Abstract
The etiologic agent Trypanosoma cruzi (Tc) has been grouped into six discrete type units (DTU I-VI); within DTU-I exists four subgroups defined Ia-Id. In Colombia, the genotype Ia is associated with human infection and domiciliated Rhodnius vector. In the Yucatan Peninsula of Mexico, the main vector involved in T. cruzi transmission is Triatoma dimidiata predominantly via sylvatic and peridomiciliated cycles. In this study, multiple sequence analysis of mini-exon intergenic regions of T. cruzi isolates obtained from T. dimidiata in the Yucatan Peninsula of Mexico revealed they belonged to Tc Ia DTU along with two additional Mexican strains located 1,570 km away from Yucatan. In conclusion Tc Ia circulates in the Yucatan peninsula in T. dimidiata vector and likewise in the northwest region of Mexico.
Keywords: Trypanosoma cruzi, Spliced leader, Discrete type unit, Chagas disease
Introduction
Trypanosoma cruzi (Tc) has been grouped into six discrete type units (DTU). In Mexico, the most prevalent DTU using mini-exon sequences was reported belonging to TcI type (Bosseno et al. 2002; Ruíz-Sánchez et al. 2005). However, using serological means and ribosomal markers, other DTU have been recognized such as TcII- TcV (Risso et al. 2011; Ibáñez-Cervantes et al. 2013; Ramos-Ligonio et al. 2012). In a recently published paper, the use of microsatellites confirmed that Mexican T. cruzi strains isolated from humans belonged to TcI and genotype 1 (Martinez et al. 2013). Because vector control programs do not exist in Mexico, the number of people infected by different routes of transmission (vector, congenital, blood transfusion, organ transplantation, or oral) is unknown (Carabarin-Lima et al. 2013). In this context, knowledge of parasitic genotypes circulating in the different cycles of transmission may help to understand the epidemiology of Chagas disease.
By analyzing the 350 bp intergenic region of the mini-exon, it is possible to detect four distinct subgroups within TcI designated as genotypes 1 through 4 where transition, transversion and insertion/deletion represent overall 17.5 % nucleotide divergence. Genotype 1 is associated with human and domiciliated vector, genotype 2 with human and sylvatic vectors, genotype 3 with human dwelling-places and genotype 4 with wild vectors and mammals respectively (Herrera et al. 2007). Using specific primers designed on this polymorphic region of mini-exon, it is possible to identify the associated vector type and transmission cycle for T. cruzi parasites. For example, genotype Ia is associated with human infection and domiciliated Rhodnius vector; genotype Ib is associated with peridomestic cycle and T. dimidiata; and genotype Id with sylvatic cycles of wild vectors and mammals in Colombia (Falla et al. 2009).
Triatoma dimidiata is the main vector recognized in the Yucatan peninsula and it is part of the sylvatic and peridomiciliated transmission cycles (Dumonteil et al. 2002; Rebollar-Tellez et al. 2009). In a recently published paper it was reported that T. dimidiata in this region is preferentially infected with T. cruzi I (Monteon et al. 2013). In this present work we extended our findings using multiple sequence analysis of mini-exon intergenic region of a set of five T. cruzi isolates from Yucatan peninsula and two additional geographically nonrelated Mexican strains.
