Abstract
We describe a comprehensive protocol for the preparation of multifunctional DNA nanostructures termed nanoflowers (NFs), which are self-assembled from long DNA building blocks generated via rolling-circle replication (RCR) of a designed template. NF assembly is driven by liquid crystallization and dense packaging of building blocks, which eliminates the need for conventional Watson-Crick base pairing. As a result of dense DNA packaging, NFs are resistant to nuclease degradation, denaturation or dissociation at extremely low concentrations. By manually changing the template sequence, many different functional moieties including aptamers, bioimaging agents and drug-loading sites could be easily integrated into NF particles, making NFs ideal candidates for a variety of applications in biomedicine. In this protocol, the preparation of multifunctional DNA NFs with highly tunable sizes is described for applications in cell targeting, intracellular imaging and drug delivery. Preparation and characterization of functional DNA NFs takes ~5 d; the following biomedical applications take ~10 d.
INTRODUCTION
Advances in nanotechnology have been accompanied by an increasing demand for nanotools that can be applied to noninvasive early diagnosis and targeted therapy. In the past few years, many different materials have been proposed as powerful biomedical tools for use in disease diagnosis and therapy. Among them are exogenous nanomaterials such as organic or inorganic nanoparticles1–5, hydrogels6,7, polymers8–10, liposomes11 and micelles12,13, which have shown to possess outstanding features such as resistance to enzymolysis, functional surface modification and excellent optical properties. However, in spite of their unique properties, most of these nanomaterials are challenged by obvious cytotoxicity, structural instability, nonspecificity and sophisticated preparation processes, which largely limit their clinical applications. In addition, when considering their range of applications, nanotools that depend on these materials may become inflexible by the different modification methods needed for new platform construction and covalent bond formation.
Compared with these exogenous materials, DNA is naturally water-soluble and biocompatible. This means that it can be used as a building block to construct various DNA nanostructures14,15 with built-in functionalities for biomedical and biotechnological applications16,17. In addition, other advantages such as sequence programmability and automated controllable synthesis also make DNA nanostructures more suitable biomedical nanotools over their exogenous counterparts. Numerous studies have reported on the remarkable potential of DNA nanostructures; representative examples include DNA tetrahedron18–20, DNA dendrimer21,22, DNA origami14,17 and hybridization chain reaction–based DNA polymers23,24. Conventional approaches to DNA nanostructure construction typically rely on Watson-Crick base pairing between short DNA building blocks25. However, when applied in complicated physiological environments, approaches based on hydrogen bonding and base stacking were proven to have some intrinsic drawbacks. First, to assemble relatively large and sophisticated nanostructures for efficient biological applications, myriad different DNA strands with complicated sequences need to be manually designed. Sometimes even software engineering is needed26,27, making construction a cumbersome process. Second, preparation of DNA nanostructures usually means bulky synthesis of DNA strands, which is an expensive and time-consuming process. Third, DNA hybridization–generated nicks in the DNA backbone typically serve as potential cleavage sites for many exonucleases, which can decrease the biostability of traditional DNA nanostructures28,29. Fourth, as a result of steric hindrance of DNA strands, hydrogen bond–based DNA nanoassemblies often have loose internal structures, which are unfavorable for nanotherapeutic and bioimaging applications. Fifth, traditional DNA nanostructures can easily lose structural integrity during denaturation or at extremely low concentrations, which is unavoidable in an in vivo circulation system30.
Development of the protocol
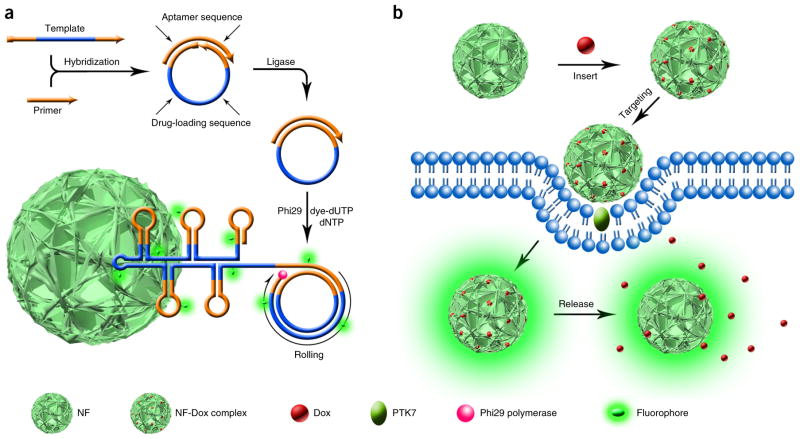
To solve these problems, our group recently reported, for the first time, RCR-based programmable and multifunctional DNA NFs with a hierarchical inner structure31,32. This kind of noncanonical DNA nanomaterial is demonstrated to have the advantages of exogenous nanomaterials, as well as DNA nanostructures, thus making it quite suitable for biomedical applications. NFs were generated from self-assembled compaction of a large amount of replicated DNA building blocks elongated through RCR33 (Fig. 1a). A benefit of the RCR-based strategy is that only one template strand and one primer strand are needed in NF assembly, thus avoiding the large number of DNA strands that are typically used in constructing previously reported DNA nanostructures. In contrast to Watson-Crick base pairing–dependent DNA nanostructure construction, NFs are self-assembled through an anisotropic DNA liquid crystallization process, which circumvents the otherwise complicated sequence design needed to construct traditional DNA nanostructures for interstrand or intrastrand hybridization. Highly concentrated DNAs in NFs are thus densely and orderly packed so that a large number of functional DNA moieties such as therapeutics and bioimaging agents could be encapsulated. The size of NFs can be tuned from ~200 nm to a few micrometers by changing the RCR reaction time. In addition to their unique construction features including long and non-nicked DNA backbones, as well as dense DNA packaging, DNA NFs showed efficient resistibility toward severe environments including nuclease treatment, heating/urea treatment–induced DNA denaturation and dilution to low concentrations. Resistance toward these factors is highly critical for many versatile applications in biomedicine.
Figure 1.
Schematic illustration of noncanonical self-assembly and biomedical applications of multifunctional DNA NFs. (a) RCR-based assembly of NFs. The linear DNA template was first hybridized with a primer and then ligated to form a circular template by T4 DNA ligase. Subsequently, phi29 DNA polymerase was used to perform RCR by polymerizing uncombined dNTP and dye-modified dUTP on the 3′-terminal end of the primer. RCR generated a large amount of elongated non-nicked concatemer DNA, and these DNAs then served as building blocks to self-assemble monodisperse, densely packed and hierarchical DNA NFs. NF sizes are tunable by changing the reaction time, with diameters ranging from ~200 nm to several micrometers. NF assembly does not rely on Watson-Crick base pairing between DNA building blocks, thus enabling tailored design of the template to carry multiple complements of functional nucleic acids. In this protocol, aptamers, drug-loading sites and dye molecules are integrated into NFs so that the prepared multifunctional NFs can act as smart nanotools for versatile biomedical applications such as targeted cancer cell recognition, bioimaging and targeted drug delivery. (b) Drug loading into NFs and targeted drug delivery. Doxorubicin (Dox) can be inserted into double-stranded 5′-GC-3′ or 5′-CG-3′ sequences by incubating it with NFs at room temperature. On the basis of the large number of generated drug-loading sites and internal hierarchical structures, drug payload capacity is extremely high when compared with other materials. The sgc8c aptamer incorporated can bind to PTK7-overexpressed cells, thereby endowing NFs with specific recognition toward target cancer cells. Dox will be released from NFs after intracellular uptake and cause selective apoptosis. The integrated dye molecules are used to give a fluorescence signal for intracellular imaging.
More importantly, the liquid crystallization and dense packaging–driven NF formation is sequence-independent, which means in theory that any sequences that conform to the basic principle of RCR can be used to prepare NFs. Through rational design, numerous nucleic acid sequences with various reported functionalities can be simply integrated into NFs through polymerase-mediated DNA elongation. For example, these can include functional moieties such as fluorescent agents for bioimaging (generated by modifying fluorophores on primers or using fluorophore-tethered deoxynucleotide triphosphates in RCR), aptamers34–36 for use in selective cell targeting and drug-associated sequences for loading drugs37–39. Multifunctional NFs can therefore be implemented in versatile biomedical applications, such as early diagnosis and targeted cancer therapy (Fig. 1b). Moreover, by simultaneously incorporating different fluorophores (fluorescein (FAM), cyanine 3 (Cy3) and 6-carboxyl-X-rhodamine (ROX)) into NFs via chemically modified deoxynucleotides (fluorescein-12-ddUTP, cyanine 3-dUTP and (6) ROX-12-ddUTP), multicolor NFs with a single excitation wavelength have also been proposed for multiplexed bioimaging with highly tunable emission wavelengths and spatial resolution32. Considering the programmability and flexibility of DNA NFs, we anticipate more potential applications for this emerging nanomaterial in the future through incorporation of nucleic acid sequences such as G-quadruplex40, i-motifs41 or even DNAzymes22,42.
In conclusion, RCR-based DNA NFs have shown exceptional promise and potential, as well as versatility, for applications in biomedicine. Specialists involved in cancer diagnosis and treatment will learn how the following protocol can be applied to their studies, whether in biosciences or medical sciences. General readers from a broad spectrum of disciplines will also appreciate an introduction to this uniquely powerful tool for molecular medicine.
Experimental design
Incorporating cDNA aptamers into templates
Aptamers35,36,43 are single-stranded oligonucleotides with a high affinity to special targets including metal ions, small organic molecules, proteins, viruses, cells and even tissues. Concatemeric ssDNA molecules containing multiple aptamers are expected to enhance the binding affinity of the resultant NFs to target cells through synergistic multivalent binding. This highlights the potential for future construction of NFs incorporated with therapeutic aptamers. Aptamers are generally 20–90 nt in length34,43–45, which makes it possible to fully incorporate their complementary sequence into the NF template with no deletion.
