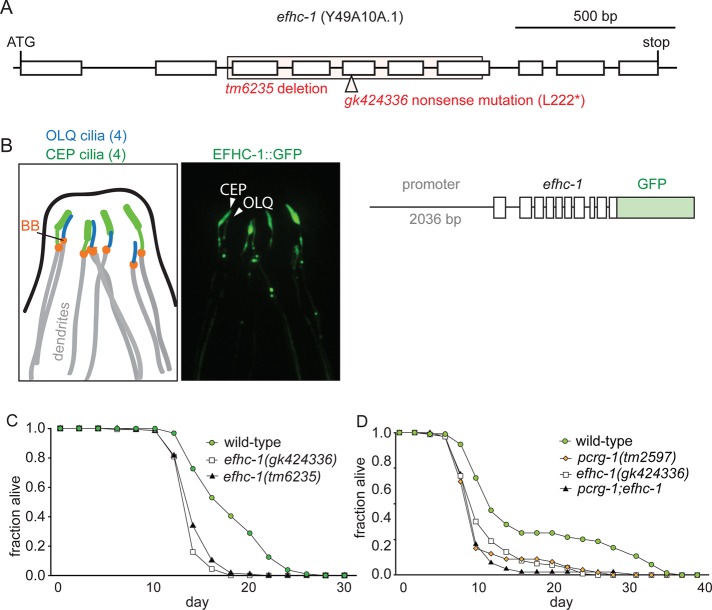
Chlamydomonas PACRG is believed to interact with Rib72 (Ikeda et al., 2007), another highly conserved ciliary protein (Ostrowski et al., 2002; Li et al., 2004; Pazour, 2005; Smith et al., 2005; Stolc et al., 2005) orthologous to human EFHC1 and commonly mutated in juvenile myoclonic epilepsy (Suzuki et al., 2004). In protist cilia, EFHC1 is a core component of the protofilament ribbon (Ikeda et al., 2003) and is also implicated in ciliary motility (Ikeda et al., 2005; Suzuki et al., 2008, 2009; Conte et al., 2009). The localization of PCRG-1 to a subset of cilia in C. elegans was surprising, given its high level of conservation in ciliated organisms. We sought to determine whether EFHC1, believed to be functionally/physically linked to PACRG (Ikeda et al., 2007), showed a similar restricted localization in a subset of ciliated neurons. We analyzed the unstudied C. elegans orthologue of EFHC1 encoded by Y49A10A.1 (efhc-1). We made a construct with the complete efhc-1 gene along with 2036 base pairs of endogenous upstream promoter fused in-frame to GFP (EFHC-1::GFP). Several independent transgenic lines consistently reveal EFHC-1::GFP localization to cilia (Figure 7B). Like PCRG-1, the fusion protein is expressed in only a subset of ciliated neurons: the four OLQ neurons like PCRG-1 and the four cephalic (CEP) neurons (Figure 7B). EFHC1::GFP appears to localize along the full length of each OLQ and CEP cilium (Figure 7B). These data suggest that although PACRG and EFHC1 play shared roles in motility in several organisms, in C. elegans, they function in only a subset of nonmotile cilia, where they can modulate specific sensory modalities.
FIGURE 7:
efhc-1 mutant alleles, localization of EFHC-1::GFP, and a reduction in lifespan in efhc-1 mutants. (A) Gene model for the Y49A10A.1 (efhc-1) gene in the nematode. Positions and size/nature of the two alleles examined in this study (gk424336 and tm6235). (B) Localization of EFHC-1::GFP to the four OLQ and the four CEP cilia of C. elegans. Basal body regions (BB) are indicated by orange ovals, cilia by green lines, and dendrites by gray lines. Scale bar, 5 μm. (C) Both efhc-1(gk424336) and efhc-1(tm6235) worms show significantly decreased lifespans compared with wild type (N2). (D) The reduced lifespan in efhc-1 mutants is similar to the lifespan of pcrg-1 mutants, as well as of double mutants, suggesting that the two genes work in the same genetic pathway to regulate lifespan. For number of animals and p values comparing differences between strains in lifespan experiments, see Supplemental Table S1.
EFHC-1 regulates lifespan likely through the same pathway as PCRG-1
Given that most known ciliary mutants show increased lifespans, the decreased lifespan seen in pcrg-1 mutants was unexpected. Considering the restricted localization patterns of PCRG-1 and EFHC-1 to a small subset of ciliated neurons in C. elegans, we queried whether disrupting EFHC-1 also influences lifespan. We tested two efhc-1 alleles in aging assays. The efhc-1(gk424336) allele contains a nonsense mutation halfway through the gene, and the efhc-1(tm6235) allele has an in-frame deletion of exons 3–7 (Figure 7A). Remarkably, both mutants show significant decreases in lifespan, similar to pcrg-1 mutants (Figure 7C). The overlapping localization of PCRG-1 and EFHC-1 in only OLQ neurons, in combination with their similar roles in lifespan extension, suggests that both proteins may act in OLQ (and perhaps not ASE or CEP) neurons to promote longevity.