Materials and methods
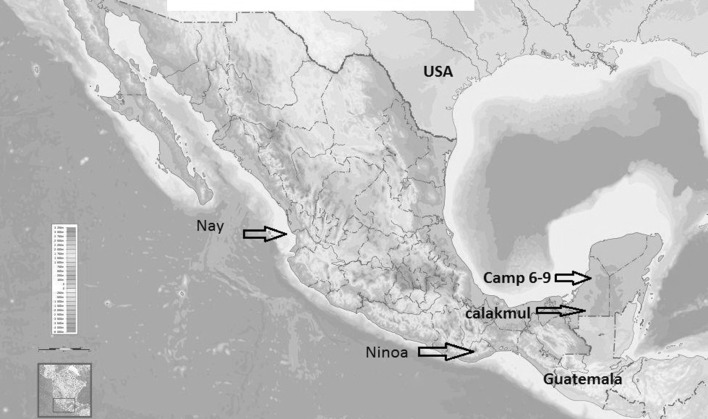
The T. cruzi parasites were isolates from T. dimidiata adults captured inside dwellings in the city of Campeche in the Yucatan Peninsula named Camp6, Camp7, Camp8 and Camp9 and one from the sylvatic region of Calakmul located near to Guatemala border. The non-geographically related strains were isolated 1) from an acute human case in Oaxaca State (Ninoa strain) 768 km away from Yucatan and 2) from Meccus picturatus in Nayarit State named “Nay” 1,570 km away from Yucatan (Fig. 1, http://es.wikipedia.org/wiki/M%C3%A9xico). In order to isolate parasites from infected triatomines captured inside dwellings, the contaminated feces were used to inoculate mice. Following 3 to 4 weeks post-inoculation a blood samples were obtained to check parasitemia and were inoculated into Liver Infusion Tryptose (LIT) culture media enriched with 10 % of fetal bovine serum. The culture was analyzed for the presence of flagellates for 2–4 weeks. Once the culture was established T. cruzi parasites were harvested and DNA was obtained using standard procedures with phenol–chloroform (Green and Sambrook 2012). The mini-exon intergenic region of T. cruzi was amplified using 3 primers, as reported (Souto et al. 1996). All Mexican isolates produce 350 bp amplicon. All PCR products were sent to Macrogen Inc., Korea for DNA purification and sequencing service. For all samples, sequencing was conducted in both forward and reverse directions. These sequences were used to generate a consensus sequence with a previous pairwise alignment using the ClustalW algorithm (Thompson et al. 1997) implemented in the Bioedit v. 7.05 (Hall 1999).
Fig. 1.
Place of isolation of Trypanosoma cruzi parasites: Camp6–9 and Calakmul from Yucatan Peninsula; Ninoa from Oaxaca State and Nay from Nayarit State in the Pacific coast
Results
A sequence of length 235–213 bp was obtained from the Mexican isolates studied. These sequences were analyzed using the ClustalW algorithm (http://www.genome.jp/tools/clustalw/) for multiple sequence alignment and compared with reference sequences deposited in the GenBank of strains cgc:AM259467 genotype Ia; FCh: AM259469 genotype Ib; X380: AM259472 genotype Ic and PALC: AM259473 genotype Id. The isolates from the Yucatan peninsula were practically identical all of which belonging to genotype Ia as well as the Mexican isolates referred to in this study as “non-geographically related showing a single transition T > C in the position 53 (Fig. 2). When comparing our data with deposited sequence in the GenBank, we observed one hundred hits with 48 of them resulting in 100 % of maximum identity and query cover of 100 %. Among these hits, we selected the Tc Ia strain reported by Cura et al. (2010). In multiple sequence alignment using the ClustalW algorithm and phylogenetic analysis using neighbor joining method of the polymorphic sites of 194 nt of miniexon gene´s intergenic region, the Yucatan T. cruzi isolates clustered together with reference strain cga Tc Ia whereas Ninoa and Nay strains presented a single transition T > C (Figs. 2, 3).
Fig. 2.
CLUSTAL 2.1 multiple sequence alignment of Mexican and Tc I references strains. Tc Ia strain cgc; Tc Ib strainFch; Tc IcX380 and Tc Id Palc. Black letter indicates site of transition T > C between Mexican Ninoa and Nay isolates. All Yucatan Mexican isolates (Camp6–9 and Calakmul) are identical with cgc Tc Ia strain. Arrows indicate the beginning and end of sequence analyzed
Fig. 3.
Phylogentic tree of Mexican isolates Camp6–9, Calakmul from Yucatan Peninsula and the Mexican not geographically related Ninoa and Nay strains compared to reference strains cgc haplotype Ia, Fchc haplotype Ib, X380 haplotype Ic and Palc haplotype Id studied by Cura et al. 2010. Data matrix was analyzed with NTSYSpc program and phylogenetic analysis was performed using neighbor-joining method. The sequences analyzed encompass 194 nt of miniexon as indicated in Fig. 2
Our results indicate that haplotype Ia is present in T. dimidiata in the Yucatan peninsula and additionally circulates in the northwest and southwest regions of Mexico as seen in the acute human case of T. cruzi Ninoa strain and vectors such as “Nay strain” in M. picturatus.