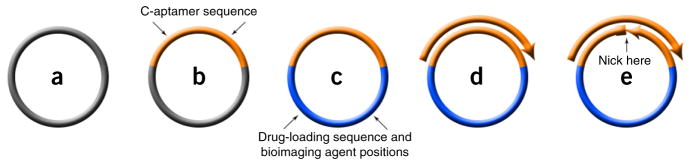
Aptamer sgc8c, which can specifically recognize protein tyrosine kinase 7 (PTK7) overexpressed on many cancer cells, was used as a model in this protocol34,46. Sgc8c was demonstrated to selectively recognize target HeLa cells and CEM cells, but not nontarget Ramos cells. Table 1 gives the details of the sgc8c sequence and, for template design, its complementary sequence C-sgc8c. This method is based on RCR. As such, template design calls for cyclization by DNA ligase with the help of a primer. Therefore, we explain conceptually the construction of the template in a step-by-step manner using Figure 2. First, the blank circular template shown in gray (Fig. 2a) is loaded with the C-aptamer targeting sequence (Fig. 2b); loading of additional sequences is discussed below.
TABLE 1.
DNA sequences used in this protocol.
| Template | 5′-Phosphate-TTCCCGGCGGCGCAGCAGTTAGATGCTGCTGCAGCGATACGCGTATCGCTATGGCATATCGTACGATATGCCGCAGCAGCATCTAACCGTACAGTATT-3′ |
| Primer | 5′-TCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA-3′ |
| Sgc8c | 5′-ATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA-3′ |
| C-sgc8c | 5′-TCTAACCGTACAGTATTTTCCCGGCGGCGCAGCAGTTAGAT-3′ |
Underlined domain: C-aptamer sequence; domain in italics: drug-loading sites and bioimaging agent positions.
Figure 2.
Designing the RCR template and primer for DNA NF preparation. (a) A blank circular sequence for RCR template design. (b) A circular template sequence integrated with C-aptamer sequence (orange domain). (c) A circular template sequence integrated with C-aptamer sequence (orange domain), drug-loading sites and bioimaging agent positions (blue domain). (d) Hybridization between linear primer and circular template. (e) A circular template nicked in the middle of the C-aptamer domain and converted to a 5′-terminal-phosphorylated linear template.
Incorporating drug-loading sites into templates
Previous studies have demonstrated that the anticancer drug doxorubicin (Dox) can be inserted into π-π stacking of double-stranded 5′-GC-3′ or 5′-CG-3′ sequences37–39. Following this principle, numerous designs have been shown in previous studies47,48. Benefiting from the easy programmability of NFs, these drug-associated sequences could be easily incorporated into templates, such that the resultant RCR products could be decorated with a series of aptamers and drug-loading sites for Dox (Fig. 2c). Oligonucleotide linkers such as poly-T or poly-A linkers are not needed between the aptamer portion and integrated drug-loading sequence, because they have no role in the construction and function of prepared DNA NFs. By incorporating drug-loading sites into templates, polymerase-mediated replication endows the generated NFs with a large number of drug-associated sequences, ultimately amplifying the drug-carrying capacity of NFs with correspondingly improved therapeutic effect.
Incorporating bioimaging agent positions into templates
To perform intracellular imaging, the nucleotide adenosine (A) should be strategically spaced along the DNA template providing regular positions to attach dye-modified dUTPs to the replicated product (Fig. 2c). Dye-modified dUTPs (fluorescein-12-ddUTP, cyanine 3-dUTP, (6) ROX-12-ddUTP or cyanine 5-dUTP in this protocol) can be recognized by many DNA polymerases, including phi29, and this enables their incorporation into NFs. When building the Förster resonance energy transfer (FRET) cascades that are required for implementation of a multifluorescent imaging platform32, dyes should be chosen on the basis of good spectral overlap as required by FRET—e.g., FAM, Cy3 and ROX. However, a comprehensive description of just one kind of fluorophore is used for the preparation of NFs in this protocol. Apart from adding the nucleotide adenosine for dye-modified dUTP attachment, it is also necessary to incorporate a sufficient number of thymine nucleobases into the template and to maintain the GC-rich domain in an intramolecular double-stranded form when positioning the nucleotide adenosine (Supplementary Fig. 1). During the design process, software such as NUPACK and Mfold can be used to simulate hybridization in order to simplify this procedure26. On the basis of the length of the aptamer and drug-associated sequences, the minimum number of nucleotides in the template should be more than 40 nt. Considering the efficiency of DNA synthesis and quality, as well as controllability, the maximum number of nucleotides is also suggested to be <120 nt.
Primer design
By following the same principle as RCR primer design, the primer should be designed such that it complements a domain of the template, which allows for subsequent elongation by DNA polymerase. The C-aptamer domain is regarded as the most suitable domain for hybridization with a primer because of its distinct and nonrepeating sequence compared with other domains, which thereby avoids nonspecific hybridization and ensures accurate intramolecular cyclization of the template (Fig. 2d). At this stage, it is possible to add a fluorophore to the 3′-terminal oligonucleotide of the primer according to the users’ own requirements (e.g., colocalization), although this is not discussed in the protocol.
Thus far, we have designed a linear primer, as well as a circular template, with three functional domains: C-aptamer domain, drug-loading sequence and bioimaging agent positions. However, the template that is finally needed in the RCR reaction is a 5′-terminal-phosphorylated single-stranded DNA that can be intramolecularly cyclized by DNA ligase after hybridization with a primer, according to the principles of RCR. Therefore, when applying this design to the final template, it should be linearized in the middle of the C-aptamer domain because of its nonrepeating sequence (Fig. 2e). The intramolecular hybridization of linear template can then be predicted by using a software program to determine the final structure and to eliminate the undesired secondary structures (Supplementary Fig. 1).
DNA preparation
Designed template and primer sequences (Table 1) can be synthesized via a mature solid-state DNA synthesizer and purified with reversed-phase HPLC on a C-18 column. After desalting, UV-visible measurements can be performed with a UV-visible spectrometer for DNA quantification. The concentration of the DNA sequence is calculated through the Beer-Lambert law: A = εbc. It is worth mentioning that commercial DNAs are available for purchase, which will greatly simplify the protocol.
Self-assembly of monodisperse, size-tunable and densely packed multifunctional DNA NFs
During RCR-based NF preparation, the template and the primer are first annealed for complete and accurate hybridization, after which the template DNA is circularized by using T4 DNA ligase. This protocol uses only T4 DNA ligase; however, Escherichia coli DNA ligase is also effective at this stage. Ligation conditions should be adapted according to the manufacturer’s instructions when using E. coli DNA ligase as an alternative. The hybridized primer is subsequently elongated by phi29 polymerase to generate periodic DNA with a myriad of aptamers, drug-loading moieties and bioimaging agents. By extending the reaction time, long and highly concentrated DNAs are produced (Table 2). NFs will gradually assemble through DNA liquid crystallization, which is an anisotropic process for orderly alignment of highly concentrated polymers. The size of NFs is strongly dependent on RCR reaction time, and hence it can be tuned by optimizing the reaction time.
TABLE 2.
The relationship between RCR time at 30 °C and NF sizes.
| RCR reaction time | NF size or description |
|---|---|
| 0.5 h | Only amorphous bulky structures are observed in SEM imaging |
| 1 h | Some crystal-like structures become visible |
| 2 h | Petal-like structures are formed on the surface |
| 3 h | Flower-like spherical structures are gradually formed on the matrices |
| 6 h | ~150 nm |
| 8 h | ~200 nm |
| 10 h | 200–300 nm |
| 15 h | ~500 nm |
| 20 h | ~1 μm |
| 30 h | 2–4 μm |
Characteristics of DNA NFs
NFs should be characterized after RCR to verify their quality. In our experiment, electrophoresis and confocal laser-scanning microscopy (CLSM) were used first to demonstrate the RCR result and to confirm the encapsulated fluorophore. Scanning electron microscopy (SEM) was then used to observe the size and surface morphology of NFs. Dynamic light scattering (DLS) was used to determine the hydrated radius of NFs. Transmission electron microscopy (TEM) and atomic force microscopy (AFM) were further used to characterize size and morphology. TEM images displayed hierarchical internal structures in NFs. Polarized light microscopy (PLM) was used to directly validate the liquid crystalline structures of NFs.
Stability of DNA NFs
As a result of the liquid crystallization and dense packaging–driven process, DNA condensation makes NFs more stable toward various environmental interferences, thus making them suitable for in vivo applications. Specifically, the exceptional stability of DNA NFs arises from their resistance to enzymolysis when incubated with 5 U/ml DNase I or 10% human serum, resistance to denaturation conditions when incubated at 170 °C or treated with 5 M urea and resistance to dissociation at low concentration (100× dilution). In our experiment, SEM imaging was used to verify the structural integrity of NFs after each treatment.
Aptamer-integrated DNA NFs for cell targeting
Aptamer-integrated DNA NFs were predicted to have similar or better recognition ability toward target cells compared with free aptamers, based on the multivalent effect of aptamers grafted onto the surface. In order to report a signal upon cancer cell binding, we integrated dye molecules into NFs through RCR. In this experiment, aptamer-integrated NFs were incubated with, and bound to, CEM and HeLa cancer cells, with Ramos cells as control. Such binding led to the endocytosis of NFs. Confocal microscopy and flow cytometry were then used to monitor the results, which showed satisfactory targeting performance of NFs toward target cells in comparison with control cells.
