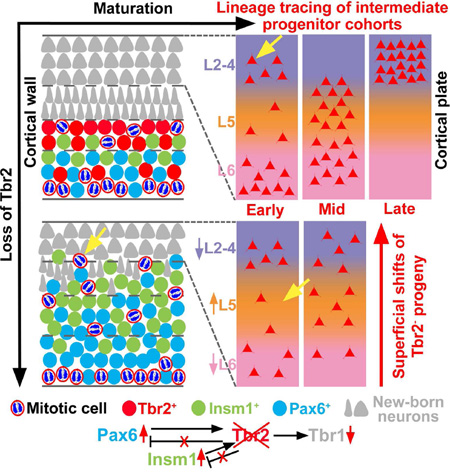
Summary
Intermediate progenitors (IPs) amplify production of pyramidal neurons, but their role in selective genesis of cortical layers or neuronal subtypes remains unclear. Using genetic lineage tracing in mice, we find that IPs destined to produce upper cortical layers first appear early in corticogenesis, by embryonic day 11.5. During later corticogenesis, IP laminar fates are progressively limited to upper layers. We examined the role of Tbr2, an IP-specific transcription factor, in laminar fate regulation using Tbr2 conditional mutant mice. Upon Tbr2 inactivation fewer neurons were produced by immediate differentiation, and laminar fates were shifted upwards. Genesis of subventricular mitoses was, however, not reduced in the context of a Tbr2 null cortex. Instead neuronal and laminar differentiation were disrupted and delayed. Our findings indicate that upper layer genesis depends on IPs from many stages of corticogenesis, and that Tbr2 regulates the tempo of laminar fate implementation for all cortical layers.
Graphical Abstract
eTOC Blurb
The role of intermediate progenitors (IPs) in corticogenesis has been unclear. Mihalas et al. show that early IP cohorts produce lower and upper layer neurons, and that Tbr2 is required to regulate differentiation, not genesis of IPs.
Introduction
Excitatory, pyramidal neurons of the cerebral cortex are generated during embryonic neurogenesis from radial glial progenitors (RGPs) both directly, and indirectly via transient-amplifying, committed neurogenic intermediate progenitors (IPs) (reviewed by Florio and Huttner, 2014; Sun and Hevner, 2014). Importantly, the generation of neurons of different cortical layers follows a general "inside-out" pattern as lower layer (LL) neurons are born first, and upper layers (ULs) last (Hevner et al., 2003).
IPs are distinguished from RGPs by short radial or multipolar morphology, mitotic division tending to occur away from the ventricular surface, and a unique molecular profile, including specific expression of the Tbr2 transcription factor (also known as Eomes; NCBI Gene Eomes) (Englund et al., 2005; Gal et al., 2006; Kawaguchi et al., 2008; Stancik et al., 2010). Given their distinct properties, IPs have been proposed to generate specific cortical neuron subtypes. However, the precise contribution of IPs to cortical neurogenesis and laminar fate specification remains poorly defined. Previously, IPs were suggested to produce mainly UL neurons (Tarabykin et al., 2001; Zimmer et al., 2004; Britanova et al., 2005), but further studies indicated that IPs produce all layers (Kowalczyk et al., 2009; Vasistha et al., 2014). More recently, the morphological and electrophysiological properties of UL neurons were reported to depend on their origins from Tbr2-negative RGPs, or from Tbr2-positive IPs (Tyler et al., 2015). Thus, one goal of the present study was to use a panel of molecular markers to more extensively define neuron subtypes produced from IPs.
The second goal of the present study was to determine if IPs generate cortical layers in a predetermined sequential order depending on the timepoint of their generation, or if individual IP cohorts may contribute to multiple layers. Previously, early IPs were observed to rapidly differentiate and produce LL neurons (Kowalczyk et al., 2009), but the possibility that some early IPs differentiate slowly and contribute to ULs has not been thoroughly investigated. The latter scenario could arise if some early IPs are restricted to UL fates, as reported for some early RGPs (Franco et al., 2012); or if some early IPs remain in the mitotic cycle for a protracted period before differentiating, as suggested for late IPs (Wu et al., 2005). We studied these possibilities by inducible genetic fate mapping using Tbr2CreER, which, together with the reporter gene Ai14, specifically labels IPs and their progeny (Pimeisl et al., 2013).
To investigate regulation of laminar fate in IPs, we studied mice lacking Tbr2. Previously, it was reported that Tbr2-deficient cortex had decreased thickness of ULs, suggesting that Tbr2 promotes UL neuron fates (Arnold et al., 2008; Sessa et al., 2008). Here, we characterized the effects of Tbr2 deficiency on laminar fates of early, middle, and late IP cohorts. We studied IP genesis, migration, differentiation, and fates using molecular markers, lineage tracing, and cell birthdating. Unexpectedly, we found that Tbr2 is not required for genesis of subventricular IPs from RGPs as reported previously (Sessa et al., 2008), but is required mainly for differentiation of IPs to projection neurons, including acquisition of LL and UL identities.
Our results show that sequential cohorts of IPs generate progressively limited sets of cortical layers, such that UL neurogenesis depends not only on late, but also on early generated IPs. Furthermore, the process of laminar neurogenesis from IPs is regulated by Tbr2, which controls the tempo of LL and UL differentiation. Thus, in addition to previously described roles to control cortical regional patterning (Elsen at al., 2013), IPs and Tbr2 play important roles in laminar differentiation of the cerebral cortex.
Results
Some early IPs produce upper layer neurons
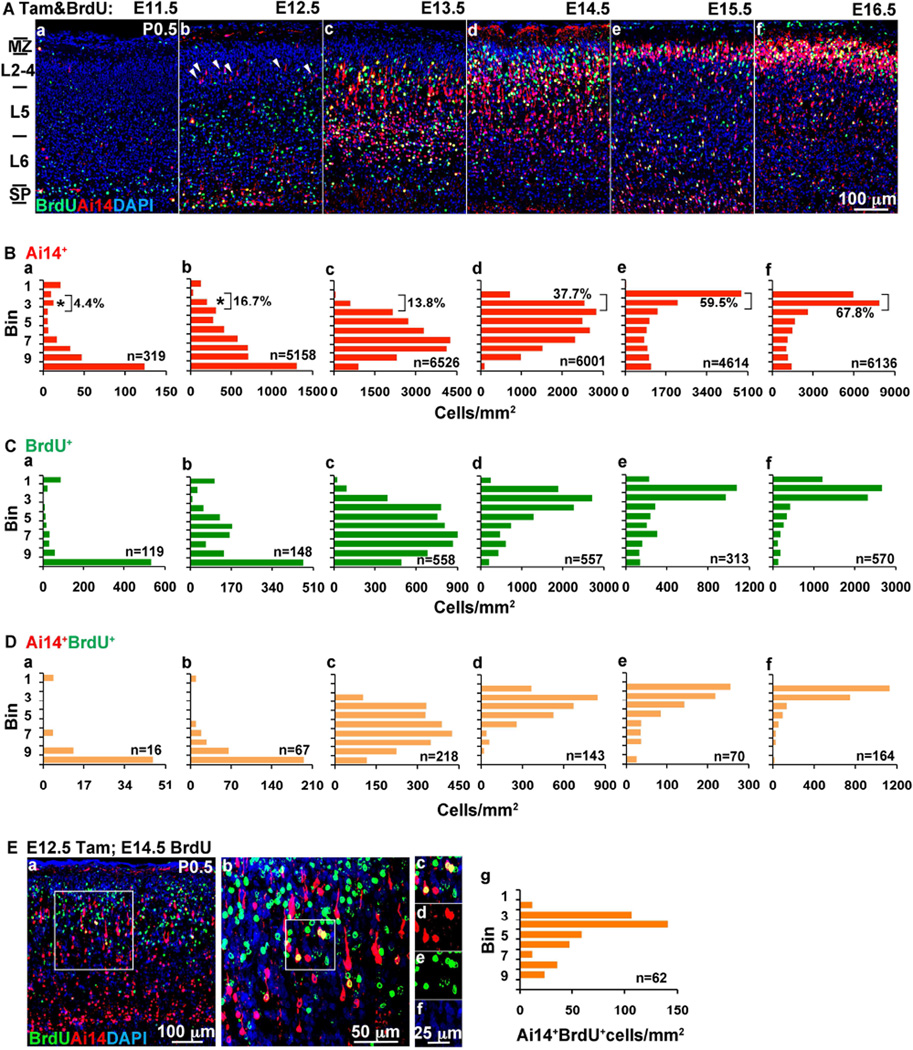
To label IP cohorts and their progeny, we used Tbr2CreER;Ai14 mice (Pimeisl et al., 2013), in which tamoxifen (Tam) administration induces permanent expression of tdTomato, a red fluorescent protein (RFP), in Tbr2-expressing cells and their progeny. Tam was administered on embryonic days 11.5 (E11.5) to E16.5, and brains were collected on postnatal day (P) 0.5 (Figure S1). To identify neurons born on the day of Tam administration, bromodeoxyuridine (BrdU) was given concurrently (Figure 1).
Figure 1. Laminar fates and birthdays of IP-derived progeny.