Discussion
Our findings are different than those observed in Colombia where Tc Ia (recently named Tc I DOM) is preferentially associated with R. prolixus and humans in domestic cycles (Falla et al. 2009; Zumaya-Estrada et al. 2012), whereas in the Yucatan Peninsula T. dimidiata is rather a visiting vector than a domiciliated participating more in peridomestic and sylvatic cycles. The human would be an accidental host in this cycle. Thus Tc Ia is being introduced into the human population through peridomestic and sylvatic vectors.
Cura et al. (2010) reported that Tc Ia (Tc I DOM) is associated with domestic cycles in southern and northern South America and sylvatic cycles in Central and North America. Yet particularly in Mexico, they studied only three T. cruzi isolate and did not provide accurate information regarding the geographical and biological origin. In our work we add new accurate information for Tc Ia suggesting it does exist in T. dimidiata in the Yucatan Peninsula but also in the pacific coast of Oaxaca in human cases and in M. picturatus in the northwest of Mexico. In a recently published paper by Zumaya-Estrada et al. 2012, using high resolution nuclear and mitochondrial genotyping tools reported that TcIDOM/Tc Ia may be as ancient as humans in South America and it is nestled among North and Central American strains.
Martinez et al. (2013) have confirmed the wide distribution of Tc I in diverse geographic areas of Mexico and transmission cycles, although subgroups within Tc I have not been identified. Our study confirms the presence of Tc Ia in Mexico. Other DTU such as Tc II to Tc V have been recognized in central parts of Mexico and Veracruz (Ibáñez-Cervantes et al. 2013; Ramos-Ligonio et al. 2012). The epidemiology of Chagas disease is influenced by several factors such as T. cruzi lineages and vectors. It would be necessary for future studies to utilize larger sample sizes.
Acknowledgments
This work was financed by Conacyt grant 153764, Mexico and Estrategia de sostenibilidad 2013 Grupo BCEI, Universidad de Antioquia.
References
- Bosseno M, Barnabé C, Magallón-Gastellum E, Lozano-Kasten F, Ramsey J, Espinoza B, Breniere S. Predominance of Trypanosoma cruzi I lineage in Mexico. J Clin Microbiol. 2002;40:627–632. doi: 10.1128/JCM.40.2.627-632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabarin-Lima A, Gonzalez-Vazquez M, Rodrigues-Morales O, Baylon-Pacheco L, Rosales-Encina J, Reyes-Lopez P, Arce-Fonseca M. Chagas disease (American trypanosomiasis) in Mexico: an update. Acta Trop. 2013;127:126–135. doi: 10.1016/j.actatropica.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Cura C, Mejia-Jaramillo A, Duffy T, Burgos J, Rodriguero M, Cardinal M, Kjos S, Gurgel-Gonçalves R, Blanchet D, de Pablos L, Tomasini N, da Silva A, Russomando G, Cuba C, Aznar C, Abate T, Levin M, Osuna A, Gürtler R, Diosque P, Solari A, Triana-Chávez O, Schijman G. Tryapanosoma cruzi I genotypes in different geographical regions and transmission cycles based on a microsatellite motif of the intergenic región spacer of spliced-leader genes. Int J Parasitol. 2010;40:1599–1607. doi: 10.1016/j.ijpara.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumonteil E, Gourbière S, Barrera-Pérez M, Rodríguez-Félix E, Ruíz-Piña H, Baños-López O, Ramírez-Sierra M, Menu F, Ravinovich J. Geographic distribution of Triatoma dimidiata and transmission dynamics in the Yucatán Peninsula of México. Am J Trop Med Hyg. 2002;67:183–189. doi: 10.4269/ajtmh.2002.67.176. [DOI] [PubMed] [Google Scholar]