The similarities between PCRG-1 and EFHC-1 also suggest a potential functional interaction between the two proteins, as was suspected but unconfirmed from Chlamydomonas studies (Ikeda et al., 2007). Indeed, pcrg-1;efhc-1 double mutants did not show a further decrease in lifespan compared with the single mutants (Figure 7D), consistent with both genes functioning in the same genetic pathway to regulate nematode lifespan. Given their colocalization to OLQ cilia, we queried whether the proteins depend on each other for ciliary localization, as implied by work in Chlamydomonas (Ikeda et al., 2007). Of interest, our results show that PCRG-1 is dispensable for EFHC-1 ciliary localization and vice versa (Supplemental Figure S3). Our findings suggest that although both proteins likely function together to exert their shared effect on lifespan, their association with the ciliary axoneme is independent.
OLQ cilia/neurons appear to be involved in longevity control in C. elegans
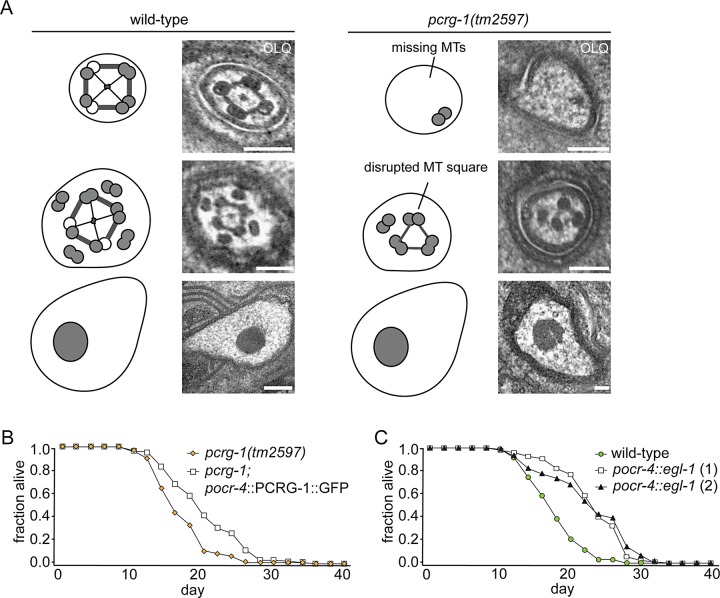
To investigate a possible role of OLQ cilia in longevity control, we first looked for structural defects in OLQ cilia of pcrg-1 mutants via transmission electron microscopy (TEM). The majority of C. elegans cilia possess a modified basal body with transition fibers, a transition zone characterized by the presence of Y-shaped axoneme–membrane linkers, a “middle segment” consisting of microtubule doublets, and a “distal segment” consisting of microtubule singlets (Perkins et al., 1986). The OLQ axoneme deviates from this, displaying nine doublet microtubules, four of which are joined into a square by thick cross-bridges and attached at the center of the axoneme with fine radial arms (Perkins et al., 1986). Our TEM analysis revealed that pcrg-1 mutant animals show variable loss of microtubule doublets and a disrupted microtubule square that frequently results in three microtubule doublets connected as a triangle (Figure 8A). Our results suggest that defects in OLQ ciliary structure in pcrg-1 mutants may lead to signaling defects that disrupt longevity control. We next investigated whether expressing PCRG-1::GFP specifically in the OLQ neurons using the ocr-4 promoter in pcrg-1 mutants increases their longevity, and indeed we found that pcrg-1 mutants carrying the pocr-4::PCRG-1::GFP construct live longer than their nontransgenic siblings (Figure 8B). To interrogate directly the involvement of OLQ neurons in longevity in C. elegans, we genetically ablated OLQ neurons by expressing the proapoptotic gene egl-1(d) (pocr-4::egl-1(d) construct). Of interest, animals carrying the ablation construct show increased longevity compared with nontransgenic, wild-type siblings (Figure 8C). This suggests that the ciliated OLQ neurons may play an important role in longevity control in C. elegans and that PCRG-1 and EFHC-1 may exert their longevity-promoting activities through their functions in OLQ.
FIGURE 8:
OLQ neurons and/or cilia appear to play a role in longevity. (A) The ultrastructure of OLQ cilia in pcrg-1 mutants shown by representative TEM images from serial cross sections. Bottom, striated rootlet at the base of OLQ cilia that is unaltered in pcrg-1 mutants. Middle, proximal region of OLQ cilia. In wild-type cilia, four outer doublets are connected in a square by thick cross-bridges and to the center with fine radial arms surrounded by additional microtubule doublets. In pcrg-1 mutants, microtubule doublets no longer form a square but frequently form a triangle instead, and radial arms are missing. Top, distal region of OLQ cilia where wild-type animals retain microtubule doublets connected in a square but lose additional doublets. pcrg-1 mutants completely lose connections between doublets and retain a small number of doublets. Scale bars, 200 nm. (B) pcrg-1 animals with PCRG-1 fused to GFP and driven by the ocr-4 promoter specifically in OLQ neurons show increased longevity compared with nontransgenic, pcrg-1 siblings. (C) Genetic ablations of OLQ neurons by expression of egl-1(d) (two lines) show increased longevity compared with nontransgenic, wild-type siblings.