DNA NF–based drug delivery
The exceptional stability of DNA NFs makes them ideal candidates for drug delivery. In this protocol, dye molecules, preferably drug-associated sequences (e.g., double-stranded 5′-GC-3′ or 5′-CG-3′ sequences for anthracycline drugs in this protocol), and aptamers (sgc8c for CEM and HeLa cell lines) were incorporated into the template sequences to functionalize NFs as ‘smart nanotherapists’, which are able to specifically deliver anticancer drugs. Compared with other delivery systems, DNA NFs have obvious advantages such as high biocompatibility, flexible programmability, stability against physiological interference, as well as high drug-loading capacity and traceability. As the preparation of NFs is sequence-independent, all reported aptamer sequences can be incorporated into NFs by easily changing the anti-aptamer sequence within the template. This could largely extend the scope of potential applications. NFs with a size of ~200 nm were used for intracellular experiments, essentially because nanomaterials of this size can be internalized by many cancer cells, according to previous reports49,50. In order to prepare drug-loaded DNA NFs, Dox was incubated with the NFs dispersed in Dulbecco’s PBS (DPBS). The loading amount could be calculated by measuring the absorption of free Dox in the supernatant on a UV-visible spectrometer. In our experiment, bioimaging of the intracellular behavior of NFs in target cells (CEM and HeLa) was performed with a Leica TCS SP5 confocal microscope in differential interference contrast (DIC) mode. An Ar laser was used for excitation of fluorescein and Dox, and a He-Ne laser was used for excitation of Cy3, ROX and Cy5. Nontarget Ramos cells were used as a control for each cell line; samples treated with no NFs were used as a control. Finally, the cytotoxicity of NFs, free drug (control) or drug-NF complexes was evaluated using the CellTiter 96 AQueous one solution cell proliferation assay.
Limitations
DNA NFs are RCR-based self-assembled nanostructures that allow the integration of polymerase-recognized monomers (e.g., dNTP and dye-modified dUTP), which can partially limit the range of possible applications. In this protocol, only aptamers can be used as a cell-targeting moiety; other targeting functional groups such as antibodies and cell-receptor ligands are difficult to incorporate into the NF structure. However, considering the large number of proposed functional nucleic acids and the various targets of aptamers, we still think that the RCR-based DNA NFs show promise for versatile biomedical applications along with applications in other fields. DNA NFs cannot carry all kinds of drugs, but it is reported that the anthracycline class of drugs (e.g., Dox and daunomycin), which are widely used in cancer treatment, can intercalate into DNA and hence can be incorporated into NFs37–39. For other drugs or large molecules, micro-encapsulation may be an accessible pathway51.
In addition, because of dense DNA packaging, NFs showed relatively slow drug-release kinetics compared with other drug carriers such as mesoporous nanoparticles4, micelles12 and hybridization-based DNA nanostructures12,17,18,22. Hence, NFs are not favorable for situations in which quick drug release is necessary. In the fields of intracellular biosensing and analyte detection, NFs do not have advantages, in contrast to previously reported biosensing platforms such as graphene oxide nanosheets52, nanoparticles53, DNA dendrimers22 and DNA tetrahedra20. This is because large quantities of DNA probes are packaged inside NFs, which means that they cannot function well toward target molecules.
It should also be noted that we have not yet found an adequate method for accurate quantification of NFs. NFs have diameters ranging from ~200 nm to a few micrometers, which are too small to be counted with a hemocytometer. Furthermore, once prepared, NFs cannot be completely dried for weighing in case of adhesion and agglomeration. To avoid this, we proposed a provisional method that measures and calculates the direct correlation between the concentration of NFs and maximum inserted Dox. Nevertheless, we still want to develop a reliable and direct method for NF quantification.
MATERIALS
REAGENTS
! CAUTION All chemical reagents used in this protocol are potentially harmful; therefore, it is necessary to use protective devices such as lab coats, gloves, goggles and mask throughout the procedure. Solid and liquid waste should be recycled or disposed of according to local or institutional regulations.
-
Trichloroacetic acid (C2HCl3O2; J&K Scientific, cat. no. T0369-500G)
! CAUTION Trichloroacetic acid is a corrosive irritant.
-
1,2-Dichloroethane (C2H4Cl2; J&K Scientific, cat. no. 353972-2.5L)
▲ CRITICAL 1, 2-Dichloroethane should be re-distilled before use, but anhydrous reagent can be used directly.
DCI activator (0.25 M; Sigma-Aldrich, cat. no. L880045-06)
Cap A (acetic anhydride:tetrahydrofuran = 9.1:90.9 (vol/vol); Sigma-Aldrich, cat. no. L840045-06) ! CAUTION This chemical is corrosive.
Cap B (tetrahydrofuran:N-methylimidazole:pyridine = 8:1:1 (vol/vol/vol); Sigma-Aldrich, cat. no. L850045-06) ! CAUTION This chemical is an irritant.
Acetonitrile (CH3CN; Sigma-Aldrich, cat. no. L810045-06)
Oxidizer (0.02 M, tetrahydrofuran:H2O:pyridine:iodine = 90.54:9.05:0.41:0.43 (vol/vol/vol/wt); Sigma-Aldrich, cat. no. L860045-06)
dA-CE phosphoramidite (Glen Research, cat. no. 10-1000-10e, 1.0g)
dmf-dG-CE phosphoramidite (Glen Research, cat. no. 10-1020-10e, 1.0g)
dT-CE phosphoramidite (Glen Research, cat. no. 10-1030-10e, 1.0g)
ac-dC-CE phosphoramidite (Glen Research, cat. no. 10-1015-10e, 1.0g)
Chemical phosphorylation reagent II (CPR II; Glen Research, cat. no. 10-1901-90, 100μmoles)
Thymidine lcaa CPG (1,000 Å, ChemGenes, cat. no. N-5104, 5g)
Deoxyadenosine (n-bz) 3′-lcaa CPG (1,000 Å, ChemGenes, cat. no. N-5101, 5g)
Ammonium hydroxide (Sinopharm Chemical Reagent, cat. no. 10002118)
Methylamine water solution (Sinopharm Chemical Reagent, cat. no. 80081228)
Absolute ethanol (General-Reagent, cat. no. P1090446)
Glacial acetic acid (Sinopharm Chemical Reagent, cat. no. 10000218)
Triethylamine ((C2H5)3N; Sigma-Aldrich, cat. no. T0886-500ML)
Sodium chloride (NaCl; Adamas, cat. no. 81793B)
Magnesium chloride hexahydrate (MgCl2·6H2O; Sinopharm Chemical Reagent, cat. no. 10012818)
Acetonitrile (CH3CN; Adamas, cat. no. 80988C)
T4 DNA ligase (400,000 units/ml; New England Biolabs, cat. no. M0202S)
DNA ligation buffer, 10× (10×(50 mM Tris-HCl, 1 mM MgCl2, 1 mM ATP and 1 mM DTT); New England Biolabs, cat. no. M0202S)
dNTP (10 mM; Takara, cat. no. 4019)
Cyanine 3-dUTP (1 mM; Enzo Life Sciences, cat. no. ENZ-42501)
Cyanine 5-dUTP (1 mM; Enzo Life Sciences, cat. no. ENZ-42502)
Fluorescein-12-ddUTP (1 mM; Enzo Life Sciences, cat. no. ENZ-42833)
(6) ROX-12-ddUTP (0.1 mM; Enzo Life Sciences, cat. no. ENZ-42857)
BSA, 10× (New England Biolabs, cat. no. M0269S)
Phi29 DNA polymerase (10,000 units/ml; New England Biolabs, cat. no. M0269S)
phi29 DNA polymerase buffer, 10× (10×(50 mM Tris-HCl, 10 mM (NH4)2SO4, 10 mM MgCl2 and 4 mM DTT); New England Biolabs, cat. no. M0269S)
Dulbecco’s PBS (1× DPBS basic; Gibco, cat. no. C14190500BT-500ml)
Agarose (HydraGene, cat. no. R9012LE-100g)
Ethidium bromide (Dingguo, cat. no. NEP028) ! CAUTION Ethidium bromide is a suspected mutagen and a possible carcinogen. Nitrile gloves and lab coats should be worn when you are handling this reagent and any gels or solutions carrying it.
Loading buffer, 6× (Takara, cat. no. 9156)
DNA ladder, 20 bp (Dye Plus; Takara Bio, cat. no. 3420A)
Tris (C4H11NO3; Scientific Research special, cat. no. AMRESCO-500G)
Boric acid (H3BO3; Sangon Biotech, cat. no. BT0897-500g)
EDTA disodium salt (EDTA-2Na; Sinopharm Chemical Reagent, cat. no. 10009717)
-
Acetone (CH3COCH3; Reagent, cat. no. G75902A-500ml)
! CAUTION Acetone is volatile.
DNase I (2,000 units/ml; New England Biolabs, cat. no. M0303S)
DNase I buffer, 10× (10×(10 mM Tris-HCl (pH 7.6 at 25 °C), 2.5 mM MgCl2 and 0.5 mM CaCl2); New England Biolabs, cat. no. M0303S)
Human serum (Asterand)
Urea (CH4N2O; Dingguo, cat. no. DH362-4.1)
-
Cell lines (CCRF-CEM (human T-cell ALL), Ramos (human B-cell Burkitt’s lymphoma) and HeLa cells (human cervical carcinoma))
! CAUTION Cell cultures are biohazardous and potentially infectious materials. Proper personal protective equipment should be provided during cell experiments.
