Abstract
We present DADA2, a software package that models and corrects Illumina-sequenced amplicon errors. DADA2 infers sample sequences exactly, without coarse-graining into OTUs, and resolves differences of as little as one nucleotide. In several mock communities DADA2 identified more real variants and output fewer spurious sequences than other methods. We applied DADA2 to vaginal samples from a cohort of pregnant women, revealing a diversity of previously undetected Lactobacillus crispatus variants.
The importance of microbial communities to human and environmental health has motivated methods for their efficient characterization. The most common and cost-effective method is the amplification and sequencing of targeted genetic elements1. Amplicon sequencing of taxonomic marker genes such as the 16S rRNA gene in bacteria, the ITS region in fungi, and the 18S rRNA gene in eukaryotes, provides a census of a community. Functional diversity can be probed by targeting functional genes2.
Disentangling biological variation from amplicon sequencing errors presents unique challenges, which prompted the development of amplicon-specific error-correction methods3,4,5,6. Most of these methods were designed for 454 pyrosequencing, and are not applicable to Illumina sequencing.
Errors in Illumina-sequenced amplicon data are currently addressed by quality filtering and the construction of Operational Taxonomic Units (OTUs): clusters of sequences that differ by less than a fixed dissimilarity threshold7,8,9 (typically 3%). Lumping together similar sequences reduces the rate at which errors are misinterpreted as biological variation (Supplementary Fig. 1), but OTUs under-utilize the quality of modern sequencing by precluding the possibility of resolving fine-scale variation5,10,11,12. Fine-scale variation can be informative about ecological niches10, temporal dynamics12, and population structure2. Fine-scale variation differentiates pathogenic from commensal strains in some cases13,14, and can contain clinically relevant information for more complex microbiome-associated diseases15.
The Divisive Amplicon Denoising Algorithm (DADA) introduced a model-based approach for correcting amplicon errors without constructing OTUs5. DADA identified fine-scale variation in 454-sequenced amplicon data while outputting few false positives2,5.
Here we present DADA2, an open-source R package (https://github.com/benjjneb/dada2) that extends and improves the DADA algorithm. DADA2 implements a new quality-aware model of Illumina amplicon errors. Sample composition is inferred by dividing amplicon reads into partitions consistent with the error model (Methods). DADA2 is reference-free, and applicable to any genetic locus. The DADA2 R package implements the full amplicon workflow: filtering, dereplication, chimera identification, and merging paired-end reads.
We compared DADA2 to four algorithms (Methods): UPARSE, an OTU construction algorithm with the best published false-positive results9; MED, an algorithm with the best published fine-scale resolution in Illumina amplicon data11; and the popular mothur (average linkage) and QIIME (uclust) OTU methods7,8.
We benchmarked these algorithms on three mock community datasets: Balanced, HMP and Extreme (Methods, Supplementary Table 1), each sequenced at a depth of over 500,000 highly overlapping paired-end Illumina MiSeq 2×250 reads. The Balanced community contained 57 bacteria and archaea at nominally equal frequencies16. The HMP community contained 21 bacteria at nominally equal frequencies17. The Extreme community contained 27 bacterial strains at frequencies spanning five orders of magnitude and differing over the sequenced region by as little as one nucleotide (Methods, Supplementary Table 2). Sequence quality varied: Balanced was higher (Mean Q = 35.9 forward/33.5 reverse), Extreme moderate (33.0/29.3), and HMP lower (32.3/28.7).
We compared output sequences to the known reference sequences present in the reference strains making up these communities. Output sequences that exactly matched a reference sequence were classified as Reference, and those differing by one mismatch or gap were classified as One Off. Contaminants were identified in the yet unclassified sequences by BLASTing against nt (Methods). Sequences with an exact BLAST hit (100% identity, 100% coverage) were classified as Exact, and those with best hits containing one mismatch or gap as One Off. Everything else was classified as Other. We evaluated sensitivity by detection of the reference strains. Note that fine-scale variation was present in all mock communities, as some reference strains contained multiple distinguishable 16S rRNA sequence variants.
