Abstract
A facile strategy has been developed to fabricate Cu(OH)2 supercages (SCs) as an artificial enzyme system with intrinsic peroxidase-mimic activities (PMA). SCs with high catalytic activity and excellent recyclability were generated via direct conversion of amorphous Cu(OH)2 nanoparticles (NPs) at room temperature. More specifically, the process that takes a single nanoparticle to a 3D supercage involves two basic steps. First, with addition of a copper–ammonia complex, the Cu2+ ions that are located on the surface of amorphous Cu(OH)2 NPs would evolve into a fine lamellar structure by coordination and migration and eventually convert to 1D nanoribbons around the NPs. Second, accompanied by the migration of Cu2+, a hollow cavity is generated in the inner NPs, such that a single nanoparticle eventually becomes a nanoribbon-assembled 3D hollow cage. These Cu(OH)2 SCs were then engineered as an artificial enzymatic system with higher efficiency for intrinsic PMA than the peroxidase activity of a natural enzyme, horseradish peroxidase.
Graphical abstract
INTRODUCTION
It is well-established that the self-assembly of nanoparticle building blocks into highly ordered superstructures results in properties superior to those of the individual building blocks, making such well-defined superstructures ideal in a wide range of technological applications, such as photonics,1 catalysis,2 biomedical diagnosis,3 sensors,4 magnetic resonance imaging,5 plasmonics,6 and surface-enhanced Raman spectroscopy.7 Up to now, colloidal assembly of nanoparticles (NPs) into designed superstructures with various geometries, such as belts, spheres, cubes, and rods, has been achieved with excellent control.8 For example, quantum belts were obtained through lamellar assembly of cadmium selenide nanoclusters.9 3D plasmonic Au nanoclusters were prepared through a polymer-assisted assembly approach.10 An assembled superstructure was obtained from anisotropic Pt nanocubes using a home-built evaporation-controlled system.11 Despite the enormous pioneering attempts, most of these superstructures have been formed by a time-consuming bottom-up approach, during which the uniformity in particle size, the properties of the solvent, and interparticle interactions must all be carefully controlled to avoid disintegration or deformation of the superstructures.12
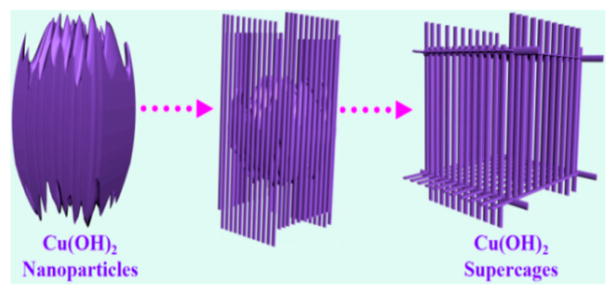
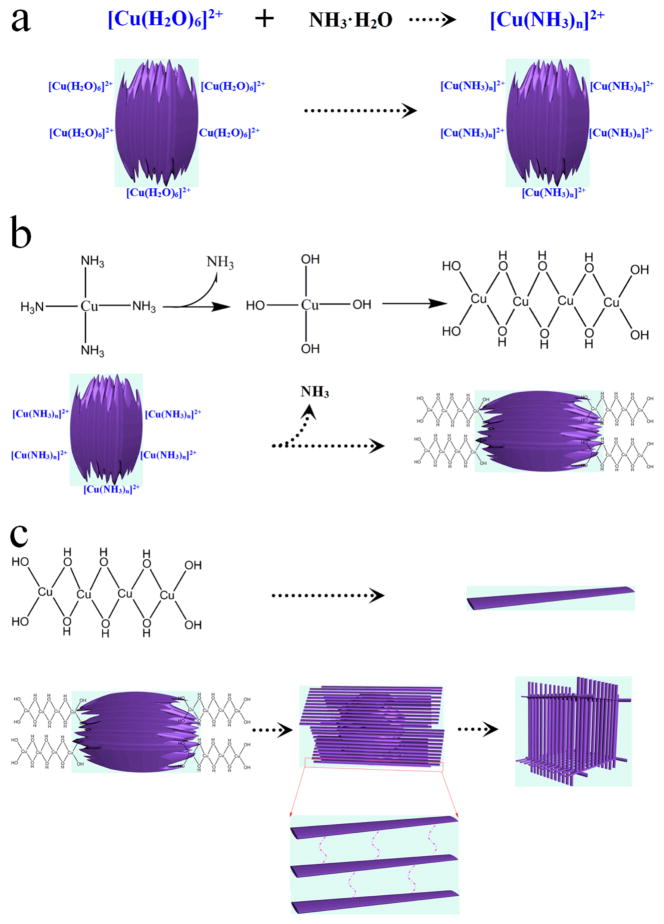
In contrast, we developed a facile top-down process for the preparation of Cu(OH)2 supercages (SCs). These Cu(OH)2 SCs, which are packed by 1D nanoribbons, are prepared by transformation of a single nanoparticle with the assistance of NH3·H2O (Scheme 1). The process that takes a single nanoparticle to a 3D supercage involves two basic steps. First, with addition of a copper–ammonia complex, the Cu2+ ions that are located on the surface of amorphous Cu(OH)2 NPs would evolve into a fine lamellar structure by coordination and migration and eventually convert to 1D nanoribbons around the NPs. Second, accompanied by the migration of Cu2+, a hollow cavity is generated in the inner NPs, such that a single nanoparticle eventually becomes a nanoribbon-assembled 3D hollow cage. Both formation and decomposition of SCs can be tuned simply by changing the concentration of NH3·H2O. More significantly, high catalytic activity can be expected from an artificial enzymatic system constructed by such SCs because (1) a large surface area results from the small size of the nanoribbons and (2) the collision probability of the active molecules can be increased when they are trapped in the cages.