RPMI 1640 medium basic (1×; Gibco, cat. no. C11875500BT-500ml)
FBS (heat-inactivated; Gibco, cat. no. 10099-141-500ml)
Penicillin-streptomycin (10,000 U/ml penicillin and 100 μg/ml streptomycin; Gibco, cat. no. 15140-122)
Ribonucleic acid, transfer from baker’s yeast (yeast tRNA; Sigma-Aldrich, cat. no. 83853-100MG)
D-(+)-Glucose (Sigma-Aldrich, cat. no. G8270-1KG)
Trypsin-EDTA, 0.25% wt/vol (1× trypsin-EDTA, Gibco, cat. no. 25200-056-100ml)
Doxorubicin (Dox; Hisun Pharmaceutical)
CellTiter 96 AQueous one solution cell proliferation assay (Promega, cat. no. G3580)
EQUIPMENT
DNA synthesizer (Polygen)
Glass beaker, 50 ml
Glass cylinder, 100 ml and 1 liter
Microcentrifuge tube, 1.5 and 2 ml
Centrifuge tube (15 ml; Santa Cruz Biotechnology, cat. no. sc-200250)
Milli-Q reference ultrapure water purification system (Millipore)
Vertical heating pressure steam sterilizer (Shanghai ShenAn, cat. no. LDZM-60KCS)
Analytical balance (Mettler Toledo, AL204)
Vortex (IKA Vortex Genius 3)
Thermostatic water bath (Jing Hong, cat. no. DKB-5015)
Timekeeper (GE Healthcare)
Micropipette with tips (Eppendorf)
Freezer (Zhongke Meiling Cryogenics Science and Technology, cat. no. YCD-EL259)
Centrifuge (Eppendorf, Centrifuge 5417R)
Micro-pore filter (0.45 μm; Jin Long, MICRO PES)
HPLC (Agilent, 1260)
C-18 reversed-phase column (5 μm, 4.6×250 mm, 100 Å; GL Sciences, Inertsil ODS-3, cat. no. 5020-01732)
Centrifugal vacuum evaporator (Labconco, CentriVAP concentrator, cat. no. 7810040)
Desalting column (GE Healthcare illustra NAP-5 column Sephadex G-25 DNA grade)
Spectrophotometer (Thermo Scientific, NanoDrop 2000)
Microscope sliders (Sail Brand, cat. no. 7101)
Microwave (Galanz)
Electrophoresis power supply (Bio-Rad, Power PAC 200)
Molecular imager (Bio-Rad, ChemiDoc XRS+ with lab imaging software)
Scanning electron microscope (Hitachi, S-4800)
Zetasizer (Malvern, Nano-ZS90)
Transmission electron microscope (JEOL, F-2010, at a working voltage of 100 kV)
Atomic force microscope (Veeco, Nanoscope IIIa)
Polarizing microscope (Nikon, Optipho-2)
Ice-making machine (XUEKE)
Parafilm (Parafilm, cat. no. PM-996)
High-temperature oven (TEST, 202-0AB)
Cell culture incubator (Thermo Scientific, series II water jacket CO2 incubator, model 3111)
TC 10 automated cell counter (Bio-Rad)
Cell culture dish (Fisher Scientific, cat. no. 310109009)
Flow cytometer (BD FACSVerse, cat. no. 651154)
UV-visible spectrophotometer (SHIMADZU, UV-2450, cat. no. 206-24301-93)
Confocal laser-scanning microscope (Olympus, FV1000)
Glass-bottom microscope dish (In Vitro Scientific, cat. no. D35-14-1-N)
Ninety-six-cell plate (Thermo Scientific, cat. no. 167008)
Microplate reader (BioTek)
Orbital shaker (Thermo Scientific, Model 420)
REAGENT SETUP
Trichloroacetic acid (TCA) deblock
To 1 liter of 1,2-dichloroethane, add 40 g of trichloroacetic acid. Store the solution at room temperature (20°C in this protocol) in a dry environment for up to 6 months.
AMA
Mix ammonium hydroxide and 40% methylamine in a 50:50 ratio (vol/vol). Store it at 4 °C for up to 6 months.
Triethylamine acetate (TEAA) solution, 2 M
To 304 ml of double-distilled water, add 57 ml of glacial acetic acid and 139 ml of triethylamine. Store the solution at 4 °C for up to 3 months. Dilute it to 0.1 M with double-distilled water before use.
TBE buffer, 10×
Add 108 g of Tris, 55 g of boric acid and 7.5 g of EDTA-2Na to 1 liter of double-distilled water. Store the buffer at room temperature for up to 3 months.
Complete medium with 10% FBS and penicillin (100 U/ml)-streptomycin (100 μg/ml)
To 500 ml of RPMI 1640 medium, add 50 ml of FBS (heat inactivated) and 5 ml of penicillin-streptomycin. Store it at −20 °C for at least 6 months or at 4 °C for up to 3 months.
NaCl solution, 3 M
To prepare 40 ml of 3 M NaCl solution, add 7.02 g of NaCl to 40 ml of double-distilled water. Store the solution at 4°C for at least 6 months.
Aqua regia
Mix concentrated nitric acid and hydrochloric acid in a volume ratio of 1:3 in a 50-ml beaker. Aqua regia should be freshly made.
Washing buffer
To 1 liter of DPBS, add 4.5 g of glucose and 5 ml of 1 M MgCl2. Store the buffer at 4 °C for up to 3 months.
Binding buffer
To 1 liter of DPBS, add 4.5 g of glucose, 100 mg of tRNA, 1 g of BSA and 5 ml of 1 M MgCl2. Store the buffer at 4 °C for up to 2 months.
Ethanol, 70% (vol/vol)
To prepare 1 liter of 70% (vol/vol) ethanol, add 700 ml of water to 300 ml of absolute ethanol.
Nonenzymatic dissociation solution
To 10 ml of DPBS, add 0.02 g of EDTA-2Na. Store the solution at 4 °C for at least 3 months.
EQUIPMENT SETUP
Setup of DNA synthesis
All phosphoramidite monomers are freshly dissolved in anhydrous acetonitrile, where the use of molecular sieves is necessary. For each conventional dA, dG, dC and dT phosphoramidite, a concentration of 0.067 M is needed. For chemical phosphorylation reagent II (CPR II), the concentration is 0.1 M. Concentrations of phosphoramidite monomers differ according to the synthesizer used. Please follow the instructions from the manufacturer when you are preparing for synthesis. To ensure high coupling efficiency, coupling time for conventional dA, dG, dC and dT phosphoramidites is suggested to be 60 s, and for CPR II 900 s is the suggested coupling time.
Setup of HPLC separation method
Eluent A is acetonitrile and eluent B is 0.1 M TEAA in double-distilled water. A C-18 reversed-phase column (250 mm × 4.6 mm, 5 μm, 100 Å) is used. Equilibrate the column with 10% acetonitrile (acetonitrile:TEAA=10:90) for more than 10 min before separation. For short sequences, use a fast gradient, and use a slow gradient for long sequences. Set the wavelength of the UV detector at 260 nm for all sequences, and use a fluorescence detector for dye-labeled sequences. For example, use a linear gradient of 10–65% eluent A (A:B = 10:90 to 65:35) in 30 min for primer (40 nt) purification, and use a linear gradient of 10–40% eluent A (A:B = 10:90 to 40:60) in 30 min for template (98 nt) purification. The flow rate is kept as 1 ml/min, unless indicated otherwise.
PROCEDURE
DNA preparation ● TIMING 30 min to 3 d
-
1|
Prepare DNA sequences according to Table 1. DNA sequences that are used in this protocol can be purchased. If you wish to make your own DNA with a synthesizer, please see Supplementary Method for our suggested steps.
-
2|
Dilute the template and primer sequences to 10 μM with ultrapure water.
-
3|
Label the concentration of each sample tube and stock it for further use.
■ PAUSE POINT Stock solutions can be stored at −20 °C for at least 1 year.
Preparation of ligated DNA template ● TIMING ~8 h
-
4|
Add the following to a 1.5-ml centrifuge tube.
Component Amount (μl) Final concentration in 100 μl Ultrapure water 69.5 10× DNA ligation buffer 10 1× Template (10 μM) 6 0.6 μM Primer (10 μM) 12 1.2 μM Total 97.5 -
5|
Mix the tube by vortexing, and heat the mixture at 95 °C for 5 min. Cool the mixture slowly to room temperature over a period of 3 h.
▲ CRITICAL STEP Heating the DNA to 95 °C and subsequent gradual cooling to room temperature are important steps for complete hybridization of cDNAs.
? TROUBLESHOOTING
-
6|
Add 2.5 μl of T4 DNA ligase (400,000 U/ml) to the annealed mixture, and mix it well with a pipette tip or by mildly vortexing it.
▲ CRITICAL STEP E. coli DNA ligase is also an effective substitute for T4 DNA ligase in this step. As noted in the discussion above, please follow the manufacturer’s instructions for ligation conditions.
-
7|
Incubate the mixture at room temperature for 4 h.
■ PAUSE POINT The ligated product can be stored at 4 °C for at least 1 month.
Self-assembly of monodisperse, size-tunable and densely packed multifunctional DNA NFs ● TIMING ~1 d
-
8|
Prepare the following mixture in a 1.5-ml centrifuge tube on ice and mix it by pipetting.
Component Amount (μl) Final concentration Circularized template-primer complex (~600 nM) 50 286 nM dNTPs (10 mM) 20 1.9 mM Dye-modified dUTPs (1 mM) 5 0.048 mM 10× BSA 10 0.95× 10× phi29 DNA polymerase buffer 10 0.95× phi29 DNA polymerase (10,000 U/ml) 10 952 units/ml Total 105 ▲ CRITICAL STEP Choose appropriate dye-modified dUTPs, according to your experiments. Fluorescein-12-ddUTP, cyanine 3-dUTP, (6) ROX-12-ddUTP and cyanine 5-dUTP are all available for NF preparation. Perform this step in an ice bath to maintain the activity of phi29 DNA polymerase.
-
9|
Seal the tube tightly with Parafilm to avoid the loss of solution by evaporation.
-
10|
Incubate the mixture at 30 °C for more than 6 h according to Table 2.
▲ CRITICAL STEP By adjusting RCR reaction time, the size of NFs can be easily tuned. The relationship between RCR reaction time at 30 °C and NF size is shown in Table 2. This protocol is adapted for the above-mentioned system. For other templates, a handy rule of thumb is that fewer base pairs correspond to a looser structure of produced NFs. NF sizes generated using a template with fewer base pairs become larger within the same reaction time.