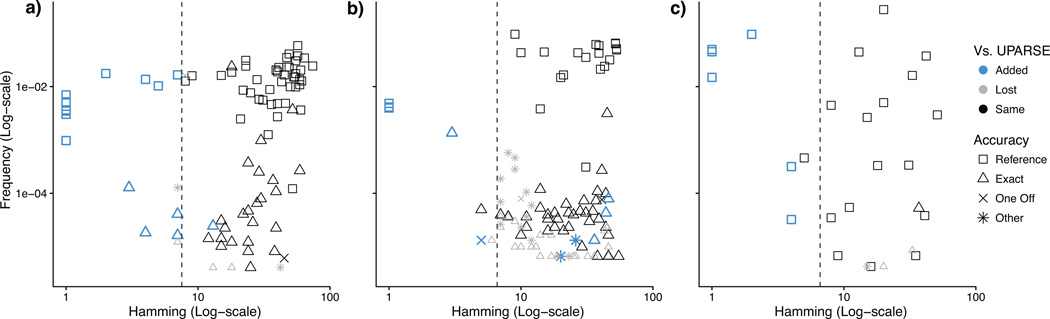
We compared the sample sequences output by DADA2 to the output of UPARSE (Fig. 1). Almost all variants with Hamming separation greater than UPARSE's OTU radius (3%, dashed line) were identified by both algorithms (black). However, DADA2 identified fine-scale variation that UPARSE did not (blue), in both the merged reads (Fig. 1) and the forward reads alone (Supplementary Fig. 2–7). DADA2 accurately resolved sequence variants differing by a single nucleotide and present in as few as two reads.
Figure 1. Sequence variants inferred by DADA2 compared to the OTUs constructed by UPARSE.
The merged sequences output by DADA2 are plotted for three Illumina amplicon datasets: (a) Balanced, (b) HMP, and (c) Extreme. Frequency is plotted on the y-axis; Hamming distance to the closest more-abundant sequence on the x-axis. Shapes represent accuracy (Methods). When variants are well separated from other members of the community the sequence variants inferred by DADA2 largely coincide with the OTUs output by UPARSE (black). However, DADA2 resolves additional variation (blue), especially within the UPARSE's OTU radius (dashed line), while outputting fewer spurious sequences (One Off and Other).
DADA2 identified more reference sequences and as many or more reference strains than UPARSE in every dataset, using merged or forward reads (Table 1). DADA2 identified every reference strain in the Balanced and HMP datasets; the Extreme reference strains it missed demonstrate its limits (Supplementary Note 1). DADA2 output fewer spurious sequences (Other and One Off) than UPARSE in every dataset (Table 1).
Table 1.
The accuracy of DADA2, UPARSE, MED, mothur and QIIME on three mock community datasets
| Output Reads(%) |
Output Sequences | Reference Strains |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Reference | Exact | One Off | Other | |||||
| Balanced | Forward | DADA2 | 99.2 | 93 | 59 | 33 | 1 | 0 | 57 |
| UPARSE | 99.1 | 81 | 48 | 29 | 2 | 2 | 53 | ||
| MED | 95.5 | 86 | 59 | 5 | 22 | 0 | 57 | ||
| Mothur | 96.3 | 249 | 44 | 25 | 15 | 165 | 49 | ||
| QIIME | 99.2 | 378 | 51 | 34 | 3 | 290 | 54 | ||
| Merged | DADA2 | 96.2 | 87 | 57 | 29 | 1 | 0 | 55 | |
| UPARSE | 94.2 | 76 | 45 | 27 | 2 | 2 | 50 | ||
| MED | 91.1 | 64 | 56 | 6 | 2 | 0 | 54 | ||
| Mothur | 94.1 | 108 | 42 | 27 | 11 | 28 | 47 | ||
| QIIME | 94.1 | 170 | 45 | 28 | 4 | 93 | 50 | ||
| HMP | Forward | DADA2 | 95.1 | 151 | 23 | 112 | 8 | 8 | 21 |
| UPARSE | 96.7 | 161 | 20 | 123 | 10 | 8 | 21 | ||
| MED | 80.9 | 83 | 23 | 2 | 58 | 0 | 21 | ||
| Mothur | 95.4 | 849 | 20 | 177 | 47 | 605 | 21 | ||
| QIIME | 97.4 | 1375 | 20 | 177 | 60 | 1118 | 21 | ||
| Merged | DADA2 | 92.3 | 67 | 23 | 40 | 2 | 2 | 21 | |
| UPARSE | 67.7 | 94 | 20 | 59 | 2 | 13 | 21 | ||
| MED | 64.8 | 32 | 23 | 3 | 6 | 0 | 21 | ||
| Mothur | 62.1 | 121 | 20 | 82 | 9 | 10 | 21 | ||
| QIIME | 67.6 | 290 | 20 | 71 | 8 | 191 | 21 | ||
| Extreme | Forward | DADA2 | 99.5 | 68 | 26 | 35 | 3 | 4 | 23 |
| UPARSE | 99.5 | 74 | 21 | 40 | 0 | 13 | 21 | ||
| MED | 86.4 | 95 | 16 | 0 | 79 | 0 | 13 | ||
| Mothur | * | * | * | * | * | * | * | ||
| QIIME | 99.5 | 3237 | 20 | 44 | 73 | 3100 | 20 | ||
| Merged | DADA2 | 97.6 | 25 | 24 | 1 | 0 | 0 | 21 | |
| UPARSE | 69.9 | 23 | 18 | 4 | 0 | 1 | 18 | ||
| MED | 67.6 | 32 | 17 | 0 | 15 | 0 | 14 | ||
| Mothur | 94.3 | 44 | 23 | 14 | 0 | 7 | 23 | ||
| QIIME | 69.9 | 36 | 19 | 8 | 1 | 8 | 19 | ||
After a common filtering step, methods were applied to the forward reads, and the merged forward and reverse reads, of the Balanced, HMP and Extreme datasets (Methods). Output sequences were classified as Reference or Exact (true positives) and One Off or Other (false positives) by comparison to the known sequences of these mock communities (reference strains) and comparison to nt to identify contaminants (Methods). DADA2 detected the most reference strains and sequences, while outputting the fewest false positives. Mothur failed to complete on the Extreme forward reads.