Given that OLQ cilia are known to function in mechanosensation at the nose of C. elegans (Kaplan and Horvitz, 1993; Hart et al., 1995), we investigated the consequences of the loss of PCRG-1 on mechanosensory responses and OLQ-related behaviors. We found that pcrg-1 mutants retain mechanosensory responses to both light (buzz stimulus) and strong (press stimulus) stimulation to the nose (Supplemental Figure S4, A and B). In addition, we found that foraging behaviors mediated by OLQ neurons are similarly unaffected in pcrg-1 mutants (Supplemental Figure S4, C and D). Together these data show that PCRG-1, and likely EFHC-1, function in OLQ to mediate signaling functions/molecules specific to longevity but are not implicated in the forms of mechanosensation and foraging behaviors tested for.
DISCUSSION
A distinction between motile and nonmotile (sensory) cilia is often made due to their well-established roles in motility and sensory transduction, respectively. Nevertheless, sensory functions have been ascribed to motile cilia (Shah et al., 2009; Bloodgood, 2010). Both forms of cilia require the same evolutionarily conserved IFT machinery for their formation, and they possess similar ultrastructures, with the exception of various appendages associated with the motile axoneme, such as the central microtubule pair, radial spokes, inner and outer dynein arms, and interdoublet links. The protofilament ribbon, located at the junction of the A- and B-tubules of outer doublet microtubules, has long been suspected of being a major attachment point for structures/proteins involved in producing movement (Linck and Norrander, 2003; Tanaka et al., 2004; Setter et al., 2006; Amos, 2008). Consistent with this, PACRG, which is specifically associated with the protofilament ribbon (Ikeda et al., 2007), functions in ciliary motility (Lorenzetti et al., 2004; Dawe et al., 2005; Brody et al., 2008; Wilson et al., 2010). Given the presence of a PACRG orthologue in C. elegans, which lacks motile cilia, we hypothesized that this protofilament ribbon protein may provide a scaffold important for sensory transduction; similar functions may also exist in motile cilia but could be masked by prominent motility phenotypes in mutants.
Our results demonstrate that C. elegans PACRG localizes specifically to cilia; however, rather than being a ubiquitous ciliary component, it appears to have been adopted for specific functions in a restricted number of ciliated sensory neurons (ASE and OLQ). In particular, whereas pcrg-1 mutants show wild-type chemotaxis to NaCl, they display defects in the learning behavior known as gustatory plasticity, in which wild-type animals avoid normally attractive NaCl concentrations when preexposed to the salt in the absence of food. pcrg-1 mutants remain attracted to NaCl after preexposure in the absence of food, a phenotype similar to that seen in mutants for the Gα subunit GPA-1 and Gγ subunit GPC-1. All three genes were found to function in the same pathway, supporting a highly intricate integration of sensory cues in the modulation of gustatory plasticity. It has been proposed that ASE neurons (which express pcrg-1) provide a signal to sensitize ADF, ADL, ASI, and ASH (many of which express gpa-1 and/or gpc-1) neurons in gustatory plasticity to increase the avoidance of normally attractive NaCl concentrations after prolonged exposure (Hukema et al., 2006). An appealing possibility is that pcrg-1 plays a role in the production of this signal.
In this work, we also find that pcrg-1 mutants display marked decreases in lifespan compared with wild-type animals. Nematode cilia collectively represent the sensory apparatus of the animal (reviewed in Inglis et al., 2007; Bae and Barr, 2008), and decreased sensation is associated with increased longevity (Kenyon, 2010). In C. elegans, mutations affecting the insulin/IGF receptor DAF-2 result in lifespan extension that depends on the FOXO transcription factor DAF-16. All investigated proteins that influence cilium formation (e.g., IFT and transition zone components) negatively regulate insulin/IGF signaling and thus reduce longevity (i.e., mutants have increased lifespans). It is therefore surprising that PCRG-1 has the opposite effect, positively regulating insulin/IGF signaling. Nonetheless, our genetic interaction studies reveal that PCRG-1 functions in a parallel pathway to the previously studied IFT and transition zone proteins, supporting the notion that its role at the cilium in modulating signaling is unique. Remarkably, EFHC1, an integral component of the protofilament ribbon, also localizes to cilia within a small, partially overlapping subset of neurons and positively regulates longevity, likely through the same pathway as PACRG. Hence both protofilament ribbon proteins carry out distinct roles in ciliary signaling compared with ciliogenic proteins.
