Abstract
Light-mediated control of protein-protein interactions to regulate metabolic pathways is an important approach of optogenetics. Here, we report the first optogenetic system based on a reversible light-induced binding between a bacterial phytochrome BphP1 and its natural partner PpsR2 from Rhodopseudomonas palustris bacteria. We extensively characterized the BphP1–PpsR2 interaction both in vitro and in mammalian cells, and then used it to translocate target proteins to specific cellular compartments, such as plasma membrane and nucleus. Applying this approach we achieved a light-control of cell morphology resulting in the substantial increase of cell area. We next demonstrated the light-induced gene expression with the 40-fold contrast in cultured cells, 32-fold subcutaneously and 5.7-fold in deep tissues in mice. The unique characteristics of the BphP1–PpsR2 optogenetic system are its sensitivity to 740–780 nm near-infrared light, ability to utilize an endogenous biliverdin chromophore in eukaryotes including mammals, and spectral compatibility with blue-light optogenetic systems.
Spatiotemporal control of biochemical processes in cells and animals should advance basic biology and biomedicine. A promising approach for this is optogenetics. Among several directions of optogenetics, the important one is a modulation of cellular protein-protein interactions (PPIs) using light. Light-dependent genetically encoded systems exhibit several advantages over pharmacologically-inducible PPI approaches due to their non-invasiveness, high PPI activation rates, reversibility and absence of side effects caused by drugs.
Several PPI optogenetic systems1 were engineered from different photoreceptor families including LOV (light/oxygen/voltage) domains2, 3, BLUF (blue light utilizing FAD) domains4, cryptochromes5, 6 and plant phytochromes7–9. Although the use of these systems in cultured mammalian cells was shown, their application in mammals can be complicated. First, a wavelengths range where light exhibits maximum depth of tissue penetration lies between 650 and 900 nm, and is called a near infra-red (NIR) tissue transparency window10. However, among the photoreceptor families only phytochromes are activated with far-red or NIR light. Second, different phytochrome subclasses incorporate different tetrapyrrole chromophores. Phytochromobilin and phycocyanobilin serve as chromophores in plant phytochromes, whereas bacterial phytochromes incorporate biliverdin IXα (BV)11,12. Among these tetrapyrroles, BV exhibits the most NIR-shifted absorbance spectra. Third, plant phytochrome-based optogenetic systems require an exogenous supply of phycocyanobilin13. In contrast, the BV chromophore, utilized by bacterial phytochromes, is abundant in eukaryotic cells14. This feature was used to engineer bacterial phytochromes into several types of NIR probes for mammalian tissues15 including constitutively fluorescent proteins16, 17, photoactivatable fluorescent proteins18 and PPI reporters19.
Bacterial phytochromes control gene expression and second messenger signaling in bacteria in response to NIR light20. They consist of a photosensory core module and an output effector module. Natural output modules can be replaced with phosphodiesterase and adenylate cyclase enabling to manipulate cyclic nucleotides in mammalian cells21–23. Spectral properties of bacterial phytochromes are defined by a photosensory core module where the protein-chromophore interaction occurs11, 24, 25. Within the chromophore-binding pocket BV can adopt two conformational states, Pr and Pfr, which absorb far-red and NIR light, respectively. Most of bacterial phytochromes undergo photoconversion from a Pr state to the Pfr state with 660–700 nm light. However, there is a small group, termed bathyphytochromes, which adopts Pfr as a ground state and undergoes Pfr→Pr photoconversion with 740–780 nm light26. Bacterial phytochromes can be converted back to a ground state either with light or through thermal relaxation in darkness.
It has been proposed that a bacterial bathyphytochrome RpBphP1 (BphP1) from Rhodopseudomonas palustris performs its signaling function by interacting with a transcriptional repressor RpPpsR2 upon NIR illumination27. The effector module of BphP1 consists of two domains, PAS/PAC (PAS domain with additional C-terminal residues) and HOS (2-helix output sensor), with no detectable enzymatic activity. While a light-induced BphP1–RpPpsR2 heterodimerization was observed by gel filtration, it was not characterized further28.
In this paper, we study the light-activatable interaction of BphP1 with RpPpsR2 in vitro and in vivo. We demonstrate recruitment of one of the interacting partners to specific cellular compartments. We utilize this feature to induce intracellular signaling and to control gene expression. We show that the BphP1–RpPpsR2 pair represents a novel PPI-inducing NIR optogenetic system, which utilizes a bacterial phytochrome, its binding partner and an endogenous BV chromophore.
RESULTS
Properties of purified BphP1
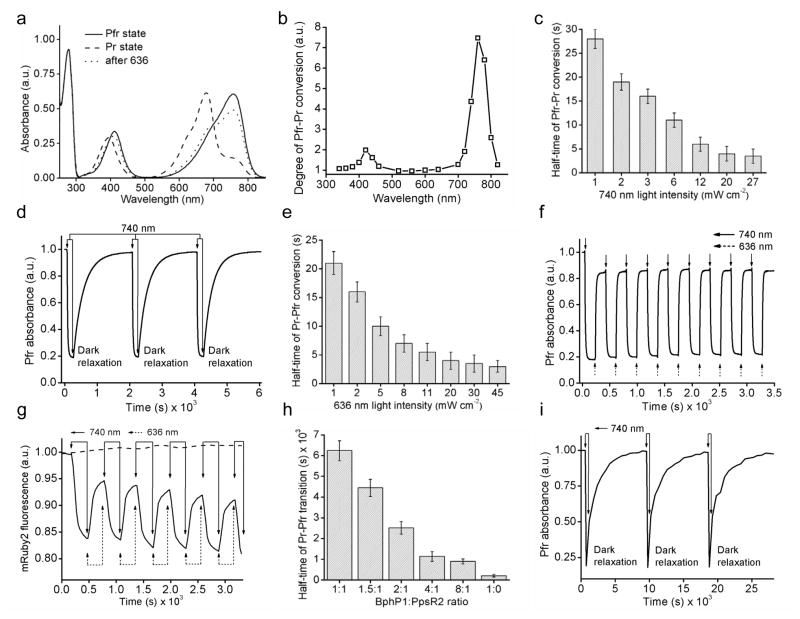
In a ground Pfr state, BphP1 has absorbance at 412 nm (Soret band) and 756 nm (Q band) (Fig. 1a). Upon NIR illumination it photoconverts into the Pr state with absorbance at 390 nm and 678 nm for Soret and Q bands, respectively. A 4.3-fold decrease in absorbance was observed at 756 nm upon 740/25 nm irradiation (Supplementary Table 1). The highest light sensitivity of the Pfr state was observed at ~740–780 nm, with notable BphP1 photoconversion at 800 nm (Fig. 1b). Kinetics of the Pfr→Pr photoconversion was monoexponential (Supplementary Fig. 1a). A half-time of the Pfr→Pr transition was 28 s at 1 mW cm−2 and then decreased to 3.5 s at 27 mW cm−2 (Fig. 1c). After the Pfr→Pr photoconversion, BphP1 returned to the ground state in darkness with a half-time of 170 s. Several cycles of the Pfr→Pr photoconversion followed by dark relaxation back to the Pfr state were shown (Fig. 1d). Irradiation with 636 nm light restored the Pfr absorbance to ~80% (Fig. 1a). The remaining ~20% were restored during dark relaxation (Supplementary Fig. 1b). A half-time of the Pfr→Pr conversion depended on the 636 nm light intensity, ranging from 21 s at 1 mW cm−2 to 3.0 s at 45 mW cm−2 (Fig. 1e). Many cycles of photoswitching without notable changes in absorbance were observed (Fig. 1f).