? TROUBLESHOOTING
-
11|
Inactivate the phi29 DNA polymerase at 75 °C for 10 min to end the reaction.
-
12|
Centrifuge the product at 14,000g for 10 min.
-
13|
Remove the supernatant with a pipette and add 100 μl of DPBS.
-
14|
Resuspend NFs by pipetting the mixture up and down or by mildly vortexing it.
-
15|
Repeat Steps 12–14 once more, and then store the solution at −20 °C.
■ PAUSE POINT The prepared stock solution of NFs can be stored at −20 °C for at least 3 months.
Confirmation of RCR by agarose electrophoresis ● TIMING ~2 h
-
16|
Prepare a 2% (wt/vol) agarose gel with ethidium bromide.
! CAUTION Ethidium bromide is a suspected mutagen and a possible carcinogen. Nitrile gloves and lab coats should be worn when you are handling this reagent and any gels or solutions carrying it. Disposal of material containing ethidium bromide should follow local institutional safety procedures.
-
17|
Prepare agarose electrophoresis samples as shown.
Lane M Lane 1 Lane 2 Lane 3 Lane 4 Loading buffer (6×, μl) — 2 2 2 2 Primer stock solution (μl) — — 2 1 — Template stock solution (μl) — 2 — 1 — NF stock solution (μl) — — — — 2 Ladder (20 bp, μl) 3 — — — — DPBS (μl) — 6 6 6 6 -
18|
Load the samples in lanes and perform electrophoresis at 100 V for 40 min.
-
19|
Stop the electrophoresis and remove the gel. Image the DNA band using a Bio-Rad molecular imager with the imaging software under UV light.
▲ CRITICAL STEP Electrophoresis here is first used to characterize the elongation result of DNA through RCR. A representative gel image is shown in Supplementary Figure 2a. The template sequence in lane 1 and the primer sequence in lane 2 should have different migration velocities for their different lengths. After ligation, more complicated structures will have formed through intramolecular and intermolecular base pairs, which, in this experiment, are shown in lane 3 as multibands and tailing. In lane 4, assembled NFs have a migration velocity close to zero because of their significantly larger molecular weight. These electrophoresis results indicate that phi29 DNA polymerase–mediated RCR was successfully performed.
Checking for integration of dye molecules by laser confocal fluorescence microscopy ● TIMING ~1 h
-
20|
Add 5 μl of DPBS to 5 μl of NF stock solution and mix it by mildly vortexing.
-
21|
Pipette 10 μl of the diluted NF solution onto a microscope slide.
-
22|
Use laser confocal fluorescence microscopy for NF imaging. Choose appropriate excitation and emission wavelengths according to the integrated fluorophore (for FAM, the maximum excitation wavelength is 496 nm and the maximum emission wavelength is 520 nm; for Cy3, the maximum excitation wavelength is 550 nm and the maximum emission wavelength is 564 nm; for ROX, the maximum excitation wavelength is 580 nm and the maximum emission wavelength is 630 nm; and for Cy5, the maximum excitation wavelength is 650 nm and the maximum emission wavelength is 662 nm).
! CAUTION High-energy lasers can cause severe thermal burns. Sufficient protective equipment and careful operation are needed.
▲ CRITICAL STEP To verify the integration of dye molecule–tethered dUTP (Cy5 is an example in this experiment), NFs are imaged in buffer conditions after washing them with ultrapure water. In this way, the fluorescence signal from free dye-molecule–tethered dUTP is eliminated, and the only signal source of remaining fluorescence is the dye-molecule–integrated NF. In this representative experiment, Cy5-integrated NFs were used as an example. The result, as shown in Supplementary Figure 2b, indicates a strong fluorescence signal emitted from each single NF, which accounts for the incorporated Cy5 molecule. For clear imaging and easy analysis, NFs at 2 μm in size (RCR for 20–30 h) were used for representative imaging.
Characterization of DNA NFs
-
23|
A number of options are available to characterize the DNA NFs produced in previous steps. Use option A for SEM characterization, option B for dynamic light-scattering characterization, option C for TEM characterization, option D for AFM characterization and option E for PLM characterization. For general users, we suggest choosing at least option A.
-
SEM imaging of NFs ● TIMING ~4 h
Soak the silicon wafer overnight in freshly prepared aqua regia.
Clean the surface of the silicon wafer twice with absolute alcohol and then once with acetone. Dry the silicon wafer for further use.
Add 10 μl of NF stock solution to the cleaned silicon wafer.
Dry the sample in an oven at 70 °C for 2 h.
Sputter-coat the sample with gold before SEM imaging.
-
Use SEM to observe the size and surface morphology of NFs.
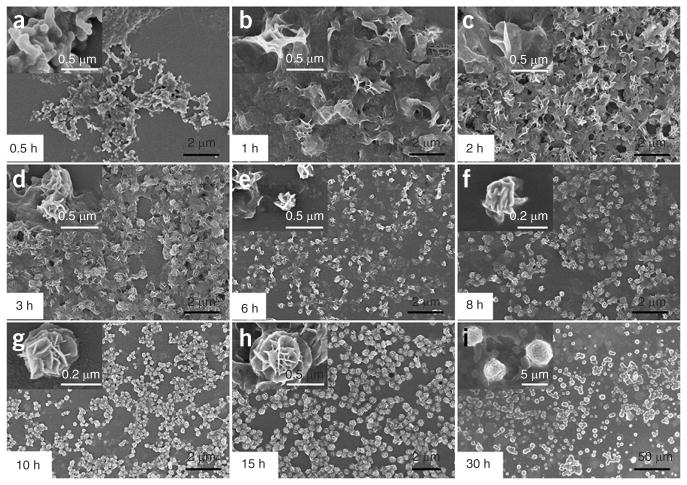
▲ CRITICAL STEP It is necessary to check the morphology and size of NFs using SEM during the growth process with a reaction time from 0.5 to 30 h to have a deeper understanding of the entire growth process. Figure 3 shows the referenced SEM results. After 0.5 h of reaction, irregular bulky structures formed, as shown in Figure 3a, and the bulky matrices then began to show some crystal-like structures on the surfaces at 1 h (Fig. 3b). Subsequently, these crystal-like structures grew further to generate ‘petal’ structures at 2 h (Fig. 3c). After 3 h, flower-like structures first appeared on the surface of matrices (Fig. 3d), and the spherical NFs gradually cleaved off and split from each other to become monodisperse (Fig. 3e). The size of NFs kept growing over time, and diameters reached ~200 nm at 8 h (Fig. 3f). After RCR for 10 h, the size of NFs reached 200–300 nm (Fig. 3g), and after 15 h the SEM image shows an NF size of ~500 nm (Fig. 3h). After a reaction time of 30 h, NFs with diameters of 2–4 μm were observed (Fig. 3i), and, in all likelihood, they did not stop growing until the substrate had been exhausted. NF self-assembly is a time-dependent process, and by adjusting the RCR reaction time NFs with diameters ranging from ~200 nm to a few micrometers can be prepared according to the needs of each application.
? TROUBLESHOOTING
-
Dynamic light-scattering measurement of NFs ● TIMING ~30 min
Dilute 10 μl of NF stock solution with 1.5 ml of ultrapure water in a centrifuge tube.
Mix it well by vortexing, and transfer the solution to a DLS cuvette.
-
Determine the hydrated radius of NFs on a Malvern Zetasizer.
▲ CRITICAL STEP The hydrated radius of NFs determined by DLS should be consistent with SEM data. NFs with 10–15 h of reaction time should have an average hydrated radius of ~150 nm, as shown in Supplementary Figure 3a.
-
TEM imaging of NFs ● TIMING ~4 h
Deposit 5 μl of NF stock solution onto a carbon-coated copper grid.
Dry the samples in an oven at 70 °C for 2 h.
-
Insert the sample into the F-2010 transmission electron microscope and image at a working voltage of 100 kV.
▲ CRITICAL STEP Ultrathin sheet sections of single NFs can be observed in TEM images (Supplementary Fig. 3b,c), as well as hierarchical surface morphology.
-
AFM imaging of NFs ● TIMING ~1 d
Prepare a clean mica sheet for sample mounting.
Add 10 μl of NF stock solution to the center of the mica sheet. The solution will spread over the whole surface.
Dry the sample naturally at room temperature.
-
Pipette 10 μl of ultrapure water onto the mica sheet and remove it after 5 s to wash away the salt crystals.
▲ CRITICAL STEP Be careful when you are adding or removing the solution with tips, to prevent damaging the mica surface. A rough mica surface may break the needle tip on the microcantilever, and it may reduce the imaging quality.
-
Image the NFs with an atomic force microscope using tapping mode in ambient air.
▲ CRITICAL STEP AFM results can corroborate the size and monodispersity of NFs. For NFs with 10–15 h of reaction time, AFM results will show a diameter of ~150 nm with great monodispersity (Supplementary Fig. 3d,e).
-
PLM imaging of NFs ● TIMING ~1 h
Add 5 μl of DPBS to 5 μl of NFs and mix it by mildly vortexing.
Pipette 10 μl of diluted NF solution on a microscope slide.
-
Use polarized light and a polarizing microscope for PLM.
▲ CRITICAL STEP When NFs are exposed between crossed polarizers, they display disc-shaped optical textures as a result of the birefringence of the liquid crystalline structure, which is an optical property of anisotropic materials (Supplementary Fig. 3f,g). The liquid crystalline structures of NFs can hence be directly illustrated.
-
Figure 3.
