Abstract
Freshwater fishes in Manipur, Northeast India frequently harbour several types of metacercariae, which based on morphological criteria were identified as Clinostomoides brieni, Euclinostomum heterostomum (Clinostomidae) and Polylekithum sp. (Allocreadiidae). Molecular techniques utilizing PCR amplification of rDNA regions of larger subunit (LSU or 28S), smaller subunit (SSU or 18S) and inter transcribed spacers (ITS1, 2) were used for molecular characterization of these types. Sequences generated from the metacercariae were compared with their related sequences available in public databases; an analysis of the identity matrices and phylogenetic trees constructed was also carried out, which confirmed their identification. Similarly, the sequences generated from Polylekithum sp. were found to be highly similar to the species of the same genus. The rDNA ITS2 secondary structure provided additional confirmation of the robustness of the molecular marker as a tool for taxon-specific characterization.
Keywords: Molecular, rDNA ITS2, 28S, 18S, Metacercaria, Fish hosts, Clinostomidae, Allocreadiidae, India
Introduction
With the advent in recent molecular techniques, the DNA-based PCR amplification methods in particular have helped resolve many taxonomic issues related to various helminth parasites, by isolating many target sequences and analysing them (Hillis and Dixon 1991). Among these sequence regions, the nuclear ribosomal DNA (rDNA) coding regions and their non-coding spacers, namely the highly conserved larger subunit (LSU or 28S), the smaller subunit (SSU or 18S) and highly variable inter transcribed spacer (ITS1 and ITS2) regions have proven usefulness in answering systematic questions at many taxonomic levels (Hillis and Davis 1986). The ITS2, flanked by more conserved 5.8S and 28S genes, evolves comparably fast and thus has a wider application for taxonomic issues at the species and genus levels relating to trematode taxa (Morgan and Blair 1995; Rinaldi et al. 2005; Goswami et al. 2009; Shylla et al. 2011; Ghatani et al. 2012). The secondary structure of this region aids in alignment of ITS2 sequences at taxonomic levels above that of the genus due to highly conserved secondary structure core (Michot et al. 1993; Morgan and Blair 1995; Coleman 2003).
Fish serves as second intermediate host for a large numbers of trematodes (Nguyen et al. 2009; Phan et al. 2010). The trematode genera Clinostomum, Euclinostomum (family Clinostomidae) and Polylekithum sp. (family Allocreadiidae) are common parasites found infecting the piscivorous birds in many parts of the globe including India as well (Verma 1936; Vidyarthi 1938; Jaiswal 1957; Ukoli 1966, Yamaguti 1971, Jhansilakshmibai and Madhavi 1997, Kanev et al. 2002). From Northeast India, Athokpam and Tandon (2013) reported the metacercariae of E. heterostomum and Polylekithum sp. from the snakehead fish, Channa punctata and Clinostomoides brieni from Heteropneustes fossilis in the state of Manipur. In the present communication we provide molecular characterization and reliable identification of these taxa using rDNA ITS2, 28S and 18S gene marker regions.
Materials and methods
Parasite material
Metacercariae representing the parasites namely, Clinostomoides brieni, Euclinostomum heterostomum and Polylekithum sp. [identified based on morphological criteria following standard literature (Yamaguti 1971; Gibson et al. 2002; Jones et al. 2005)] were recovered from the snakehead Channa punctata and the singhi fish Heteropneustes fossilis from various localities in Manipur and made to excyst by artificial digestion as reported earlier (Athokpam and Tandon 2013). The excysted metacercariae were fixed in 70 % ethanol for further molecular study.
Molecular study
DNA isolation, amplification and sequencing
Total genomic DNA was extracted from the fixed specimens using a QIAamp DNA Mini Kit (Qiagen, GmbH, Hilden, Germany) following the manufacturer’s instructions. The rDNA genes under consideration were amplified with different sets of primers as detailed below (Table 1).
Table 1.
Primers used for the amplification of rDNA marker regions
| Primer sequences | References | |
|---|---|---|
| ITS2 | 3S (forward):5′-GTACCGGTGGATCACTCGGCTCGTG-3′ A28 (reverse): 5′-GGGATCCTGGTTAGTTTCTTTTCCTCCGC-3′ |
Bowles et al. (1995) |
| 28S | dig12 (forward): 5′-AAGCATATCACTAAGCGG-3′ 1500R (Reverse): 5′-GCTATCCTGAGGGAAACTTCG-3′ |
Tkach et al. (2000) |
| 18S | EukA (forward): 5′-AACCTGGTTGATCCTGCCAGT-3′ EukB (reverse): 5′-TGATCCTTCTGCAGGTTCACCTAC-3′ |
Diez et al. (2001) |
The PCR-amplified products were purified using Genei Quick PCR purification kit for DNA and sequenced using automated sequencing services provided by Macrogen Service Center, Seoul, Korea.