Our genetic epistasis studies reveal that PCRG-1 modulates lifespan upstream of the longevity-promoting FOXO transcription factor DAF-16; this is like other ciliary (IFT, transition zone) proteins, but PCRG-1 exerts an opposite effect on lifespan compared with other ciliary proteins. Although ciliary mutants most often display increased longevity, the ablation of specific gustatory neurons, and likely that of other ciliated sensory neurons, can either inhibit or promote longevity (Alcedo and Kenyon, 2004). The longevity-promoting nature of both PCRG-1 and EFHC-1 and the overlapping localization of the two proteins in OLQ neurons suggest the existence of a lifespan-modulating signal derived from OLQ neurons. This putative signal is independent of the G-protein signaling network implicated in gustatory plasticity. A role for OLQ cilia/neurons in longevity control is further supported by the discovery that ciliary structure is abrogated in pcrg-1 mutants, which could lead to impairment of signaling pathways that rely on intact cilia. Furthermore, we find that expression of PCRG-1::GFP specifically in OLQ neurons results in lifespan extension of pcrg-1 mutants. Of interest, genetic ablations of OLQ neurons result in increased longevity, suggesting that OLQ is responsible for producing a lifespan-reducing signal and that ciliary dysfunction in pcrg-1 mutants may increase the production of this signal and reduce lifespan.
In conclusion, our work has identified roles for the C. elegans PACRG orthologue in regulating signaling functions associated with nonmotile, sensory cilia. It is possible that the proteins have comparable roles in signaling within motile cilia—which have known sensory-signaling functions (Shah et al., 2009; Bloodgood, 2010). Uncovering such signaling functions may be difficult, given the prominent roles for PACRG in maintaining axonemal structure integrity and motility. The other possibility is that the signaling roles of PACRG are innovations in metazoans; the protein was adopted by nonmotile cilia to regulate sensory signal transduction. Furthermore, in this work, we show that additional ciliary motility–signaling connections may exist, since EFHC1 (a core protofilament ribbon protein associated with ciliary motility) also positively regulates lifespan. Future studies aimed at dissecting the roles of mammalian PACRG and EFHC1 in primary cilia–mediated sensory processes and insulin/IGF (and potentially other) signaling pathways should prove to be valuable, as they may provide insights into lifespan control, as well as into other physiological and developmental processes.
MATERIALS AND METHODS
C. elegans strains and genetic constructs
Unless otherwise noted, all strains were cultured and maintained at 20°C. The pcrg-1(tm2597) and efhc-1(tm6235) strains used in this work were obtained from National Bioresource Project (shigen.lab.nig.ac.jp/c.elegans/index.jsp). The pcrg-1(gk480) strain was obtained from the C. elegans Gene Knockout Consortium (www.zoology.ubc.ca/~dgmweb/research1.htm). The efhc-1(gk424336) strain was obtained from the Million Mutation Project (genome.sfu.ca/mmp/). These strains were outcrossed to wild-type (N2) worms at least six times. Other strains used in this work include gpa-1(pk15), gpa-11(pk349), gpa-11XS(pkIs539), gpc-1(pk298), odr-3(n1605), osm-5(p813), che-11(e1810), daf-16(mu86), daf-2(e1370), nphp-1(ok500), nphp-4(tm925), eat-2(ad465), clk-1(e2519), dpy-5(e907);Ex[gcy-5p::gfp + dpy5(+)], dpy-5(e907);Ex[gcy-5p::osm-6::gfp) + dpy5(+)], N2;osm-9::gfp5, and the chameleon strain AQ1490 ljEx130[pocr-4::YCD3 OLQ] (Kindt et al., 2007). The wild-type C. elegans used was the strain Bristol N2.
Construction of transgenic C. elegans strains
Two translational PCRG-1::GFP constructs (A and B) and a translational EFHC-1::GFP construct were generated as described previously (Blacque et al., 2004). The 5′ upstream sequence included in the PCRG-1::GFP constructs also contain the predicted mitochondrial RNA processing noncoding RNA mprp-1. The 5′ upstream sequence of PCRG-1::GFP (construct B) also contains the E04F6.6 gene, which codes for a protein of unknown function. All constructs contain the entire genomic coding regions along with 991 base pairs (PCRG-1::GFP construct A), 3182 base pairs (PCRG-1::GFP construct B), or 2036 base pairs (EFHC-1::GFP) of promoter sequence 5′ of the start codon fused upstream of and in-frame with the GFP coding sequence. To obtain ocr-4::egl-1(d) genetic ablations, the ocr-4::egl-1(d) construct was injected at 90 ng/µl with 10 ng/μl of rol-6. The PCRG-1::GFP construct was driven in OLQ neurons using 3200 base pairs of the upstream promoter of ocr-4.
Behavioral assays
The response to 25 mM NaCl, with or without preexposure to 100 mM NaCl, was assessed as described before (Wicks et al., 2000; Jansen et al., 2002). A chemotaxis index was calculated as (A − C)/(A + C), where A is the number of animals at the quadrants with NaCl, and C is the number of animals at the quadrants without attractant. The behavior of animals was always compared with controls performed on the same day(s). From 100 to 200 animals were tested in each assay, and assays were performed at least four times. Statistical significance was determined using an analysis of variance, followed by a Bonferroni post hoc test. Error bars represent SEM.