SEM imaging of the morphology and size of NFs during the growth process with reaction time from 0.5 to 30 h. (a) RCR-produced amorphous bulky structures were observed at 0.5 h. (b) RCR products then began to display some crystal-like structures on the surfaces of bulky matrices after incubation for 1 h. (c) Petal-like structures are subsequently formed on the surfaces of crystal-like structures after incubation for 2 h. (d) Flower-like spherical structures were first formed on the matrices after 3 h of reaction. (e–i) The size of NFs kept growing over time. (e) SEM imaging of NFs after 6 h with diameters of ~150 nm. (f) SEM imaging of NFs after 8 h with diameters of ~200 nm. (g) SEM imaging of NFs after 10 h with diameters of 200–300 nm. (h) SEM imaging of NFs after 15 h with diameters of ~500 nm. (i) With a reaction time of 30 h, NFs grew to a diameter of 2–4 μm. Adapted with permission from Zhu et al.31 (© 2013, American Chemical Society).
Examining resistance to nuclease degradation ● TIMING 1–2 d
-
24|
Dilute DNase I to 50 U/ml with 1× DNase I buffer.
▲ CRITICAL STEP Perform this step in an ice bath.
-
25|
Add the following components to a 200-μl centrifuge tube.
Component Amount (μl) Final concentration NF stock solution 8 10× DNase I buffer 1 1× DNase I (50 U/ml) 1 5 U/ml Total 10 -
26|
Mix with a pipette tip.
-
27|
Seal the tube tightly with Parafilm.
-
28|
Incubate the mixture in a 37 °C water bath for 24 h.
-
29|
Deactivate DNase I by heating it at 75 °C for 10 min.
-
30|
Use SEM to verify the structural integrity of NFs. Operations are defined in Step 23A.
Examining resistance to human serum ● TIMING 1–2 d
-
31|
To a 200-μl centrifuge tube, add 9 μl of DPBS dispersed NFs (from Step 15) and 1 μl of human serum.
-
32|
Mix with a pipette tip.
-
33|
Seal the tube tightly with Parafilm.
-
34|
Incubate the mixture in a 37 °C water bath for 24 h.
-
35|
Use SEM to verify the structural integrity of NFs. Operations are defined in Step 23A.
Examining thermal stability ● TIMING ~5 h
-
36|
Pipette 10 μl of NF stock solution (from Step 15) on a silicon wafer.
-
37|
Dry the samples at 70 °C for 2 h and subsequently incubate them at 170 °C in a high-temperature oven for 0.5 h.
-
38|
Use SEM to verify the structural integrity of NFs. Operations are defined in Step 23A.
Examining stability against urea ● TIMING ~5 h
-
39|
To a 200-μl centrifuge tube, add 10 μl of NF stock solution (from Step 15) and 3 mg of urea (final concentration of 5 M).
-
40|
Mix with a pipette tip or by mildly vortexing.
-
41|
Incubate the mixture at room temperature for 30 min.
-
42|
Centrifuge the mixture for 5 min at 14,000g. Discard the supernatant with a pipette tip.
-
43|
Add 10 μl of DPBS to disperse the sample.
-
44|
Repeat Steps 43 and 44 3–5 times.
-
45|
Use SEM to verify the structural integrity of NFs. Operations are defined in Step 23A.
Examining resistance to dissociation at low concentration ● TIMING ~4 h
-
46|
To a 200-μl centrifuge tube, dilute 1 μl of NF stock solution (from Step 15) with 99 μl of DPBS.
-
47|
Use SEM to verify the structural integrity of NFs. Operations are defined in Step 23A.
Cell culture and cell counting ● TIMING 2–3 d
-
48|
Culture CCRF-CEM, Ramos and HeLa cells in RPMI 1640 medium supplemented with 10% (vol/vol) FBS (heat-inactivated) and penicillin (100 U/ml)-streptomycin (100 μg/ml) in a cell culture incubator at 37 °C with 5% CO2. When necessary, use a hemocytometer to determine cell density. For adherent cell lines (e.g., HeLa), follow the steps given in Box 1 for cell dissociation and counting.
! CAUTION Cell cultures are biohazardous and potentially infectious materials. Proper personal protective equipment should be provided during cell experiments.
Box 1. Dissociation of adherent cells and cell counting before cell targeting ● TIMING ~1 h.
For adherent cell lines (HeLa), split cells 24 h before dissociation treatment.
Discard the culture medium with a pipette and wash the cells with 3 ml of DPBS.
-
Dissociate the cells from the culture flask or dish by adding 1 ml of nonenzymatic dissociation solution or by short-term (30 s to 1 min) trypsin treatment.
▲ CRITICAL STEP Long-term treatment with trypsin will cause undesirable cell damage. Therefore, restrict trypsin treatment to 5 min. In cell targeting experiments (Step 49), do not use trypsin in this step. Trypsin would damage membrane proteins and hence affect the binding between the aptamer and the target cells. In addition, thorough mixing will ensure an accurate count.
Add 3 ml of complete culture medium to end the dissociation reaction after nonenzymatic dissociation solution or trypsin treatment.
Transfer the cells dissociated from culture flask or dish into a 15-ml centrifuge tube.
Count the number of cells with a hemocytometer.
Test cell targeting of aptamer-integrated DNA NFs ● TIMING 3–4 h
-
49|
Count the number of target (CEM and HeLa) and control (Ramos) cells according to Step 48. In total, 4 × 105 cells are needed for each cell line (duplicate samples of 2 × 105 cells each).
-
50|
Add the cell volume that yields the desired number of cells to a 15-ml centrifuge tube.
-
51|
Centrifuge the cells at 150g for 3 min at 4 °C.
-
52|
Remove the supernatant and add 3 ml of washing buffer.
-
53|
Resuspend the cells by pipetting up and down, by tapping the bottom of the centrifuge tube or by mildly vortexing.
-
54|
Repeat Steps 51–53.
-
55|
Finally, centrifuge the cells at 150g for 3 min at 4 °C and resuspend each cell line in 100 μl of binding buffer to a concentration of 4 × 105 cells per 100 μl.
▲ CRITICAL STEP Avoid strong vortexing, as it can cause cell breakage.
-
56|
Seed each cell line into two flow cytometry tubes labeled 0 and 1 for target CEM cells, 0′ and 1′ for target HeLa cells and 0″ and 1″ for nontarget Ramos cells. Each tube contains 50 μl of cells.
-
57|
Add 5 μl of binding buffer to 0, 0′ and 0″ tubes and 5 μl of NF stock solution to 1, 1′ and 1″ tubes individually.
-
58|
Incubate the tubes on ice for 30 min.
-
59|
Centrifuge and wash the cells according to Steps 51 through 54.
-
60|
Resuspend the cells in 200 μl of binding buffer.
-
61|
Use flow cytometry to determine the fluorescence signal intensity of the integrated fluorophore in each tube for target and control cells.
-
62|
Analyze the data with FlowJo software. An obvious fluorescence signal shift will appear with target cells but not with nontarget cells.
? TROUBLESHOOTING
Drug loading into NFs ● TIMING 1–2 d
-
63|
Prepare 2 mM Dox in DPBS.
-
64|
Centrifuge 100 μl of NF stock solution in a 1.5-ml centrifuge tube at 14,000g for 15 min, and discard the supernatant.
-
65|
Resuspend the NFs with the prepared 100 μl of 2 mM Dox.
-
66|
Incubate the mixture at room temperature for 24 h.
-
67|
Centrifuge the mixture at 14,000g for 15 min. NFs will precipitate out, whereas free Dox remains in the supernatant.
-
68|
Collect the supernatant and measure the absorbance of Dox at 480 nm on a UV-visible spectrometer. Mark the value as Asupernatant.
-
69|
Measure the absorbance of 1 mM Dox in DPBS at 480 nm on a UV-visible spectrometer. Mark the value as Atotal.
-
70|The Dox loading amount into NFs is calculated as shown in Equation 1:
(1) where x and y denote the dilution factor of total Dox amount and Dox amount in the supernatant.
▲ CRITICAL STEP Define 1 U as a NF concentration that can load 10 μM Dox in the above-mentioned experimental condition. This unit will be used in cytotoxicity experiments.
-
71|
Disperse the precipitate (NF-Dox complexes) in 100 μl of DPBS.
▲ CRITICAL STEP After loading drugs, NF-Dox complexes should be used as soon as possible to avoid drug leakage.
Bioimaging of intracellular behavior of NFs and Dox delivered by NFs ● TIMING ~6 h
-
72|
Prepare fresh cells according to Step 48, and count the number of target CCRF-CEM cells and control Ramos cells; 6 × 105 cells, in total, are needed for each cell line. For adherent cells (HeLa), follow the steps given in Box 2 to perform the intracellular imaging experiment, and continue the procedure from Step 81.
-
73|
Centrifuge the mixture at 150g and wash the cells twice with washing buffer.
-
74|
Resuspend each cell line in 600 μl of binding buffer with a concentration of 1 × 105 cells per 100 μl.
-
75|
Seed each suspended cell line into three tubes with 200 μl in each tube. Label the tubes of the target cell line as 0, 1 and 2, and label the tubes of the nontarget cell line as 0′, 1′ and 2′.
-
76|
Add NFs (10 μl equivalent NF stock solution from Step 15) to 1 and 1′ and NF-Dox complexes (2 μM Dox equivalent) to 2 and 2′. Keep 0 and 0′ tubes as controls.
-
77|
Incubate 0, 0′, 1, 1′, 2 and 2′ in a cell culture incubator at 37 °C for 2 h.
-
78|
Centrifuge the tubes at 150g and wash the cells twice with 1 ml of washing buffer
-
79|
Resuspend the cells with 200 μl of DPBS using pipette tips, and transfer the cells into glass-bottomed microscope dishes individually. Label the dishes according to the tubes.
-
80|
By using CLSM in DIC mode, perform bioimaging of intracellular behavior of NFs and Dox. Choose appropriate excitation and emission wavelengths according to the integrated fluorophore (see Step 22 for maximum excitation wavelength and maximum emission wavelength of different fluorophores; for Dox, maximum excitation wavelength is 480 nm and maximum emission wavelength is 580 nm.).