Minimum Entropy Decomposition (MED) is a method to distinguish fine-scale diversity in amplicon data11. Like DADA2, MED divides amplicon reads into partitions within which variation is supposed to be artefactual. MED effectively uses a single-site minor-allele-frequency threshold to distinguish real variation, and prevents false positives with a minimum abundance threshold. Thus while MED identified fine-scale variation, it output more false positives and couldn’t detect rare variants (Table 1, Supplementary Fig. 2–7).
Mothur and QIIME output significantly more spurious sequences than the other methods, although this deficiency was reduced when merging reads (Table 1, Supplementary Fig. 2–7). UPARSE was the most accurate OTU method tested by all measures save the number of reference strains identified in the merged Extreme dataset. The spurious output of mothur and QIIME included chimeric and non-chimeric errors (Supplementary Table 3).
The residual error rates in the output of DADA2 were very low. For the Balanced dataset, DADA2's residual error rate of 2.46 × 10−8 (forward) and 2.53 × 10−8 (merged), compares favorably to the best error rates of 5.9 × 10−3 and 5.0 × 10−4 previously reported16. For the HMP dataset, DADA2's residual error rate of 1.66 × 10−5 (forward) and 2.74 × 10−6 (merged) compares favorably to the previously reported error rates of 9.2 × 10−4 and 4.6 × 10−4 in [18], and 2 × 10−4 (merged) in [17]. DADA2 discarded relatively few reads compared to the other methods (Table 1).
DADA2’s core denoising algorithm was slower but comparable to UPARSE, and DADA2 easily processed Illumina samples on a laptop. For the filtered Balanced forward reads (33,516 unique sequences) UPARSE ran in 9s, QIIME/uclust in 17s, DADA2 in 21s, mothur in 2m26s and MED in 2m34s on a 2013 MacBook Pro (Methods).
We further evaluated DADA2 on two longitudinal, Illumina-sequenced datasets: 142 vaginal samples from 42 pregnant women19, and 360 mouse fecal samples17.
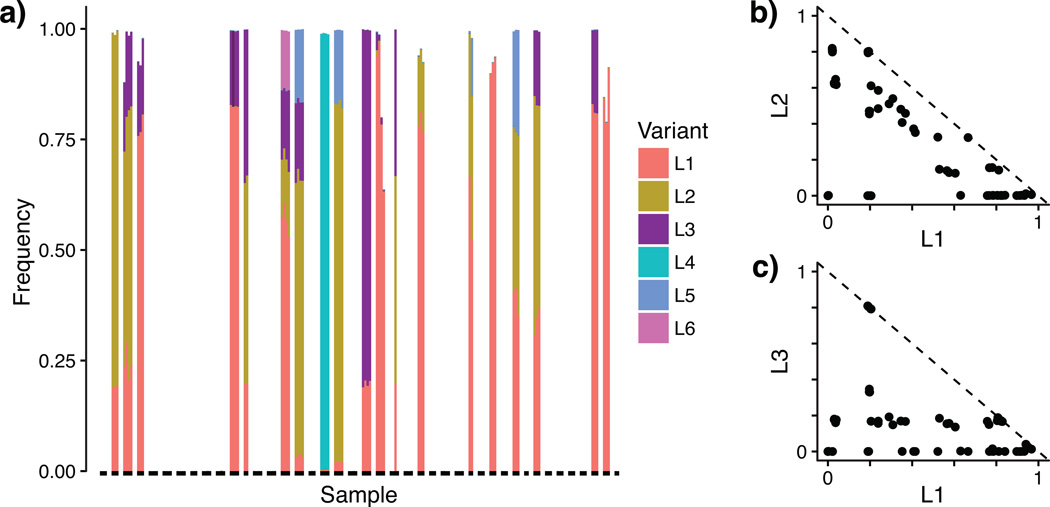
The vagina is the least diverse human body habitat1, often dominated by a single Lactobacillus OTU20. Lactobacillus crispatus is the most common species, and L. crispatus dominated communities have been associated with good health and stability20. DADA2 revealed that L. crispatus communities are more complex than generally recognized: six distinct L. crispatus sequence variants were present in multiple samples and substantial abundance (Fig. 2a). This variation is imperceptible to OTU methods, as these variants differ by just 1–2 nucleotides over the sequenced region.