Cilium length measurements
ASE(R) amphid sensory neurons were visualized by expressing the transcriptional reporter gcy-5p::GFP (Blacque et al., 2006). In this construct, GFP diffuses freely throughout the neuron to stain the cell bodies, axons, dendrites, transition zone, and cilia. Fifty animals were measured per assay. Assays were repeated at least twice, with consistent results. Cilium length was measured from the distal tip of the dendrite (base of cilium) to the distal tip of the cilium.
Lifespan assays
Lifespan assays were performed similar to those described in Apfeld and Kenyon (1999). Worms were grown at 20°C for one generation before the eggs were collected through treatment with sodium hypochlorite. At the L4 molt, the animals were transferred to nematode growth medium (NGM) plates seeded with Escherichia coli strain OP50 and containing 16 μM fluorodeoxyuridine to prevent progeny growth. The assay plates were kept at 20°C for the duration of the assay. One hundred animals were picked for each of the indicated strains, with 10 worms on each individual plate. Plates were scored every 1–2 d for the presence of live and dead worms. Individual worms were considered dead once they no longer responded to harsh touch (prodding with a platinum wire). Worms that crawled off of the plate or exploded were censored. All assays were performed at least twice with consistent results. Lifespan analyses of many mutant strains, together with wild-type controls, were conducted in parallel; some of these results are split into separate graphs to simplify the data presented and show more clearly the relevant comparisons.
Microscopy and intraflagellar transport assays
Transgenic animals harboring GFP-tagged proteins were mounted on 1% agarose pads and immobilized with 100 mM levamisole. Amphid and phasmid cilia were examined with a 100×/1.35 numerical aperture objective and an ORCA AG charge-coupled device camera (Hamamatsu Photonics Systems, Hamamatsu City, Japan) mounted on a Zeiss (Oberkochen, Germany) Axioskop 2 mot plus microscope, with time-lapse images acquired at 300–500 ms/frame, depending on the specific marker used. Images and movies were obtained in Openlab, version 5.02 (ImproVision). Kymographs were generated using the MultipleKymograph ImageJ plug-in (www.embl-heidelberg.de/eamnet/html/body_kymograph.html). Rates from middle and distal segments were obtained essentially as described (Ou et al., 2005). For transgenic strains, at least 50 animals were observed per strain, with consistent results.
Transmission electron microscopy
Wild-type or mutant worms were washed directly into a primary fixative of 2.5% glutaraldehyde in 0.1 M Sorensen phosphate buffer. To facilitate rapid ingress of fixative, worms were cut in half under a dissection microscope using a razor blade, transferred to Eppendorf tubes, and fixed for 1 h at room temperature. Samples were then centrifuged at 3000 rpm for 2 min, and supernatant was removed and washed for 10 min in 0.1 M Sorensen phosphate buffer. Worms were then postfixed in 1% osmium tetroxide in 0.1 M Sorensen phosphate buffer for 1 h at room temperature. After washing in buffer, specimens were processed for electron microscopy by standard methods; briefly, they were dehydrated in ascending grades of alcohol to 100%, infiltrated with Epon, and placed in aluminum planchettes oriented in a longitudinal aspect and allowed to polymerize at 60°C for 24 h. A Leica (Wetzlar, Germany) UC6 ultramicrotome was used to section individual worms in cross section from anterior tip at 1 μm until the area of interest was located by using by light microscopy to examine the sections stained with toluidine blue. Then serial ultrathin sections of 80 nm were taken for electron microscopic examination. These were picked up onto 100-mesh copper grids and stained with uranyl acetate and lead citrate. Using a Tecnai Twin (FEI) electron microscope (Hillsboro, OR), sections were examined to locate, first, the most distal region of the ciliary region and then from that point to the more proximal regions of the ciliary apparatus. At each strategic point, distal segment, middle segment, and transition zone/fiber regions were tilted using the Compustage of the Tecnai to ensure that the axonemal microtubules were imaged in an exact geometrical normalcy to the imaging system. All images were recorded at an accelerating voltage (120 kV) and objective aperture of 10 μm using a MegaView 3 digital recording system (Munster, Germany).
Calcium imaging of OLQ neurons
Calcium imaging of nose-touch stimulation of glued animals was essentially as described previously, using a 1-s “press” or a 3.7-s “buzz” stimulus (Kindt et al., 2007). Images were recorded at 10 Hz using an iXon EM camera (Andor Technology, Belfast, United Kingdom), captured using IQ1.9 software (Andor Technology), and analyzed using a Matlab (MathWorks, Natick, MA) program custom written by Ithai Rabinowitch. A rectangular region of interest (ROI) was drawn around the cell body, and for every frame, the ROI was shifted according to the new position of the center of mass. Fluorescence intensity, F, was computed as the difference between the sum of pixel intensities and the faintest 10% pixels (background) within the ROI. Fluorescence ratio R = Fyellow/Fcyan (after correcting for bleedthrough) was used for computing ratio change, expressed as a percentage of R0, the average R within the first 3 s of recording (Rabinowitch et al., 2013).