▲ CRITICAL STEP All of the imaging parameters should be kept the same in order to compare the signal in each image.
Box 2. Bioimaging of intracellular behavior of NFs and Dox delivery by NFs in adherent cells ● TIMING 1–2 d.
For adherent cells, refer to Box 1 for cell dissociation and counting. Seed 6×104 cells in 200 μl of culture medium into glass-bottomed microscope dishes. Incubate the dishes at 37 °C for 24–36 h to ensure that each dish has reached 60–80% confluence before adding NFs or DNAs.
Discard the medium with a pipette. Wash it twice with 200 μl of washing buffer. Finally, immerse the cells in 200 μl of binding buffer.
Add NFs (10 μl equivalent NF stock solution) and NF-Dox complex (2 μM Dox equivalent) to different microscope dishes. Keep another dish as control.
Incubate the cells at 4 °C for 30 min.
Discard the supernatant with a pipette. Wash the cells twice with 200 μl of washing buffer.
Perform bioimaging with CLSM in DIC mode as described in Step 80.
DNA NF–based high-performance targeted drug delivery ● TIMING 3–4 d
-
81|
Prepare fresh cells according to Step 48. For cytotoxicity experiments, count the number of target and control cells with a hemocytometer; 5 × 104 CEM or Ramos, or 5 × 103 HeLa, cells are used for one sample.
-
82|
Seed cells in a 96-well plate in FBS-free medium. For adherent cells, incubate the cells at 37 °C for 24–36 h to ensure good adherence. For suspension cells, no incubation is needed before the next step.
-
83|
Incubate the cells with a concentration gradient of NFs, free Dox and Dox-NF complexes for 2 h in a cell culture incubator at 37 °C. See the following table for a guide.
Samples incubated with nanoflowers 1 2 3 4 5 6 7 8 Nanoflower (U) 0.01 0.02 0.04 0.08 0.16 0.32 0.64 1.28 Free Dox — — — — — — — — Nanoflower-Dox complex — — — — — — — — Samples incubated with free Dox 1′ 2′ 3′ 4′ 5′ 6′ 7′ 8′ Nanoflower — — — — — — — — Free Dox (μM) 0.1 0.2 0.4 0.8 1.6 3.2 6.4 12.8 Nanoflower-Dox complex — — — — — — — — Samples incubated with nanoflower-Dox complex 1″ 2″ 3″ 4″ 5″ 6″ 7″ 8″ Nanoflower — — — — — — — — Free Dox — — — — — — — — Nanoflower-Dox complex (U) 0.01 0.02 0.04 0.08 0.16 0.32 0.64 1.28 See Step 70 for the definition of U. -
84|
For suspension cells, centrifuge the cells at 150g for 5 min and then remove the supernatant medium with a pipette. For adherent cells, remove the supernatant medium directly with a pipette.
-
85|
Add 200 μl of fresh medium (with 10% FBS) into each well and incubate for 48 h in a cell culture incubator at 37 °C for further cell growth.
-
86|
For suspension cells, centrifuge the cells at 150g for 5 min and then remove the medium. For adherent cells, remove the medium directly.
-
87|
Dilute every 20 μl of CellTiter reagent in 100 μl of fresh FBS-free medium. Add 100 μl of the diluted reagent to each well and incubate for 1–2 h in a cell culture incubator at 37 °C.
-
88|
Use a microplate reader to record the absorbance at 490 nm.
-
89|
Determine cell viability according to the manufacturer’s description.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 3.
TABLE 3.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 5 | RCR proceeded slowly. No obvious monodisperse NFs are observed after hours of polymerization | Imperfect hybridization can result from fast cooling during the annealing step | Cool the mixture gradually to room temperature over 3 h after incubation at 95 °C |
| 10 | Size of NFs does not correspond with the given value | Phosphorylation efficiency of the template and activity of the polymerase vary between batches | Optimize the size of NFs at different time points, depending on your own reagents. The given values are suggested for reference and may be changed according to the specific source |
| 23A | The SEM imaging quality is poor | NF samples are not completely dry | Prevent this by slightly increasing the drying time in the oven |
| 62 | No obvious shift is observed between target and nontarget cells | Cell state is poor | Change to a new cell source and culture with caution |
| Trypsin is used to dissociate the adherent cells, and membrane protein is damaged | Use nonenzymatic dissociation solution for adherent cell dissociation |
● TIMING
Steps 1–3, DNA preparation: 30 min to 3 d
Steps 4–7, preparation of ligated DNA template: ~8 h
Steps 8–15, self-assembly of monodisperse, size-tunable and densely packed multifunctional DNA NFs: ~1 d
Steps 16–19, use of agarose electrophoresis to check the RCR result: ~2 h
Steps 20–22, use of laser confocal fluorescence microscopy to check the integration of the dye molecule: ~1 h
Step 23A, SEM imaging of NFs: ~4 h
Step 23B, dynamic light-scattering measurement of NFs: ~30 min
Step 23C, TEM imaging of NFs: ~4 h
Step 23D, AFM imaging of NFs: ~1 d
Step 23E, PLM imaging of NFs: ~1 h
Steps 24–30, examining resistance to nuclease degradation: 1–2 d
Steps 31–35, examining resistance to human serum: 1–2 d
Steps 36–38, examining thermal stability: ~5 h
Steps 39–45, examining stability against urea: ~5 h
Steps 46 and 47, examining resistance to dissociation at low concentration: ~4 h
Step 48, cell culture and cell counting: 2–3 d
Steps 49–62, aptamer-integrated DNA NFs for cell targeting: 3–4 h
Steps 63–71, drug loading into NFs: 1–2 d
Steps 72–80, bioimaging of intracellular behavior of NFs and Dox delivery by NFs: ~6 h
Steps 81–89, DNA NF–based high-performance targeted drug delivery: 3–4 d
Box 1, dissociation of adherent cells and cell counting before cell targeting: ~1 h
Box 2, bioimaging of intracellular behavior of NFs and Dox delivery by NFs in adherent cells: 1–2 d
ANTICIPATED RESULTS
Characterization of DNA NFs
In our experiment, NFs with 10–15 h of reaction time were chosen for further characterization using SEM (Fig. 3), DLS (Supplementary Fig. 3a), TEM (Supplementary Fig. 3b,c), AFM (Supplementary Fig. 3d,e) and PLM (Supplementary Fig. 3f,g). These results provide clear evidence of the internal hierarchical structures and the dense DNA packaging in NFs, as well as the great monodispersity and the liquid crystalline structures of NFs.
Stability of DNA NFs
By using SEM, the exceptional resistance of NFs to enzymolysis, denaturation conditions and dissociation at low concentration was demonstrated. The results are shown in Supplementary Figure 4. DNase I is a ubiquitous endonuclease that can nonspecifically cleave both single- and double-stranded DNAs and is responsible for at least 90% of the deoxynuclease activity in human plasma. After treatment with 5 U/ml DNase I (a concentration considerably higher than that in human blood (<1 U/ml); Supplementary Fig. 4a,b) or 10% human serum for 24 h, the NFs remain in flower structures, as displayed in SEM images (Supplementary Fig. 4c), thus demonstrating high stability toward enzymolysis. Most of the hydrogen bond–based DNA nanostructures will denature under severe conditions such as high temperature and high urea concentrations. However, in contrast to traditional hydrogen bonding, DNA NFs benefit from a unique stacking force, which can maintain structural integrity even at 170 °C (Supplementary Fig. 4d) or with 5 M urea treatment (Supplementary Fig. 4e). Resistance to dissociation at low concentration was also studied through diluting the NF reaction solution 100 times in water. No dissociation was observed in SEM imaging results (Supplementary Fig. 4f).
Aptamer-integrated DNA NFs for cell targeting
Aptamer sgc8c can specifically recognize PTK7, which is overexpressed on many cancer cells. By incorporating sgc8c aptamer into NFs by RCR, DNA NFs are endowed with high affinity and selectivity toward the target cells, HeLa and CEM, compared with nontarget Ramos cells. A representative flow cytometry result is shown in Supplementary Figure 5. In this experiment, FAM-integrated NFs were used for cell targeting and signal reporting, and these NFs showed high recognition capability toward HeLa and CEM cells according to experimental result. No clear fluorescence signal shift was observed with nontarget Ramos cells. In conclusion, the conjugated aptamer preserves its binding affinity and specificity, making aptamer-integrated DNA NFs powerful nanotools for cell targeting.
DNA NF–based high-performance drug delivery
By incubating NFs with Dox in DPBS at room temperature for 24 h, Dox was loaded into the drug-loading sites of NFs. Drug-loaded NFs and free Dox can be separated through centrifugation at 14,000g for 5 min. After centrifugation, the supernatant is light-colored as a result of having lost part of Dox, whereas the NF concentrate turns dark red upon drug loading, as shown in Supplementary Figure 6a. According to the described method, every 100 μl of prepared NF stock solution could load 66 nmol of Dox, and here we can calculate that the defined 1 U corresponds to an ~66-fold dilution of the original stock solution.
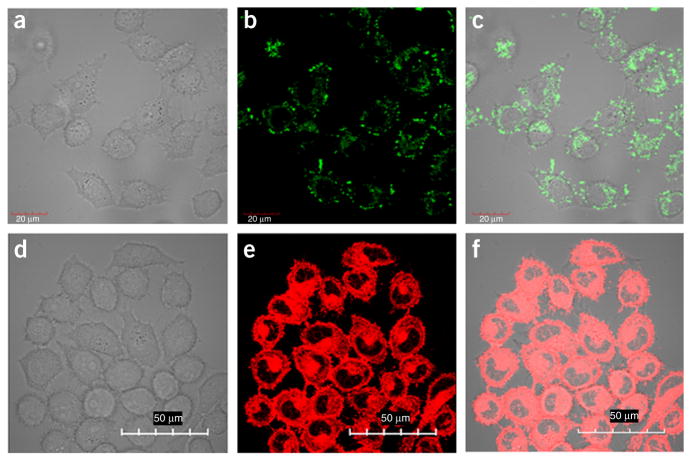
Bioimaging of intracellular behavior of NFs and Dox delivery by NFs is shown in Figure 4. The green and red signals show that FAM-integrated NFs were taken up into the target cells and that drug molecules were released with high performance. In contrast, such fluorescence signals are difficult to visualize in control cells.