Figure 1. Spectral properties of BphP1 and characterizations of BphP1–PpsR2 interaction in vitro.
(a) Absorbance spectra in Pfr state before (solid line), after photoconversion to Pr state with 740/25 nm (dashed line) and after conversion back to Pfr state with 636/20 nm (dotted line). (b) Action spectrum of Pfr→Pr photoconversion measured as the relative decrease of Pfr absorbance detected at 780 nm upon irradiation with light of specific wavelength. (c) Dependence of half-time of light induced Pfr→Pr photoconversion on intensity of 740/25 nm light (n=3, error bars are s.e.m.). (d) Reversible absorbance BphP1 in Pfr state with 740/25 nm light followed by dark relaxation. (e) Dependence of half-time of light induced BphP1 Pr→Pfr photoconversion on intensity of 636/20 nm light (n=3, error bars are s.e.m.). (f) Reversible absorbance BphP1 in Pfr state turned off with 740/25 nm light and then turned on with 636/20 nm light. Absorbance in (b), (d) and (f) was measured at 780 nm. (g) Time-course of FRET changes during BphP1 photoswitching between the Pr and Pfr states either together with PpsR2-mRuby2 (sold line) or together with mRuby2 control (dashed line). Solid arrows correspond to 740/25 nm illumination. Dashed arrows correspond to 636/20 nm illumination. (h) Half-time of dark relaxation of BphP1 (Pr→Pfr transition) in presence of various quantities of PpsR2-mRuby2 (n=3, error bars are s.e.m.). (i) Reversible dark relaxation cycles from the Pr to the Pfr state of the BphP1 and PpsR2 mixture with 8:1 ratio. Arrows correspond to 740/25 nm illumination.
BphP1–PpsR2 interaction in vitro
RpPpsR2 has a single cysteine residue in position 439 and forms a noncovalent homodimer. To exclude possibility of a disulfide bond formation, cysteine 439 was substituted with serine. This RpPpsR2/C439S variant was termed PpsR2. We used a FRET to characterize the BphP1–PpsR2 interaction. For this we fused mRuby2 to a C-terminus of PpsR2. mRuby2 emission and BphP1 absorbance in the Pr state have a good overlap (Supplementary Fig. 2a), resulting in quenching of mRuby2 fluorescence (Supplementary Fig. 2b). 740 nm light caused an increase in the Pr state and up to 12% decrease in mRuby2 fluorescence resulted from the BphP1–PpsR2-mRuby2 binding. A 27-fold change in 740 nm light intensity did not cause changes in the kinetics of the mRuby2 fluorescence decrease (Supplementary Fig. 2c). For all intensities the fluorescence quenching exhibited half-time of ~60 s, which was larger than observed for BphP1 alone (Fig. 1c). This suggested that PpsR2 did not bind BphP1 in the Pfr state, and that BphP1–PpsR2 interaction was not limited by rate of Pfr→Pr photoconversion but rather by BphP1 and/or PpsR2 protein structural changes involved in the interaction.
Using 740 nm for Pfr→Pr and 636 nm for Pr→Pfr photoconversion we observed several cycles of FRET changes (Fig. 1g) with half-times of ~60 s and ~30 s, respectively (Supplementary Table 1). The observed incomplete Pr→Pfr switching with 636 nm light possibly was caused by overlap of the Pr and Pfr spectra at this wavelength (Fig. 1a). Dark relaxation in the BphP1–PpsR2 complex was slower than photoswitching with 636 nm. It depended on the BphP1:PpsR2 ratio, with shorter half-times of Pr→Pfr dark relaxation detected using Pfr absorbance corresponded to the larger BphP1 concentrations (Fig. 1h). The dark relaxation half-time was 900 s (Supplementary Table 1). This suggested that the PpsR2 binding to BphP1 substantially slowed the Pr→Pfr relaxation and BphP1–PpsR2 dissociation in darkness. After complete dark relaxation the complexes can be formed again by 740 nm light illuminating (Fig. 1i).
BphP1–PpsR2 interaction in mammalian cells
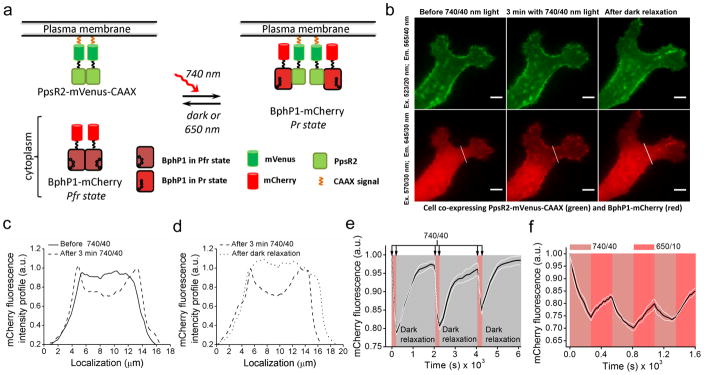
To study the BphP1–PpsR2 interaction in mammalian cells we used a translocation assay (Fig. 2a). PpsR2 was fused with mVenus followed by a -CAAX plasma membrane localization signal, and BphP1 was fused with mCherry for cytoplasmic expression. Excitation of these fluorescent proteins did not photoconvert BphP1 (Supplementary Fig. 3). Illumination of HeLa cells with 740 nm light caused a translocation of BphP1-mCherry to plasma membrane and decrease of mCherry cytoplasmic intensity (Fig. 2b,c). With epifluorescence microscope we observed ~25% decrease of mCherry fluorescence in cytoplasm after 3 min of 740 nm illumination (Fig. 2c). Subsequent dark incubation resulted in restoring of BphP1-mCherry fluorescence in cytoplasm, reaching the original level in ~24 min (Fig. 2b,d). Reversibility of the BphP1 translocation to plasma membrane and back to cytoplasm was shown for 3 cycles of 3 min of 740 nm illumination followed by 30 min of dark relaxation (Fig. 2e), with recovering of ~95% of the initial mCherry cytoplasmic intensity. Then we studied effect of 650 nm light on acceleration of the BphP1 dissociation from plasma membrane. 650 nm illumination resulted in 1.6-fold higher initial rates of the BphP1-mCherry cytoplasmic signal recovery as compared to that in darkness (Supplementary Fig. 4). However, the complete dissociation of the BphP1–PpsR2 complexes required darkness. Illumination with 740 nm and 650 nm light for 3 min each resulted in ~8% reversible changes of the BphP1-mCherry cytoplasmic signal after the initial 25% decrease (Fig. 2f), indicating that one third of the BphP1–PpsR2 complexes could be modulated with light.
Figure 2. Re-localization of BphP1 to plasma membrane induced by light.