For the 28S and 18S genes, as the full length could not be obtained through one direction sequencing, the contigs were created assembling both the forward and reversed sequences in DNA Baser v3.5.3 (http://www.dnabaser.com/). The sequences generated were submitted to GenBank (NCBI) and their accession numbers acquired.
Sequence and phylogenetic analyses
For the sequence similarity search the Basic Local Alignment Search Tool (BLAST), available at http://www.ncbi.nlm.nih.go v/blast, was used. Multiple sequence alignment was done and sequence identity matrices were constructed using Bioedit software version 7.0.9.0 (Hall 1999) for each of the amplified markers for all metacercaria species under study, using sequences of their closely related taxa as available in the public domain.
The output fasta format files from Bioedit were then entered into MEGA5 (Tamura et al. 2011) for phylogenetic tree construction deducing the distance-based Neighbour Joining (NJ) and the character-based Maximum likelihood (ML) methods.
Predicted ITS2 RNA secondary structures
The ITS2 sequences generated were annotated using the Hidden Markov Models (HMM)-based annotation (available at http://its2.bioapps.biozentrum.uni-wuerzburg.de/) to retrieve the exact sequence of the region (Keller et al. 2009); their secondary structures were constructed using MFOLD software version 3.2, using free energy folding algorithms (Zuker 2003). The output Vienna format was then imported to 4SALE to view the secondary structure (Seibel et al. 2006).
Results
The sequences generated from the three metacercaria species were analysed for their sequence identity with their closely related sequences from the families Clinostomidae and/or Allocreadiidae as retrieved from NCBI (Tables 2, 3, 4). For phylogenetic analysis the trees constructed with ML and NJ methods showed almost the same topology of taxa; therefore, only the ML tree is shown herein. In all trees Fasciolopsis buski was used as the outgroup. The species-wise results of analyses are detailed below.
Table 2.
List of Clinostomidae taxa included in the sequence analysis along with their host and accession numbers of marker genes
| Sr. No. | Parasite name | Host name | Locality | Genes | ||
|---|---|---|---|---|---|---|
| ITS2 | 28S | 18S | ||||
| 1. | Clinostomum complanatum | Squalius cephalus | Italy | FJ609420.1 | FJ609420.1 | FJ609420.1 |
| 2. | C. complanatum | Squalius cephalus | Croatia | – | JX235337.1 | – |
| 3. | C. complanatum | Squalius cephalus | Israel | – | – | AY245701.1 |
| 4. | C. marginatum | Pimephales notatus | Canada | JF718643.1 | – | – |
| 5. | C. marginatum | Perca flavescens | Canada | JF718642.1 | – | – |
| 6. | C. marginatum | Perca flavescens | USA | – | – | AY245760.1 |
| 7. | C. marginatum | Perca flavescens | Canada | JF718641.1 | – | – |
| 8. | C. marginatum | Ardea herodias | Canada | JF718636.1 | – | – |
| 9. | C. tataxumui | Petenia splendida | Mexico | JX631140.1 | – | – |
| 10. | C. phalacrocoracis | Oreochromis niloticus | Kenya | FJ609422.1 | FJ609422.1 | – |
| 11. | C. piscidium | Colisa fasciata | India | – | – | FJ970655.1 |
| 12. | C. giganticum | Channa punctatus | India | – | – | FJ970654.1 |
| 13. | Clinostomum sp. | Hypeseleotris galii | Australia | – | AY222175.1 | AY222094.1 |
| 14. | Clinostomum sp. | Barbus barbus | Italy | JF718621.1 | – | – |
| 15. | Clinostomum sp. | Rana catesbeiana | USA | – | AY222176.1 | – |
| 16. | Clinostomid sp. | Rana catesbeiana | USA | – | AY858877.1 | AY829252.1 |
Table 3.
List of the Allocreadiidae taxa and details of their host and gene accession numbers used in the study
| Sr. No. | Parasite name | Host name | Locatity | Genes | |
|---|---|---|---|---|---|
| ITS2 | 28S | ||||
| 1. | Allocreadium isoporum | Alburnus alburnus | Russia | FJ874921.1 | GU462126.1 |
| 2. | A. lobatum | Semotilus corporalis | USA | – | EF032693.1 |
| 3. | A. neotenicum | Hydroporus rufifrons | UK | – | JX977132.1 |
| 4. | Bunodera luciopercae | Sphaerium rivicola | Lithuania | FJ874916.1 | GU462116.1 |
| 5. | B. luciopercae | Sphaerium rivicola | Ukraine | FJ874915.1 | GU462111.1 |
| 6. | Crepidostomum illinoiense | Hiodon alosoides | USA | – | HQ833705.1 |
| 7. | C. cornutum | Lepomis gulosus | USA | – | EF032695.1 |
| 8. | Creptotrema funduli | Fundulus notatus | USA | JQ425256.1 | JQ425256.1 |
| 9. | Paracreptotrematina limi | Umbra limi | USA | HQ833706.1 | HQ833706.1 |
| 10. | Polylekithum ictaluri | Ictalurus punctatus | Canada | – | DQ189999.1 |
| 11. | P. catahoulensis | Ictalurus furcatus | USA | EF032698.1 | EF032698.1 |
| 12. | P. ictaluri | Ictalurus furcatus | USA | EF032697.1 | EF032697.1 |
Table 4.