Scheme 1.
Schematic Illustration of the Synthesis Process of Cu(OH)2 Supercages
EXPERIMENTAL SECTION
Synthesis of Amorphous Cu(OH)2 Nanoparticles
The synthesis of amorphous Cu(OH)2 nanoparticles (Figure 3a) followed the approach developed by our group based on a previous strategy,13 which involves reacting CuCl2 with NaBH4 in ethanol. In our experiment (Scheme S1), CuCl2·2H2O (17 mg), polyvinylpyrrolidone (PVP) (molecular weight = 55 000) (130.5 mg), and ethanol (40 mL) were added into a 100 mL flat-bottomed flask one by one. After 10 min of ultrasonication and 30 min of stirring, 7.7 mg of NaBH4 was dissolved in 10 mL of ethanol and quickly added to the solution under vigorous stirring. After 72 h, the products were collected by centrifugation and washed with ethanol three times.
Figure 3.
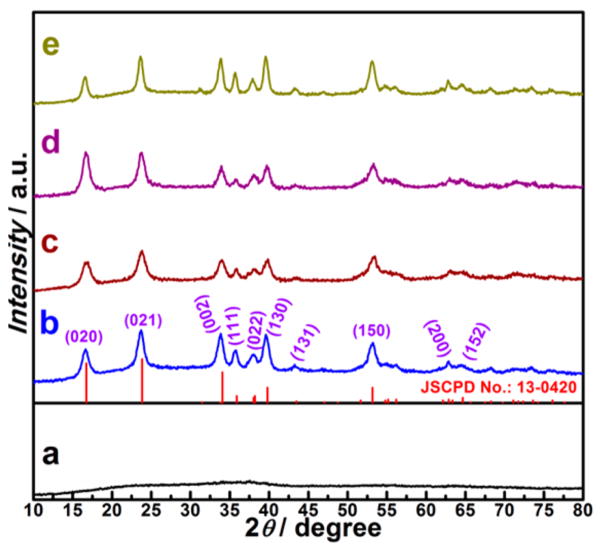
XRD patterns of the corresponding samples from the synthesis with different amounts of NH3 reacted in copper–ammonia complex: (a) 0 μL, (b) 400 μL, (c) 200 μL, (d) 600 μL, and (e) 800 μL.
Synthesis of 3D Cu(OH)2 Supercages
In a typical experiment, amorphous Cu(OH)2 nanoparticles (9.7 mg) and PVP (molecular weight = 1.3 × 106) (20 mg) were dispersed in 20 mL of DI water by ultrasonication for 15 min in a 100 mL flat-bottomed flask. The solution was vigorously stirred for 15 min. A copper–ammonia complex solution was prepared from 5 mL of Cu(NO3)2·3H2O (24.16 mg) aqueous solution plus 15 mL of NH3·H2O solution (400 μL of NH3·H2O (29%) plus 14.6 mL of DI-water). Then, 20 mL of copper–ammonia complex solution was added to the above mixture of amorphous Cu(OH)2 nanoparticles in 1 min and stirred for 10 min. The products were collected by centrifugation and washed with acetone and methanol three times.
Using the same strategy, Cu(OH)2 supercages were disassembled by the addition of 200, 600, and 800 μL of NH3·H2O (29%) to form solutions of copper–ammonia complex.
Catalyzed Oxidation
Unless otherwise stated, steady-state kinetic assays were carried out at 25 °C in a 1.5 mL tube with 30 μg of Cu(OH)2 SCs (3.5 × 109 supercages) or 300 ng HRP (4.1 × 1012 enzyme molecules) in 500 μL of reaction buffer (0.2 M NaAc, pH 4.5) in the presence of 530 μM H2O2 for Cu(OH)2 SCs or HPR using 800 μM TMB as the substrate. For experiments at different pHs (1–12) at 25 °C, 30 μL of H2O2 (30%) was added to 400 μL of reaction buffer and vortexed for 4 min. Then, 40 μL of TMB (10 mM) was added into the mixture and vortexed for another 4 min. Finally, 30 μL of Cu(OH)2 SCs (1 mg/mL) was quickly added to the mixture. Immediately after addition of Cu(OH)2 SCs, color changes were observed. All reactions were monitored in time-scan mode at 652 nm using a Cary Bio-100 UV/vis spectrometer (Varian).