Figure 4.
Bioimaging of intracellular behavior of NFs and Dox delivered by NFs. (a–c) Confocal microscopy images show that NFs were internalized into target HeLa cells after incubation at 37 °C for 2 h. (a) The bright-field image of HeLa cells. (b) The fluorescence channel of FAM. (c) The merged bright-field image and fluorescence channel for confocal images. Images were captured by confocal microscopy after extensive washing of cells with PBS. Scale bars, 20 μm. NFs with a size of 200–300 nm (RCR for 10–15 h) were used for this experiment, and fluorescein-12-ddUPT was integrated into NFs. (d–f) Confocal microscopy images show Dox distribution in HeLa cells treated with NF-Dox (2 μM Dox equivalent) for 2 h. Scale bars, 50 μm Adapted with permission from Zhu et al.31 (© 2013, American Chemical Society) and Hu et al.32.
By treating target cells and nontarget cells with NFs, free Dox and NF-Dox complex in equivalent drug concentrations or NF concentrations, we further present the results of cell viabilities using an MTS assay. On the basis of the outstanding bio-compatibility of DNA molecules, NFs showed no cytotoxicity toward either CEM or Ramos cells (Supplementary Fig. 6b,c). For target CEM and HeLa cells, the inhibition of Dox delivered by NFs on cell proliferation is similar to that of free Dox (Supplementary Fig. 7a,b). For nontarget Ramos cells, in contrast to free Dox, NF-Dox complexes induced distinctly less inhibition of cell proliferation (Supplementary Fig. 7c). These results distinctly demonstrated the selective cytotoxicity of NF-Dox complex toward target cells over nontarget cells, as well as the great potential of multifunctional NFs for targeted drug delivery and intracellular bioimaging.
Supplementary Material
Acknowledgments
This work was supported by the National Key Scientific Program of China (2011CB911000), NSFC grants (21325520, 21327009, 21221003, J1210040, 21177036 and 21135001) and the China National Instrumentation Program 2011YQ03012412. It is also supported by the US National Institutes of Health (GM079359 and CA133086).
Footnotes
AUTHOR CONTRIBUTIONS G.Z., R.H., X.Z. and W.T. originated the method of preparing DNA NFs; Y.L., G.Z., R.H., X.Z., L.M., Q.L. and W.T. were responsible for the content of the protocol. Y.L. and W.T. wrote the manuscript, and all authors contributed to the revision of the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Rosi NL, et al. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 2.Wu P, Yan XP. Doped quantum dots for chemo/biosensing and bioimaging. Chem Soc Rev. 2013;42:5489–5521. doi: 10.1039/c3cs60017c. [DOI] [PubMed] [Google Scholar]
- 3.Fan Q, et al. Transferring biomarker into molecular probe: melanin nanoparticle as a naturally active platform for multimodality imaging. J Am Chem Soc. 2014;136:15185–15194. doi: 10.1021/ja505412p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan Q, et al. Photon-manipulated drug release from a mesoporous nanocontainer controlled by azobenzene-modified nucleic acid. ACS Nano. 2012;6:6337–6344. doi: 10.1021/nn3018365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng J, et al. A spherical nucleic acid platform based on self-assembled DNA biopolymer for high-performance cancer therapy. ACS Nano. 2013;7:6545–6554. doi: 10.1021/nn402344v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn SS, et al. Reductively responsive siRNA-conjugated hydrogel nanoparticles for gene silencing. J Am Chem Soc. 2012;134:7423–7430. doi: 10.1021/ja300174v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang H, et al. Near-infrared light-responsive core–shell nanogels for targeted drug delivery. ACS Nano. 2011;5:5094–5099. doi: 10.1021/nn201171r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, et al. Engineering polymeric aptamers for selective cytotoxicity. J Am Chem Soc. 2011;133:13380–13386. doi: 10.1021/ja201285y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Jin X, Ma PX. Nanofibrous hollow microspheres self-assembled from star-shaped polymers as injectable cell carriers for knee repair. Nat Mater. 2011;10:398–406. doi: 10.1038/nmat2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shim MS, Xia Y. A reactive oxygen species (ROS)-responsive polymer for safe, efficient, and targeted gene delivery in cancer cells. Angew Chem Int Ed. 2013;52:6926–6929. doi: 10.1002/anie.201209633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovell JF, et al. Porphysome nanovesicles generated by porphyrin bilayers for use as multimodal biophotonic contrast agents. Nat Mater. 2011;10:324–332. doi: 10.1038/nmat2986. [DOI] [PubMed] [Google Scholar]
- 12.Wu Y, et al. DNA aptamer–micelle as an efficient detection/delivery vehicle toward cancer cells. Proc Natl Acad Sci USA. 2010;107:5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T, et al. DNA micelle flares for intracellular mRNA imaging and gene therapy. Angew Chem Int Ed. 2013;125:2066–2070. doi: 10.1002/anie.201209440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 15.Yin P, Choi HM, Calvert CR, Pierce NA. Programming biomolecular self-assembly pathways. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 16.Bath J, Turberfield AJ. DNA nanomachines. Nat Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Q, et al. DNA origami as a carrier for circumvention of drug resistance. J Am Chem Soc. 2012;134:13396–13403. doi: 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- 18.Lee H, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman RP, et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 20.Pei H, et al. Reconfigurable three-dimensional DNA nanostructures for the construction of intracellular logic sensors. Angew Chem Int Ed. 2012;51:9020–9024. doi: 10.1002/anie.201202356. [DOI] [PubMed] [Google Scholar]
- 21.Lee JB, et al. Multifunctional nanoarchitectures from DNA-based ABC monomers. Nat Nanotechnol. 2009;4:430–436. doi: 10.1038/nnano.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng HM, et al. DNA dendrimer: an efficient nanocarrier of functional nucleic acids for intracellular molecular sensing. ACS Nano. 2014;8:6171–6181. doi: 10.1021/nn5015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci USA. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi HM, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, Nangreave J, Liu Y, Yan H. Structural DNA nanotechnology: state of the art and future perspective. J Am Chem Soc. 2014;136:11198–11211. doi: 10.1021/ja505101a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zadeh JN, et al. NUPACK: analysis and design of nucleic acid systems. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 27.Douglas SM, et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009;37:5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamkovich SN, et al. Circulating DNA and DNase activity in human blood. Ann NY Acad Sci. 2006;1075:191–196. doi: 10.1196/annals.1368.026. [DOI] [PubMed] [Google Scholar]
- 29.Hamblin GD, et al. Rolling circle amplification-templated DNA nanotubes show increased stability and cell penetration ability. J Am Chem Soc. 2012;134:2888–2891. doi: 10.1021/ja2107492. [DOI] [PubMed] [Google Scholar]
- 30.Shu D, et al. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu G, et al. Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J Am Chem Soc. 2013;135:16438–16445. doi: 10.1021/ja406115e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu R, et al. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew Chem Int Ed. 2014;53:5821–5826. doi: 10.1002/anie.201400323. [DOI] [PubMed] [Google Scholar]
- 33.Johne R, et al. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol. 2009;17:205–211. doi: 10.1016/j.tim.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Shangguan D, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 36.Xiong X, et al. Nucleic acid aptamers for living cell analysis. Annu Rev Anal Chem. 2014;7:405–426. doi: 10.1146/annurev-anchem-071213-015944. [DOI] [PubMed] [Google Scholar]
- 37.Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer–doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 38.Chaires JB, Herrera JE, Waring MJ. Preferential binding of daunomycin to 5′TACG and 5′TAGC sequences revealed by footprinting titration experiments. Biochemistry. 1990;29:6145–6153. doi: 10.1021/bi00478a006. [DOI] [PubMed] [Google Scholar]
- 39.Frederick CA, et al. Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin. Biochemistry. 1990;29:2538–2549. [PubMed] [Google Scholar]
- 40.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modi S, et al. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat Nanotechnol. 2013;8:459–467. doi: 10.1038/nnano.2013.92. [DOI] [PubMed] [Google Scholar]
- 42.Wu P, Hwang K, Lan T, Lu Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J Am Chem Soc. 2013;135:5254–5257. doi: 10.1021/ja400150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sefah K, et al. Development of DNA aptamers using cell-SELEX. Nat Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 44.Dua P, et al. Alkaline phosphatase ALPPL-2 is a novel pancreatic carcinoma-associated protein. Cancer Res. 2013;73:1934–1945. doi: 10.1158/0008-5472.CAN-12-3682. [DOI] [PubMed] [Google Scholar]
- 45.Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc Chem Res. 2009;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shangguan D, et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu G, et al. Self-assembled aptamer-based drug carriers for bispecific cytotoxicity to cancer cells. Chem Asian J. 2012;7:1630–1636. doi: 10.1002/asia.201101060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu G, et al. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc Natl Acad Sci USA. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peer D, et al. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 50.Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 51.Sun W, et al. Cocoon-like self-degradable DNA nanoclew for anticancer drug delivery. J Am Chem Soc. 2014;136:14722–14725. doi: 10.1021/ja5088024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, et al. Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J Am Chem Soc. 2010;132:9274–9276. doi: 10.1021/ja103169v. [DOI] [PubMed] [Google Scholar]
- 53.Qian R, Ding L, Ju H. Switchable fluorescent imaging of intracellular telomerase activity using telomerase-responsive mesoporous silica nanoparticle. J Am Chem Soc. 2013;135:13282–13285. doi: 10.1021/ja406532e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.