- Falla A, Herrera C, Fajardo A, Montilla M, Vallejo G, Guhl F. Haplotype identification within Trypanosoma cruzi I in Colombian isolates from several reservoirs, vectors, and humans. Acta Trop. 2009;110:15–21. doi: 10.1016/j.actatropica.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Green M, Sambrook J. Molecular cloning: a laboratory manual. 4. New York: Cold Spring Harbor Laboratory Press; 2012. Isolation and quantification of DNA; pp. 90–93. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Herrera C, Bargues D, Fajardo A, Montilla M, Triana O, Vallejo G, Guhl F. Identify four Trypanosoma cruzi haplotypes from different geographic regions in Colombia. Infect Genet Evol. 2007;7:535–539. doi: 10.1016/j.meegid.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Ibáñez-Cervantes G, Martínez-Ibarra A, Nogueda-Torres B, López-Orduña E, Alonso AL, Perea C, Maldonado T, Hernández JM, León-Avila G. Identification by Q-PCR of Trypanosma cruzi lineage and determination of blood meal sources in triatomine gut samples in Mexico. Parasitol Int. 2013;62:36–43. doi: 10.1016/j.parint.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Martinez I, Nogueda B, Martinez-Hernandez F, Espinoza B. Microsatellite and Mini-Exon analysis of Mexican human DTU I Trypanosoma cruzi strains and their susceptibility to nifurtimox and benznidazole. Vector Borne Zoonotic Dis. 2013;13:181–187. doi: 10.1089/vbz.2012.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteon V, Alducin C, Hernández J, Ramos-Ligonio A, Lopez R. High frequency of human blood in Triatoma dimidata captured inside dwellings in a rural community in the Yucatan Peninsula, Mexico, but low antibody seroprevalence and electrocardiographic findings compatible with Chagas disease in humans. Am J Trop Med Hyg. 2013;88:566–571. doi: 10.4269/ajtmh.12-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Ligonio A, Torres-Montero J, López-Monteon A, Dumonteil E. Extensive diversity of Trypanosoma cruzi discrete typing units circulating in Triatoma dimidiata from central Veracruz, Mexico. Infect Genet Evol. 2012;12:1341–1343. doi: 10.1016/j.meegid.2012.04.024. [DOI] [PubMed] [Google Scholar]
- Rebollar-Tellez EA, Reyes-Villanueva F, Escobedo-Ortegon J, Balam-Briceño P, May-Concha I. Abundance and nigthly activity behavior of a sylvan population of Triatoma dimidiata (Hemíptera:Reduviidae:Triatominae) from the Yucatan. Mexico Vector Ecol. 2009;34:304–310. doi: 10.1111/j.1948-7134.2009.00038.x. [DOI] [PubMed] [Google Scholar]
- Risso G, Sartor P, Burgos J, Briceño L, Rodríguez E, Guhl F, Triana-Chavez O, Espinoza B, Monteón V, Russomando G, Schijman A, Bottasso O, Leguizamón Maria Susana. Immunological identification of Trypanosoma cruzi lineages in human infection along the endemic area. Am J Trop Med Hyg. 2011;84:78–84. doi: 10.4269/ajtmh.2011.10-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruíz-Sánchez R, de León P, Matta V, Reyes PA, López R, Jay D, Monteón VM. Trypanosoma cruzi isolates from Mexican and Guatemalan acute and chronic chagasic cardiopathy patients belong to Trypanosoma cruzi I. Mem Inst Oswaldo Cruz. 2005;100:281–283. doi: 10.1590/S0074-02762005000300012. [DOI] [PubMed] [Google Scholar]
- Souto R, Fernandes O, Macedo A, Campbell D, Zingales B. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol Biochem Parasitol. 1996;83:141–152. doi: 10.1016/S0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: xexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumaya-Estrada FA, Messenger LA, Lopez-Ordonez T, Lewis MD, Flores-Lopez CA, Martínez-Ibarra AJ, Pennington PM, Cordon-Rosales C, Carrasco HV, Segovia M, Miles MA, Llewellyn MS. North American import? Charting the origins of an enigmatic Trypanosoma cruzi domestic genotype. Parasit Vectors. 2012;5:226. doi: 10.1186/1756-3305-5-226. [DOI] [PMC free article] [PubMed] [Google Scholar]