(Aa–f) Representative images of Ai14+ and BrdU+ cells from P0.5 pups injected with Tam and BrdU on different days from E11.5 – E16.5. DAPI counterstain (blue). Cells were counted in bins from marginal zone (MZ) to subplate (SP). Typically, bin 1 represents the MZ, bins 2–3 are layers 2–4, bins 4–6 are layer 5, bins 7–9 are layer 6, and bin 10 is SP. (Ba–f) Bin analysis of Ai14+ cells. (Ca–f) Bin analysis of BrdU+ cells. (Da–f) Bin analysis of Ai14+/BrdU+ double-positive neurons, born from IPs that last divided on the BrdU/Tam injection day. The n represents the total number of cells included in the analysis. N=2–4 embryos for each time point of Tam induction. Scale bar: 100 µm (Aa–f). (E) Diverse latency to last division. While many E12.5 IPs (RFP labeled by Tam injection on E12.5) exited the cell cycle on E12.5 (Figure 1Db), others did not divide finally until E14.5, as shown by E14.5 BrdU labeling with survival to P0.5. (Ea–f) Double-labeled Ai14+/BrdU+ neurons seen by confocal microscopy. (Eg) Bin analysis of Ai14+/BrdU+ cells showed these neurons occupied mainly upper layers. (Some Ai14+/BrdU+ cells in lower layers were still migrating.) N=2 embryos. Also see Figure S1.
Lineage tracing revealed that IP cohorts contributed differentially to cortical layers (Figure 1A,B). Early IP cohorts (E11.5–E12.5) generated not only LL neurons as expected from BrdU birthdating (Figure 1B–D), but also substantial numbers of UL neurons. Indeed, neurons derived from E11.5–E12.5 IPs were distributed bimodally in LLs and ULs, and as many as 17% of the RFP+ neurons generated from E12.5 IPs settled in ULs (Figure 1Ba,b). While LL neurons derived from early IPs were early-born, the UL neurons derived from early IPs were not early-born, as they did not incorporate BrdU given concurrently with Tam (Figure 1Da,b).
IPs from mid-neurogenesis (E13.5–E14.5) contributed widely to LLs and ULs, while late IPs (E15.5–E16.5) produced ULs selectively (Figure 1A–D).
Early IPs produce diverse molecular subtypes of cortical projection neurons
Since UL neurons are generally born later in neurogenesis (Hevner et al., 2003), we considered the possibility that some early IPs (E12.5) do not undergo final mitosis until much later in neurogenesis. To test this hypothesis, we administered Tam on E12.5 and BrdU on E14.5, followed by survival to P0.5. With this schema, many double-labeled (RFP+/BrdU+) neurons were detected in ULs (but not LLs), confirming that some early IPs persisted for at least two embryonic days before final mitosis (Figure 1E). However, we do not know if the early IPs divided only once, or repeatedly during this period.
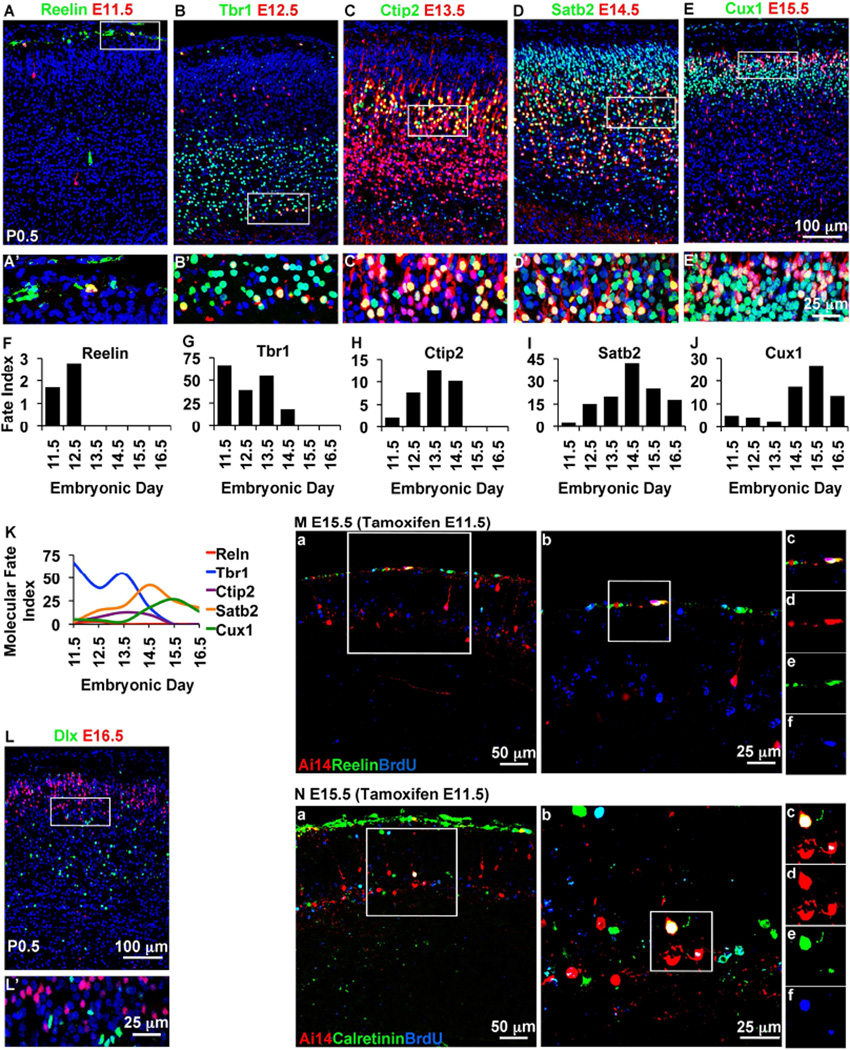
Cortical projection neurons are defined by not only laminar position, but also molecular expression (Hevner et al., 2003; Molyneaux et al., 2007). To further identify the neuron subtypes produced from E12.5 IPs, we evaluated their expression of Reelin, a Cajal-Retzius (C-R) cell marker; Tbr1, expressed at high levels in layer 6 (L6); Ctip2 (L5); Satb2 (callosal projection neurons); Cux1 (L2–4); and Dlx (interneuron precursors) (Figure S2). Remarkably, E12.5 IPs produced all five subtypes of projection neurons (Figure S3). As expected, no IP-derived neurons expressed Dlx (data not shown; see also Figure 2L,L').
Figure 2. IP cohorts produce all types of projection neurons in overlapping sequence.
(A–E) Different IP cohorts showed distinct profiles of neurogenesis and different predominant fates. In these images, neurons from different IP cohorts are shown with selected colocalized molecular markers in P0.5 cortex: (A) E11.5 Tam, Reelin; (B) E12.5 Tam, Tbr1; (C) E13.5 Tam, Ctip2; (D) E14.5 Tam, Satb2; (E) E15.5 Tam, Cux1. Scale bar: 100 µm. (A’–E’) Higher magnification of boxed areas in A–E. Scale bar: 25µm. (F–J) Different neuron types were produced with different frequency from different IP cohorts. For each molecular type of neuron, the fate index (% of Ai14+ cells that express a marker) changed with each sequential IP cohort, revealing a profile of neurogenesis. (K) Summary of molecular fate index profiles from different IP cohorts. (L) IPs did not produce Dlx+ interneurons on E16.5 (shown) or any other ages. (M, N) Some Cajal-Retzius and SP neurons were produced from IPs. Tam and BrdU were administered on E11.5, with survival to E15.5. IP-derived Cajal-Retzius neurons (Reelin+/BrdU+/Ai14+) were located in the MZ, and SP neurons (Calretinin+/BrdU+/Ai14+) in the morphological SP. Scale bar: (a) 50µm; (b–f) 25µm. Also see Figure S2–S4.
Progressive limitation of IP laminar and molecular fates
To molecularly characterize the subtypes of projection neurons produced by sequential IP cohorts, we extended our analysis to include E11.5–E16.5 IP cohorts (Figure 2). Cajal-Retzius cells (Reelin+) were produced from early IPs (E11.5–E12.5) only (Figure 2A,A’,F). LL neurons (Tbr1+ or Ctip2+) were produced from early to middle (E11.5–E14.5) IPs only. In contrast, UL neurons (Cux1+) and callosal projection neurons (Satb2+) were produced from all IP cohorts (Figure 2). Some RFP+ progeny of late IPs (E15.5–E16.5) were located in LLs on P0.5 (Figures 1Ae,f, 2E,L), but these appeared to be migrating neurons destined for ULs, as they had elongated morphologies and did not express LL markers (Figure S4). Thus, at the population level, early IPs exhibit diverse laminar and molecular fates, while later IP cohorts have progressively limited fates.
The interpretation of fates from E11.5–E12.5 IPs was complicated by the fact that at these ages, Tbr2 is expressed by not only IPs, but also postmitotic C-R and subplate (SP) neurons (Englund et al., 2005). Thus, C-R and SP neurons may be labeled by Tam treatment on E11.5 or E12.5, even if these neurons were not derived from IPs. To verify the origins of C-R and SP neurons from IPs, we injected Tam and BrdU on E11.5, and studied cortex on E15.5. Triple-label immunofluorescence (IF) demonstrated that many Reelin+ C-R cells, and Calretinin+ SP neurons, were labeled with RFP and BrdU, confirming their origins from proliferating Tbr2+ IPs (Figure 2M,N). Thus, IPs produce preplate (C-R and SP), as well as LL and UL projection neuron subtypes.