(a) Chart depicting a light-induced interaction between cytoplasmic BphP1-mCherry and membrane-bound PpsR2-mVenus-CAAX. (b) Fluorescence images of the representative HeLa cell co-expressing PpsR2-mVenus-CAAX (green) and BphP1-mCherry (red) before illumination (left), after 3 min of 740/40 nm illumination with 0.9 mW cm−2 (middle), and after 20 min of dark relaxation (right). Bars, 10 μm. (c) Intensity profile of BphP1-mCherry fluorescence of the cell in (b) marked with a white line before (solid line) and after (dashed line) 3 min of 740/40 nm illumination. (d) Intensity profile of BphP1-mCherry fluorescence of the cell in (b) marked with a white line after 3 min of 740/40 nm illumination (dashed line) and after subsequent 24 min in darkness (dotted line). (e) Time-course of BphP1-mCherry fluorescence intensity in cytoplasm during three cycles of 3 min of 740/40 nm irradiation with 0.2 mW cm−2 followed by 30 min in darkness (n=5; white lines represent mean ±s.e.m.). mCherry fluorescence was measured every 15 s during 740/40 nm light illumination and every 180 s during dark relaxation. (f) Time-course of the BphP1-mCherry fluorescence intensity in cytoplasm during three cycles of 3 min of 740/40 nm irradiation with 0.2 mW cm−2 followed by 3 min of 650/10 nm irradiation with 0.35 mW cm−2 (n=5; white lines represent mean ±s.e.m.). mCherry fluorescence was measured every 15 s. All imaging was performed at 37°C using an epifluorescence microscope. Light intensities were measured at the back focal plane of a 60×1.35 NA objective lens.
Light-induced activation of signaling pathway
We next utilized the BphP1–PpsR2 interaction for recruitment of a DHPH domain of intersectin-129 to plasma membrane for activation of small GTPase Cdc4230, 31. DHPH domain was fused to a C-terminus of BphP1-mCherry and co-expressed in HeLa cells with PpsR2-mVenus-CAAX (Fig. 3a). 740 nm illumination caused changes of cell morphology and gradual increase of a cell area with a plateau after ~30 min (Fig. 3b). No notable changes were detected in cells co-transfected with BphP1-mCherry-DHPH and mVenus-CAAX (Fig. 3c). The area increase reached 50% in some cells, with the average increase of ~25% (Fig. 3d). The initial relative decrease of BphP1-mCherry-DHPH in cytoplasm was 2-fold higher than the cell area increase, indicating that BphP1-mCherry-DHPH was translocated mainly during the first 5 min of 740 nm irradiation. The further changes were rather similar for mCherry fluorescence and cell area, suggesting that they were caused by an increase in the cell area (Supplementary Fig. 5).
Figure 3. Light-induction of cellular cytoskeletal re-arrangements.
(a) Chart depicting a light-induced recruitment of cytoplasmic BphP1-mCherry-DHPH to membrane-bound PpsR2-mVenus-CAAX that results in cytoskeletal rearrangements of mammalian cells. (b, c) Fluorescence images of the representative HeLa cells co-expressing either (b) BphP1-mCherry-DHPH (red) and PpsR2-mVenus-CAAX (green) or (c) BphP1-mCherry-DHPH (red) and mVenus-CAAX control (green) before (top) and after (bottom) 30 min irradiation with 740/40 nm light of 0.2 mW cm−2 (first 3 min continuous irradiation with 740/40 with following 27 min of pulse illumination 15 s On 45 s Off). Bars, 10 μm. (d) Time dependent changes in cellular areas of HeLa cells, which co-express either BphP1-mCherry-DHPH and PpsR2-mVenus-CAAX (n=5; error bars are s.e.m.), or BphP1-mCherry-DHPH and mVenus-CAAX control (n=5; error bars are s.e.m.), during irradiation with 740/40 nm light of 0.2 mW cm−2 (first 3 min continuous irradiation with 740/40 with following 27 min of pulsed illumination 15 s On and 45 s Off). Fluorescent images were taken every 15 s during continuous and every 60 s during pulsed irradiation with 740/40 nm. All imaging was performed at 37°C using an epifluorescence microscope. The light power densities were measured at the back focal plane of a 60×1.35 NA objective lens.
BphP1 recruitment to cell nucleus
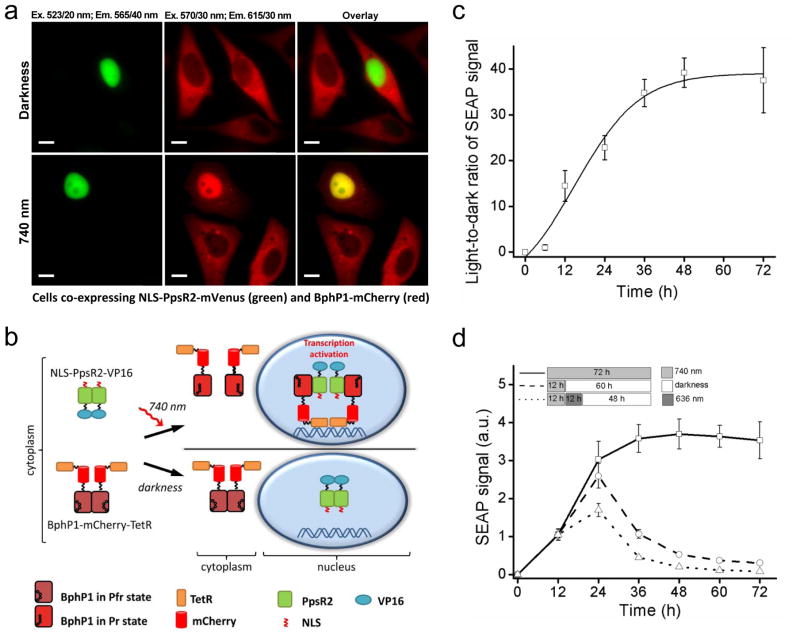
HeLa cells stably expressing BphP1-mCherry were transfected with PpsR2-mVenus having a nuclear localization signal (NLS). In darkness NLS-PpsR2-mVenus was localized to nucleus and BphP1-mCherry stayed in cytoplasm (Fig. 4a), with a mCherry intensity ratio between the nucleus and cytoplasm of ~0.5 (Supplementary Fig. 6). 740 nm illumination caused an increase of BphP1-mCherry fluorescence in nucleus (Fig. 4a), resulting in the nucleus/cytoplasm ratio of 1.8. Cells expressing BphP1-mCherry only had the nucleus/cytoplasm ratios in darkness and in light not exceeding 0.5 (Supplementary Fig. 6). Thus, the light-induced BphP1–PpsR2 interaction caused ~3.5-fold increase of the BphP1-mCherry signal in nuclei of the illuminated cells.
Figure 4. Recruitment of BphP1 to nucleus and light-induced transcription activation.