To study the effect of different temperatures (22–65 °C) at pH 4.5, 400 μL of reaction buffer was held at the desired temperature for 5 min. Then, 30 μL of H2O2 (30%) was added to the reaction buffer and vortexed for 1 min, and the mixture was held at that temperature for 4 min. Then, 40 μL of TMB (10 mM) was added to the mixture and vortexed for 1 min, and the mixture was held at that temperature for another 4 min. Finally, 30 μL of Cu(OH)2 SCs (1 mg/mL) was quickly added to the mixture. The reaction was monitored in time-scan mode at 652 nm using a Cary Bio-100 UV/vis spectrometer (Varian).
RESULTS AND DISCUSSION
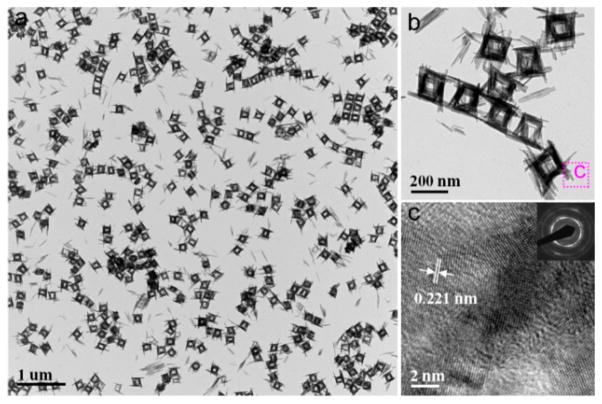
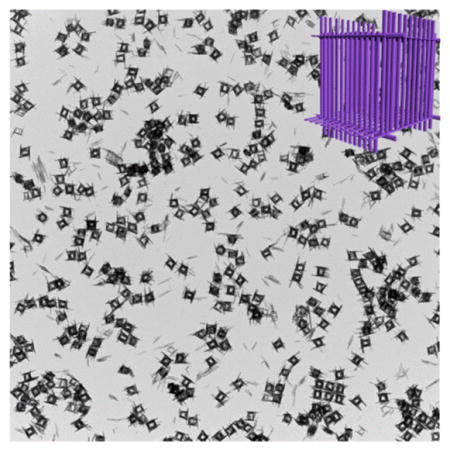
In a typical procedure, Cu(OH)2 NPs (Figures S1 and S2a) were employed as a starting materials in a top-down process. The Cu(OH)2 SCs were obtained after a copper–ammonia complex ([Cu(NH3)n](NO3)2) was added to the mixture of Cu(OH)2 NPs and PVP with vigorous stirring at room temperature. The resulting products were collected by centrifugation after stirring for 10 min. Low-magnification transmission electron microscopy (TEM) showed that uniform hollow nanocages with an average edge length of ~200 nm were formed (Figure 1a). High-magnification TEM (Figure 1b) images further revealed that these hollow nanocages were composed of packed nanoribbons. The wall thickness of the hollow supercages was about 30 nm, and the length of the nanoribbons ranged from 150 to 250 nm. The crystal phase of the hollow nanocages was determined to be orthorhombic Cu(OH)2 (Powder Diffraction File no. 13-0420, International Centre for Diffraction Data, [year]) from the corresponding powder X-ray diffraction (XRD) pattern shown in Figure 3b. The observed lattice fringe of 0.221 nm in the high-resolution TEM (HRTEM) image (Figure 1c) corresponds to the d spacing of the (130) lattice planes in Cu(OH)2. The ring-type selected-area electron diffraction (SAED) pattern (inset in Figure 1c) indicates the polycrystalline nature of these Cu(OH)2 SCs. To determine the specific surface area, full nitrogen sorption isotherms of these supercages were measured. According to the Brunauer–Emmett–Teller (BET) model and the data in Figure S3, the specific surface area of the supercages was 172 m2 g−1.
Figure 1.
Characterization of Cu(OH)2 supercages (SCs): (a) low-magnification and (b) high-magnification TEM images; (c) HRTEM image (inset, SAED).