Figure 2. Lactobacillus crispatus sequence variants in the human vaginal community during pregnancy.
DADA2 identified six Lactobacillus crispatus 16S rRNA sequence variants present in multiple samples and a significant fraction of all reads (L1: 19.7%, L2: 11.1%, L3: 6.5%, L4: 3.1%, L5: 1.3%, L6: 0.4%). (a) The frequency of L1–L6 in each sample. Black bars at the bottom link samples from the same subject. The frequency of (b) L1 vs. L2, and (c) L1 vs. L3, by sample. The dashed line indicates a total frequency of 1.
The joint distribution of L. crispatus variants strongly suggests they represent multiple bacterial strains. The distribution of sequence variants was stable over time, but differed substantially between women, and distinct ecological relationships appear to exist between variants. L1 and L2 showed a pattern of mutual exclusion consistent with competition for a common niche (Fig. 2b). L1 and L3 showed a pattern more consistent with an absence of direct competition (Fig. 2c): The frequency of L1 was independent of the frequency of L3, which strongly tended towards 20%.
The fecal community is more diverse than the vaginal community or mock communities, which could present different bioinformatic challenges. When applied to the mouse fecal dataset, DADA2 output 389 sequences and UPARSE 327 OTUs, of which 257 were shared. Output sequences were BLASTed against nt: 247/257 of the shared sequences, 123/132 of the DADA2-only sequences, and 26/70 of the UPARSE-only sequences were Exact matches. DADA2-only sequences were typically close to other sample sequences: the median Hamming distance of DADA2-only sequences to a more abundant output sequence was 2, for shared sequences it was 15. Sample richness was highly correlated between the methods (Pearson correlation = 0.99), but DADA2 identified more variants (mean ratio = 1.21). These results are consistent with the mock community benchmarking: DADA2 identified more biological variants, especially within UPARSE's OTU radius, while outputting fewer spurious sequences.
These comparisons show that DADA2 is more accurate than other methods. DADA2 resolves fine-scale variation better than the current best method for that task, while outputting fewer incorrect sequences than the most robust OTU method. The precision of DADA2 improves downstream measures of diversity and dissimilarity, and potentially allows amplicon methods to probe strain-level variation.
The output of DADA2 can be clustered into OTUs, but that often eliminates biological information present in the data. OTUs are not species, and their construction is not necessitated by amplicon errors. DADA2 enhances the study of microbial communities by allowing researchers to accurately reconstruct amplicon-sequenced communities at the highest resolution.
Online Methods
The Divisive Amplicon Denoising Algorithm
The core denoising algorithm in the DADA2 R package is built on a model of the errors in Illumina-sequenced amplicon reads. This error model quantifies the rate λji at which an amplicon read with sequence i is produced from sample sequence j as a function of sequence composition and quality. A Poisson model for the number of repeated observations of the sequence i, parameterized by the rate λji, is then used to calculate the p-value of the null hypothesis that the number of amplicon reads (the abundance) of sequence i is consistent with the error model. These p-values are used as the division criteria for an iterative partitioning algorithm, which continues dividing sequencing reads until all partitions are consistent with being produced from their central sequence5. We now describe each of these steps in detail.
Sequence comparison
Pairwise sequence alignments are performed by a vectorized implementation of the Needleman-Wunsch algorithm with ends-free gapping. As alignment dominates the computational costs of the algorithm, two heuristics are enabled by default. The first heuristic is the use of a kmer-distance screen prior to alignment. If the kmer-distance between i and j exceeds a user-settable parameter (KDIST_CUTOFF), no alignment is performed21. The default value of this parameter was chosen to exclude only pairs of reads with >10% nucleotide mismatch. The second heuristic is banded alignment, which forgoes calculation of potential alignments in which the net number of gaps of one sequence relative to the other exceeds a user-settable parameter (BAND_SIZE). The default value of this parameter was chosen to minimally impact the alignment of sequences with few indels, such as ribosomal RNA genes. Both heuristics can be disabled by the user, and the default values should be re-examined if the algorithm is applied to genetic regions with significantly different characteristics, such as the indel-rich ITS region.