Sequence identity matrix (%) among the ITS2 (a), 28S (b) and 18S (c) sequences of the various Clinostomidae taxa as retrieved from GenBank
| (a) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequences | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| 1. | gb|KF781298|Clinostomoides brieni, India* | ID | |||||||||
| 2. | gb|KF781297|Euclinostomum heterostomum, India* | 91.0 | ID | ||||||||
| 3. | gb|JF718641.1|Clinostomum marginatum, Canada | 78.7 | 72.9 | ID | |||||||
| 4. | gb|JF718636.1|C. marginatum, Canada | 78.0 | 72.0 | 98.5 | ID | ||||||
| 5. | gb|FJ609420.1|C. complanatum, Italy | 95.3 | 88.5 | 77.3 | 76.5 | ID | |||||
| 6. | gb|JF718621.1|Clinostomum sp., Italy | 23.1 | 22.8 | 28.0 | 28.4 | 23.5 | ID | ||||
| 7. | gb|JF718643.1|C. marginatum, Canada | 76.3 | 70.5 | 96.1 | 97.6 | 74.8 | 29.1 | ID | |||
| 8. | gb|JF718642.1|C. marginatum, Canada | 51.7 | 48.9 | 64.0 | 64.9 | 52.4 | 43.7 | 66.5 | ID | ||
| 9. | gb|JX631140.1|C. tataxumui, Mexico | 85.8 | 80.0 | 88.2 | 87.1 | 83.4 | 25.6 | 85.7 | 57.3 | ID | |
| 10. | gb|FJ609422.1|C. phalacrocoracis, Kenya | 96.3 | 88.8 | 77.9 | 77.0 | 96.0 | 23.2 | 75.3 | 51.3 | 83.8 | ID |
| (b) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Sequences | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| 1. | gb|KF781299|Clinostomoides brieni, India* | ID | ||||||
| 2. | gb|FJ609420.1|Clinostomum complanatum, Italy | 32.7 | ID | |||||
| 3. | gb|JX235337.1|C. complanatum, Croatia | 16.1 | 40.1 | ID | ||||
| 4. | gb|FJ609422.1|C. phalacrocoracis, Kenya | 32.3 | 98.1 | 39.2 | ID | |||
| 5. | gb|AY222175.1|Clinostomum sp., Australia | 46.6 | 18.6 | 32.3 | 18.5 | ID | ||
| 6. | gb|AY222176.1|Clinostomum sp., USA | 46.9 | 18.9 | 32.1 | 18.8 | 96.5 | ID | |
| 7. | gb|AY858877.1|Clinostomid sp., USA | 45.1 | 18.0 | 33.8 | 17.8 | 95.2 | 92.1 | ID |
| (c) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequences | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| 1. | gb|KF781300|Clinostomoides brieni, India* | ID | |||||||||
| 2. | gb|FJ609423.1|Clinostomum phalacrocoracis, Kenya | 95.8 | ID | ||||||||
| 3. | gb|AY245701.1|C. complanatum, Israel | 95.5 | 98.2 | ID | |||||||
| 4. | gb|FJ609422.1|C. phalacrocoracis, Kenya | 95.9 | 99.9 | 98.1 | ID | ||||||
| 5. | gb|FJ970655.1|C. piscidium, India | 21.0 | 22.1 | 22.4 | 22.1 | ID | |||||
| 6. | gb|FJ970654.1|C. giganticum, India | 29.9 | 30.8 | 31.0 | 30.8 | 67.3 | ID | ||||