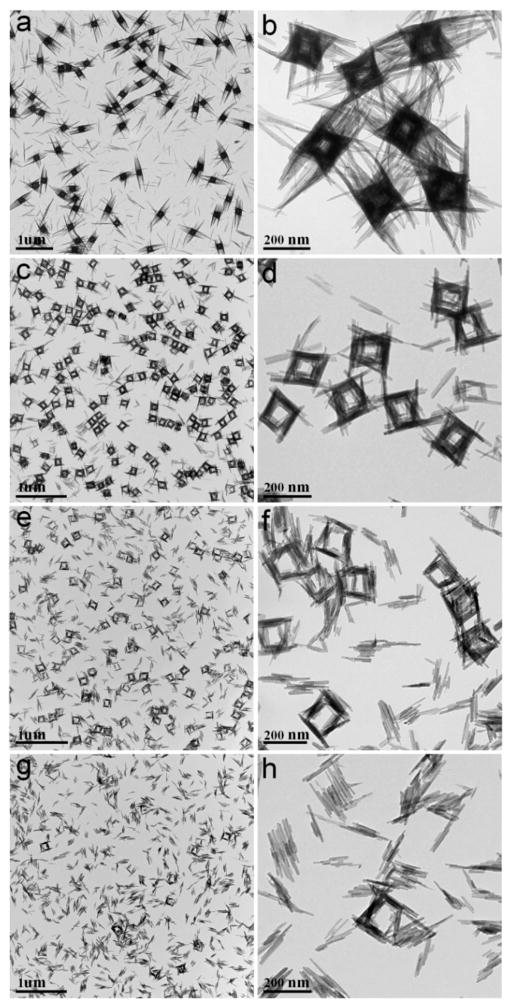
To investigate the growth process of the supercages, aliquots of reaction products were prepared by adding different amounts of 29% NH3·H2O (200, 400, 600, and 800 μL) to the copper–ammonia complex. As revealed by TEM images in Figure 2, relatively dense cages were formed in the aliquot with 200 μL of NH3·H2O added (Figure 2a, b). They were piled up by 1D nanoribbons with lengths in the range of 200–250 nm, and importantly, cavities were faintly visible at the center of the cages. When the amount of NH3·H2O was increased to 400 μL, nearly perfect supercages with side lengths around 200 nm could be observed (Figure 2c, d). However, a further increase of NH3·H2O solution to 600 μL caused slight disassembly of the supercages, in which some nanoribbons were dissociated (Figure 2e, f). Full disintegration of the supercages was observed when 800 μL of NH3·H2O was added, leaving well-dispersed nanoribbons (Figure 2g, h). It is worth mentioning that the entire process was also monitored by XRD, which showed (Figure 3) that all the above products are orthorhombic Cu(OH)2.
Figure 2.
TEM images of nanoribbon assembly for Cu(OH)2 supercages. Samples were prepared from synthesis using different amounts of NH3 in copper–ammonia complex: (a and b) 200 μL, (c and d) 400 μL, (e and f) 600 μL, and (g and h) 800 μL.
On the basis of the above results, a scheme can be proposed for the formation of SCs (Scheme 1 and Figure 4). Before the reaction, a protective layer of PVP is generated by mixing PVP with Cu(OH)2 NPs in order to control the reaction rate.14 With addition of the copper–ammonia complex ([Cu(H2O)6]2+ + nNH3·H2O ⇌ [Cu(NH3)n]2+ + (n + 6)H2O, NH3·H2O ⇌ NH4+ + OH−), Cu2+ ions ([Cu(H2O)6]2+) on the surface of the amorphous Cu(OH)2 NPs (Figure S1) first coordinate with NH3·H2O to generate [Cu(NH3)n]2+ (Figure 4a). [Cu(NH3)n]2+ tends to coordinate in a square planar manner with OH−,15 leading to an extended complex chain structure on the particle surface, i.e., [Cu(NH3)n]2+ → [Cu(NH3)n−1(OH)]+ → [Cu(NH3)n−2(OH)2] → … → [Cu(OH)n](n−2)− (Figure 4b).16 These chains can be connected through the coordination of OH− and Cu2+, growing into a lamellar structure (nanoribbons) on the surface of the original NPs (Figure 4b).17 As a result, Cu2+ migrates from the inner NPs to the tips of nanoribbons in the form of [Cu(NH3)n−m(OH)m](2−m)+,16 generating, in turn, a hollow cavity in the original NPs (Figures 2a, b and 4c). Because nanoribbons are stacked around the particles through hydrogen bonds,18 3D hollow cages are eventually formed (Figures 2c, d and 4c). It is interesting that the obtained SCs have size similar to that of amorphous Cu(OH)2 NPs (200–250 nm, Figure S1). Further addition of NH3·H2O was shown to weaken the hydrogen bonds between the nanoribbons, thus causing partial breakdown of the cages (NH3·H2O ⇌ NH4+ + OH−, Cu(OH)2 + 2OH− ⇌ [Cu(OH)4]2−; Figure 2e, f). Finally, by increasing the concentration of NH3·H2O, the supercages can be fully disassembled into well-dispersed nanoribbons (Figure 2g, h). It is worth noting that all assembly and disassembly processes occur at room temperature, indicating that this is a green procedure with low energy cost.