Tbr2-deficient early IPs produce fewer rapidly differentiating neurons
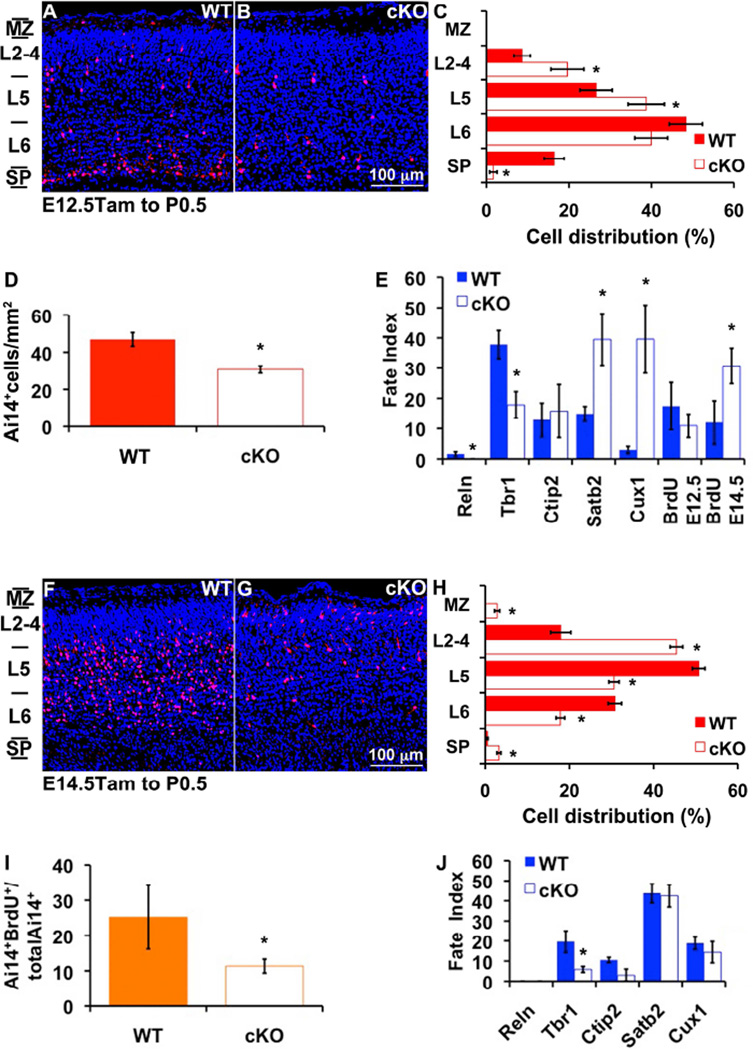
To test if Tbr2 regulates laminar fate in IPs cell-autonomously, we utilized Tbr2CreER to inactivate floxed Tbr2 (Tbr2FL; Intlekofer et al., 2008) in IP cohorts. Administration of Tam to Tbr2CreER/FL;Ai14 mice resulted in RFP labeling of Tbr2-deficient IPs and their progeny. (However, IPs presumably expressed functional Tbr2 transiently before Tbr2FL was recombined.) We labeled cohorts of Tbr2-deficient IPs on E12.5 or E14.5, and evaluated RFP+ progeny on P0.5. Control mice were treated identically, but lacked the Tbr2FL allele (i.e., Tbr2CreER/+;Ai14).
Tbr2-deficient E12.5 IPs produced significantly fewer total neurons than did control E12.5 IPs (control=49±4 cells/mm2, cKO=31±2 cells/mm2; p=0.0004; Figure 3A–D). Among Tbr2-deficient IPs, fewer became SP neurons (WT=16.4±2%, cKO=1.6±1%; p=7.1*10−7), while more became L5 neurons (WT=27±3%, cKO=39±4%; p=0.043) and L2–4 neurons (WT=9±2%, cKO=20±4%; p=0.0145; Figure 3A–C). The upward laminar shift of neurons from Tbr2-deficient IPs was matched by genesis of fewer Tbr1+ SP neurons (WT=38±5%, cKO=18±4%; p=0.0154), but increased numbers of Satb2+ callosal projection neurons (WT=15±2%, cKO=40±8%; p=0.0209), and Cux1+ UL neurons (WT=3±1%, cKO=40±11%; p=0.0104). Also, Tbr2-deficient E12.5 IPs were more likely to delay final division until E14.5 (WT=12±7%, cKO=30.7±6%; p=0.0493; Figure 3E). Thus, E12.5 IPs require Tbr2 mainly for genesis of early-born LL neuron subtypes, but not for later-born callosal and UL neuron subtypes.
Figure 3. Tbr2-deficient IP cohorts shift to more superficial laminar fates.
Tbr2CreER was used for both lineage tracing and Tbr2 inactivation in IP cohorts. (A–E) Neurogenesis of RFP+ neurons from E12.5 IPs was reduced and shifted after Tbr2 inactivation. (A) Control experiment showing E12.5 IP-derived, Ai14+ neurons in P0.5 cortex. (B) Tbr2-deficient IPs produced fewer neurons overall, with marked loss of LL fates and relatively sparing of UL fates. (C) Cortical layer analysis (assignment of cotical layers was rendered from 10 equidistant bins spanning from pial surface to SP, as described in Figure 1 legend) showed that Tbr2-deficient E12.5 IPs produced Ai14+ neurons that were shifted superficially in the CP (*, p≤0.043). (D) Tbr2-deficient E12.5 IPs produced approximately 35% fewer neurons overall (p=3.9*10−4). (E) The molecular fates and cell birthdays of neurons derived from Tbr2-deficient E12.5 IPs were shifted. Tbr2-deficient IPs produced fewer (none) Reelin+ (p=0.006) Cajal-Retzius neurons and fewer Tbr1+ L6 neurons (p=0.015), but relatively more Cux1+ (p=0.01) and Satb2+ (p=0.02) neurons. Neurons from Tbr2-deficient IPs were also more likely to be born later, on E14.5 (p=0.049). N=4 WT and 4 cKO embryos. (F–J) Neurogenesis from E14.5 IPs was reduced and superficially shifted after Tbr2 inactivation (survival to P0.5). Compared to controls (F), Tbr2-deficient E14.5 IPs (G) produced fewer LL neurons, and more neurons in the superficial CP. (H) Cortical layer analysis confirmed the gain of UL fates (L2–4 p=6.7*10−13) and loss of LL fates (L5, 6, p≤1.4*10−8). (I) E14.5 Tam and BrdU administration. Less than half as many Tbr2-deficient E14.5 IPs had theit last division at E14.5 (p=0.035). (J) Molecular fate analysis showed that Tbr2-deficient E14.5 IPs produced fewer Tbr1+ L6 neurons (p=0.014). N=2 WT and 3 cKO embryos. Scale bar: 100 µm (all images). Error bars indicate standard error of the mean (SEM).
The effects of Tbr2 deficiency on E14.5 IPs were broadly similar as on E12.5 IPs. Laminar fates were shifted upwards (Figure 3F–H), with more IP-derived neurons distributed in ULs 2–4 (WT=18±2%, cKO=45.4±2%; p=6.7*10−13), and fewer in L5 (WT=50.7±2%, cKO=30.6±1%; p=2.7*10−12) and L6 (WT=30.8±2%, cKO=17.9±1%; p=1.4*10−8). Also, Tbr2-deficient E14.5 IPs produced fewer Tbr1+ (SP/L6) neurons (WT=19.7±5%, cKO=5.9±1%; p=0.0144), and were less likely to differentiate rapidly on E14.5 (WT=25.2±9%, cKO=11.4±2%; p=0.0357; Figure 3I, J).
Together, these results suggest that Tbr2 is necessary for differentiation of IPs to produce rapidly-generated neuron types. The shift to genesis of later-generated neuron types from Tbr2-deficient IPs may result from delayed differentiation related to molecular dysregulation (see below).
Shifts of neurogenesis and laminar fates in Tbr2-deficient cortex
We next investigated the effects of complete Tbr2 deficiency on cortical neurogenesis. To produce mice lacking Tbr2 in the nervous system, we recombined Tbr2FL using Nes11Cre (Tronche et al., 1999). For simplicity, we designated the Nes11Cre;Tbr2FL/FL mice as Tbr2 conditional knockout (cKO) mutants.