| 7. | gb|AY245760.1|C. marginatum, USA | 96.3 | 98.3 | 98.0 | 98.3 | 22.0 | 30.8 | ID | |||
| 8. | gb|FJ609420.1| C. complanatum, Italy | 96.0 | 98.9 | 99.2 | 98.9 | 22.3 | 30.8 | 98.5 | ID | ||
| 9. | gb|AY222094.1|Clinostomum sp., Australia | 95.6 | 98.2 | 99.1 | 98.2 | 23.0 | 31.3 | 97.9 | 98.5 | ID | |
| 10. | gb|AY829252.1|Clinostomid sp., USA | 86.6 | 88.7 | 89.0 | 88.7 | 25.1 | 34.2 | 88.4 | 89.0 | 89.5 | ID |
* Query sequence generated from the study; highest value marked in bold
Clinostomoides brieni
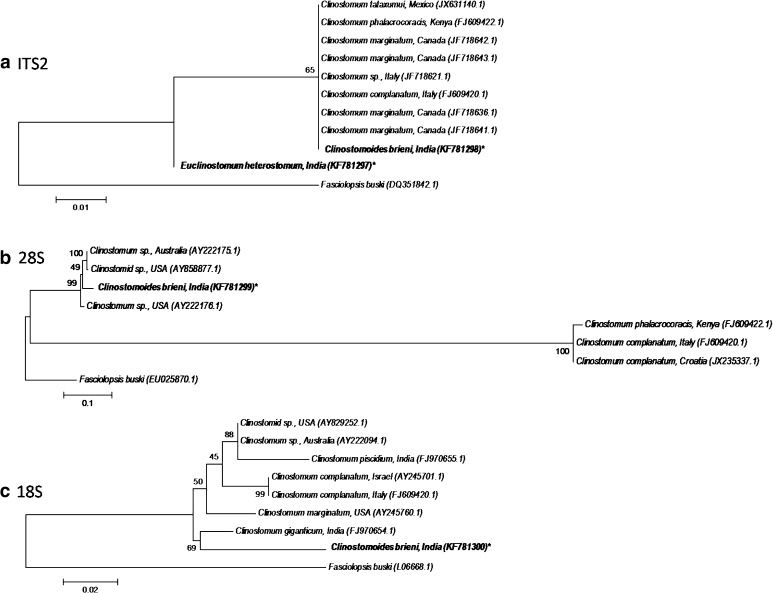
The rDNA ITS2, 28S and 18S sequences deposited in GenBank (under accession numbers: KF781298, KF781299 and KF781300) are 430, 2546 and 1907 bp in length, respectively. For sequence analysis sequences of the various Clinostomidae taxa available in the public domain were used (Table 2). The metacercaria under the present study comes close to Clinostomum spp, showing 96.3 % sequence similarly for ITS2 (with C. phalacrocoracis) as well as 18S (with C. marginatum); however, with regard to 28S the sequence similarity with Clinostomum sp. was less than 50 % (Table 4a–c). In phylogenetic analysis, the query sequence C. brieni claded with clinostomid species, but with bootstrap values of 65–99 % (Fig. 1a–c). In all trees, Fasciolopsis buski, the outgroup, formed a separate clade.
Fig. 1.
Phylogenetic trees constructed for Clinostomidae: a ITS2; b 28S; c 18S (Asterisk Query sequence generated from the study)
Euclinostomum heterostomum
The ITS2 sequence (accession number: KF781297) has a length of 415 bp. There being no sequence available for Euclinostomum species in public database, the sequence analysis was done comparing with Clinostomum spp. (Table 2). In sequence identity matrix, the metacercaria under the present study stands very close to Clinostomoides brieni sequence, with a sequence identity of 91.0 % (Table 4a); in phylogenetic tree it forms a separate branch from other Clinostomidae taxa (Fig. 1a).
Polylekithum sp.
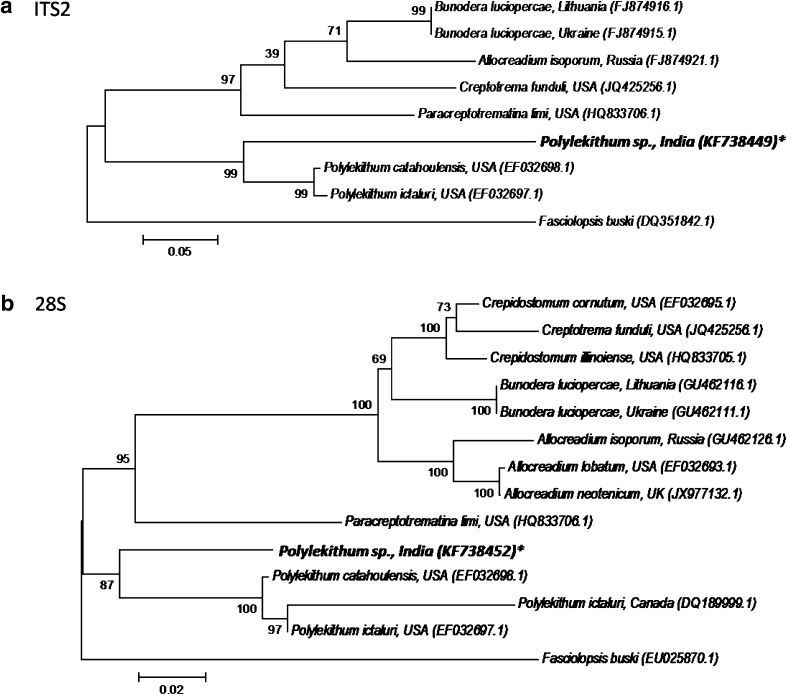
The rDNA ITS2 and 28S sequences (accession numbers: KF738449 and KF738452, respectively) deposited in GenBank have a length of 459 and 985 bp, respectively. The metacercaria under the present study stands very close to Polylekithum catahoulensis, with sequence identities of 73.1 % (ITS2) and 91.1 % (28S) (Table 5). In phylogenetic analysis Polylekithum sp. claded within the genus Polylekithum, with significant bootstrap value of 87–99 % (Fig. 2).