Error model
DADA2 models errors as occurring independently within a read, and independently between reads. Under this model, the rate at which an amplicon read with sequence i is produced from sample sequence j is reduced to the product over the transition probabilities between the L aligned nucleotides:
The transition probability between aligned nucleotides is allowed to depend on the original nucleotide, substituting nucleotide, and associated quality score, e.g. p(A→C, 35).
After sequence alignment, the error rate λji is calculated and stored. If sequences i and j were not aligned because they exceeded the kmer-distance cutoff λji is set to 0.
The abundance p-value
The abundance p-value quantifies the notion that sequence i is too abundant for it to be explained by errors in amplicon sequencing. If sequencing errors are independent across reads, the number of amplicon reads with sequence i that will be produced from sample sequence j is Poisson distributed with expectation equal to an error rate λji multiplied by the expected reads of sample sequence j. Let unique sequence i with abundance ai be in partition j containing nj reads. Then, conditional on i being read at least once, the abundance p-value is the probability of seeing that many or more identical reads (ρpois is the Poisson density function):
A low pA indicates that there are more reads of sequence i than can be explained by errors introduced during the amplification and sequencing of nj copies of sample sequence j.
Note that the abundance p-value is calculated conditional on at least one sequence being observed. As a result, all singleton sequences have an abundance p-value of 1, and are never judged inconsistent with the error model. This means that singletons cannot form their own partitions, and DADA2 will not infer singleton sequences. The effect of this is similar in practice to the UPARSE developer's recommendation to remove singleton sequences prior to picking OTUs, and in both cases is driven by the difficulty in robustly differentiating singleton errors from real singleton sequences.
The divisive partitioning algorithm
First, amplicon reads with the same sequence are grouped into unique sequences with an associated abundance and consensus quality profile (or dereplicated). The divisive partition algorithm is initialized by placing all unique sequences into a single partition, and assigning the most abundant sequence as the center of that partition. All unique sequences are then compared to the center of their partition, error rates are calculated and stored, and the abundance p-value is calculated for each unique sequence. If the smallest p-value, after bonferroni correction, falls below the user-settable threshold OMEGA_A, a new partition is formed with the unique sequence with the smallest p-value as its center, and all unique sequences are compared to the center of that new partition.
After a new partition is formed, every unique sequence is allowed to join the partition most likely to have produced it (i.e. that produces the highest expected number of that unique sequence). At that point the division procedure iterates, with each iteration consisting of identifying the unique sequence with the smallest p-value, forming a new partition with that sequence as its center, and reshuffling sequences to their most likely partition.
Division continues until all abundance p-values are greater than OMEGA_A, i.e. all unique sequences are consistent with being produced by amplicon sequencing the center of their partition. The inferred composition of the sample is then the set of central sequences and the corresponding total abundances of those partitions (alternatively: each read is denoised by replacing it with the central sequence of its partition).
A detailed description of the original version of this divisive algorithm is available in Rosen et al. 20125. The DADA2 implementation has been slightly simplified for computational speed, in particular there is no longer any construction of “indel families”.
Error model parameterization
DADA2 depends on a parameterized error model (the 16 × 41 transition probabilities, e.g. p(A→C, 35) but if those parameters are not known a priori then DADA2 can estimate them from the data.
Given an inferred partition of the amplicon sequences, DADA2 records the mismatches between every sequence and the center of its partition, and counts each type of mismatch (e.g. the number of A→C mismatches where Q=35). That table of observed mismatches represents the errors inferred by DADA2, and can be used to estimate the parameters of the error model. DADA2's default parameter estimation method is to perform a weighted loess fit to the regularized log of the observed mismatch rates as a function of their quality, separately for each transition type (e.g. A→C mismatches are fit separately from A→G mismatches). However, the error rate estimation function is a modular part of the algorithm, and the user is able to provide their own R function to estimate the parameters of the error model from the observed mismatches if they prefer a different method.
Alternating estimation until consistency
DADA2's selfConsist mode alternates sample inference (conditional on the parameters of the error model) with parameter estimation (conditional on the inferred sample composition) until convergence, at which point jointly consistent estimates of the error parameters and sample composition are reported. This improves the accuracy of DADA2: while Illumina quality scores are informative, they do not exactly match their textbook definition (Supplementary Fig. 8), and we have observed significant variation in this relationship between different runs. The plotErrors function in the DADA2 R package produced Supplementary Fig. 8, and is a useful tool to visualize the observed and estimated error rates in various datasets.