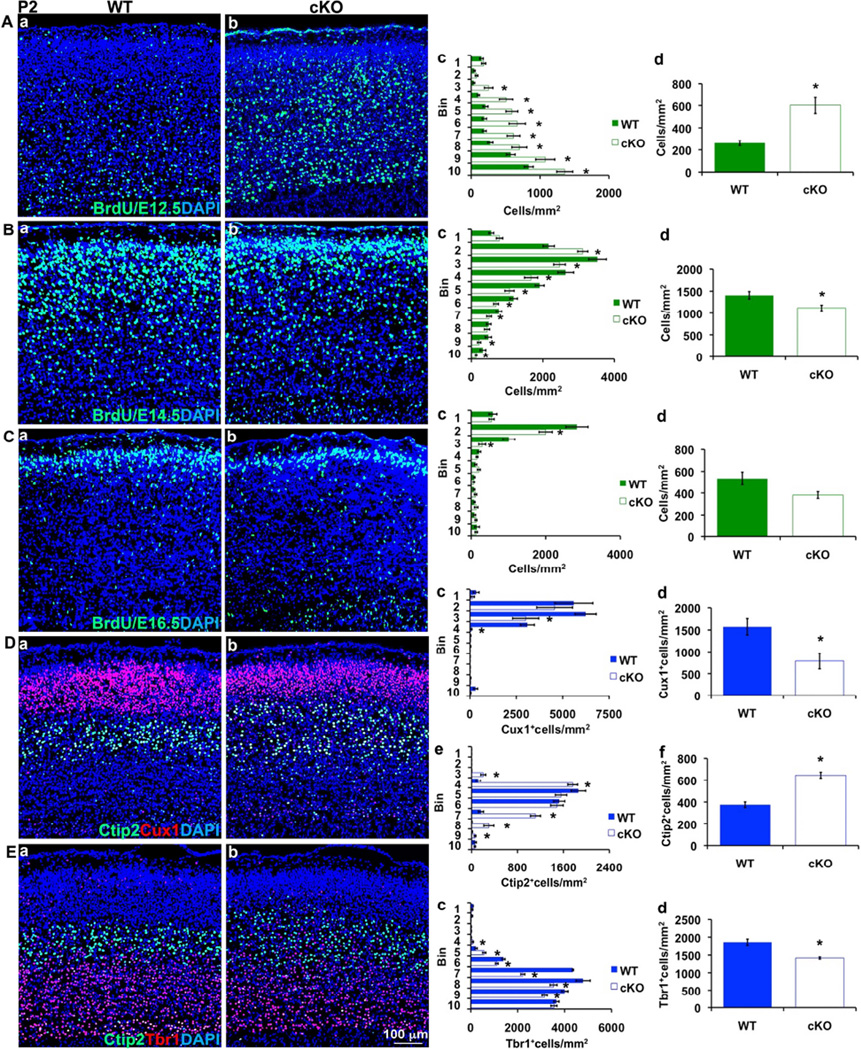
To profile the timing of neurogenesis in control and Tbr2 cKO cortex, we labeled early- (E12.5), middle- (E14.5), or late-born (E16.5) neurons with BrdU, and studied the distribution of BrdU+ cells after survival to P2. As expected from lineage tracing of Tbr2-deficient IPs (Figure 3), neurogenesis was moderately decreased in Tbr2 cKO cortex on E14.5 (79% of control; p=0.016) and E16.5 (71% of control; p=0.06), while the laminar fates of these cells were unchanged or shifted slightly upwards (Figure 4B,C). In contrast, the genesis of E12.5-born neurons was significantly increased by 2.3-fold (p=0.0001) in Tbr2 cKO cortex (Figure 4A).
Figure 4. Tbr2 cKO cortex shows precocious neurogenesis and increased layer 5 thickness.
(A–C) BrdU birthdating (with survival to P2) showed that, compared to controls (Aa, Ba, Ca), Tbr2 cKO mice (Ab, Bb, Cb) had increased early neurogenesis (A, E12.5; p=1.4*10−4), followed by decreased middle (B, E14.5; p=0.016) and late (C, E16.5; p=0.061) neurogenesis. Bin analysis (Ac, Bc, Cc) showed shifts of laminar fate (*, p<0.045), and cell counts confirmed changes in overall neurogenesis (Ad, Bd, Cd). Interestingly, early neurogenesis in Tbr2 cKO mice was shifted to increased genesis of middle bins, corresponding to layer 5 of P2 cortex (Ac). (D, E) Tbr2 cKO mice have increased layer 5 thickness, at the expense of layers 6 and 2–4 (P2). (Da,b–Ea,b) Analysis by IF to detect Cux1 (L2–4), Ctip2 (L5) and Tbr1 (L6) showed that Ctip2+ L5 appeared thicker in Tbr2 cKO cortex. Quantitatively, Cux1+ cells were overall reduced (Dd; p=0.012) and restricted to more superficial bins (Dc) in Tbr2 cKO cortex. In contrast, Ctip2+ cells were greatly increased (Df; p=3.2*10−8) and distributed within an expanded L5 (De) of Tbr2 cKO mutants. Tbr1+ neurons were moderately reduced (Ed; p=7.7*10−4) and shifted. (*, p≤0.042 in bin analyses). N= 3 WT and 3 cKO pups. Scale bar: 100 µm (all images). All error bars represent SEM. Also see Figure S5.
The increased genesis of E12.5-born neurons in Tbr2 cKO cortex suggested that SP and L6 thickness might be increased postnatally, as SP and L6 are the major neuron types born on E12.5 in normal mice (Figure 1C). Instead, we found significant expansion of L5 (Ctip2+) neurons (1.7 fold; p=3.2*10−8), at the expense of SP/L6 (Tbr1+, 76%; p=0.0008) and L2–4 (Cux1+, 50%; p=0.0124) neurons (Figure 4D,E). Thus, approximately twice as many E12.5-born cells differentiated as Ctip2+ L5 neurons (p=0.0139) in Tbr2 cKO mutants as in controls (Figure S5B). These results indicated that the relation between cell birthday and laminar fate was perturbed in Tbr2 cKO cortex. Interestingly, one previous study also reported slight expansion of L5 in Tbr2 cKO cortex, although not statistically significant (Arnold et al., 2008).
Tbr2 cKO mice have small brains but no deficits of simple motor skills
Tbr2 cKO mice survived to adulthood, but had 20–30% reduced body and brain mass (Figure S5F–J). (The causes of reduced body mass in Tbr2 cKO mice remain uncertain, but could include reduced feeding, hyperactivity, or hormonal changes.)
To evaluate motor development, we used rotarod and balance beam tests. Paradoxically, Tbr2 cKO mice had enhanced performance on both tests, although the enhancement declined in older mutants (Figure S5C,E). Importantly, body size did not correlate with motor performance (R2<0.02, Figure S5D), so the enhancement in Tbr2 cKO mutants cannot be attributed to lower body mass.
Other brain abnormalities in Tbr2 cKO mice included severe olfactory bulb hypoplasia, and reduced cortical surface area (Figure S5H,J; see also Arnold et al., 2008). In contrast, cortical thickness was not significantly reduced (Figure 4). Also, the anterior commissure was absent, although the corpus callosum and hippocampal commissure showed no obvious defects (Figure S6G,H; see also Hodge et al., 2013).
Gene dysregulation in Tbr2 cKO IPs and neuronal progeny
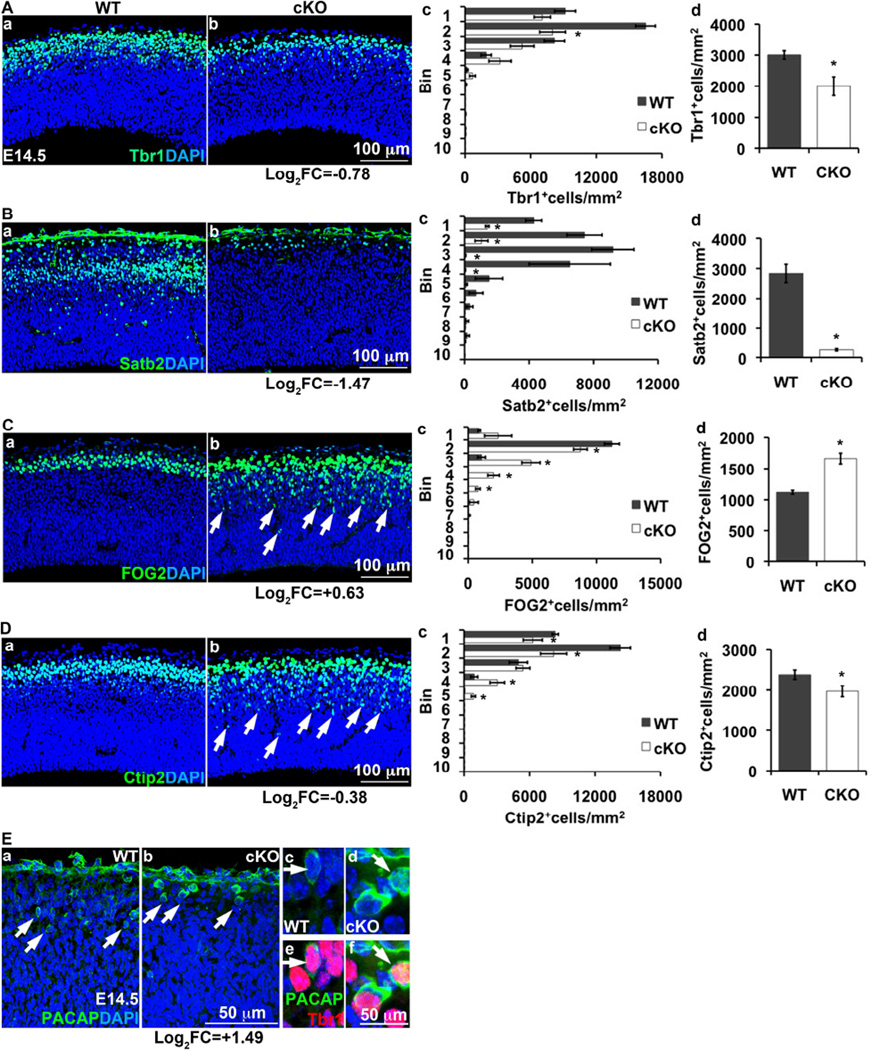
To investigate molecular defects in Tbr2 cKO mutants, we analyzed microarray data comparing E14.5 WT and Tbr2 cKO neocortex from our previous study, which focused on rostrocaudal identity (Elsen et al., 2013). Here, we focused on critical genes in neurogenesis and laminar fate acquisition. Genes up- or down-regulated in Tbr2 cKO neocortex were identified by positive or negative log2FC (log2 of the fold change) values, indicating significant differences (p<0.05) between Tbr2 cKO and control cortex.