Table 5.
Sequence identity matrix (%) among the ITS2 (a) and 28S (b) gene sequences of the various Allocreadiidae taxa as retrieved from GenBank
| (a) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sequences | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 1. | Polylekithum sp., India* | ID | |||||||
| 2. | P. catahoulensis, USA | 73.1 | ID | ||||||
| 3. | P. ictaluri, USA | 72.7 | 98.0 | ID | |||||
| 4. | Allocreadium isoporum, Russia | 49.0 | 50.1 | 49.8 | ID | ||||
| 5. | Bunodera luciopercae, Lithuania | 57.7 | 59.4 | 58.9 | 80.0 | ID | |||
| 6. | B. luciopercae, Ukraine | 58.2 | 59.8 | 59.3 | 79.5 | 99.4 | ID | ||
| 7. | Creptotrema funduli, USA | 61.2 | 61.0 | 60.7 | 71.1 | 79.5 | 80.0 | ID | |
| 8. | Paracreptotrematina limi, USA | 60.7 | 61.8 | 61.3 | 66.6 | 77.0 | 77.5 | 80.3 | ID |
| (b) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequences | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| 1. | Polylekithum sp., India* | ID | ||||||||||||
| 2. | P. ictaluri, Canada | 67.0 | ID | |||||||||||
| 3. | P. catahoulensis, USA | 91.1 | 72.6 | ID | ||||||||||
| 4. | P. ictaluri, USA | 90.9 | 73.2 | 98.9 | ID | |||||||||
| 5. | Allocreadium isoporum, Russia | 82.0 | 59.7 | 82.3 | 82.2 | ID | ||||||||
| 6. | A. lobatum, USA | 83.0 | 60.2 | 82.9 | 83.0 | 95.9 | ID | |||||||
| 7. | A. neotenicum, UK | 83.1 | 60.3 | 83.0 | 83.1 | 95.9 | 99.7 | ID | ||||||
| 8. | Bunodera luciopercae, Lithuania | 83.0 | 60.7 | 83.6 | 83.4 | 92.1 | 92.5 | 92.7 | ID | |||||
| 9. | B. luciopercae, Ukraine | 83.0 | 60.7 | 83.6 | 83.4 | 92.1 | 92.5 | 92.7 | 100.0 | ID | ||||
| 10. | Crepidostomum illinoiense, USA | 83.6 | 60.6 | 82.9 | 82.9 | 92.1 | 93.1 | 93.3 | 94.1 | 94.1 | ID | |||
| 11. | C. cornutum, USA | 83.9 | 61.1 | 83.4 | 83.4 | 92.2 | 93.1 | 93.3 | 94.2 | 94.2 | 97.8 | ID | ||
| 12. | Creptotrema funduli, USA | 82.3 | 60.1 | 82.2 | 82.2 | 90.9 | 92.2 | 92.2 | 92.8 | 92.8 | 95.9 | 96.9 | ID | |
| 13. | Paracreptotrematina limi, USA | 87.3 | 63.6 | 86.7 | 86.8 | 82.5 | 83.8 | 83.8 | 83.7 | 83.7 | 84.2 | 84.3 | 83.3 | ID |
* Query sequence generated from the study; highest value marked in bold
Fig. 2.