Figure 4.
Schematic of (a) [Cu(HO2)6]2+ coordinating with NH3·H2O to generate [Cu(NH3)n]2+, (b) coordination growth of the lamellar structure, and (c) formation of nanoribbons and Cu(OH)2 supercage.
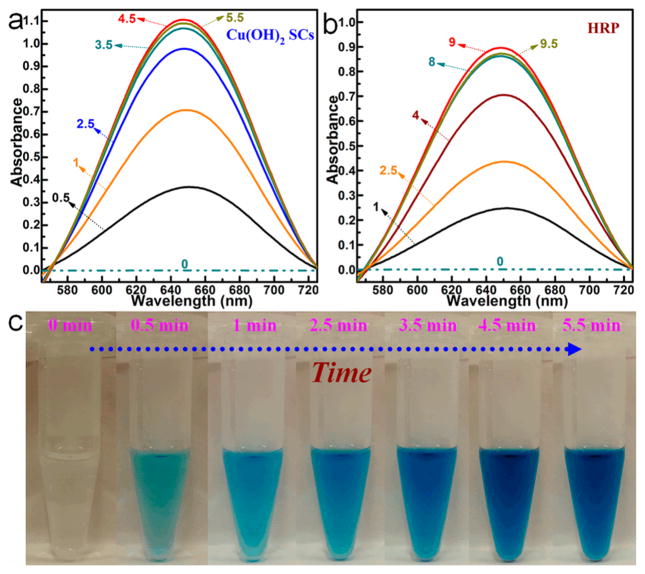
Artificial enzymes are very important components of biomimetic chemistry, which aims to imitate the general principles of natural enzymes using alternative materials.19 Compared with natural enzymes, artificial enzyme systems display high operational stability, low cost, facile preparation, and tunable catalytic activity,19,20 and they are promising in such applications as biosensors, immunoassays, cancer diagnostics, neuroprotection, stem cell growth, and pollutant removal.19,21 For our first set of experiments, we investigated if Cu(OH)2 SCs would exhibit peroxidase-mimic activity (PMA) for the substrate 3,3′,5,5′-tetramethylbenzidine (TMB). For comparison, the activity of horseradish peroxidase (HRP), which is one of the most utilized natural enzymes for biocatalysis,22 was also investigated. After addition of Cu(OH)2 SCs into the TMB–H2O2 solution (pH 4.5) (Figure S4–S5), the solution changed from colorless to a deep blue within 5 min at room temperature, as shown by the photo and UV/vis absorption curves (Figure 5). This indicates that the reaction of H2O2 and TMB could be catalyzed by Cu(OH)2 SCs (Figure S6). The characteristic absorption peak at 652 nm was chosen as the parameter to monitor the catalysis process.23 However, for HRP, a relatively slow reaction rate was observed (Figure 5b), even though the concentration of HRP was 2000 times higher than that of Cu(OH)2 SCs. (See the calculation sections in the Supporting Information.) In addition, the catalytic ability of Cu(OH)2 SCs is better than that of amorphous Cu(OH)2 NPs (Figure S7). This experiment confirmed that Cu(OH)2 SCs could serve as an efficient artificial enzymatic system for intrinsic PMA.21
Figure 5.
Absorption spectra for the oxidation catalysis process (min) of (a) Cu(OH)2 SCs and (b) HRP, where TMB was used as the substrate in the presence of H2O2, pH was 4.5, and temperature was 25 °C. (c) Photographs of Cu(OH)2 SCs-catalyzed oxidation of TMB in the presence of H2O2 at different times.