DADA2 pipeline
The DADA2 R package implements a complete pipeline to turn paired-end fastq files from the sequencer into merged, denoised, chimera-free, inferred sample sequences. Parts of this pipeline can be substituted with outside methods, but there are some structural differences between the DADA2 pipeline and most others. One such difference is that the DADA2 pipeline performs merging of paired-end reads after denoising. This is because the core denoising algorithm uses the empirical relationship between the quality score and the error rates. When reads are merged, this relationship will differ between the forward-only, overlapping, and reverse-only portions of the merged read. That variation interferes with the denoising algorithm, and therefore greater accuracy can be achieved by denoising before merging, albeit at some computational cost.
Filtering
fastqFilter() implements filtering of fastq files that largely recapitulates the usearch fastq_filter command (http://www.drive5.com/usearch/manual/cmd_fastq_filter.html). In short, this function trims sequences to a specified length, removes sequences shorter than that length, and filters based on the number of ambiguous bases, a minimum quality score, and the expected errors in a read18. fastqPairedFilter() implements the same trimming and filtering, but applies it to paired reads jointly, only outputting reads where both the forward and reverse reads pass the filter.
Dereplication
derepFastq() imports a fastq file and outputs a dereplicated list of unique sequences and their abundances. derepFastq() also outputs consensus positional quality scores for each unique sequence by taking the average (mean) of the positional qualities of the component reads. These consensus scores are used by the error model of the dada() function.
Denoising
dada() implements the core denoising algorithm described above.
Chimeras
isBimeraDenovo() identifies sequences that are exact bimeras (two-parent chimeras) of more abundant output sequences. Bimeras are identified by performing a Needleman-Wunsch global alignment of each sequence to all more abundant sequences, and then searching for combinations of a left-parent and a right-parent that cover the child sequence without any mismatches or internal indels. Child sequences that differ by a single mismatch or indel from the chimeric model are also flagged if the left-parent and right-parent are both at least 4-away from the child sequence.
isBimeraDenovo() is intended to be used after denoising, and on exactly inferred sample sequences, rather than on noisy input reads or fuzzy OTUs. It was necessary to implement isBimeraDenovo() because most current stand-alone chimera identification programs are intentionally conservative about identifying chimeras that are relatively close to other more abundant sequences, because such sequences are expected to be later joined together in the same OTU (Supplementary Note 2). DADA2 does not create OTUs, and does differentiate closely related sequence variants, therefore we implemented this simple, but more sensitive, chimera detection method.
Merging
mergePairs() performs a global ends-free alignment between paired forward and reverse reads, and merges them together if they exactly overlap. mergePairs() requires that the input forward and reverse reads read in by derepFastq() were in the same order, a feature which is maintained by fastqPairedFilter(). Note that merging in the DADA2 pipeline happens after denoising, hence the strict requirement of exact overlap since it is expected that nearly all substitution errors have already been removed.
Benchmarking
Test datasets
The Balanced community consists of 57 bacteria and archaea from a broad range of habitats. The 16S rRNA gene sequences of most of these strains were well separated (>3%) over the region sequenced. However, the sequences of 5 strains were identical to other more abundant strains, while 5 strains had a total of 7 additional distinguishable sequence variants in their genomes that differed by 1 or 2 nucleotides. There were also two strains that were less than 3% different from more abundant strains. The Balanced dataset was downloaded from http://www.ebi.ac.uk/ena/data/view/PRJEB6244, and its construction was described in [16] where it is identified as dataset DS 35.
The HMP community consists almost entirely of strains well separated (>3%) over the region sequenced (S. epidermis and S. aureus are indistinguishable), most of which colonize the human body. The HMP dataset was downloaded from http://www.mothur.org/MiSeqDevelopmentData.html, and its construction was described in [17] where it is identified as the MOCK1 sample in run 130403. This dataset was also analyzed in [18].
The Extreme dataset was generated for this study. The organisms for the Extreme community include human gastrointestinal tract bacterial isolates (Supplementary Table 2). The Extreme dataset was intended to include more fine-scale variation than the other mock communities, the members of which were chosen in part for their well-separated 16S rRNA gene sequences. The Extreme strains are all distinguishable over the sequenced region of the 16S rRNA gene, but some pairs of strains differ by as little as 1 nucleotide. The Extreme dataset is available under SRA accession number SRX1478507.