Interestingly, transcription factor genes "upstream" of Tbr2 were up-regulated in Tbr2 cKO cortex, including Pax6 (log2FC= +0.36; p=0.001) and Insm1 (log2FC= +0.46; p=0.001). In contrast, "downstream" markers of laminar differentiation were mixed, with down-regulation of Tbr1 (log2FC= −0.78; p=0.00004), Bcl11b/Ctip2 (log2FC= −0.38; p=0.009) and Satb2 (log2FC= -1.47; p=0.00008), but up-regulation of Zfpm2/FOG2 (log2FC= +0.63; p=0.0005) and Adcyap1/PACAP (log2FC= +1.49; p<10−7). Interestingly, both Tbr1 and FOG2 are L6 markers (Bedogni et al., 2010), but they were regulated in opposite directions on E14.5, as were L5 markers Ctip2 and PACAP. These data suggested that differentiation of postmitotic neurons was severely dysregulated in Tbr2 cKO cortex.
All stages of cortical differentiation are abnormal in Tbr2 cKO cortex
To further investigate the differentiation defects inferred from microarray analysis, we studied the expression of neuronal differentiation markers by immunofluorescence. This approach allowed us to define not only quantitative changes in gene expression, but also qualitative changes in zonal differentiation patterns (Bystron et al., 2008).
Patterns of neuron differentiation were profoundly disturbed in E14.5 Tbr2 cKO cortex (Figure 5). The cortical plate (CP) and intermediate zone (IZ) appeared thin, and fewer cells expressed markers of postmitotic LL (Tbr1+, 67%; p=0.0126) and callosal (Satb2+, 9%; p=5.9*10−6) differentiation (Figure 5A,B). In contrast, FOG2 (L6, 1.5 fold increase; p=0.0038) and Ctip2 (L5, 83% decrease; p=0.0441), which are restricted to the CP on E14.5 in WT embryos, showed ectopic expression in the IZ of Tbr2 cKO mutants (Figure 5C,D). PACAP+ cells, representing a subset of L5 neurons (Lodato et al., 2014), were more abundant and more immunoreactive in the Tbr2 cKO CP (Figure 5E). C-R neurons (Reelin+) appeared slightly increased in Tbr2 cKO mice (data not shown), similarly as in Pax6 mutants (Stoykova et al., 2003). Together, these results demonstrated that projection neuron differentiation was severely disorganized and dysregulated in E14.5 Tbr2 cKO mutants.
Figure 5. Zonal expression patterns of TFs that regulate projection neuron differentiation are altered in E14.5 Tbr2 cKO cortex.
(Aa,b–Da,b) Expression of (A) Tbr1, (B) Satb2, (C) FOG2, and (D) Ctip2 in E14.5 control (Aa, Ba, Ca, Da) and Tbr2 cKO (Ab, Bb, Cb, Db) cortex. (Ac, Bc, Cc, Dc) Bin analysis of cells expressing each marker in WT and Tbr2 cKO cortex, in bins spanning VZ to MZ (*, p≤0.049). Note ectopic expression of FOG2 in the IZ (arrows, Cb). (Ad, Bd, Cd, Dd) Cell counts showed decreased Tbr1+ (p=0.012), Satb2+ (p=5.8*10−6) and Ctip2+ (p=0.044) cells in Tbr2 cKO cortex, while FOG2+ cells were increased (p=0.003). Despite overall reduction, Ctip2+ cells were located not only in the very thin CP, but also ectopically in the IZ (D, arrows). (E) PACAP+ cells were larger and expressed higher levels of PACAP in Tbr2 cKO (b) than in control cortex (a), and co-expressed Tbr1 (in E14.5 neocortex, a marker of all projection neurons) in both genotypes (c–f). Numbers under the (b) column indicate changes (log2FC) in the expression of mRNA for each marker (all p<0.05) in Tbr2 cKO cortex. Positive numbers indicate increased, and negative numbers decreased expression. N≥ 3 WT and 3 cKO embryos. Error bars represent SEM. Also see Figure S6.
To further characterize the trajectory of cortical differentiation in Tbr2 cKO cortex, we studied E12.5 and E16.5 time points (Figure S6). In E12.5 mutants, the preplate appeared thicker than normal, due to an abundance of NeuroD+/Tbr1− immature neurons, along with approximately normal numbers of Tbr1+ neurons (Figure S6A,B). Interestingly, the boundary between VZ (Sox2+) and preplate (Tbr1+) appeared irregular due to the accumulation of immature neurons (Figure S6B). These results suggested that differentiation of preplate neurons was impaired in E12.5 Tbr2 cKO mutants, and early neurogenesis was increased (Figure 4A) in compensation.
In E16.5 Tbr2 mutants, the numbers of Tbr1+, Ctip2+, and PACAP+ LL neurons were strikingly increased over WT, and CP thickness was increased (Figure S6C,D,E). PACAP+ neurons also expressed Ctip2 (Figure S6E,F), supporting their identity as a subset of L5 neurons (Lodato et al., 2014). In contrast to increased LL thickness, UL thickness was decreased in E16.5 Tbr2 cKO cortex compared to WT (Figure S6C,D), and ULs remained thin on P2 (Figure 4D,E).
Together with BrdU birthdating (Figure 4A–C), these data indicated that early, middle, and late phases of cortical differentiation were severely abnormal in Tbr2 cKO cortex. The delayed differentiation of early-born neurons (Figure S6A,B) may account for the shift from L6 to L5 fates (Figure 4D,E), and for the compensatory burst of early neurogenesis (Figure 4A) in Tbr2 cKO mutants. In turn, excessive early neurogenesis may have depleted progenitors, leading to decreased neurogenesis by E14.5–E16.5 (Figure 4B,C) and consequent thinning of ULs (Figure 4D,E).
Basal progenitors are dysregulated and increased in Tbr2 cKO cortex
The defects of neuronal differentiation in Tbr2 cKO neocortex might be attributable to defective genesis and/or differentiation of IPs. To identify IPs and distinguish them from RGPs, we studied abventricular (basal) mitoses (AVMs) and ventricular (apical) mitoses (VMs) by phospho-histone H3 (pH3) IF (Kowalczyk et al., 2009).