Phylogenetic trees constructed for Allocreadiidae: a ITS2; b 28S (Asterisk Query sequence generated from the study)
ITS2 rDNA secondary structures
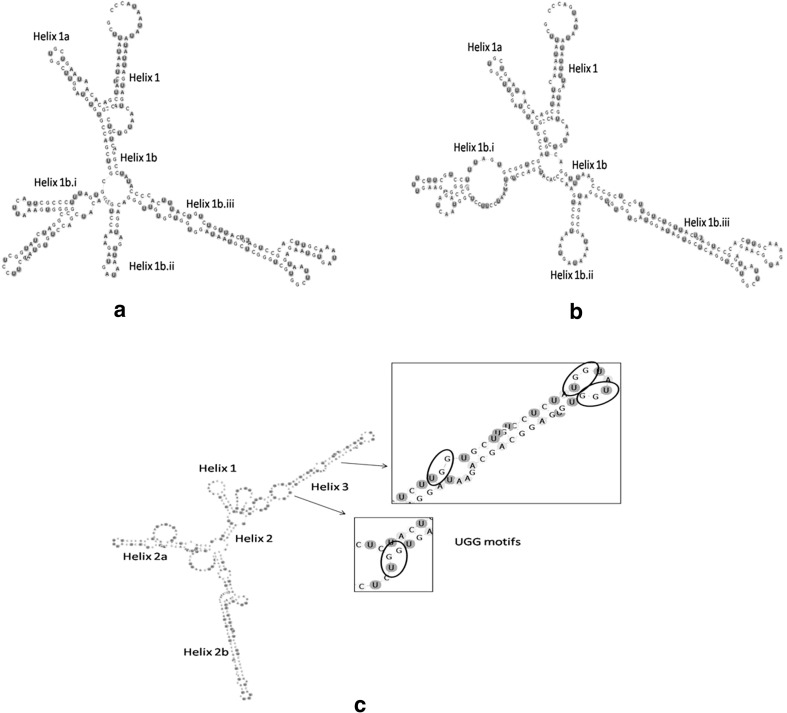
The secondary structure of ITS2 of both C. brieni and E. heterostomum shows deviation from the 4-helical structure model and consists of only one helix having two branches: Helix 1a and 1b, the latter (Helix 1b) again showing three sub branches—Helix 1b.i, ii and iii (Fig. 3a, b). However, in contrast to clinostomatids the ITS2 secondary structure of Polylekithum sp. comprises three helices—Helix 1, Helix 2 (with two sub-branches—2a and 2b) and Helix 3; Helix 2 is the longest and Helix 3 shows UGG motifs (Fig. 3c).
Fig. 3.
Inferred secondary structure of ITS2 rDNA region (based on minimum free energy modelling using Mfold); a C. brieni India: Northeast, dG = −94.80 kcal/mol; b E. heterostomum India: Northeast, dG = −90.00 kcal/mol; c Polylekithum sp. India: Northeast, dG = −107 kcal/mol
Discussion
The sequences of the rDNA marker regions have been successfully used, as in the present study, for molecular characterization of the trematode taxa including the clinostomids and allocreadiids. The 18S gene was used for validating C. marginatum (Dzikowski et al. 2004). Caffara et al. (2011) also supplemented the studies in morphological difference among various Clinostomum spp. utilizing the rDNA ITS and 18S regions, and mitochondrial cytochrome oxidase 1 (mtCO1) regions. From India, only three sequences from the genus Clinostomum are available in the public domain—28S partial sequence (GQ925911.1) (Singh and Chaudhary, unpublished) and two 18S partial sequences (FJ970655.1 and FJ970654.1, derived from Clinostomum piscidium found infecting the host Colisa fasciatus and Clinostomum giganticum found infecting the fish Channa punctatus, respectively (Agrawal et al., unpublished). Our present study provides for the first time the sequence data for C. brieni and E. heterostomum. With regard to the various Allocreadiidae taxa, Curran et al. (2006) characterized Polylekithum sp. utilizing the rDNA (ITS2 and 28S) marker regions. Petkevičiūtė et al. (2010) also used these marker sequences for discrimination of host-specific subspecies of the allocreadiid trematode species, and to clarify species relationships among these taxa. Our results of the phylogenetic analysis indicating a high bootstrap value also supported the sequence analysis; as generally accepted a bootstrap value of ≥70 % for a given interior branch of a phylogenetic tree is indicative of an authentic analysis (Hillis and Bull 1993; Tandon et al. 2007).
The ITS2 secondary structure can provide additional informations related to taxonomic characters, which otherwise cannot be explained by primary sequence alone (Caetano-Anollés 2002; Grajales et al. 2007; Krüger and Gargas 2008). Schultz et al. (2005) showed a common core of secondary structure of ITS2 throughout the Eukaryota. High sequence variation with conservedness in its secondary structure have made ITS2 useful in various evolutionary comparison studies (Coleman 2003). The secondary structures generated from the ITS2 regions of E. heterostomum and C. brieni in our study do not display the typical four-helix model. Generally RNA secondary structure prediction is based on the minimum free energy at 37 °C and variations in topology are attributed to differences in their nucleotide length (Prasad et al. 2009). Only 17.9 % of the available ITS2 sequences in the public databases show the four-domain structure (Wolf et al. 2005), though three-helix models are also available (Subbotin et al. 2005). Schultz et al. (2005) attributed several reasons for the absence of the four-helix structure—firstly, the chosen consensus structure may be very conservative to avoid false positives; secondly, the low abundance of the consensus structure could be caused by the fold prediction itself as only the minimal energy structure was considered and this might not represent the biological active form.
In conclusion, the present study is the first to provide the molecular characterization of the clinostomatiid/allocreadiid metacercariae that occur in freshwater fishes in the region. The study also corroborates the utility of molecular tools in validating the species identification.
Acknowledgments
VDA acknowledges the award of Research Fellowship for meritorious students by University Grants Commission (UGC) and Senior Research Fellowship by Council of Scientific & Industrial Research (CSIR), Government of India.