Extreme strains were grown overnight in liquid broth with the medium recommended from the source culture collection for each respective strain (Table 1). An aliquot of the bacterial culture was used to directly amplify the 16S rRNA gene. One microliter of the bacterial culture was used as template to amplify the V4 region of the 16S rRNA gene using fusion gene primers (515f/806r) that incorporate Illumina adapter sequences and indexing barcodes22. The PCR reaction was carried out in a 25 uL mixture containing 1× HotMaster Mix with 2.5 mM Mg2+ (5 PRIME, Gaithersburg, MD), 400 nM forward primer, 400 nM reverse primer, along with the bacterial culture template. The following cycling parameters were used: initial cell lysis and DNA denaturing at 95$^\circ$C for 10 minutes, followed by 30 cycles of 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 30 seconds, ending with a final annealing step at 72°C for 10 min. PCR amplicons were cleaned using Agencourt AMPure XP beads (Beckman Coulter, Pasadena, CA) following the manufacturer's instructions. Cleaned PCR amplicons were analyzed and quantified using an Agilent 2100 Bioanalyzer.
Strains were grouped into two taxonomic groups, Firmicutes and Bacteroidetes. Within each taxonomic group, strains were designated for one of six 10-fold dilution groups (Supplementary Table 2). PCR amplicons for each strain were first normalized to the same concentration. From there, each amplicon was individually diluted to its respective dilution level and then all amplicons were pooled. The concentration of the pooled library was quantified using the Quant-iT PicoGreen dsDNA Assay kit (Life Technologies, Carlsbad, CA) and analyzed on an Agilent 2100 Bioanalyzer. The pooled library was diluted to 4 nM and then Illumina's protocol for preparing libraries for sequencing on the MiSeq was followed. The final concentration of the library was diluted to 6 pM with ~20% PhiX spiked in to account for the low base-diversity library. The final pooled library was sequenced on an Illumina MiSeq with a MiSeq Sequencing Reagent Kit v3 to obtain 250 bp paired end reads utilizing custom sequencing primers as described in [22].
Workflow on test data
A common filtering and trimming was performed before applying each method: The DADA2 fastqPairedFilter (paired reads) and fastqFilter (forward reads only) functions were used to remove sequences with Ns or more than two expected errors18, and to trim the first 20 nucleotides and the last 10 nucleotides (forward reads) or 10–50 nucleotides (reverse reads) depending on the quality profile of the data.
The usearch command fastq_mergepairs with a minimum overlap of 20 bases and maximum differences of 1 was used to merge the filtered forward and reverse reads for further analysis by UPARSE, MED and QIIME. Mothur used its native read merging function make.contigs. DADA2 denoised the forward and reverse reads independently, and then merged them with its mergePairs function.
Chimeras were removed from the denoised output of DADA2 and MED by isBimeraDenovo in the DADA2 R package, as this tool is intended for the exactly inferred sequences output by these methods. UPARSE has built-in chimera removal. The uchime method included in mothur and QIIME was used to remove chimeras for those pipelines23.
A list of output sequences and associated abundances was obtained for each algorithm. For DADA2 these were the inferred sample sequences, for UPARSE, mothur and QIIME the representative OTU sequences, and for MED the representative sequences of its “nodes”.
We also removed singleton OTUs from the outputs of mothur and QIIME. The DADA2, UPARSE and MED pipelines all decline to call singleton variants, so removing singletons from mothur and QIIME allows a cleaner comparison between methods. Additionally, nearly all of the singleton variants output by mothur and QIIME were spurious, so removing singletons improved their reported accuracy.
Software versions: DADA2 version 0.99.8, usearch version 8.1.1831 (implements UPARSE), MED version 2.0, mothur version 1.36.1, QIIME version 1.9.1.
Specificity
Output sequences were first compared to the known 16S rRNA gene reference sequences of the members of each mock community. If an output sequence matched a reference sequences, it was classified as Reference, and if it had one mismatch or gap to a reference sequence it was classified as One Off. Output sequences that were at least Hamming distance 2 from any reference sequence were then BLASTed against the nr/nt database. If the best hit was an exact match covering the full output sequence, it was classified Exact. If there was a single mismatch or indel, it was classified One Off. Output sequences that remained unclassified to this point were classified Other.
We included the BLAST against nr/nt step because even amplicon sequencing data from communities with a putatively known reference composition will contain contaminant sequences. Contaminants are real, albeit unwanted, biological variation, and should be identified when correcting amplicon errors. While the nr/nt database is imperfect, it is reasonable to expect that Exact matches are far more likely to be real variants than are Others. Output sequences classified as Other, and output sequences classified as One Off that differed by one substitution from a more abundant output sequence, were considered a proxy for false positives. Output sequences classified as Reference or Exact were considered true positives.
Sensitivity
We compiled the 16S rRNA gene sequences (reference sequences) for the intended members of each mock community (reference strains). The presence of each reference strain was confirmed by checking that at least one read matching one of its 16S rRNA gene sequences was present in the filtered dataset. If no such read existed, that strain was removed from the reference list.