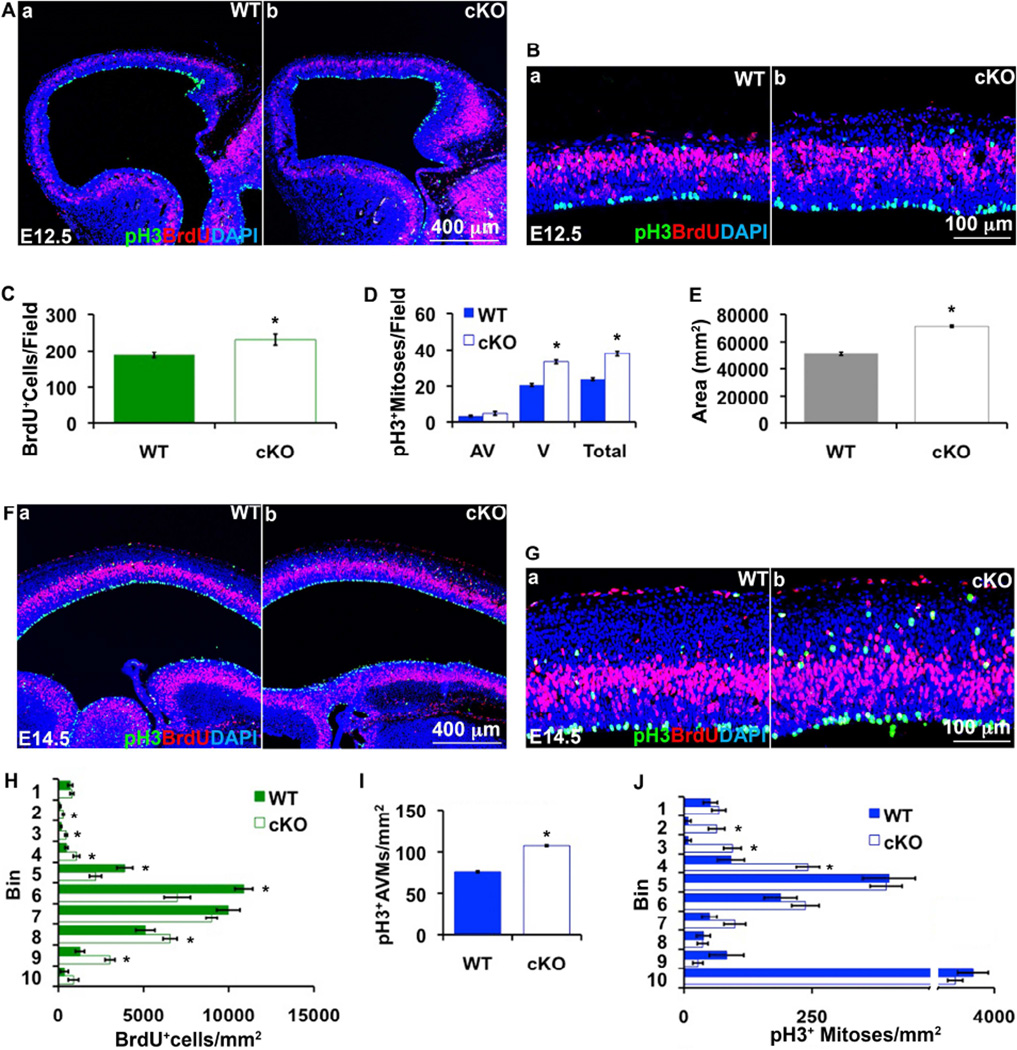
In E12.5 Tbr2 cKO mutants, the number of AVMs was unchanged from controls, but ventricular mitoses (VMs) were increased to 1.7-fold (p=4.4*10−9), and S-phase cells (acute BrdU+) to 1.2-fold (p=0.0245) of control values (Figure 6A–E). By E14.5, VMs normalized, but AVMs increased to significantly exceed control numbers (1.4-fold; p=4.7*10−5; Figure 6F,G). Moreover, AVMs in Tbr2 cKO cortex were not confined to the VZ/SVZ as in controls, but were also found ectopically in the IZ and CP (Figure 6G,J). Furthermore, acute BrdU+ (S-phase) cells were also observed in the Tbr2 cKO IZ and CP, and BrdU+ cells in the VZ were disorganized (Figure 6G,H). Nevertheless, the total numbers of BrdU+ cells were similar in E14.5 mutants and controls. These results indicated that basal IPs were not diminished, but were actually increased in Tbr2 cKO mutants, contradicting previous reports (Arnold et al., 2008; Sessa et al., 2008).
Figure 6. Tbr2 cKO cortex has increased numbers of basal/abventricular mitoses on E14.5.
(A, B) E12.5 WT (a) and Tbr2 cKO (b) cortex labeled with acute BrdU (red) and pH3 (green). Ba and Bb show higher magnifications of Aa and Ba, respectively. (C) The number of BrdU+ cells was increased in E12.5 Tbr2 cKO cortex (p=0.024), as was the total number of pH3+ mitoses (D), due to an increase of VMs (p<1.2*10−8) only. (VMs can be RGP or IP mitoses.) (E) Cortical thickness (measured as area over a defined length of ventricular surface) was increased in the E12.5 Tbr2 cKO cortex (p=8.4*10−7), due mainly to preplate expansion. (F, G) E14.5 WT (a) and Tbr2 cKO (b) cortex labeled with acute BrdU (red) and pH3 (green). Ga and Gb show higher magnifications of Fa and Fb, respectively. (H) The distribution of BrdU+ cells was altered in E14.5 Tbr2 cKO cortex, with increased numbers in abventricular (bins 2–4) and adventricular (bins 8–9) zones (*, p<0.048). (I) The number of AVMs was overall increased in Tbr2 cKO cortex (p=4.6*10−5). (J) pH3+ AVMs were also shifted superficially, with increased numbers in bins 2–4 (IZ and CP) (*, p≤0.005), N≥ 3 WT and 3 cKO embryos. Error bars represent SEM.
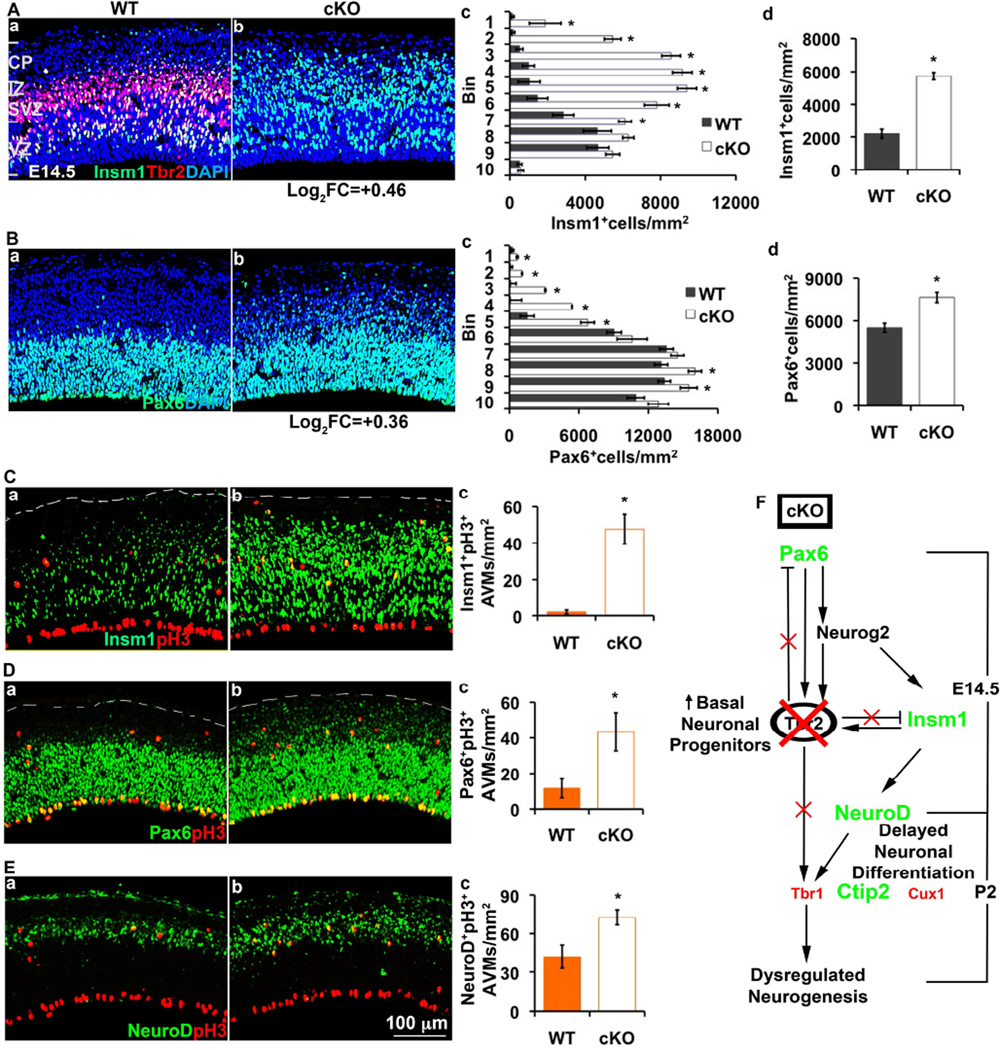
We next studied expression of Pax6 and Insm1, transcription factors upstream of Tbr2 that promote genesis of Tbr2+ IPs (Quinn et al., 2007; Farkas et al., 2008; Sansom et al., 2009). Strikingly, Pax6+ and Insm1+ cells were not only increased 1.4-fold (p=0.0011) and 2.6-fold (p=4.3*10−10), respectively, but were also located ectopically in the IZ and CP of Tbr2 mutants (Figure 7A,B). Moreover, pH3+ AVMs were more likely to express Pax6 (3.6-fold; p=0.024) and Insm1 (24-fold; p=3*10−5) in mutant than in control cortex (Figure 7C,D), suggesting that Tbr2-deficient IPs failed to down-regulate these transcription factors (Englund et al., 2005; Farkas et al., 2008).
Figure 7. Tbr2-deficient basal IPs express Insm1 and Pax6 ectopically, and NeuroD at increased levels.
(A) Two-color IF to detect Tbr2 (red) and Insm1 (green) in E14.5 control (Aa) and Tbr2 cKO (Ab) cortex showed increased numbers of Insm1+ cells in Tbr2 cKO cortex (Ad; p=4.3*10−10), especially in superficial bins (Ac) representing IZ and CP (*, p≤0.041). (B) Pax6 expression in E14.5 WT (Ba) and Tbr2 cKO (Bb) cortex. Pax6+ cells were found ectopically in the IZ and CP (Bc), and were overall increased (Bd; p=0.001). Numbers under column (b) indicate changes in mRNA expression (log2FC) between Tbr2 cKO and control cortex from microarray analysis. (C–E) Insm1+, Pax6+ and NeuroD+ AVMs were increased in E14.5 Tbr2 cKO cortex (b) relative to control (a), as confirmed by cell counting (c, Insm1+ AVMs; p=3.0*10−5, Pax6+ AVMs, p=0.024; NeuroD+ AVMs, p=0.01). N≥ 3 WT and 3 cKO embryos. (F) Diagram illustrating the dysregulated TF network in E14.5 Tbr2 cKO cortex, and altered laminar fates in P2 Tbr2 cKO cortex. Green labeled TFs show increased, whereas red labeled TFs show decreased expression in Tbr2 cKO. Scale bar: 100 µm (all images). Error bars represent SEM. Also see Figure S7.