Single-worm tracking of C. elegans behavior
Single-worm tracking was essentially as described previously (Yemini et al., 2013). Worms were maintained under strictly controlled conditions up until the point of tracking. Animals were staged by picking late L4 animals the evening before tracking. On the day of tracking, animals were transferred to 3.5-cm plates with low-peptone NGM and seeded with 20 μl of E. coli OP50 that was allowed to dry for at least 30 min. A 30-min wait was implemented before tracking to allow worms to habituate after being picked and moved to their tracking plate. To ensure sufficient phenotypic sampling, at least 20 young-adult hermaphrodites per strain were filmed for 15 min. The camera magnification was to set between 3.5 and 4.5 μm/pixel (a corresponding field of view of ∼2.5 × 2 mm at 640 × 480 resolution) and the frame rate to 20–30 frames/s to ensure a high-resolution analysis. To avoid potential room conditions that might bias measurement, recording was randomized across multiple trackers. Strains were matched to all wild-type animals (N2) recorded alongside the mutants.
Supplementary Material
Acknowledgments
M.R.L. acknowledges funding by the Canadian Institutes of Health Research (grant MOP 82870) and a senior salary award from the Michael Smith Foundation for Health Research (MSFHR). Work in the lab of G.J. was funded by the Royal Netherlands Academy of Sciences and ALW/NOW. O.E.B. acknowledges principal investigator funding from Science Foundation Ireland (11/PI/1037). W.R.S. received funding from the Medical Research Council (MC_A023_5PB91) and Wellcome Trust (WT103784MA). C.M.L. was supported by a CIHR Frederick Banting and Charles Best Canada Graduate Scholarship. Some of the nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources.
Abbreviations used:
- CEP
cephalic
- Che
chemosensory
- EFHC1
EF-hand containing 1
- FOXO
class O forkhead box transcription factor
- IFT
intraflagellar transport
- IGF
insulin/insulin-like growth factor
- OLQ
outer labial quadrant
- Osm
osmosensory
- PACRG
parkin coregulated gene
- TEM
transmission electron microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0490) on May 18, 2016.
REFERENCES
- Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Amos LA. The tektin family of microtubule-stabilizing proteins. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- Apfeld J, O’Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Bae Y-K, Barr MM. Sensory roles of neuronal cilia: cilia development, morphogenesis, and function in C. elegans. Front Biosci. 2008;13:5959–5974. doi: 10.2741/3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. Chemosensation in C. elegans. WormBook. 2006:2006(Oct 25), 1–29. doi: 10.1895/wormbook.1.123.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- Bialas NJ, Inglis PN, Li C, Robinson JF, Parker JDK, Healey MP, Davis EE, Inglis CD, Toivonen T, Cottell DC, et al. Functional interactions between the ciliopathy-associated Meckel syndrome 1 (MKS1) protein and two novel MKS1-related (MKSR) proteins. J Cell Sci. 2009;122:611–624. doi: 10.1242/jcs.028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque OE, Reardon MJ, Li C, McCarthy J, Mahjoub MR, Ansley SJ, Badano JL, Mah AK, Beales PL, Davidson WS, et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque OE, Li C, Inglis PN, Esmail MA, Ou G, Mah AK, Baillie DL, Scholey JM, Leroux MR. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol Biol Cell. 2006;17:5053–5062. doi: 10.1091/mbc.E06-06-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloodgood RA. Sensory reception is an attribute of both primary cilia and motile cilia. J Cell Sci. 2010;123:505–509. doi: 10.1242/jcs.066308. [DOI] [PubMed] [Google Scholar]
- Brody KM, Taylor JM, Wilson GR, Delatycki MB, Lockhart PJ. Regional and cellular localisation of Parkin co-regulated gene in developing and adult mouse brain. Brain Res. 2008;1201:177–186. doi: 10.1016/j.brainres.2008.01.050. [DOI] [PubMed] [Google Scholar]
- Christensen ST, Pedersen LB, Schneider L, Satir P. Sensory cilia and integration of signal transduction in human health and disease. Traffic. 2007;8:97–109. doi: 10.1111/j.1600-0854.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- Conte FF, Ribeiro PAO, Marchesini RB, Pascoal VDB, Silva JM, Oliveira AR, Gilioli R, Sbragia L, Bittencourt JC, Lopes-Cendes I. Expression profile and distribution of Efhc1 gene transcript during rodent brain development. J Mol Neurosci. 2009;39:69–77. doi: 10.1007/s12031-009-9179-6. [DOI] [PubMed] [Google Scholar]
- Dawe HR, Farr H, Portman N, Shaw MK, Gull K. The Parkin co-regulated gene product, PACRG, is an evolutionarily conserved axonemal protein that functions in outer-doublet microtubule morphogenesis. J Cell Sci. 2005;118:5421–5430. doi: 10.1242/jcs.02659. [DOI] [PubMed] [Google Scholar]