(a) Fluorescence images of representative HeLa cells co-expressing NLS-PpsR2-mVenus and BphP1-mCherry incubated either in darkness or under irradiation with 740/25 nm pulsed light (30 s On and 180 s Off) of 0.2 mW cm−2. Images were acquired at 37°C using an epifluorescence microscope. Bars, 10 μm. (b) Chart depicting the proposed systems for light-inducible transcription activation. NIR light converts BphP1 into Pr state and induces heterodimerization with PpsR2. Nuclear localization signal (NLS) fused with PpsR2 facilitates translocation of the heterodimer to nucleus where BphP1 fusions interact with tetO DNA repeats via fused TetR. VP16 fused with PpsR2 recruits transcription initiation complex and triggers transcription of a reporter gene. (c) Kinetics of a light-to-dark ratio of SEAP signal detected in culture media of HeLa cells bearing BphP1-mCherry-TetR co-transfected with NLS-PpsR2-VP16 producing plasmid and pTRE-Tight-SEAP (7×tetO) reporter plasmid after 48 h (n=3; error bars are s.e.m.). (d) Termination of SEAP transcription in HeLa cells with the same constructs as in (c) illuminated with 740/25 nm followed by 60 h of darkness or followed by 12 h of 636/25 nm illumination and then by 48 h of darkness. Data were normalized to SEAP signal of cells irradiated with 740/25 nm for 72 h; the signal from non-irradiated cells was subtracted (n=3; error bars are s.e.m.).
Light-inducible TetR-tetO transcription system
We applied light-induced recruitment of BphP1 to a nucleus to activate gene transcription using a tetracycline repressor-based system (Fig. 4b). We fused VP16 to a C-terminus of NLS-PpsR2 and tetracycline repressor (TetR) to BphP1-mCherry. HeLa cells stably expressing BphP1-mCherry-TetR were co-transfected with a plasmid encoding NLS-PpsR2-VP16 and a pTRE-Tight-SEAP plasmid containing 7×tetO upstream of the SEAP gene. 740 nm illumination increased the SEAP level and resulted in ~40-fold contrast over the darkness after 48 h (Supplementary Fig. 7a,b). Time course of SEAP production revealed ~3-fold SEAP increase after 12 h of 740 nm illumination comparing to dark-treated cells (Supplementary Fig. 7c). SEAP accumulation exhibited a half-time of ~18 h and reached a plateau after 48 h (Fig. 4c, Supplementary Fig. 7c). SEAP expression level depended on the power of activating light with SEAP signal observed in cells irradiated with as low as 0.05 mW cm−2 (Supplementary Fig. 8). Analogously, flow cytometry of light-induced EGFP expression from a pTRE-Tight-EGFP plasmid resulted in more than 27-fold higher EGFP signal in illuminated over dark-treated cells (Supplementary Fig. 9).
We next studied how fast the light-induced transcription could be terminated. We illuminated cells with 740 nm light for 12 h and then kept them in darkness. The SEAP reporter production increased ~2.6-fold during the first 12 h in darkness, likely due to pre-accumulation of SEAP mRNA, followed by its subsequent decrease with a half-time of ~8 h (Fig. 4d). We tested whether 636 nm light would accelerate the transcription termination. Indeed, 12 h illumination with 636 nm, following 12 h with 740 nm light, decreased the SEAP production ~2.3-fold comparing to the cells kept in darkness. Moreover, the SEAP decrease in the subsequent darkness also was ~2-fold faster (Fig. 4d). Similar to cell membrane re-localization (Fig. 2), the termination of gene transcription can be accelerated with 636 nm illumination, which causes dissociation of the BphP1-TetR and PpsR2-VP16 complexes in a nucleus.
Light-activation of gene expression in vivo
For activation of gene expression in vivo, stably expressing BphP1-mCherry-TetR HeLa cells were injected subcutaneously in the interscapulum area of FVB mice 24 h after co-transfection with NLS-PpsR2-VP16 and pTRE-Tight-Rluc8 plasmids. Then animals were either illuminated with 740 nm light or kept in darkness. After 48 h we detected a substantial increase of the Rluc8 signal in the illuminated mice (Fig. 5a) as compared to those kept in darkness. The observed 32-fold activation contrast (Fig. 5b) was rather similar to that obtained in cell culture experiments (Supplementary Fig. 7b).
Figure 5. Light-induced transcription activation in mice.
(a) Rluc8 bioluminescence detected in mice with subcutaneously injected HeLa cells stably expressing BphP1-mCherry-TetR and co-transfected with the NLS-PpsR2-VP16 producing plasmid and pTRE-Tight-Rluc8 reporter plasmid kept either in darkness (top) or illuminated with 740/25 nm light of 1 mW cm−2 (bottom) for 48 h. (b) Rluc8 signals detected in dark treated animals and in illuminated animals shown in (a) (n=3; error bars are s.e.m.). (c) Rluc8 signals in mice with subcutaneously injected HeLa cells co-transfected with pGAVPO plasmid encoding GAL4(65)-VVD-VP16 and pU5-Rluc8 reporter plasmid kept in darkness (top) or illuminated with 470/15 nm light of 1 mW cm−2 (bottom) for 48 h. (d) Rluc8 signals detected in the dark treated and illuminated animals shown in (c) (n=3; error bars are s.e.m.). (e) Rluc8 signals detected in mice after hydrodynamic co-transfection with pKA-207I10 (encoding NLS-PpsR2-VP16-IRESv10-BphP1-mCherry-VP16) (50 μg) and pTRE-Tight-Rluc8 (5 μg) plasmids. Mice kept in darkness (top) or illuminated with 740/25 nm light of 5 mW cm−2 (bottom) for 48 h. (f) Kinetics of the Rluc8 expression in mice shown in (e) kept in darkness or illuminated for 72 h (n=3; error bars are s.e.m.). (g) Rluc8 signals detected in mice after hydrodynamic co-transfection with pGAVPO (50 μg) and pU5-Rluc8 (5 μg) plasmids. Mice kept in darkness (top) or illuminated with 470/15 nm light of 5 mW cm−2 (bottom) for 24 h. (h) Kinetics of Rluc8 expression in mice shown in (g) kept in darkness or illuminated for 72 h (n=3; error bars are s.e.m.).
We next compared the BphP1 PpsR2 system with a blue light-activated LightON system32. In cultured cells, LightON activation with 470 nm light resulted in 42-fold increase of the Rluc8 signal over the dark treated cells (Supplementary Fig. 10). However, experiments with subcutaneously injected cells in FVB mice showed only 15-fold Rluc8 activation contrast (Fig. 5c,d). This 2.8-fold drop in the Rluc8 production in LightON system was likely caused by higher absorbance of 470 nm light than 740 nm light by ~1 mm thick tissue.
Activation of both systems in cultured cells with 470 nm and 740 nm light revealed their low cross-activation. We detected 12-fold higher activation of the LightON system with 470 nm light and 18-fold higher activation of the BphP1–PpsR2 system with 740 nm light (Supplementary Fig. 11).