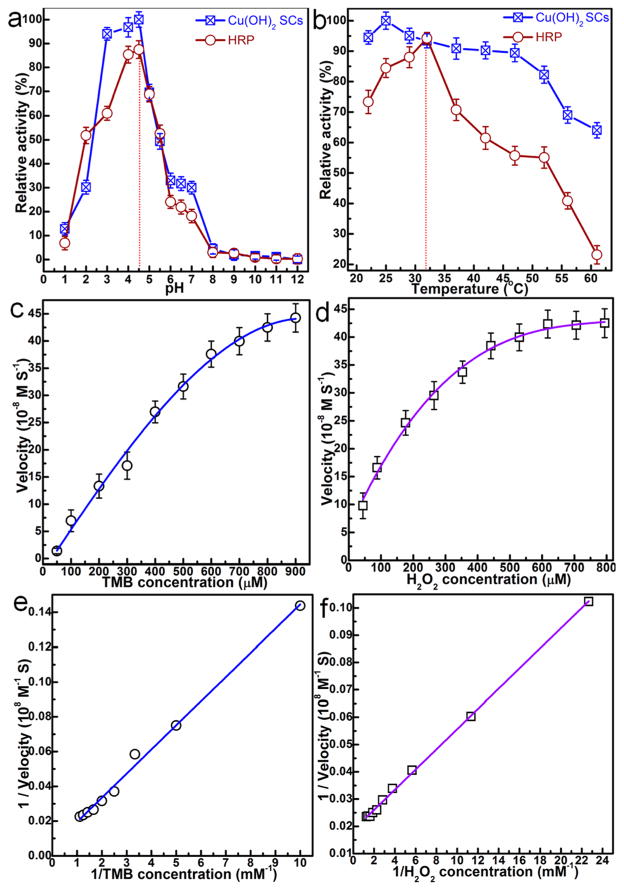
Next, the catalytic activity of Cu(OH)2 SCs was systematically investigated at different pHs and temperatures. To accomplish this, PMA of Cu(OH)2 SCs was measured while varying the pH from 1 to 12 and the temperature from 22 to 65 °C. For comparison, the activity of HRP using the same parameters was also studied. The results in Figure 6a show the high PMA of Cu(OH)2 SCs in the pH range of 3–5, with catalytic efficiency reaching above 90%. Cu(OH)2 SCs also exhibited excellent peroxidase catalytic activity over a broad range of temperatures (Figure 6b). The optimum pH and temperature for Cu(OH)2 SCs catalysis are approximately 4.5 and 25 °C, respectively, which are very close to those of HRP (Figure 6a, b). To test the reusability of the SCs, we used fresh and recovered Cu(OH)2 SCs (artificial enzyme, 1.5 mg) to catalyze the oxidation of TMB (800 μM TMB) by H2O2 (530 μM) in 25 mL of NaAc buffer (0.2 M NaAc, pH 4.5). The absorbance was measured after 4.5 min reaction (round 1). The recovered Cu(OH)2 SCs were separated from the reaction mixture by centrifuging, then washed with 20 mL of ethanol two times and with 20 mL of ether solvent one time to remove the product. The recycled Cu(OH)2 SCs were mixed with a fresh reaction mixture, and the absorbance at 4.5 min was measured (round 2). The procedure was repeated once more (round 3). The results (Figure S8) show that the catalytic efficiency (ratio of the absorbance at 4.5 min to the absorbance of the fresh SCs at 4.5 min) was 87.6% for round 2 and 75.2% for round 3.
Figure 6.
(a and b) pH- and temperature-dependent peroxidase-mimic activity of Cu(OH)2 SCs and HRP. (a) Cu(OH)2 SCs and HRP show an optimal pH of 4.5; (b) Cu(OH)2 SCs and HRP show an optimal temperature around 25 and 32 °C, respectively. Experiments were carried out using 30 μg of SCs or 300 ng of HRP in a reaction volume of 0.5 mL of 0.2 M NaAc buffer, with 800 μM TMB as substrate. H2O2 concentration was 530 μM for SCs and HRP. The maximum point in each curve (a and b) was set as 100%. Steady-state kinetic assays and catalytic mechanism of Cu(OH)2 SCs were carried out under the following conditions: (c) The concentration of H2O2 was 530 μM, and TMB concentration was varied. (d) The concentration of TMB was 800 μM, and H2O2 concentration was varied. (e and f) Double reciprocal plots for Cu(OH)2 SCs with the concentrations of (e) H2O2 fixed and TMB varied and (f) TMB fixed and H2O2 varied.
To provide further insight, the catalytic activity of the Cu(OH)2 SCs was studied by enzyme kinetics theory and methods.23,24 Typical Michaelis–Menten curves (Figure 6c, d) were obtained for a range of TMB or H2O2 concentrations, and fitted by the Lineweaver–Burk equation (Figure 6e, f).22 Important enzyme kinetic parameters, such as the Michaelis–Menten constant (Km) and maximum initial velocity (Vmax), were obtained and are listed in Table 1. Km is an indicator of enzyme affinity to substrate,25 with a high Km value representing a weak affinity and vice versa. Although the apparent Km value of Cu(OH)2 SCs with TMB as the substrate was significantly higher than that of HRP,26 the apparent Km value of Cu(OH)2 SCs with H2O2 as the substrate was about 20 times lower than that of HRP. Thus, Cu(OH)2 SCs showed better affinity to H2O2 compared to HRP. This is ascribed to the high surface area-to-volume ratio in Cu(OH)2 SCs, leading to more active sites for H2O2, which, in turn, results in a lower Km and a higher Vmax, giving an overall kcat (Vmax/[E]) of approximately the same value for SCs and HRP, which is around two times higher than the values of irregularly shaped platinum nanoparticles in the previous report.27 In addition, Cu(OH)2 SCs would have strong ability to convert H2O2 into hydroxyl radicals (·OH) and thus exhibit excellent peroxidase-like activity.28
Table 1.