- de Bono M, Tobin DM, Davis MW, Avery L, Bargmann CI. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature. 2002;419:899–903. doi: 10.1038/nature01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewbank JJ, Barnes TM, Lakowski B, Lussier M, Bussey H, Hekimi S. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Sengupta P, McIntire SL. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Hukema RK, Rademakers S, Dekkers MPJ, Burghoorn J, Jansen G. Antagonistic sensory cues generate gustatory plasticity in Caenorhabditis elegans. EMBO J. 2006;25:312–322. doi: 10.1038/sj.emboj.7600940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukema RK, Rademakers S, Jansen G. Gustatory plasticity in C. elegans involves integration of negative cues and NaCl taste mediated by serotonin, dopamine, and glutamate. Learn Mem. 2008;15:829–836. doi: 10.1101/lm.994408. [DOI] [PubMed] [Google Scholar]
- Ikeda T. Parkin-co-regulated gene (PACRG) product interacts with tubulin and microtubules. FEBS Lett. 2008;582:1413–1418. doi: 10.1016/j.febslet.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Brown JA, Yagi T, Norrander JM, Hirono M, Eccleston E, Kamiya R, Linck RW. Rib72, a conserved protein associated with the ribbon compartment of flagellar A-microtubules and potentially involved in the linkage between outer doublet microtubules. J Biol Chem. 2003;278:7725–7734. doi: 10.1074/jbc.M210751200. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Ikeda K, Enomoto M, Park MK, Hirono M, Kamiya R. The mouse ortholog of EFHC1 implicated in juvenile myoclonic epilepsy is an axonemal protein widely conserved among organisms with motile cilia and flagella. FEBS Lett. 2005;579:819–822. doi: 10.1016/j.febslet.2004.12.070. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Ikeda T, Morikawa K, Kamiya R. Axonemal localization of Chlamydomonas PACRG, a homologue of the human Parkin-coregulated gene product. Cell Motil Cytoskeleton. 2007;64:814–821. doi: 10.1002/cm.20225. [DOI] [PubMed] [Google Scholar]
- Inglis PN, Ou G, Leroux MR, Scholey JM. The sensory cilia of Caenorhabditis elegans. WormBook. 2007;2007(Mar 8):1–22. doi: 10.1895/wormbook.1.126.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RH. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet. 1999;21:414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- Jansen G, Weinkove D, Plasterk RHA. The G-protein gamma subunit gpc-1 of the nematode C.elegans is involved in taste adaptation. EMBO J. 2002;21:986–994. doi: 10.1093/emboj/21.5.986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J-LF, Leroux MR. cAMP and cGMP signaling: sensory systems with prokaryotic roots adopted by eukaryotic cilia. Trends Cell Biol. 2010;20:435–444. doi: 10.1016/j.tcb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Kaplan JM, Horvitz HR. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1993;90:2227–2231. doi: 10.1073/pnas.90.6.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR, Marshall WF. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kindt KS, Viswanath V, Macpherson L, Quast K, Hu H, Patapoutian A, Schafer WR. Caenorhabditis elegans TRPA-1 functions in mechanosensation. Nat Neurosci. 2007;10:568–577. doi: 10.1038/nn1886. [DOI] [PubMed] [Google Scholar]
- Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Gleeson JG. The primary cilium as a cellular signaling center: lessons from disease. Curr Opin Genet Dev. 2009;19:220–229. doi: 10.1016/j.gde.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Jansen G. Multiple sensory G proteins in the olfactory, gustatory and nociceptive neurons modulate longevity in Caenorhabditis elegans. Dev Biol. 2007;303:474–482. doi: 10.1016/j.ydbio.2006.11.028. [DOI] [PubMed] [Google Scholar]
- Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Linck R, Fu X, Lin J, Ouch C, Schefter A, Steffen W, Warren P, Nicastro D. Insights into the structure and function of ciliary and flagellar doublet microtubules: tektins, Ca2+-binding proteins and stable protofilaments. J Biol Chem. 2014;289:17427–17444. doi: 10.1074/jbc.M114.568949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linck RW, Norrander JM. Protofilament ribbon compartments of ciliary and flagellar microtubules. Protist. 2003;154:299–311. doi: 10.1078/143446103322454086. [DOI] [PubMed] [Google Scholar]
- Lorenzetti D, Bishop CE, Justice MJ. Deletion of the Parkin coregulated gene causes male sterility in the quakingviable mouse mutant. Proc Natl Acad Sci USA. 2004;101:8402–8407. doi: 10.1073/pnas.0401832101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh EC, Katsanis N. Cilia in vertebrate development and disease. Development. 2012;139:443–448. doi: 10.1242/dev.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- Pazour GJ. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Rabinowitch I, Chatzigeorgiou M, Schafer WR. A gap junction circuit enhances processing of coincident mechanosensory inputs. Curr Biol. 2013;23:963–967. doi: 10.1016/j.cub.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI. The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20:55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Satir P, Guerra C, Bell AJ. Evolution and persistence of the cilium. Cell Motil Cytoskeleton. 2007;64:906–913. doi: 10.1002/cm.20238. [DOI] [PubMed] [Google Scholar]