For deep tissues, we studied kinetics of light-induced Rluc8 expression in mice after hydrodynamic transfection (Fig. 5e–h, Supplementary Fig. 12). After 24 h we detected an increase of the Rluc8 signal in livers of the 740 nm illuminated mice in comparison with the dark treated animals (Fig. 5f). After 48 h the Rluc8 signal reached its maximum with a 5.7-fold light-to-dark signal ratio (Fig. 5e,f). The large difference of the Rluc8 expression between the illuminated and dark-treated animals was observed up to 72 h after the transfection (Fig. 5f). To test the LightON system we hydrodynamically transfected mice with the high amounts of the LightON and reporter plasmids used in the original paper32 (Supplementary Fig. 12) or with the amounts similar to those of the BphP1–PpsR2 system (Fig. 5g,h). In the former case, after 470 nm illumination of mice for 24 h we observed 1.7-fold increase of the Rluc8 production as compared to the dark treated animals. In the latter case, the signal increase was 2.8-fold (Fig. 5g,h), which was twice lower than that detected for the BphP1–PpsR2 system (Fig. 5e,f). The Rluc8 ‘bell-shaped’ kinetics of the LightON activation in liver observed in both conditions was similar to that reported33.
DISCUSSION
We have developed a novel optogenetic system based on the light-inducible interaction of bacterial phytochrome BphP1 and its binding partner PpsR2 from R. palustris. This system takes an advantage of the high-sensitivity of bacterial phytochromes to NIR light and their unique feature to incorporate heme-derived BV as a chromophore.
The interaction of the BphP1–PpsR2 occurs upon Pfr→Pr photoconversion of BphP1. This photoconversion demonstrates high sensitivity to NIR 740–780 nm light and high rates of the Pfr→Pr transition. Characterization of BphP1–PpsR2 interaction in vitro and in mammalian cells showed its complete reversibility in darkness. Because far-red 636 nm light is absorbed by both the Pr and Pfr states it caused just a partial BphP1–PpsR2 dissociation (~80%), likely due to the incomplete Pr→Pfr photoconversion (Fig. 1a).
The BphP1–PpsR2 optogenetic system can be used for activation of several types of cellular processes. We showed the light-induced recruitment of cytoplasmic BphP1 to membrane-bound PpsR2. We utilized this approach to activate Cdc42 signaling pathway by translocating the DHPH domain of intersectin-129 to membrane that resulted in the increase of cell area and changes in cell morphology (Fig. 3). This increase was likely caused by the sustained activation of Cdc42 at lamellipodia and membrane ruffles34,35.
We extended the light-induced targeting approach to the recruitment of BphP1 to a nucleus and developed the transcription activation system. The light-induced PpsR2-VP16–BphP1-TetR interaction was used for transcription activation of reporter genes downstream of the 7×tetO. The system enabled the 40-fold increase in the reporter gene expression. The light-induced transcription was reversible in darkness. Far-red illumination accelerated the transcription termination 2-fold, allowing its more precise control. The NIR light-inducible transcription system should be transferable to model organisms, genetically constructed to utilize TetR-tetO36 interaction, to enable protein expression in spatiotemporal manner. This light-inducible TetR-based system should allow to avoid drawbacks associated with the tetracycline consumption by experimental animals37.
For in vivo applications, optogenetic systems sensing NIR light are preferable due to its deep-tissue penetration and low phototoxicity. Majority of the far-red optogenetic systems are based on a PPI between plant phytochrome PhyB and PIF6 factor8, 13, 38. They are activated with far-red light peaked at 660 nm, whereas BphP1–PpsR2 interaction is activated with NIR 740–780 nm light (Fig. 1b). Moreover, a use of the PhyB–PIF6 systems is limited by the requirement to either supply an exogenous phycocyanobilin chromophore11, 12 or to produce it by co-expressing several bacterial enzymes39, thus, affecting cellular metabolism.
We compared two similar TetR-tetO transcription activation systems, one based on the BphP1–PpsR2 pair and another on the PhyB–PIF6 pair13, using the same reporter plasmid. The reporter expression was 2-fold higher for the BphP1–PpsR2 than for PhyB–PIF6 interaction (Supplementary Fig. 13). Comparison of light propagation for 660 nm and 740 nm wavelengths in various mammalian tissues revealed a substantially more effective penetration of NIR light (Supplementary Note, Supplementary Fig. 14). This results in the slower decrease of the reporter expression level with the tissue depth for the BphP1–PpsR2 system as compared to the PhyB–PIF6 system (Supplementary Fig. 15).
In vivo comparison of the transcription activation by the blue-light driven LightON system and NIR-light activated BphP1 PpsR2 system revealed the higher efficiency of the latter one at light intensities of ≤5 mW cm−2 (Fig. 5), which compliant to safety regulations for permissible for eyes visible light power density40. These light intensities caused a limited increase of the Rluc8 signal in a deep-seated liver in the 470 nm illuminated animals as compared to the Rluc8 signal resulted from 90 mW cm−2 in the original LightON paper32.
In summary, we have characterized the BphP1–PpsR2 light-controllable PPI in vitro, in cells and animals and demonstrated the use of this light-sensitive pair as an optogenetic system for the subcellular protein targeting, induction of intracellular enzymatic activity, and activation of gene expression. The BphP1–PpsR2 system is orthogonal to mammalian cells and minimally interferes with their metabolism. Our results form a basis for development of the new generation of optogenetic systems, which can be used either solely or in combination with blue-light optogenetic tools (Supplementary Fig. 11). Since we used practically unmodified BphP1 and PpsR2 proteins, we expect that light sensitivity, kinetics and reversibility of their interaction will be further optimized using molecular engineering approaches.
ONLINE METHODS
Design of bacterial and mammalian plasmids
A RpBphP1 gene of R. palustris (NCBI Gene rpa1537) was kindly provided by E. Giraud (Institute for Research and Development, France). A RpPpsR2 gene of R. palustris (NCBI Gene rpa1536) was kindly provided by M. Papiz (Liverpool University, UK) and T. Beatty (University of British Columbia, Canada). A DHPH domain of human intersectin-1 was PCR amplified from a pAL189 plasmid (Addgene #22278)8. An mRuby2 gene was PCR amplified from a pcDNA3-mRuby2 plasmid (Addgene #40260)41 A transactivation domain of transactivating tegument protein VP16 from Herpes simplex was PCR amplified from a pGV-2ER plasmid (Systasy). A Rluc8 gene encoding modified luciferase from Renilla reniformis was PCR amplified from a Nano-lantern/pcDNA3 plasmid (Addgene #51970). A SEAP gene was PCR amplified from a pKM611 plasmid kindly provided by W. Weber (University of Freiburg, Germany). A plasmid pKM022 encoding PhyB (1-650) was obtained by mutagenesis, using the pKM020 plasmid as a template13. A TetR gene (residues 1-207) that binds DNA in the absence of tetracycline/doxycycline was PCR amplified from a pTet-Off vector (Clontech).
The reporter plasmids pTRE-Tight-EGFP, pTRE-Tight-SEAP and pTRE-Tight-Rluc8 were obtained by cloning of the EGFP, SEAP and Rluc8 genes, respectively, into a pTRE-Tight2 vector (Addgene #19407). The LightON system plasmids, pGAVPO (encoding GAL4(65)-VVD-VP16) and pU5-mCherry, were kindly provided by Y. Yang (East China University of Science and Technology, China)32. A pU5-Rluc8 reporter plasmid was generated by substituting of a mCherry gene in the pU5-mCherry plasmid with the Rluc8 gene. To develop stable preclonal cell mixtures of HeLa cells, plasmids encoding a transposase SB100X, pCMV(CAT)T7-SB100 (Addgene #34879), and transposon bearing plasmids pT2/SVNeo and pT2/BH (Addgene #26553 and #26556)42 were utilized. An IRESv10 with ~3-fold reduced expression strength43 was obtained from IRES using mutagenesis.