We next studied expression of NeuroD, a transcription factor expressed in basal IPs and newly generated neurons (Hevner et al., 2006). In E14.5 Tbr2 cKO cortex, NeuroD was expressed by increased numbers of cells (1.2-fold; p=0.033), including many in the IZ, SP and CP, demonstrating ectopic expression of NeuroD in neuronal maturation zones (Figures 7E and S7A–D). The fraction of AVMs that expressed NeuroD was also increased in Tbr2 cKO cortex relative to controls (1.7-fold; p=0.0105; Figure 7E), consistent with protracted IP differentiation despite active NeuroD expression. Many NeuroD+ cells aberrantly co-expressed Pax6 in Tbr2 mutant cortex (25-fold more than in WT; Figure S7D,E). Thus, the differentiation of IPs and new neurons was disorganized and prolonged in Tbr2 cKO cortex.
Together, these findings indicate that Tbr2 is not necessary for IP genesis, but is required to promote the transition from IPs to postmitotic neurons (Figure 7F). In the absence of Tbr2, IP genesis continues, AVMs accumulate, and differentiation of IPs to neurons is profoundly abnormal.
Discussion
In the present study, we found that IP cohorts make complex contributions to cortical layers, including an unexpected contribution from early IPs to upper cortical layers. We also showed that Tbr2 regulates laminar organization of the cortex, by facilitating the transition from IP to neuron, and promoting the timely acquisition of laminar identity.
The finding that some early IPs produce UL neurons (Figure 1A) suggested two possible interpretations. First, if laminar fate is specified in RGPs and some early RGPs have restricted UL fates (Franco et al., 2012), then early IPs inherit UL fates from parent RGPs. Alternatively, if IPs are initially multipotent with regard to laminar identities, then daughter neuron fates may be determined by the timing of final mitosis, and limited by progressive fate restriction (Desai and McConnell, 2000). The latter possibility is favored by previous evidence that IPs can divide asymmetrically (with respect to laminar fate) to produce multiple layers (Wu et al., 2005). To resolve this issue, clonal analysis of IP lineages will be necessary.
Our findings challenge the previous conclusion that Tbr2 is required primarily for IP genesis (Sessa et al., 2008). Specifically, we found that basal mitoses were not reduced, but were actually increased in E14.5 Tbr2 cKO neocortex (Figure 6F,G,I). In contrast, previous studies reported that basal mitoses were significantly depleted in Tbr2 cKO neocortex (Arnold et al., 2008; Sessa et al., 2008). We attribute these discrepancies to different Cre drivers and floxed Tbr2 alleles. The previous study implicating Tbr2 in IP genesis (Sessa et al., 2008) used Foxg1Cre, a knockin allele that itself causes IP depletion (Siegenthaler et al., 2008). Another previous study (Arnold et al., 2008) used Sox1Cre, likewise a knockin allele (Takashima et al., 2007) that causes defects of brain development and function (Malas et al., 2003). In contrast, Nes11Cre, used in the present study, is a transgene that does not directly interfere with gene expression, or with brain development and function (Tronche et al., 1999).
The aberrant and protracted differentiation of neurons in embryonic Tbr2 cKO cortex can be traced to gene dysregulation in IPs. The ectopic expression of Pax6 and Insm1 in Tbr2 cKO IZ and CP (Figure 7) suggests that Tbr2 is required to downregulate these transcription factors, possibly by direct transcriptional repression in IPs. Indeed, Tbr2 binding sites are found near the Pax6 and Insm1 promoters (Teo et al., 2011). The persistent expression of Pax6 and Insm1 may interfere with neuronal differentiation in cortex, as demonstrated for ectopic Pax6 in the spinal cord (Bel-Vialar et al., 2007). Thus, Tbr2 appears to facilitate neuronal differentiation in part by repressing molecules that are normally expressed only in progenitor cells.
Conversely, Tbr2 may direct neuronal maturation by activating transcription of molecules expressed in differentiating neurons, such as Tbr1 and Satb2. Consistent with this possibility, both Satb2 and Tbr1 are initially detected in basal IPs, albeit at low levels (Britanova et al., 2005; Nelson et al., 2013). On the other hand, some important neuronal differentiation factors (such as NeuroD) are clearly not dependent on Tbr2. Also, despite abnormal gene expression in the Tbr2 cKO cortex, most projection neurons ultimately differentiated successfully. Indeed, the Tbr2 cKO cortex underwent a marked change between E14.5, when the CP was thin with a paucity of Tbr1+ neurons (Figure 5A), and E16.5, when the CP was thick with abundant Tbr1+ and Ctip2+ neurons (Figure S6C,D). The number of Satb2+ neurons likewise recovered substantially after E14.5 (Figure S6I). In the absence of Tbr2, upregulation of other molecules, such as NeuroD (Figure S7A–D), may compensate to ensure neuronal differentiation.
Tbr2 appears to regulate laminar fate by multiple mechanisms. Inactivation of Tbr2 in IP cohorts (E12.5 and E14.5) led to reduced genesis of rapidly-generated neuron subtypes (deeper in cortex), and relatively increased genesis of later-generated subtypes (more superficial) (Figure 3). Since Tbr2-deficient cohorts were sparse, these results indicated that Tbr2 is required cell-autonomously for rapid IP differentiation and neurogenesis. Extrapolating from these findings, Tbr2 cKO (throughout cortex) may have delayed the differentiation of all IP cohorts, causing an overall shift away from early-born neuron subtypes, towards increased genesis of later-born subtypes. Indeed, Tbr1+ neurons and L6 thickness were decreased in Tbr2 cKO mice (Figure 4E). However, Tbr2 may also regulate the balance of L6 and L5 fates directly: E12.5-born (BrdU+) cells were more likely to differentiate as Ctip2+ neurons in Tbr2 cKO cortex (Figure S5Bc), and L5 markers PACAP and Ctip2 were markedly increased by E16.5 in Tbr2 cKO cortex (Fig. S6D–F). Thus, Tbr2 appears to directly regulate both the rate of IP differentiation, and the balance of L6 and L5 fates during early neurogenesis.
Despite the delayed differentiation of IPs, neurogenesis was initially accelerated in E12.5 Tbr2 cKO cortex, but decreased subsequently on E14.5 and E16.5 (Figure 4A–C). The acceleration of early neurogenesis may represent a non-autonomous effect of altered IP differentiation. Such effects are anticipated because IPs interact with RGPs, for example, by Delta-Notch signaling (Nelson et al., 2013). We speculate that deficient Delta-Notch signaling in early Tbr2 cKO cortex caused RGPs to respond by increasing direct neurogenesis (Figure 4A) and overproducing preplate neurons (Figure S6A,B). In turn, the RGP pool may have been depleted prematurely in Tbr2 cKO mutants, thus accounting for reduced genesis of late-born UL neurons, and decreased UL thickness (Figure 4B–D). In sum, laminar defects in Tbr2 cKO cortex reflect a complex system of differentiation and feedback.
Interestingly, the reduction of late neurogenesis and UL thickness occurred despite ample production of basal progenitors in Tbr2 cKO cortex (Figure 6I). Previous studies have shown that IP genesis is driven by low Notch signaling in RGPs (Nelson et al., 2013), and by neurogenic transcription factors including Pax6, Insm1, and Neurog2 (Sun and Hevner, 2014). Those "upstream" mechanisms of IP specification occur in RGPs prior to the expression of Tbr2, so our finding that IP genesis was spared in Tbr2 cKO cortex is logical. Rather, Tbr2 deficiency perturbed gene expression in new IPs, and impaired their ability to differentiate as cortical projection neurons with well-defined laminar subtype identities. Our findings indicate that Tbr2 plays an important transitional role in neurogenesis, by both suppressing RGP identity and promoting specific features of cortical layers. Ultimately, neuronal differentiation and layer formation were delayed in Tbr2 cKO mice, but proceeded to completion due to compensatory mechanisms.
Remarkably, major motor skills were not impaired in Tbr2 cKO mice (Figure S5C,E), although previous studies detected hyperactivity and weakness (Arnold et al., 2008). The small olfactory bulb and rudimentary dentate gyrus in Tbr2 mutants (Hodge et al., 2013) presumably impair olfaction and memory, but those functions have not been tested. Interestingly, Nlgn3 mutant mice also show improved performance on repetitive motor tasks (Rothwell et al., 2014).