- Schafer JC, Winkelbauer ME, Williams CL, Haycraft CJ, Desmond RA, Yoder BK. IFTA-2 is a conserved cilia protein involved in pathways regulating longevity and dauer formation in Caenorhabditis elegans. J Cell Sci. 2006;119:4088–4100. doi: 10.1242/jcs.03187. [DOI] [PubMed] [Google Scholar]
- Setter PW, Malvey-Dorn E, Steffen W, Stephens RE, Linck RW. Tektin interactions and a model for molecular functions. Exp Cell Res. 2006;312:2880–2896. doi: 10.1016/j.yexcr.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JC, Northey JGB, Garg J, Pearlman RE, Siu KWM. Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J Proteome Res. 2005;4:909–919. doi: 10.1021/pr050013h. [DOI] [PubMed] [Google Scholar]
- Snow JJ, Ou G, Gunnarson AL, Walker MRS, Zhou HM, Brust-Mascher I, Scholey JM. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- Stolc V, Samanta MP, Tongprasit W, Marshall WF. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc Natl Acad Sci USA. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C-H, Leroux MR. The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat Cell Biol. 2013;15:1387–1397. doi: 10.1038/ncb2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Delgado-Escueta AV, Aguan K, Alonso ME, Shi J, Hara Y, Nishida M, Numata T, Medina MT, Takeuchi T, et al. Mutations in EFHC1 cause juvenile myoclonic epilepsy. Nat Genet. 2004;36:842–849. doi: 10.1038/ng1393. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miyamoto H, Nakahari T, Inoue I, Suemoto T, Jiang B, Hirota Y, Itohara S, Saido TC, Tsumoto T, et al. Efhc1 deficiency causes spontaneous myoclonus and increased seizure susceptibility. Hum Mol Genet. 2009;18:1099–1109. doi: 10.1093/hmg/ddp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Inoue I, Yamagata T, Morita N, Furuichi T, Yamakawa K. Sequential expression of Efhc1/myoclonin1 in choroid plexus and ependymal cell cilia. Biochem Biophys Res Commun. 2008;367:226–233. doi: 10.1016/j.bbrc.2007.12.126. [DOI] [PubMed] [Google Scholar]
- Swoboda P, Adler HT, Thomas JH. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, Kaseda K, Maekawa M, Nishimune Y. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol Cell Biol. 2004;24:7958–7964. doi: 10.1128/MCB.24.18.7958-7964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumberger T, Hagenlocher C, Tisler M, Beyer T, Tietze N, Schweickert A, Feistel K, Blum M. Ciliary and non-ciliary expression and function of PACRG during vertebrate development. Cilia. 2012;1:13. doi: 10.1186/2046-2530-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin DM, Madsen DM, Kahn-Kirby A, Peckol EL, Moulder G, Barstead R, Maricq AV, Bargmann CI. Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron. 2002;35:307–318. doi: 10.1016/s0896-6273(02)00757-2. [DOI] [PubMed] [Google Scholar]
- Wicks SR, de Vries CJ, van Luenen HG, Plasterk RH. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol. 2000;221:295–307. doi: 10.1006/dbio.2000.9686. [DOI] [PubMed] [Google Scholar]
- Wilson GR, Wang HX, Egan GF, Robinson PJ, Delatycki MB, O’Bryan MK, Lockhart PJ. Deletion of the Parkin co-regulated gene causes defects in ependymal ciliary motility and hydrocephalus in the quakingviable mutant mouse. Hum Mol Genet. 2010;19:1593–1602. doi: 10.1093/hmg/ddq031. [DOI] [PubMed] [Google Scholar]
- Winkelbauer ME, Schafer JC, Haycraft CJ, Swoboda P, Yoder BK. The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J Cell Sci. 2005;118:5575–5587. doi: 10.1242/jcs.02665. [DOI] [PubMed] [Google Scholar]
- Wolf MTF, Lee J, Panther F, Otto EA, Guan K-L, Hildebrandt F. Expression and phenotype analysis of the nephrocystin-1 and nephrocystin-4 homologs in Caenorhabditis elegans. J Am Soc Nephrol. 2005;16:676–687. doi: 10.1681/ASN.2003121025. [DOI] [PubMed] [Google Scholar]
- Yeh C, Li A, Chuang J-Z, Saito M, Cáceres A, Sung C-H. IGF-1 activates a cilium-localized noncanonical Gβγ signaling pathway that regulates cell-cycle progression. Dev Cell. 2013;26:358–368. doi: 10.1016/j.devcel.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yemini E, Jucikas T, Grundy LJ, Brown AEX, Schafer WR. A database of C. elegans behavioral phenotypes. Nat Methods. 2013;10:877–879. doi: 10.1038/nmeth.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Shi S, Wang H, Liao K. Growth arrest induces primary-cilium formation and sensitizes IGF-1-receptor signaling during differentiation induction of 3T3-L1 preadipocytes. J Cell Sci. 2009;122:2760–2768. doi: 10.1242/jcs.046276. [DOI] [PubMed] [Google Scholar]
- Zwaal RR, Ahringer J, van Luenen HG, Rushforth A, Anderson P, Plasterk RH. G proteins are required for spatial orientation of early cell cleavages in C. elegans embryos. Cell. 1996;86:619–629. doi: 10.1016/s0092-8674(00)80135-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.