Comparison of the Kinetic Parameters of Cu(OH)2 SCs and HRPa
| [E] (M) | substrate | Km (mM) | Vmax (Ms−1) | kcat (s1−) | |
|---|---|---|---|---|---|
| Cu(OH)2 SCs | 11.63 × 10−12 | TMB | 2.448 | 44.83 × 10−8 | 3.83 × 104 |
| Cu(OH)2 SCs | 11.63 × 10−12 | H2O2 | 0.199 | 42.51 × 10−8 | 3.66 × 104 |
| HRPM26 | 2.5 × 10−11 | TMB | 0.434 | 10 × 10−8 | 4.00 × 104 |
| HRP26 | 2.5 × 10−11 | H2O2 | 3.700 | 8.71 × 10−8 | 3.48 × 104 |
[E] is the enzyme (or SC) concentration, Km is the Michaelis constant, Vmax is the maximal reaction velocity, and kcat is the catalytic constant, where kcat = Vmax/[E].
CONCLUSIONS
We have developed a facile strategy to fabricate 3D Cu(OH)2 SCs via direct conversion of amorphous Cu(OH)2 NPs at room temperature. The SCs displayed lengths in the range of 150–200 nm and an average wall thickness of around 30 nm. To further understand the mechanism of SCs formation, an assembly/disassembly process for the specific shape was also studied. The artificial enzyme system composed of these SCs exhibited high catalytic activity and excellent reusability as mimics of HRP. By leveraging the color changes caused by the artificial enzymatic system, these SCs can also be utilized for the detection of biomolecules. The successful demonstration of this work may offer researchers engaged in materials science guidelines toward the construction of various superstructures into organized functional systems.
Supplementary Material
Acknowledgments
This work is supported by grants awarded by the National Institutes of Health (GM079359, GM111386 and CA133086), the National Key Scientific Program of China (2011CB911000), NSFC grants (NSFC 21221003 and NSFC 21327009), and the China National Instrumentation Program 2011YQ03012412.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b09337.
Detailed experimental procedures and additional data. (PDF)
References
- 1.Talapin DV, Lee JS, Kovalenko MV, Shevchenko EV. Chem Rev. 2010;110:389–458. doi: 10.1021/cr900137k. [DOI] [PubMed] [Google Scholar]
- 2.Chen C, Nan C, Wang D, Su Q, Duan H, Liu X, Zhang L, Chu D, Song W, Peng Q, Li Y. Angew Chem, Int Ed. 2011;50:3725–3729. doi: 10.1002/anie.201007229. [DOI] [PubMed] [Google Scholar]
- 3.Zhu G, Hu R, Zhao Z, Chen Z, Zhang X, Tan W. J Am Chem Soc. 2013;135:16438–16445. doi: 10.1021/ja406115e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Y, Thai T, Reineck P, Qiu L, Guo Y, Bach U. Adv Funct Mater. 2013;23:1519–1526. [Google Scholar]
- 5.Chen O, Riedemann L, Etoc F, Herrmann H, Coppey M, Barch M, Farrar CT, Zhao J, Bruns OT, Wei H, Guo P, Cui J, Jensen R, Chen Y, Harris DK, Cordero JM, Wang Z, Jasanoff A, Fukumura D, Reimer R, Dahan M, Jain RK, Bawendi MG. Nat Commun. 2014;5:5093–5105. doi: 10.1038/ncomms6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, Shao L, Li Q, Wang J. Chem Soc Rev. 2013;42:2679–2724. doi: 10.1039/c2cs35367a. [DOI] [PubMed] [Google Scholar]
- 7.Lee A, Andrade GFS, Ahmed A, Souza ML, Coombs N, Tumarkin E, Liu K, Gordon R, Brolo AG, Kumacheva E. J Am Chem Soc. 2011;133:7563–7570. doi: 10.1021/ja2015179. [DOI] [PubMed] [Google Scholar]
- 8.(a) Wang T, LaMontagne D, Lynch J, Zhuang J, Cao YC. Chem Soc Rev. 2013;42:2804–2823. doi: 10.1039/c2cs35318k. [DOI] [PubMed] [Google Scholar]; (b) Zhang SY, Regulacio MD, Han MY. Chem Soc Rev. 2014;43:2301–2323. doi: 10.1039/c3cs60397k. [DOI] [PubMed] [Google Scholar]; (c) Singh G, Chan H, Baskin A, Gelman E, Repnin N, Král P, Klajn R. Science. 2014;345:1149–1153. doi: 10.1126/science.1254132. [DOI] [PubMed] [Google Scholar]; (d) Wang T, Wang X, LaMontagne D, Wang Z, Cao YC. J Am Chem Soc. 2013;135:6022–6025. doi: 10.1021/ja402539f. [DOI] [PubMed] [Google Scholar]