For bacterial expression of the BphP1, BphP1-mRuby2 and PpsR2-mRuby2 proteins, a pBAD/His-D, a pBAD/His-B (Life Technologies-Invitrogen) and a pET22b (Novagen) vectors were used, respectively. In the pET22b vector an N-terminal pelB signal was replaced with a Strep-tag-II.
Mammalian expression plasmids were based either on a pEGFP-N1 vector (Clontech), having either a standard CMV promoter or a truncated to CMVd1 promoter, or on a pFC15K vector (Promega) with the truncated CMVd1 promoter. The flexible linkers of 10 (-DSAGSAGSAG-), 16 (–SAGGSAGGSAGGSAGG-), 20 (-SAGGSAGGSAGGSAGGSAGG-) or 24 (-SGGGSGGGSGGGSGGGSGGGSGGG-) amino acids, a C-terminal membrane-localization -CAAX signal from Kras4B (-KKKKKKSKTKCVIM), a nuclear localization signal of nuclear cap-binding protein subunit 1 (MSRRRHSYENDGGQPHKRRK-), and a Strep-tag-II (-WSHPQFEK-) were added by oligonucleotide annealing. The designed plasmids are summarized in Supplementary Table 2.
Protein expression and purification
BphP1 and BphP1-mRuby2 proteins with polyhistidine tags on the N-terminus were expressed in LMG194 bacterial cells (Life Technologies-Invitrogen) containing a pWA23h plasmid encoding heme oxygenase for biliverdin synthesis in Escherichia coli18. The bacterial cells were grown in RM medium supplemented with ampicillin, kanamycin and 0.02% rhamnose for 6-8 h followed by an induction of the protein expression by adding of 0.002% arabinose. The proteins were purified using a Ni-NTA agarose (Qiagen). The PpsR2 and PpsR2-mVenus proteins with a Strep-tag-II at the N-terminus and a polyhistidine tag at the C-terminus was expressed in BL21(DE3) bacterial cells grown in LB medium supplemented with ampicillin for 6 h, followed by an induction of a protein expression with 250 μM IPTG. The proteins were first purified with a Ni-NTA agarose (Qiagen) followed by purification with a Strep-Tactin sepharose (IBA Lifesciences).
In vitro characterizations of BphP1 properties
For absorbance measurements, a Hitachi U-2000 spectrophotometer was used. For fluorescence measurements of mRuby2 fusions, a FluoroMax-3 spectrofluorometer (Horiba-Jobin Yvon) equipped with a 600 nm shortpass filter before detector was used. A photoconversion of BphP1 containing proteins was performed with 740/25 nm and 636/20 nm custom assemble LED sources in quartz microcuvettes (Starna Cells). A determination of action spectrum was performed by measurement of changing in absorbance of Pfr state BphP1 at 780 nm upon illumination with photoconversion light. As a source of light the FluoroMax-3 spectrofluorometer was used. The illumination time was normalized to total amount of irradiated light energy, measured with a PM100 optical powermeter equipped with a S130A sensor (ThorLabs) at the respective wavelength. A half-time of Pr→Pfr transition (or dark relaxation) was measured by registering of absorbance at 780 nm after 5 min illumination of samples with 740/25 nm. Samples containing fixed quantity of BphP1 (5 μM) and various quantities of PpsR2 (0-5 μM) were pre-incubated in darkness for 30 min. Reversible dark relaxation cycles were obtained by registration 780 nm absorbance of protein mixture at BphP1:PpsR2 molar ratio of 8:1. To study changes in mRuby2 fluorescence due to FRET between the PpsR2-mRuby2 fusion and the Pr state of BphP1, the proteins (2.5 μM each) were pre-incubated in darkness for 30 min and then transferred to a microcuvette. An mRuby2 fluorescence intensity was registered each 30 s with excitation 545/2 nm and emission 585/10 nm. All spectroscopic measurements were performed at a room temperature in PBS.
Mammalian cell culture
HeLa cells were purchased from ATCC (CCL-2 line) and were not additionally authenticated or tested for mycoplasma contamination. The cells were grown in DMEM medium supplemented with 10% FBS, penicillin-streptomycin mixture and 2 mM glutamine (all from Life Technologies-Invitrogen) at 37°C. For imaging, 105 cells were plated on a pre-coated with ECL mixture (EMD-Millipore) 35 mm glass-bottom culture dishes (MatTek). Transient cell transfections were performed using an Effectene reagent (Qiagen).
Preclonal mixtures of HeLa cells were obtained using the plasmid-based Sleeping Beauty transposon system. For this, the desired for integration into genome sequences were cloned into the transposon bearing plasmids pT2/BH or pT2/SVNeo and co-transfected with a plasmid encoding a hyperactive transposase SB100X. Cells were further selected with 700 μg/ml of G418 antibiotic for two weeks and enriched using a FACSAria sorter (BD Biosciences), resulting in the preclonal HeLa cell mixtures stably expressing BphP1-mCherry-TetR.
For light-induced re-localization to plasma membrane, HeLa cells were transiently co-transfected with pKA-140 or pKA-142 and pKA-141 plasmids in 1:1 ratio. For light-induced cytoskeletal rearrangement, cells were co-transfected with pKA-142 (or pKA-147) and pKA-144 plasmids in 1:2 ratio. The cell light-activation and imaging were typically performed 48 h after the transfection. For light-induced nuclear re-localization, HeLa cells stably expressing BphP1-mCherry-TetR were transiently transfected with pCMV-160. To study light-induced transcription activation in TetR-based system HeLa cells stably expressing BphP1-mCherry-TetR were co-transfected with pCMV-104 and either pTRE-Tight-EGFP, pTRE-Tight-SEAP or pTRE-Tight-Rluc8 plasmids in 5:1 ratio. To compare the PhyB–PIF6 system with the BphP1–PpsR2 system, HeLa cells were transfected with pKM022 and pTRE-Tight-SEAP plasmids in 2:1 ratio. All procedures after the transfection were performed as described in the PhyB–PIF6 paper13.
Cell light-activation and imaging
Imaging was performed using an Olympus IX81 inverted epifluorescence microscope equipped with a 200 W metal halide arc lamp (Lumen 220PRO, Prior) and a 60×1.35 NA oil immersion objective lens (UPlanSApo, Olympus). During imaging HeLa cells were incubated in a cell imaging medium (Life Technologies-Invitrogen) and kept at 37°C. Yellow (523/20 nm exciter and 565/40 nm emitter) and red (570/30 nm exciter and 615/30 nm emitter) channel filter sets (Chroma) were used for detection of mVenus and mCherry fluorescence, respectively. The Pfr→Pr photoconversion of BphP1 was done by illuminating with 740/40 nm filter (Chroma).