References
- Athokpam VD, Tandon V. A survey of metacercarial infections in commonly edible fish and crab hosts prevailing in Manipur, Northeast India. J Parasit Dis. 2013 doi: 10.1007/s12639-013-0360-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Blair D, Mcmanus DP. A molecular phylogeny of the human schistosomes. Mol Phylogenet Evol. 1995;4:103–109. doi: 10.1006/mpev.1995.1011. [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés G. Tracing the evolution of RNA structure in ribosomes. Nucleic Acids Res. 2002;30:2575–2587. doi: 10.1093/nar/30.11.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffara M, Locke SA, Gustinelli A, Marcogliese DJ, Fioravanti ML. Morphological and molecular differentiation of Clinostomum complanatum and Clinostomum marginatum (Digenea: Clinostomidae) metacercariae and adults. J Parasitol. 2011;97(5):884–891. doi: 10.1645/GE-2781.1. [DOI] [PubMed] [Google Scholar]
- Coleman AW. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 2003;19:370–375. doi: 10.1016/S0168-9525(03)00118-5. [DOI] [PubMed] [Google Scholar]
- Curran SS, Tkach VV, Overstreet RM. A review of Polylekithum Arnold, 1934 and its familial affinities using morphological and molecular data, with description of Polylekithum catahoulensis sp. nov. Acta Parasitolog. 2006;51(4):238–248. doi: 10.2478/s11686-006-0037-1. [DOI] [Google Scholar]
- Diez B, Pedros-Alio C, Massana R. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R, Levy MG, Poore MF, Flowers JR, Paperna I. Clinostomum complanatum and Clinostomum marginatum (Rudolphi, 1819) (Digenea: Clinostomidae) are separate species based on differences in ribosomal DNA. J Parasitol. 2004;90:413–414. doi: 10.1645/GE-159R. [DOI] [PubMed] [Google Scholar]
- Ghatani S, Shylla JA, Tandon V, Chatterjee A, Roy B. Molecular characterization of pouched amphistome parasites (Trematoda: Gastrothylacidae) using ribosomal ITS2 sequence and secondary structures. J Helminthol. 2012;86:117–124. doi: 10.1017/S0022149X11000125. [DOI] [PubMed] [Google Scholar]
- Gibson DI, Jones A, Bray RA. Keys to the Trematoda. CABI: Natural History Museum; 2002. [Google Scholar]
- Goswami LM, Prasad PK, Tandon V, Chatterjee A. Molecular characterization of Gastrodiscoides hominis (Platyhelminthes: Trematoda: Digenea) inferred from ITS rDNA sequence analysis. Parasitol Res. 2009;104:1354–1358. doi: 10.1007/s00436-009-1354-8. [DOI] [PubMed] [Google Scholar]
- Grajales A, Aguilar C, Sánchez JA. Phylogenetic reconstruction using secondary structures of internal transcribed spacer 2 (ITS2, rDNA): finding the molecular and morphological gap in Caribbean gorgonian corals. BMC Evol Biol. 2007;7:90. doi: 10.1186/1471-2148-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;4:95–98. [Google Scholar]
- Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42(2):182–192. doi: 10.1093/sysbio/42.2.182. [DOI] [Google Scholar]
- Hillis DM, Davis SK. Evolution of ribosomal DNA; fifty million years of recorded history in the frog genus Rana. Evolution. 1986;40:1275–1288. doi: 10.2307/2408953. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- Jaiswal GP. Studies on the trematode parasites of fishes and birds found in Hyderabad State. Zool Jahrb Abt Syst Part I-IV. 1957;85(12):1–72. [Google Scholar]
- Jhansilakshmibai K, Madhavi R. Euclinostomum heterostomum (Rudolphi, 1809) (Trematoda): life-cycle, growth and development of the metacercaria and adult. Syst Parasitol. 1997;38:51–64. doi: 10.1023/A:1005829625739. [DOI] [Google Scholar]
- Jones A, Bray RA, Gibson DI. Keys to the Trematoda. CABI: Natural History Museum; 2005. [Google Scholar]
- Kanev I, Radev V, Fried B. Family Clinostomidae Lühe, 1901. In: Gibson DI, Jones A, Bray RA, editors. Keys to the Trematoda. London: CABI Publishing and the Natural History Museum; 2002. pp. 113–120. [Google Scholar]
- Keller A, Schleicher T, Schultz J, Müller T, Dandekar T, Wolf M. 5.8S–28S rRNA interaction and HMM-based ITS2 annotation. Gene. 2009;430:50–57. doi: 10.1016/j.gene.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Krüger D, Gargas A. Secondary structure of ITS2 ribosomal RNA provides taxonomic characters for systematic studies—a case in Lycoperdaceae (Basidiomycota) Mycol Res. 2008;112:316–330. doi: 10.1016/j.mycres.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Michot B, Despres L, Bonhomme F, Bachellerie JP. Conserved secondary structures in the ITS2 of trematode pre-rRNA. FEBS Lett. 1993;316:247–252. doi: 10.1016/0014-5793(93)81301-F. [DOI] [PubMed] [Google Scholar]