In sum, we have shown that IPs can persist in the cortex for prolonged periods, and that early IP cohorts contribute to multiple cortical layers. The pace of laminar neurogenesis, and the identities of projection neurons, are regulated by Tbr2 although the genesis of IPs is not. In future studies, it will be interesting to conduct clonal analysis of IP progeny, and determine if individual IPs contribute to multiple layers, as well as the size and distribution of IP-derived clones.
Experimental Procedures
Animals and tissue collection
C57BL/6 mice used in this study were kept in a 12 hour light/dark cycle, with food and water ad-libitum, in Seattle Children’s Research Institute’ vivarium. All animal experimental procedures were performed with Institutional Animal Care and Use Committee approval. The following previously described mouse transgenic alleles were used: Ai14 reporter (Madisen et al., 2010), EomesCreER (Tbr2CreER) (Pimeisl et al., 2013), Nestin-Cre (Nes11Cre) (Tronche et al., 1999) (stock 003771, Jackson Labs), Tbr2-Flox (Tbr2FL) (Intlekofer et al., 2008). Also see Supplemental Experimental Procedures.
Tamoxifen and bromodeoxyuridine administration
Pregnant dams were administered Tamoxifen (Sigma, T5648; 5mg/kg), Progesterone (Sigma, P3972; 2.5mg/kg) and BrdU (Sigma, B5002; 50mg/kg), by intraperitoneal injection, at the indicated embryonic ages. Acute BrdU treatment was done 30 minutes before brain collection.
Immunofluorescence
The IF procedure was previously described (Englund et al., 2005). Also see Supplemental Experimental Procedures.
Image acquisition, cell counting and statistics
Single plane optical sections and stacks were acquired with Zeiss LSM-710 confocal microscope. Cell counts were reported either as absolute number or density per area (mm2), or as distribution per bin/cortical zone. Also see Supplemental Experimental Procedures. Data was reported as mean ± SEM, from at least 3 sections from an animal, and 2–4 animals per condition/data point. Statistical analysis used 2-tailed, unpaired Student’s t test, and the confidence threshold chosen is p<0.05.
Rotarod
Male mice between 6 weeks and 6 months of age were tested on the Rotamex-5 (Columbus Instruments) for rotarod performance to assess their motor skills as described (Hsu et al., 2014). Also see Supplemental Experimental Procedures.
Balance Beam
Tbr2 cKO and littermate control male mice, 3–4 months of age, were used in the balance beam test, as described (Hsu et al., 2014). Also see Supplemental Experimental Procedures.
Microarrays
Data from our previous microarray experiment (www.ncbi.nlm.nih.gov/geo, accession no. GSE43387; Elsen et al., 2013) was analyzed in the present study. Also see Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Intermediate progenitors (IPs) generate all types of cortical projection neurons.
A subset of early IPs produce upper layer neurons.
Projection neuron laminar fates are dysregulated in the Tbr2-deficient cortex.
Tbr2 is mainly necessary for differentiation, not genesis, of IPs.
Acknowledgments
We thank E. Young and L. Honican for assistance with experiments and analysis. We thank Dr. C. Birchmeier (Max-Delbrück-Center for Molecular Medicine, Berlin) for Insm1 antibody, Dr. J. Kohtz (Northwestern Univ., Chicago) for Dlx antibody and Dr. J. Hannibal for PACAP antibody (University of Copenhagen). We thank Dr. T. Bammler, Dr. F. Farin, and Dr. D. Beyer (University of Washington Center for Ecogenetics and Environmental Health) for assistance with microarray experiments, and the UW Center on Human Development and Disability for partial support of microarray experiments (supported by grant U54 HD083091 from the National Institute of Child Health and Human Development to the University of Washington’s Center on Human Development and Disability). This study was supported by National Institutes of Health Grants R01 NS085081 and R01 NS092339 to RFH, German Research Foundation (DFG) Emmy Noether Programme (AR732/1-1) to SJA, Lejeune Foundation and ProRett Italia to FB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
ABM and RFH designed the study and wrote the manuscript. ABM, RAD, KAR, GEE and FB performed experiments. SJA provided research materials and manuscript editing/commenting. ABM and RFH analyzed data.
References
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, Molnar Z, Robertson EJ, Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, Bammler TK, Rubenstein JL, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci U S A. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel-Vialar S, Medevielle F, Pituello F. The on/off of Pax6 controls the tempo of neuronal differentiation in the developing spinal cord. Dev Biol. 2007;305:659–673. doi: 10.1016/j.ydbio.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21:658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–2872. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- Elsen GE, Hodge RD, Bedogni F, Daza RA, Nelson BR, Shiba N, Reiner SL, Hevner RF. The protomap is propagated to cortical plate neurons through an Eomes-dependent intermediate map. Proc Natl Acad Sci U S A. 2013;110:4081–4086. doi: 10.1073/pnas.1209076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas LM, Haffner C, Giger T, Khaitovich P, Nowick K, Birchmeier C, Paabo S, Huttner WB. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron. 2008;60:40–55. doi: 10.1016/j.neuron.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, Muller U. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Daza RA, Rubenstein JL, Stunnenberg H, Olavarria JF, Englund C. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Hodge RD, Garcia AJ, 3rd, Elsen GE, Nelson BR, Mussar KE, Reiner SL, Ramirez JM, Hevner RF. Tbr2 expression in Cajal-Retzius cells and intermediate neuronal progenitors is required for morphogenesis of the dentate gyrus. J Neurosci. 2013;33:4165–4180. doi: 10.1523/JNEUROSCI.4185-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YW, Wang SD, Wang S, Morton G, Zariwala HA, de la Iglesia HO, Turner EE. Role of the dorsal medial habenula in the regulation of voluntary activity, motor function, hedonic state, and primary reinforcement. J Neurosci. 2014;34:11366–11384. doi: 10.1523/JNEUROSCI.1861-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, Molyneaux BJ, Zuccaro E, Goff LA, Chen HH, Yuan W, Meleski A, Takahashi E, Mahony S, Rinn JL, et al. Gene co-regulation by Fezf2 selects neurotransmitter identity and connectivity of corticospinal neurons. Nat Neurosci. 2014;17:1046–1054. doi: 10.1038/nn.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malas S, Postlethwaite M, Ekonomou A, Whalley B, Nishiguchi S, Wood H, Meldrum B, Constanti A, Episkopou V. Sox1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability. Neuroscience. 2003;119:421–432. doi: 10.1016/s0306-4522(03)00158-1. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Hodge RD, Bedogni F, Hevner RF. Dynamic interactions between intermediate neurogenic progenitors and radial glia in embryonic mouse neocortex: potential role in Dll1-Notch signaling. J Neurosci. 2013;33:9122–9139. doi: 10.1523/JNEUROSCI.0791-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimeisl IM, Tanriver Y, Daza RA, Vauti F, Hevner RF, Arnold HH, Arnold SJ. Generation and characterization of a tamoxifen-inducible Eomes(CreER) mouse line. Genesis. 2013;51:725–733. doi: 10.1002/dvg.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JC, Molinek M, Martynoga BS, Zaki PA, Faedo A, Bulfone A, Hevner RF, West JD, Price DJ. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev Biol. 2007;302:50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell PE, Fuccillo MV, Maxeiner S, Hayton SJ, Gokce O, Lim BK, Fowler SC, Malenka RC, Sudhof TC. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 2014;158:198–212. doi: 10.1016/j.cell.2014.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler JA, Tremper-Wells BA, Miller MW. Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: effect of increased p21 expression. Cereb Cortex. 2008;18:1865–1875. doi: 10.1093/cercor/bhm209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30:7028–7036. doi: 10.1523/JNEUROSCI.6131-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A, Hatano O, Gruss P, Gotz M. Increase in reelin-positive cells in the marginal zone of Pax6 mutant mouse cortex. Cereb Cortex. 2003;13:560–571. doi: 10.1093/cercor/13.6.560. [DOI] [PubMed] [Google Scholar]
- Sun T, Hevner RF. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci. 2014;15:217–232. doi: 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Era T, Nakao K, Kondo S, Kasuga M, Smith AG, Nishikawa S. Neuroepithelial cells supply an initial transient wave of MSC differentiation. Cell. 2007;129:1377–1388. doi: 10.1016/j.cell.2007.04.028. [DOI] [PubMed] [Google Scholar]
- Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- Teo AK, Arnold SJ, Trotter MW, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Tyler WA, Medalla M, Guillamon-Vivancos T, Luebke JI, Haydar TF. Neural precursor lineages specify distinct neocortical pyramidal neuron types. J Neurosci. 2015;35:6142–6152. doi: 10.1523/JNEUROSCI.0335-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasistha NA, Garcia-Moreno F, Arora S, Cheung AF, Arnold SJ, Robertson EJ, Molnar Z. Cortical and Clonal Contribution of Tbr2 Expressing Progenitors in the Developing Mouse Brain. Cereb Cortex. 2015;25:3290–3302. doi: 10.1093/cercor/bhu125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SX, Goebbels S, Nakamura K, Nakamura K, Kometani K, Minato N, Kaneko T, Nave KA, Tamamaki N. Pyramidal neurons of upper cortical layers generated by NEX-positive progenitor cells in the subventricular zone. Proc Natl Acad Sci U S A. 2005;102:17172–17177. doi: 10.1073/pnas.0508560102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.