For Pfr→Pr re-localization assay, HeLa cells were exposed to 740/40 nm for 3 min while imaged every 15 s. An intensity of activation light was 0.9 mW cm-2. To quantify mCherry fluorescence in cell cytoplasm during BphP1 dark relaxation, HeLa cells were imaged every 3 min during 24 min, starting immediately after the Pfr→Pr photoconversion. Intensity profile of mCherry fluorescence through cell was determined using ImageJ v.1.46f software. Intensity of mCherry fluorescence in cytoplasm and nucleus was measured using SlideBook v.4.2.09.
To study light-induced cytoskeletal rearrangements, the Pfr→Pr photoconversion was done by illuminating cells using 740/40 nm filter for 3 min and imaged every 15 s, followed by further maintaining of BphP1 in the Pr state by illuminating with 740/40 nm for 15 s every minute and imaged every 60 s. An intensity of activation light was 0.2 mW cm-2. The total time of cells imaging was 30 min. The light power densities were measured at a back focal plane of a 60×1.35 NA objective lens. Membrane-localized mVenus was used for determination of cell area. The data were analyzed using ImageJ v.1.46f software.
Unless otherwise indicated, a light-induced transcription activation HeLa cells was performed with 740/25 nm LED source at 1.0 mW cm-2 using the 30 s On and 180 s Off cycle in CO2 incubator at 37°C. Time of illumination varied in different experiments. For nuclear localization and EGFP or SEAP transcription activation experiments, cells were illuminated for 24 and 48 h, respectively. For kinetic studies of SEAP accumulation in culture media, 3 different illumination regimes were used: 72 h of 740/25 nm; 12 h of 740/25 nm followed by 60 h of darkness; and 12 h of 740/25 nm followed by 12 h of 636/20 nm followed by 38 h of darkness. To measure light sensitivity of activation of SEAP expression, HeLa cells were irradiated with either 740/25 (with BphP1–PpsR2) or 660/20 nm (with PhyB–PIF6) light of various power densities (5-1000 μW cm-2). Light power densities and durations of illumination were converted to photons counts.
Light-induced LightON transcription activation of Rluc8 in HeLa cells was performed as described in the original paper32. In brief, HeLa cells were transfected with pGAVPO and pU5-Rluc8 plasmids in 1:1 ratio. Then cells were kept in darkness for 10 h. After change of culture medium, the cells were continuously illuminated with a 470/15 nm LED array (LuxeonStar) of 1 mW cm-2 or remained in a darkness for 48 h before analysis. For an analysis, the cells were resuspended in PBS and disrupted by freezing-thawing. An Rluc8 bioluminescence signal was measured in supernatants after adding of 5 μM h-coelenterazine (NanoLight Technology). A signal was detected using an IVIS Spectrum instrument (PerkinElmer/Caliper Life Sciences) and analyzed using Living Image 3.0 software (PerkinElmer/Caliper Life Sciences).
Flow cytometry analysis
Flow cytometry analysis of a light-induced EGFP expression was performed 48 h after the HeLa cell transfection using a LSRII flow cytometer (BD Biosciences) equipped with a 488 nm laser and a 530/40 nm emission filter and with a 561 nm laser and 610/20 nm emission filter. To calculate an efficiency of the light-induced EGFP expression, a mean fluorescent intensity of EGFP positive cells was multiplied by a number of the positive cells, resulting in the total amount of synthesized proteins. Gates for counting of EGFP positive cells were set using non-transfected cells as a negative control. Typically, the cell samples were triplicated. Minimally, 5x104 cells were analyzed per a sample. The data were analyzed using FACSDiva v. 6.1.3 and FlowJo v. 7.6.2 software.
Secreted alkaline phosphatase assay
For SEAP detection in culture media, a Great EscApe fluorescent SEAP Assay kit (Clontech) was used. 25 μl aliquots of cell culture media from wells of a 12-well plate were collected at each time point and stored at -20°C. For kinetics studies, a culture medium was changed with a fresh medium at each time point. A fluorescence intensity of the SEAP reaction product was measured using the SpectraMax-M2 plate reader (Molecular Devices).
Light-activation and imaging in mice
The FVB 2-3 month old female mice (National Cancer Institute, NIH) of 20-25 g body weight were used for in vivo comparison of the LightON and BphP1 PpsR2 systems. To compare an efficiency of the light-induced transcription activation, HeLa cells bearing either the LightON system or the BphP1 PpsR2 system were injected subcutaneously in the interscapular area of FVB mice. For better illumination the fur on the back was removed using a depilatory cream. For the LightON system, HeLa cells were co-transfected with the pGAVPO and pU5-Rluc8 plasmids in a 1:1 ratio, and for the BphP1 PpsR2 system, HeLa cells stably expressing BphP1-mCherry-TetR were co-transfected with the plasmid encoding NLS-PpsR2-VP16 and the pTRE-Tight-Rluc8 reporter plasmid in a 5:1 ratio. For both systems, the 3×106 HeLa cells in 100 μl of RPMI-1640 media supplemented with 2 mM L-glutamine were injected subcutaneously 24 h after the transfection. All manipulations with cells before and during the injection were performed under a 640 nm safelight for the LightON system and under a 530 nm safelight for the BphP1 PpsR2 system. After cells injection, mice were placed in transparent cages with the top illumination of either the 470/15 nm LED array or the 740/25 nm LED array, respectively. Intensities of 470/15 nm light and 740/25 nm light were the same and equal to 1 mW cm-2. Control mice after injection of the cells were kept in darkness in conventional cages. Animals were continuously illuminated or kept in darkness for 48 h, and every 12 h were released and fed for 30 min. Each experimental group contained 3 mice.
For hydrodynamic transfection into liver44, the Swiss Webster 2-3 month old female mice (National Cancer Institute, NIH) with body weight of 22-25 g were used. For the BphP1 PpsR2 system, 50 μg of the pKA-207I10 plasmid and 5 μg of the pTRE-Tight-Rluc8 reporter plasmid in 2.5 ml of PBS were intravenously injected through a tail vein. For hydrodynamic transfection by the LightON system, we used the plasmid amounts either indicated in the original paper32 (10 μg of the pGAVPO plasmid and 300 μg of the pU5-Rluc8 plasmid) or similar to those used for the BphP1–PpsR2 system (50 μg of the pGAVPO plasmid and 5 μg of the pU5-Rluc8 plasmid). The mice were placed in transparent cages and illuminated from the bottom with the 470/15 nm LED array and the 740/25 nm LED array, respectively. Intensity of the both activation light was the same and equal to 5 mW cm-2. For better illumination and imaging the belly fur was removed using a depilatory cream. Control animals were kept in conventional cages in complete darkness. Animals were continuously illuminated or kept in darkness for 72 h, and every 12 h were released and fed for 30 min. Each experimental group contained 3 mice.
For bioluminescence detection, 48 h after the HeLa cells injection or every 24 h after the hydrodynamic transfection of livers the animals were imaged using an IVIS Spectrum instrument (Perkin Elmer/Caliper Life Sciences) in luminescence mode with an open emission filter. Throughout the imaging, animals were maintained under anesthesia with 1.5% vaporized isoflurane. Prior to imaging, 80 μg of Inject-A-Lume CTZ native (NanoLight Technology) were intravenously injected through a retro-orbital vein. Data were analyzed using Living Image 3.0 software (Perkin Elmer/Caliper Life Sciences).