- Morgan JAT, Blair D. Nuclear rDNA ITS sequence variation in the trematode genus Echinostoma: an aid to establishing relationships within the 37-collar spine group. Parasitology. 1995;111:609–615. doi: 10.1017/S003118200007709X. [DOI] [PubMed] [Google Scholar]
- Nguyen TLA, Nguyen TP, Johansen MV, Murrell KD, Phan TV, Dalsgaard A. Prevalence and risks for fish borne zoonotic trematode infections in domestic animals in a highly endemic area of North Vietnam. Acta Trop. 2009;112:198–203. doi: 10.1016/j.actatropica.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Petkevičiūtė R, Stunžėnas V, Stanevičiūte G, Sokolov SG. Comparison of the developmental stages of some European allocreadiid trematode species and a clarification of their life-cycles based on ITS2 and 28S sequences. Syst Parasitol. 2010;76:169–178. doi: 10.1007/s11230-010-9249-8. [DOI] [PubMed] [Google Scholar]
- Phan VT, Ersboll KA, Nguyen VK, Madsen H, Dalsgaard A. Farm-level risk factors for fishborne zoonotic trematode infection in integrated small-scale fish farms in North Vietnam. PLoS Negl Trop Dis. 2010;4:e742. doi: 10.1371/journal.pntd.0000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad PK, Tandon V, Biswal DK, Goswami LM, Chatterjee A. Phylogenetic reconstruction using secondary structures and sequence motifs of ITS2 rDNA of Paragonimus westermani (Kerbert, 1878) Braun, 1899 (Digenea: Paragonimidae) and related species. BMC Genom. 2009;10(Suppl 3):S25. doi: 10.1186/1471-2164-10-S3-S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi L, Perugini AG, Capuano F, Fenizia D, Musella V, Veneziano V, Cringoli G. Characterization of the second internal transcribed spacer of ribosomal DNA of Calicophoron daubneyi from various hosts and locations in southern Italy. Vet Parasitol. 2005;131:247–253. doi: 10.1016/j.vetpar.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Schultz J, Maisel S, Gerlach D, Müller T, Wolf M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA. 2005;11(4):361–364. doi: 10.1261/rna.7204505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel PN, Müller T, Dandekar T, Schultz J, Wolf M. 4SALE—a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinform. 2006;7:498. doi: 10.1186/1471-2105-7-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shylla JA, Ghatani S, Chatterjee A, Tandon V. Secondary structure analysis of ITS2 in the rDNA of three Indian paramphistomid species found in local livestock. Parasitol Res. 2011;108:1027–1032. doi: 10.1007/s00436-010-2148-8. [DOI] [PubMed] [Google Scholar]
- Subbotin SA, Madani M, Krali E, Sturhan D, Moens M. Molecular diagnostics, taxonomy, and phylogeny of the stem nematode Ditylenchus dipsaci species complex based on the sequences of the internal transcribed spacer rDNA. Phytopathology. 2005;95:1308–1315. doi: 10.1094/PHYTO-95-1308. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon V, Prasad PK, Chatterjee A, Bhutia PT. Surface fine topography and PCR-based determination of metacercaria of Paragonimus sp. from edible crabs in Arunachal Pradesh, Northeast India. Parasitol Res. 2007;102:21–28. doi: 10.1007/s00436-007-0715-4. [DOI] [PubMed] [Google Scholar]
- Tkach VV, Pawlowski J, Sharpilo VP. Molecular and morphological differentiation between species of the Plagiorchis vespertilionis group (Digenea, Plagiorchiidae) occurring in European bats, with a re-description of P. vespertilionis (Müller, 1780) Syst Parasitol. 2000;47:9–22. doi: 10.1023/A:1006358524045. [DOI] [PubMed] [Google Scholar]
- Ukoli FMA. On Euclinostomum heterostomum (Rudolphi, 1809) J Helminthol. 1966;40:227–234. doi: 10.1017/S0022149X00034210. [DOI] [PubMed] [Google Scholar]
- Verma SC. Notes on trematode parasites of Indian birds. Part. I. Allahabad Univ Studies. 1936;12:147–188. [Google Scholar]
- Vidyarthi RD. New avian trematodes (family: Diplostomidae) from Indian birds. Proc Nat Acad Sci India. 1938;8(3):76–84. [Google Scholar]
- Wolf M, Achtziger M, Schultz J, Dandekar T, Mueller T. Homology modeling revealed more than 20,000 rRNA internal transcribed spacer 2 (ITS2) secondary structures. RNA. 2005;11:1616–1623. doi: 10.1261/rna.2144205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguti S. Synopsis of the digenetic trematodes of vertebrates. Japan: Keigaku Publishers; 1971. p. 1074. [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]