- 9.Liu YH, Wang F, Wang Y, Gibbons PC, Buhro WE. J Am Chem Soc. 2011;133:17005–17013. doi: 10.1021/ja206776g. [DOI] [PubMed] [Google Scholar]
- 10.Urban AS, Shen X, Wang Y, Large N, Wang H, Knight MW, Nordlander P, Chen H, Halas NJ. Nano Lett. 2013;13:4399–4403. doi: 10.1021/nl402231z. [DOI] [PubMed] [Google Scholar]
- 11.Quan Z, Xu H, Wang C, Wen X, Wang Y, Zhu J, Li R, Sheehan CJ, Wang Z, Smilgies DM, Luo Z, Fang J. J Am Chem Soc. 2014;136:1352–1359. doi: 10.1021/ja408250q. [DOI] [PubMed] [Google Scholar]
- 12.(a) Ross MB, Ku JC, Vaccarezza VM, Schatz GC, Mirkin CA. Nat Nanotechnol. 2015;10:453–458. doi: 10.1038/nnano.2015.68. [DOI] [PubMed] [Google Scholar]; (b) Wang Y, Chen G, Yang M, Silber G, Xing S, Tan LH, Wang F, Feng Y, Liu X, Li S, Chen H. Nat Commun. 2010;1:87. doi: 10.1038/ncomms1089. [DOI] [PubMed] [Google Scholar]; (c) Zhang J, Luo Z, Martens B, Quan Z, Kumbhar A, Porter N, Wang Y, Smilgies DM, Fang J. J Am Chem Soc. 2012;134:14043–14049. doi: 10.1021/ja304108n. [DOI] [PubMed] [Google Scholar]
- 13.LaGrow AP, Sinatra L, Elshewy A, Huang KW, Katsiev K, Kirmani AR, Amassian A, Anjum DH, Bakr OM. J Phys Chem C. 2014;118:19374–19379. [Google Scholar]
- 14.Shahmiri M, Ibrahim NA, Shayesteh F, Asim N, Motallebi N. J Mater Res. 2013;28:3109–3118. [Google Scholar]
- 15.Hou H, Zhu Y, Hu Q. J Nanomater. 2013;2013:797082. [Google Scholar]
- 16.Wen X, Zhang W, Yang S, Dai ZR, Wang ZL. Nano Lett. 2002;2:1397–1401. [Google Scholar]
- 17.Gao P, Zhang M, Niu Z, Xiao Q. Chem Commun. 2007:5197–5199. doi: 10.1039/b713154b. [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Varghese OK, Ruan C, Paulose M, Grimes CA. J Mater Res. 2003;18:2756–2759. [Google Scholar]
- 19.Fu Y, Zhao XY, Zhang JL, Li W. J Phys Chem C. 2014;118:18116–18125. [Google Scholar]
- 20.Lin Y, Ren J, Qu X. Acc Chem Res. 2014;47:1097–1105. doi: 10.1021/ar400250z. [DOI] [PubMed] [Google Scholar]
- 21.Wei H, Wang E. Chem Soc Rev. 2013;42:6060–6093. doi: 10.1039/c3cs35486e. [DOI] [PubMed] [Google Scholar]
- 22.Deng H, Shen W, Peng Y, Chen X, Yi G, Gao Z. Chem - Eur J. 2012;18:8906–8911. doi: 10.1002/chem.201200643. [DOI] [PubMed] [Google Scholar]
- 23.Marquez LA, Dunford HB. Biochemistry. 1997;36:9349–9355. doi: 10.1021/bi970595j. [DOI] [PubMed] [Google Scholar]
- 24.Tao Y, Ju E, Ren J, Qu X. Adv Mater. 2015;27:1097–1104. doi: 10.1002/adma.201405105. [DOI] [PubMed] [Google Scholar]
- 25.Guo Y, Deng L, Li J, Guo S, Wang E, Dong S. ACS Nano. 2011;5:1282–1290. doi: 10.1021/nn1029586. [DOI] [PubMed] [Google Scholar]
- 26.Gao L, Zhuang J, Nie L, Zhang J, Zhang Y, Gu N, Wang T, Feng J, Yang D, Perrett S, Yan X. Nat Nanotechnol. 2007;2:577–583. doi: 10.1038/nnano.2007.260. [DOI] [PubMed] [Google Scholar]
- 27.Gao Z, Xu M, Hou L, Chen G, Tang D. Anal Chim Acta. 2013;776:79–86. doi: 10.1016/j.aca.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 28.(a) Sun H, Zhao A, Gao N, Li K, Ren J, Qu X. Angew Chem, Int Ed. 2015;54:7176–7180. doi: 10.1002/anie.201500626. [DOI] [PubMed] [Google Scholar]; (b) Sun H, Gao N, Dong K, Ren J, Qu X. ACS Nano. 2014;8:6202–6210. doi: 10.1021/nn501640q. [DOI] [PubMed] [Google Scholar]; (c) Lin SS, Gurol MD. Environ Sci Technol. 1998;32:1417–1423. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.