All animal experiments were performed in an AAALAC approved facility using protocols approved by the Albert Einstein College of Medicine Animal Usage Committee.
Supplementary Material
Acknowledgments
We thank E. Giraud (Institute for Research and Development, France), M. Papiz (Liverpool University, UK), T. Beatty (University of British Columbia, Canada), W. Weber (University of Freiburg, Germany), P. Hackett (University of Minnesota, USA), Z. Izsvak (Max Delbrück Center for Molecular Medicine, Germany) and Y. Yang (East China University of Science and Technology, China) for the plasmids and D. Shcherbakova and K. Chernov for the useful suggestions. This work was sponsored by the National Institutes of Health grants GM073913, GM108579 and CA164468 to V.V.V.
Footnotes
AUTHOR CONTRIBUTIONS
A.A.K. and A.A.S. characterized the proteins in vitro, in mammalian cells culture and in vivo. V.V.V. planned and directed the project and together with A.A.K. and A.A.S. designed the experiments, analyzed the data, and wrote the manuscript.
COMPETING INTEREST STATEMENT
There are no competing financial interests.
References
- 1.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu Rev Biochem. 2015;84:519–550. doi: 10.1146/annurev-biochem-060614-034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motta-Mena LB, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawano F, Suzuki H, Furuya A, Sato M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun. 2015;6:6256. doi: 10.1038/ncomms7256. [DOI] [PubMed] [Google Scholar]
- 4.Stierl M, et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem. 2011;286:1181–1188. doi: 10.1074/jbc.M110.185496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taslimi A, et al. An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun. 2014;5:4925. doi: 10.1038/ncomms5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S, et al. Reversible protein inactivation by optogenetic trapping in cells. Nat Methods. 2014;11:633–636. doi: 10.1038/nmeth.2940. [DOI] [PubMed] [Google Scholar]
- 7.Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 8.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez EJ, Gerhardt K, Judd J, Tabor JJ, Suh J. Light-Activated Nuclear Translocation of Adeno-Associated Virus Nanoparticles Using Phytochrome B for Enhanced, Tunable, and Spatially Programmable Gene Delivery. ACS Nano. 2016;10:225–237. doi: 10.1021/acsnano.5b05558. [DOI] [PubMed] [Google Scholar]
- 10.Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 11.Ulijasz AT, Vierstra RD. Phytochrome structure and photochemistry: recent advances toward a complete molecular picture. Curr Opin Plant Biol. 2011;14:498–506. doi: 10.1016/j.pbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Piatkevich KD, Subach FV, Verkhusha VV. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chemical Society reviews. 2013;42:3441–3452. doi: 10.1039/c3cs35458j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller K, et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 2013;41:e77. doi: 10.1093/nar/gkt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran MT, et al. In vivo image analysis using iRFP transgenic mice. Exp Anim. 2014;63:311–319. doi: 10.1538/expanim.63.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shcherbakova DM, Baloban M, Verkhusha VV. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr Opin Chem Biol. 2015;27:52–63. doi: 10.1016/j.cbpa.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nature methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shcherbakova DM, et al. Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins. Chem Biol. 2015;22:1540–1551. doi: 10.1016/j.chembiol.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piatkevich KD, Subach FV, Verkhusha VV. Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nature Commun. 2013;4:2153. doi: 10.1038/ncomms3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filonov GS, Verkhusha VV. A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chemistry & biology. 2013;20:1078–1086. doi: 10.1016/j.chembiol.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auldridge ME, Forest KT. Bacterial phytochromes: more than meets the light. Crit Rev Biochem Mol Biol. 2011;46:67–88. doi: 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- 21.Ryu MH, Gomelsky M. Near-infrared Light Responsive Synthetic c-di-GMP Module for Optogenetic Applications. ACS Synth Biol. 2014;3:802–810. doi: 10.1021/sb400182x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu MH, et al. Engineering adenylate cyclases regulated by near-infrared window light. Proc Natl Acad Sci U S A. 2014;111:10167–10172. doi: 10.1073/pnas.1324301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasser C, et al. Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc Natl Acad Sci U S A. 2014;111:8803–8808. doi: 10.1073/pnas.1321600111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner JR, Zhang J, Brunzelle JS, Vierstra RD, Forest KT. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. The Journal of biological chemistry. 2007;282:12298–12309. doi: 10.1074/jbc.M611824200. [DOI] [PubMed] [Google Scholar]
- 25.Rockwell NC, Lagarias JC. A brief history of phytochromes. Chemphyschem. 2010;11:1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rottwinkel G, Oberpichler I, Lamparter T. Bathy phytochromes in rhizobial soil bacteria. J Bacteriol. 2010;192:5124–5133. doi: 10.1128/JB.00672-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojadinovic M, et al. Dual role for a bacteriophytochrome in the bioenergetic control of Rhodopseudomonas palustris: enhancement of photosystem synthesis and limitation of respiration. Biochim Biophys Acta. 2008;1777:163–172. doi: 10.1016/j.bbabio.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Bellini D, Papiz MZ. Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure. 2012;20:1436–1446. doi: 10.1016/j.str.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Hussain NK, et al. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat Cell Biol. 2001;3:927–932. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- 30.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 31.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Li T, Wang X, Yang Y. A light-switchable bidirectional expression module allowing simultaneous regulation of multiple genes. Biochem Biophys Res Commun. 2015;465:769–776. doi: 10.1016/j.bbrc.2015.08.085. [DOI] [PubMed] [Google Scholar]
- 34.Kurokawa K, Itoh RE, Yoshizaki H, Nakamura YO, Matsuda M. Coactivation of Rac1 and Cdc42 at lamellipodia and membrane ruffles induced by epidermal growth factor. Mol Biol Cell. 2004;15:1003–1010. doi: 10.1091/mbc.E03-08-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guntas G, et al. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci U S A. 2015;112:112–117. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schönig K, Bujard H, Gossen M. The power of reversibility regulating gene activities via tetracycline-controlled transcription. Methods Enzymol. 2010;477:429–453. doi: 10.1016/S0076-6879(10)77022-1. [DOI] [PubMed] [Google Scholar]
- 37.Albanese C, Hulit J, Sakamaki T, Pestell RG. Recent advances in inducible expression in transgenic mice. Semin Cell Dev Biol. 2002;13:129–141. doi: 10.1016/s1084-9521(02)00021-6. [DOI] [PubMed] [Google Scholar]
- 38.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller K, et al. Synthesis of phycocyanobilin in mammalian cells. Chemical communications. 2013;49:8970–8972. doi: 10.1039/c3cc45065a. [DOI] [PubMed] [Google Scholar]
- 40.IEC 60825-1:2014, Safety of laser products, Part 1: Equipment classification and requirements, Edn. 3.0., International Electrotechnical Comission (2014).
- 41.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 43.Koh EY, et al. An internal ribosome entry site (IRES) mutant library for tuning expression level of multiple genes in mammalian cells. PLoS One. 2013;8:e82100. doi: 10.1371/journal.pone.0082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.