Output sequences were compared to the list of reference sequences. If any output sequence matched any 16S rRNA gene sequence present in a strain, that reference strain was considered to have been identified.
Time Benchmarking
When benchmarking computational time, we attempted to isolate the core sample inference algorithms for each pipeline as much as possible. Thus, for the time benchmarking we applied each algorithm to an identically prepared set of sequences: the filtered forward-only reads from the Balanced dataset including singletons. We did not include chimera identification in this benchmarking. The specific commands benchmarked are listed at the end of the Balanced workflow.
Note that preprocessing steps, such as discarding singletons and reference-based chimera removal, can substantially reduce subsequent computation time for all of these methods.
Analysis of Vaginal and Fecal Samples
The 16S rRNA gene amplicon data from human vaginal samples in [19] (2.13M paired-end Illumina Miseq reads in 157 samples) and from mouse feces in [17] (3.65M paired-end Illumina Miseq reads in 362 samples) were analyzed with the DADA2 pipeline outlined above. First the demultiplexed fastq files were filtered and trimmed in the same manner as the test datasets. Each sample was dereplicated, a portion of the dataset was used to estimate the error parameters, and dada() was applied to the full pooled dataset using those inferred error parameters. isBimeraDenovo() was used to remove chimeras.
For the human vaginal samples, output sequences that appeared in at least two samples and at least 0.3% of the total reads were taxonomically identified by BLAST. Further analysis focused on the six L. crispatus sequence variants identified by this procedure.
Supplementary Material
Acknowledgments
We thank M Schirmer and D MacIntyre for productive correspondence. This work was supported by the NSF (DMS-1162538 to SH), the NIH (R01AI112401 to SH) and the Samarth Foundation (Stanford Microbiome Seed Grant to BC and SH).
Footnotes
Accession Codes
The Extreme dataset is available from the SRA under accession number SRX1478507 (http://www.ncbi.nlm.nih.gov/sra/SRX1478507).
Author Contributions
BJC and SPH designed the research; BJC, PJM and MJR implemented the algorithm; BJC performed the analysis; BJC, PJM, MJR and SPH wrote the paper; AWH and AAJ generated the Extreme dataset designed by BJC, PJM, and AWH.
Code availability
The DADA2 R package is open source and available on github (https://github.com/benjjneb/dada2). Package binaries are available through Bioconductor as of its 3.3 release. R markdown files implementing the benchmarking on the Balanced, HMP and Extreme datasets, as well as the analyses of the human vaginal and mouse fecal samples are available from the Stanford Digital Repository: https://purl.stanford.edu/mh194vj6733.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Human Microbiome Project Consortium. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosen MJ, Davison M, Bhaya D, Fisher DS. Science. 2015;348:1019–1023. doi: 10.1126/science.aaa4456. [DOI] [PubMed] [Google Scholar]
- 3.Reeder J, Knight R. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. BMC Bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosen MJ, Callahan BJ, Fisher DS, Holmes SP. BMC Bioinformatics. 2012;13:283. doi: 10.1186/1471-2105-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bragg L, Stone G, Imelfort M, Hugenholtz P, Tyson GW. Nat Methods. 2012;9:425–426. doi: 10.1038/nmeth.1990. [DOI] [PubMed] [Google Scholar]
- 7.Schloss PD, et al. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporaso JG, et al. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar RC. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 10.Eren AM, Borisy GG, Huse SM, Welch JLM. Proc Natl Acad Sci USA. 2014;111:E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. ISME J. 2015;9:968–979. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tikhonov M, Leach RW, Wingreen NS. ISME J. 2015;9:68–80. doi: 10.1038/ismej.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Genome Research. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McElroy K, Zagordi O, Bull R, Luciani F, Beerenwinkel N. BMC Genomics. 2013;14:501. doi: 10.1186/1471-2164-14-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarner F. Nat Rev Gastroenterol Hepatol. 2014;11:647–649. doi: 10.1038/nrgastro.2014.156. [DOI] [PubMed] [Google Scholar]
- 16.Schirmer M, et al. Nucleic Acids Res. 2015;43:e37. doi: 10.1093/nar/gku1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC, Flyvbjerg H. Bioinformatics. 2015;31:3476–3482. doi: 10.1093/bioinformatics/btv401. [DOI] [PubMed] [Google Scholar]
- 19.MacIntyre DA, et al. Sci Rep. 2015;5:8988. doi: 10.1038/srep08988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravel J, et al. Proc Natl Acad Sci USA. 2011;108(Supplement 1):4680–4687. [Google Scholar]
Methods-only References
- 21.Sun Y, et al. ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res. 2009;37:e76. doi: 10.1093/nar/gkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, et al. ISME J. 2012;6:1621. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.